Abstract
Selection of cells having the most osteogenic potential is a strategy used in bone tissue engineering. Preclinical studies using murine bone marrow cells must consider the large amount of hematopoietic cells in the adherent fraction. The aim of this study was to enrich a murine bone marrow cell population with osteoprogenitors by using a simple and reliable method. Bone marrow from C57Bl/6 mice was extracted and cells which adhered onto plastic were expanded in primary culture for 14 days. Immunolabeling of the CD11b surface antigen was performed and the CD11b− cell fraction was isolated by FACS. Sorted and unsorted populations were analyzed for gene expression of osteoblast differentiation, alkaline phosphatase (AlkP) activity and matrix mineralization capacities. Selection of CD11b− cells increased the number of AlkP+ cells from the plastic adherent fraction from 6.3% ± 0.8 to 56% ± 3.3 with a sevenfold increase in AlkP activity. mRNA analysis revealed a significant increase in the CD11b− fraction for Osterix (41-fold), RANKL (17-fold), M-CSF (8-fold) and Runx-2 (8-fold). An osteogenic population was obtained with improved capacities to produce a mineralized extracellular matrix in vitro, independently of the presence of glucocorticoids in the culture medium.
Keywords: Osteoprogenitor, Immunodepletion, CD11b, Alkaline phosphatase, mRNA profiles
Introduction
Bone marrow contains a large variety of stem cells comprising multipotent and restricted progenitors. The bone marrow stroma contains cells which have the capacity to form bone when transplanted in vivo in diffusion chambers. They are usually described as “plastic-adherent cells” or “colony forming unit-fibroblasts” (CFU-F) (Friedenstein et al. 1987), or are also referred to as mesenchymal stem cells (MSC) (Prockop 1997; Bianco et al. 2001). Several preclinical and clinical studies have suggested that transplantation of progenitors of aspirated bone marrow improves bone-healing (Werntz et al. 1996; Hernigou et al. 2005). However, methods using whole bone marrow are limited by the low number of osteogenic donor cells that can be harvested from the patient. For large bone defects or non-union fractures, it may be insufficient to obtain a complete repair and recovery of the skeletal function (Muschler et al. 2004). Methods may be required to increase the population of osteogenic cells through ex vivo expansion techniques (Bianco and Gehron Robey 2000; Dennis et al. 2007). MSC from culture-expanded marrow have been shown to repair segmental bone defects in animal models (Kadiyala et al. 1997; Ringe et al. 2002). It is now evident that such plastic adherent cells are not exclusively stem cells and osteoprogenitors (Ohgushi and Caplan 1999; Baddoo et al. 2003). A prolonged culture time of MSC seems sufficient to increase the number of osteoprogenitors in human and rat (Kassem 2004). On the contrary, isolation of murine osteoprogenitors from bone marrow necessitates the removal of contaminant adherent cells from hematopoietic lineages (Phinney et al. 1999). Various protocols have been developed to circumvent this problem, including immuno-selection or -depletion of cells based on specific surface markers (Van Vlasselaer et al. 1994; Baddoo et al. 2003; Tropel et al. 2004; Shiota et al. 2007).
The aim of this study was to enrich the murine plastic adherent cell population in stromal cells by removing adherent hematopoietic cells that express the CD11b antigen. In the mouse, CD11b (Mac-1 α; integrin αM chain) is expressed on monocytes/macrophages and to a lower extent on granulocytes, natural killer cells and a subset of dendritic cells (Stewart et al. 1995). We found that selection of plastic adherent CD11b− cells correlates with the selection of MSC expressing alkaline phosphatase (AlkP+ cells), an osteoblastic marker (Komori 2006). CD11b− cells were identified as mature osteoprogenitors because they expressed a mRNA profile typical of osteoblasts.
Materials and methods
Mouse bone marrow stromal cells isolation and culture
Six to eight-week-old C57BL/6NHsd (Harlan, Gannat, France) were used to harvest bone marrow. The Animal Care and Use committee at the University of Angers approved all procedures. Bone marrow from the femoral, tibial, and humeral shafts was flushed with Dubbleco’s-modified Eagle’s medium (DMEM) supplemented with 2 mM glutamine, 100 U/mL penicillin, 100 mg/mL streptomycin (Eurobio, Les Ulis, France) and filtered through a 70 μm cell strainer (BD Biosciences, Pont-de-Claix, France). The whole marrow content was plated into two 75 cm2 culture flasks (Becton Dickinson, Franklin Lakes, NJ) containing DMEM, supplemented with 20% heat-inactivated fetal calf serum (FCS, Seromed Biochrom, Berlin, Germany). Cells were cultured at 37 °C in a humidified atmosphere and 5% CO2. Confluent layers of adherent cells were formed in 14–16 days and were harvested for immuno depletion and subculture using trypsin-EDTA (for 15 min at 37 °C) and scraping.
Immunolabeling, cell sorting and subculture
For a single cell sorting assay, cultured bone marrow cells from two mice were pooled for immunolabeling. Trypsinized cells washed with phosphate buffer saline (PBS), were incubated for 30 min with phycoerythrin (PE)-conjugated IgG2b rat anti-mouse CD11b (Milteney Biotec, Paris, France) on ice. Isotype controls were run in parallel using the same concentration. After washing twice with PBS containing 1% FCS, cells were filtered through a 70 μm cell strainer (BD Biosciences, Pont-de-Claix, France) and analyzed on a fluorescent activating cell sorter (FACSAria, BD Biosciences) for measurements of cell size (forward scatter) and complexity or granularity (side scatter). The non-fluorescent CD11b negative population (CD11b− cells) were sorted with sterile PBS in the sheath fluid and recovered in a separated tube containing 500 μL of FCS. Cells were maintained under rotation and at 4 °C during the sort. 50,000 CD11b− or unsorted cells were seeded on glass slides in 24 wells plates (day 0) and subcultured for 21 days in the culture medium used for the primary culture supplemented with ascorbic acid-2 phosphate (100 μM) and β-glycerophosphate (10 mM) (Sigma–Aldrich, Saint-Quentin Fallavier, France).
Alkaline phosphatase (AlkP) cytochemical staining
Twenty hours after seeding (day 1), cells were incubated 20 min with Naphtol AS-MX 0.5% (w/v) and Fast Red TR 1% (Sigma) in Tris–HCl, 0.2 M, pH 8.9. A precipitate appeared at the site of AlkP activity, it is fluorescent in red under green light (540 nm). After washing with PBS, cells were fixed with 4% paraformaldehyde in PBS and counterstained with Harris’ hematoxylin. Observations were done under transmitted and fluorescent lights using a DMR-B microscope (Leica Microsystems, Rueil-Malmaison, France) with a N2.1 filter cube (515–560 nm excitation filter and 590 nm suppression filter). Microphotographs were taken at ×100 magnification; at least five fields per slide (2 slides per condition) were analyzed using the cell counter plugin of the Image J software (NIH, USA) to evaluate the percentage of AlkP positive cells (fluorescent). Analyses were done on six different experiments.
AlkP activity assay
Cells were lysed by sonication in Tris–HCl, 0.1 M, pH 7.5, 0.1% Triton X 100. AlkP activity was quantified by reaction with 2.5 mM paranitrophenyl phosphate in 0.1 M 2-amino-1-methyl-1-propanol buffer, pH 9.8, supplemented with 2 mM MgCl2. The reaction was stopped with NaOH 0.5 N and paranitrophenol (pNP) formed was measured at 410 nm on a spectrophotometer (Shimadsu, Kyoto, Japan). Results were normalized for protein content, determined using a protein assay kit (Biorad, Hercules, CA, USA). Samples from ten experiments were analyzed.
Quantitative PCR
Total RNA from unsorted or CD11b− cell populations were extracted at day 1 using RNeasy mini kit (Qiagen, Courtaboeuf, France) following the manufacturer’s procedure. After elution, total RNA were aliquoted and stored at −80 °C until use. The quality of the RNA samples was examined on a denaturing agarose gel and the RNA concentration was determined by spectrometry.
Reverse transcription
About 1 μg of total RNA sample was mixed with 3 μg of random hexamers (Invitrogen, Cergy Pontoise, France) in 10 μL final volume, incubated at 70 °C for 5 min and chilled on ice. Then, the reaction was performed at 25 °C for 10 min, 42 °C for 1 h and 70 °C for 10 min with 10 mM dNTPs, 40 units of RNase Inhibitor (Invitrogen), 0.1 M of DTT and 200 units of SuperScript II reverse transcriptase (SSII) (Invitrogen) in a 10 μL final volume of buffer as recommended by the supplier. For each sample, a reaction without SSII was done as negative control. cDNA was purified using an Qiaquick PCR Purification Kit (Qiagen), aliquoted and stored at −20 °C.
Real time PCR
RT-PCR analyses were done using a Chromo 4™ (Bio-Rad, Marnes-la-Coquette, France) and SYBR Green detection. The primers were designed using Primer3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). Studied genes were Runx2 (cfba-1), Osterix, Msx2, RANKL, osteoprotegerin (OPG) and M-CSF. Sequences of primers used are given in Table 1. Amplification was done in a final volume of 15 μL containing 5 μL of cDNA in sterile distilled water (sample or standard) and 10 μL of iQ SYBR Supermix (Biorad) containing 5 μM of each primer. The following protocol was used: (1) denaturation program (95 °C for 10 min), (2) amplification and quantification program repeated 40 cycles (95 °C for 15 s, 55 °C for 11 s, 72 °C for 22 s, with a single fluorescence measurement of SYBR green I at each end of cycle), (3) melting curve program (65–99 °C with a heating rate of 0.1 °C and continuous fluorescence measurement). The difference of expression level was determined by normalization to the expression of housekeeping genes (HPRT1, B2M and ACTB) in parallel runs and quantification was made using a standard curve assay.
Table 1.
Sequences of primers used for real time PCR
| Gene | Sense | Anti-sense |
|---|---|---|
| Runx2 | 5′-GTG GC CACT TAC CAC AGA GC | 5′-GTT CTG AGG CGG GAC ACC |
| Osterix | 5′-TGA GGA AGA AGC CCA TTC AC | 5′-GTG GTC GCT TCT GGT AAA GC |
| Msx2 | 5′-CTC GGT CAA GTC GGA AA | 5′-GGC TAG AAG CTG GGA TGT GG |
| RANKL | 5′-TGT ACT TTC GAG CGC AGA TG | 5′-CCC ACA ATG TGT TGC AGT TC |
| OPG | 5′-GAA CTG CAG TCC GTG AAG C | 5′-CAA ACT GTG TTT CGC TCT GG |
| M-CSF | 5′-GGG CTC TTC AGC CAC TAG C | 5′-CAC CTC CTT GGC AAT ACT CC |
Extracellular mineralization assay
At day 21 of subculture in medium supplemented with ascorbic acid and β-glycerophosphate, unsorted and CD11b− cell layers were incubated 2 h in 1 mL of a solution of calcein in DMEM (25 μg/mL, Sigma) to reveal extracellular mineralization. After two washes with PBS, slides were fixed with 4% paraformaldehyde in PBS and observed under fluorescent microscopy with an I3 filter cube (550–490 nm excitation filter and 515 nm suppression filter).
Statistical analysis
Statistical study was performed using SYSTAT statistical software (release 11.0). All data were reported as mean ± standard error of the mean (SEM). Significant differences between groups were assessed by pairwise comparisons of variance (non parametric Mann and Whitney U test). Differences were considered significant at p < 0.05.
Results
FACS analysis of plastic adherent cells (Fig. 1)
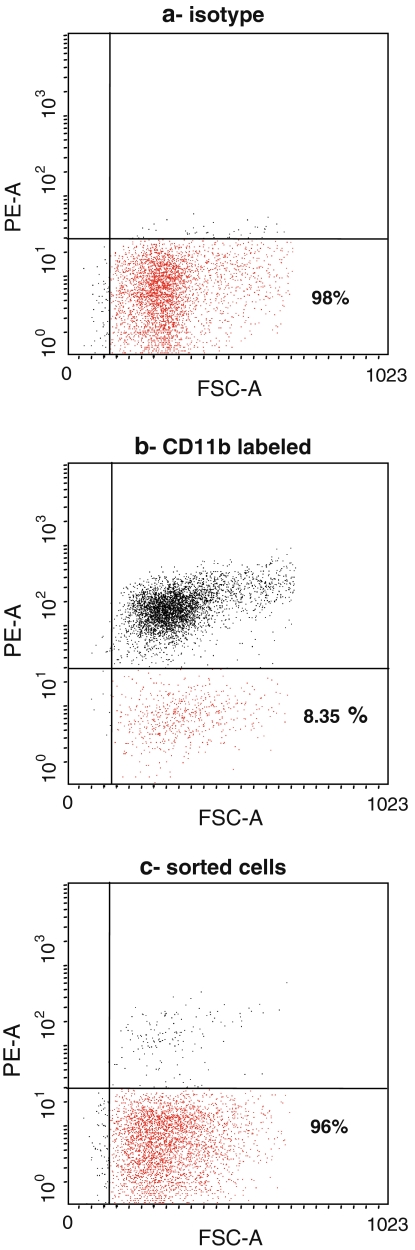
Fig. 1.
FACS analysis of plastic adherent murine marrow cells prior to (a, isotype control and b, CD11b labeled cells) and after immunodepletion (c). CD11b− cells are in the lower area. Only 8.2% of plastic adherent bone marrow cells were CD11b−. This percentage reached 96% after immunodepletion
After 14 days of primary culture, the CD11b− population represented 8.35 % ± 0.72 of the plastic adherent cells. They were clearly distinguishable from positive cells by the fluorescence parameter, allowing CD11b− cell sorting (Fig. 1b). Sorted population was more than 96 % negative for the CD11b (Fig. 1c). Positive and negative populations differed neither in size nor in granularity, according to the FACS analysis.
CD11b− cell population and AlkP
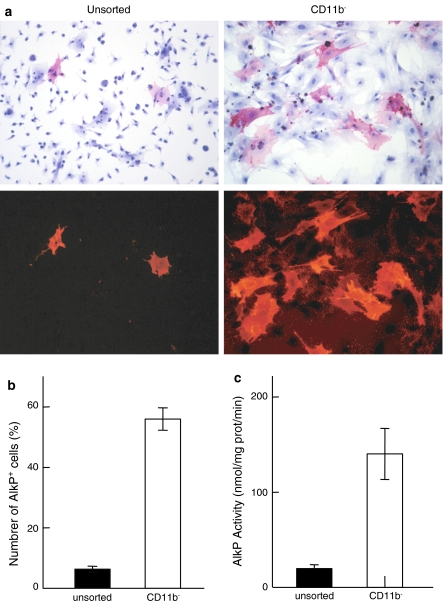
One day after cell sorting and seeding, AlkP cytochemistry showed a marked heterogeneity of the unsorted cell population (Fig. 2a). AlkP+ cells (red stained and fluorescent) were exclusively represented by large flat cells. They were counted and expressed as a percentage of all cells (Fig. 2b). In unsorted cells, 6.3% ± 0.8 were AlkP+. The immunodepletion of CD11b+ cells considerably enriched the population in AlkP+ cells (56% ± 3.3 were AlkP+) and all appeared with a widespread shape. The biochemical AlkP activity was 19.7 ± 3.9 nmol of pNP formed/mg of protein/min for the unsorted cells and was sevenfold increased after immunodepletion (140.2 ± 25.3 for the CD11b− cells) (Fig. 2c). When experiments were repeated, the percentage of CD11b− cells (determinated by FACS) and AlkP+ cells (counted after a 1 day culture) appeared highly correlated (r = 0.908).
Fig. 2.
a Microphotographs of unsorted and CD11b− cells, at day 1 of subculture after cytochemical staining of the AlkP. Same cultures were observed at ×100 magnification under transmitted and fluorescent light. Note the selection of large flattened and AlkP+ cells in the CD11b− population. b Number of AlkP+ cells among the unsorted or CD11b− cell populations, at day 1 of subculture, counted after cytochemical staining. c AlkP activity assay of unsorted and CD11b− cell populations at day 1 of subculture
mRNA expression profile
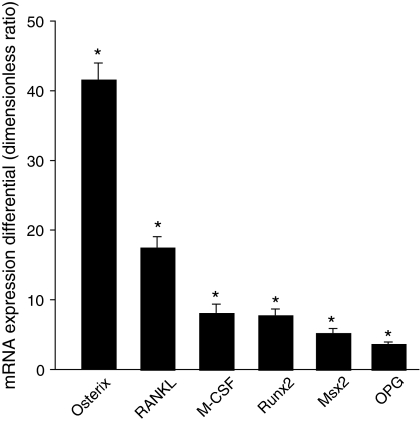
Genes of the osteoblastic differentiation were identified in unsorted and CD11b− cell populations at day 1 (Fig. 3). Expression level of all analyzed genes was higher in the CD11b− population than the unsorted population. Osterix expression was 41-fold increased whereas the one of others transcription factors Runx2 and Msx2 were eight and fivefold increased, respectively. Expression levels of RANKL, M-CSF and OPG, proteins expressed by bone marrow stromal cells and osteoblasts, were significantly increased in the CD11b− population relative to the unsorted population (resp. 17-fold, 8-fold and 4-fold).
Fig. 3.
mRNA expression profile of the CD11b− cells compared to the unsorted population. Values are ratios of mRNA levels (CD11b− cells/unsorted cells) as determined by real-time PCR, averaged ± SEM for three independent experiments. * p < 0.05 from unsorted cells (value = 1)
Functional characterization
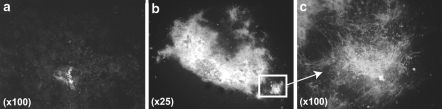
Both unsorted and CD11b− cell populations were seeded at relatively high density and subcultured during 21 days in presence of ascorbic acid and β-glycerophosphate to produce extra cellular mineralization. The unsorted cells remained in layer; nodules were small and scarcely disposed and only a few of them appeared mineralized (Fig. 4a). On the contrary, CD11b− cells tended to aggregate and formed large nodules covering most of the culture surface with prominent mineralized areas (Fig. 4b, c).
Fig. 4.
Calcein labeling of calcium deposits in unsorted (a, at magnification ×100) and CD11b− cell populations (b, at magnification ×25 and c, at magnification ×100), after 21 days of subculture in medium containing ascorbic acid and β-glycerophosphate
Discussion
Cells derived from the marrow stroma exhibit a spectrum of differentiation levels in the osteogenic lineage ranging from MSC, early and late osteoprogenitors to mature osteoblasts, responsible for synthesis, deposition and mineralization of the extracellular matrix of bone (Friedenstein et al. 1987; Owen and Friedenstein 1988; Aubin 1998). Mouse strains, either wild type or genetically altered, provide effective experimental models to study both the cell biology and to evaluate the therapeutic potential of bone marrow-derived cells. Engineering strategies for bone repair exist but technical difficulties of isolating and expanding mouse stromal cells have limited the number of experimentations. Since results depend of the osteogenic properties of used cells (Yoshikawa et al. 2000; Ringe et al. 2002), isolation of progenitors from murine bone marrow must be investigated.
This study describes an easy and reliable method to enrich murine MSC in osteoprogenitors. CD11b immunodepletion allowed the selection of AlkP+ cells. CD11b is a cell adhesion molecule (CAM) which mediates signals regulating intracellular processes (Hebert 2000; Shur et al. 2005). It is in majority expressed on granulo-monocytic cells but is also described at a relatively low level on murine cells from the marrow stromal cell line MBA-15 (Marom et al. 2005). Previous studies have shown that most round cells in murine plastic adherent cultures express CD11b (Phinney et al. 1999). These cells were identified as lymphohematopoietic cells according to their co-expression of CD45. CD11b has been used for depletion of granulo-monocytic cells from C57Bl/6 mouse adherent bone marrow cells (Tropel et al. 2004). The resultant fibroblastic cell population CD11b− was subsequently grown on fibronectin-coated dishes in presence of growth factors and exhibited a MSC phenotype according to their differentiation along osteoblastic, adipocytic and chondrocytic pathways. The cell isolation strategy developed in the present study was less sophisticated. No additional growth factor known to increase the immunogenic properties of cells (Kruyt et al. 2004) was used and the experimental cost was reduced. Others have used CAMs as surface antigen to select fibroblastic cells, notably the CD49a protein (α1-integrin subunit) (Gindraux et al. 2007; Rider et al. 2007). But CD49a+ cells from murine bone marrow appeared contaminated by CD45+ hematopoietic cells. CD11b appears to be a more powerful CAM for discrimination of MSC and hematopoietic cells. This marker can be used after cell proliferation in culture while most known CD markers are only useful for selection before plating the cells. Other methods used to discard hematopoietic cells such as leucine methyl ester (LME-a lysosomotropic agent) followed by Percoll gradient were efficient to purify multipotent progenitors (Seshi et al. 2000). The use of more differentiated osteoprogenitor cells seems more relevant in bone engineering strategies.
In the present study, selection of CD11b− cells showed significant enrichment in AlkP+ cells since AlkP was not expressed by MSC. Its expression is associated with osteoblastic differentiation and formation of the bone extracellular matrix (Owen et al. 1990; Aubin 1998). AlkP+ cells formed mineralized nodules in culture without addition of dexamethasone. Early and late osteoprogenitors respond differently to exogenous glucocorticoid stimulation. Early AlkP− progenitors are dependent of dexamethasone whereas mature AlkP+ osteoprogenitors form mineralized nodules in vitro without dexamethasone (Turksen and Aubin 1991; Aubin 1999; Atmani et al. 2003). Our results in C57Bl/6 mice confirmed the bone forming capacities of CD11b− AlkP+ bone marrow adherent cells without dexamethasone in vitro. Because they clearly proliferate in culture, CD11b− AlkP+ cells were not osteoblasts but mature osteoprogenitors. The osteoblast characteristics of the CD11b− cells were also confirmed by the higher expression levels in transcription factors and proteins of osteoblast differentiation, compared to the unsorted population. The expression level of Osterix was more increased than that of Runx2. These findings evidence that CD11b− cells are mature osteoprogenitors because Osterix is expressed after Runx2 during osteoblast differentiation (Komori 2006). In primary cultures of osteoblastic cells, glucocorticoids reduce the number of bone forming cells by decreasing cell replication and preventing terminal differentiation into mature functioning osteoblasts. In addition, glucocorticoids enhance apoptosis of mature osteoblasts (Pereira et al. 2001; Canalis and Delany 2002). Addition of 10−8 M dexamethasone in the culture medium was evaluated and our results confirmed the well known inhibitory actions of glucocorticoids: dexamethasone did not enhance bone nodule formation in the unsorted cells cultures and induced CD11b− cells death (data not shown). The differentiation of CD11b− cells into other lineages was not investigated.
Limitation in the use of such isolated osteoprogenitors for bone forming experiments is their low frequency in the plastic adherent fraction after 14 days of primary culture. In the present work, the fraction of CD11b+ cells constituted 91.7% of the adherent cell population; nonhematopoietic cells represented only 8.3%. The contamination rate vary extensively between bone marrow cultures of different mouse strains, influencing their proliferation rate and differentiation potentials (Peister et al. 2004). Phinney et al. (1999) have compared heterogeneity of plastic adherent culture from five mouse strains. Culture from bone marrow of C57BL/6 mice was the most contaminated by CD11b+ and CD45+ cells. The yield of the CD11b− immunodepletion could be enhanced with another mouse strain, like FVB/N whose non hematopoietic cells constitute up to 20% of the adherent cell population.
In conclusion, mature osteoprogenitors can be isolated from murine plastic adherent bone marrow cells by negative selection using the surface antigen CD11b. The negative fraction was greatly enriched in osteogenic cells that were able to form mineralized nodules without dexamethasone stimulation. This immunodepletion method provided an osteogenic cell population, suitable for bone engineering experiments.
Acknowledgments
This work was made possible by grants from Contrat de Plan Etat—Région “Pays de la Loire”, INSERM and the Bioregos Program. We thank Laurence Preisser and Marie-Hélène Guilleux from SCCAN Service Commun de Cytométrie et d’Analyses Nucléotidiques, IFR132 for their assistance with RT-PCR quantification.
Abbreviations
- AlkP
Alkaline phosphatase
- MSC
Mesenchymal stem cells
- FACS
Fluorescent activating cell sorting
- CAM
Cell adhesion molecule
References
- Atmani H, Chappard D, Baslé MF (2003) Proliferation and differentiation of osteoblasts and adipocytes in rat bone marrow stromal cell cultures: effects of dexamethasone and calcitriol. J Cell Biochem 89:364–372. doi:10.1002/jcb.10507 [DOI] [PubMed]
- Aubin JE (1998) Bone stem cells. J Cell Biochem Suppl 30–31:73–82. doi:10.1002/(SICI)1097-4644(1998)72:30/31+<73::AID-JCB11>3.0.CO;2-L [DOI] [PubMed]
- Aubin JE (1999) Osteoprogenitor cell frequency in rat bone marrow stromal populations: role for heterotypic cell–cell interactions in osteoblast differentiation. J Cell Biochem 72:396–410. doi:10.1002/(SICI)1097-4644(19990301)72:3<396::AID-JCB9>3.0.CO;2-6 [DOI] [PubMed]
- Baddoo M, Hill K, Wilkinson R et al (2003) Characterization of mesenchymal stem cells isolated from murine bone marrow by negative selection. J Cell Biochem 89:1235–1249. doi:10.1002/jcb.10594 [DOI] [PubMed]
- Bianco P, Gehron Robey P (2000) Marrow stromal stem cells. J Clin Invest 105:1663–1668. doi:10.1172/JCI10413 [DOI] [PMC free article] [PubMed]
- Bianco P, Riminucci M, Gronthos S et al (2001) Bone marrow stromal stem cells: nature, biology, and potential applications. Stem Cells 19:180–192. doi:10.1634/stemcells.19-3-180 [DOI] [PubMed]
- Canalis E, Delany AM (2002) Mechanisms of glucocorticoid action in bone. Ann N Y Acad Sci 966:73–81 [DOI] [PubMed]
- Dennis JE, Esterly K, Awadallah A et al (2007) Clinical-scale expansion of a mixed population of bone-marrow-derived stem and progenitor cells for potential use in bone-tissue regeneration. Stem Cells 25:2575–2582. doi:10.1634/stemcells.2007-0204 [DOI] [PubMed]
- Friedenstein AJ, Chailakhyan RK, Gerasimov UV (1987) Bone marrow osteogenic stem cells: in vitro cultivation and transplantation in diffusion chambers. Cell Tissue Kinet 20:263–272 [DOI] [PubMed]
- Gindraux F, Selmani Z, Obert L et al (2007) Human and rodent bone marrow mesenchymal stem cells that express primitive stem cell markers can be directly enriched by using the CD49a molecule. Cell Tissue Res 327:471–483. doi:10.1007/s00441-006-0292-3 [DOI] [PubMed]
- Hebert E (2000) Endogenous lectins as cell surface transducers. Biosci Rep 20:213–237. doi:10.1023/A:1026484722248 [DOI] [PubMed]
- Hernigou P, Poignard A, Beaujean F et al (2005) Percutaneous autologous bone-marrow grafting for nonunions. Influence of the number and concentration of progenitor cells. J Bone Joint Surg Am 87:1430–1437. doi:10.2106/JBJS.D.02215 [DOI] [PubMed]
- Kadiyala S, Young RG, Thiede MA et al (1997) Culture expanded canine mesenchymal stem cells possess osteochondrogenic potential in vivo and in vitro. Cell Transplant 6:125–134. doi:10.1016/S0963-6897(96)00279-5 [DOI] [PubMed]
- Kassem M (2004) Mesenchymal stem cells: biological characteristics and potential clinical applications. Cloning Stem Cells 6:369–374. doi:10.1089/clo.2004.6.369 [DOI] [PubMed]
- Komori T (2006) Regulation of osteoblast differentiation by transcription factors. J Cell Biochem 99:1233–1239. doi:10.1002/jcb.20958 [DOI] [PubMed]
- Kruyt MC, Dhert WJ, Oner C et al (2004) Optimization of bone-tissue engineering in goats. J Biomed Mater Res 69B:113–120. doi:10.1002/jbm.b.10073 [DOI] [PubMed]
- Marom R, Shur I, Solomon R et al (2005) Characterization of adhesion and differentiation markers of osteogenic marrow stromal cells. J Cell Physiol 202:41–48. doi:10.1002/jcp.20109 [DOI] [PubMed]
- Muschler GF, Nakamoto C, Griffith LG (2004) Engineering principles of clinical cell-based tissue engineering. J Bone Joint Surg Am 86-A:1541–1558 [DOI] [PubMed]
- Ohgushi H, Caplan AI (1999) Stem cell technology and bioceramics: from cell to gene engineering. J Biomed Mater Res 48:913–927. doi:10.1002/(SICI)1097-4636(1999)48:6<913::AID-JBM22>3.0.CO;2-0 [DOI] [PubMed]
- Owen M, Friedenstein AJ (1988) Stromal stem cells: marrow-derived osteogenic precursors. Ciba Found Symp 136:42–60 [DOI] [PubMed]
- Owen TA, Aronow M, Shalhoub V et al (1990) Progressive development of the rat osteoblast phenotype in vitro: reciprocal relationships in expression of genes associated with osteoblast proliferation and differentiation during formation of the bone extracellular matrix. J Cell Physiol 143:420–430. doi:10.1002/jcp.1041430304 [DOI] [PubMed]
- Peister A, Mellad JA, Larson BL et al (2004) Adult stem cells from bone marrow (MSCs) isolated from different strains of inbred mice vary in surface epitopes, rates of proliferation, and differentiation potential. Blood 103:1662–1668. doi:10.1182/blood-2003-09-3070 [DOI] [PubMed]
- Pereira RM, Delany AM, Canalis E (2001) Cortisol inhibits the differentiation and apoptosis of osteoblasts in culture. Bone 28:484–490. doi:10.1016/S8756-3282(01)00422-7 [DOI] [PubMed]
- Phinney DG, Kopen G, Isaacson RL et al (1999) Plastic adherent stromal cells from the bone marrow of commonly used strains of inbred mice: variations in yield, growth, and differentiation. J Cell Biochem 72:570–585. doi:10.1002/(SICI)1097-4644(19990315)72:4<570::AID-JCB12>3.0.CO;2-W [DOI] [PubMed]
- Prockop DJ (1997) Marrow stromal cells as stem cells for nonhematopoietic tissues. Science 276:71–74. doi:10.1126/science.276.5309.71 [DOI] [PubMed]
- Rider DA, Nalathamby T, Nurcombe V et al (2007) Selection using the alpha-1 integrin (CD49a) enhances the multipotentiality of the mesenchymal stem cell population from heterogeneous bone marrow stromal cells. J Mol Histol 38:449–458. doi:10.1007/s10735-007-9128-z [DOI] [PubMed]
- Ringe J, Kaps C, Burmester GR et al (2002) Stem cells for regenerative medicine: advances in the engineering of tissues and organs. Naturwissenschaften 89:338–351. doi:10.1007/s00114-002-0344-9 [DOI] [PubMed]
- Seshi B, Kumar S, Sellers D (2000) Human bone marrow stromal cell: coexpression of markers specific for multiple mesenchymal cell lineages. Blood Cells Mol Dis 26:234–246. doi:10.1006/bcmd.2000.0301 [DOI] [PubMed]
- Shiota M, Heike T, Haruyama M et al (2007) Isolation and characterization of bone marrow-derived mesenchymal progenitor cells with myogenic and neuronal properties. Exp Cell Res 313:1008–1023. doi:10.1016/j.yexcr.2006.12.017 [DOI] [PubMed]
- Shur I, Zilberman M, Benayahu D et al (2005) Adhesion molecule expression by osteogenic cells cultured on various biodegradable scaffolds. J Biomed Mater Res A 75:870–876. doi:10.1002/jbm.a.30507 [DOI] [PubMed]
- Stewart M, Thiel M, Hogg N (1995) Leukocyte integrins. Curr Opin Cell Biol 7:690–696. doi:10.1016/0955-0674(95)80111-1 [DOI] [PubMed]
- Tropel P, Noel D, Platet N et al (2004) Isolation and characterisation of mesenchymal stem cells from adult mouse bone marrow. Exp Cell Res 295:395–406. doi:10.1016/j.yexcr.2003.12.030 [DOI] [PubMed]
- Turksen K, Aubin JE (1991) Positive and negative immunoselection for enrichment of two classes of osteoprogenitor cells. J Cell Biol 114:373–384. doi:10.1083/jcb.114.2.373 [DOI] [PMC free article] [PubMed]
- Van Vlasselaer P, Falla N, Snoeck H et al (1994) Characterization and purification of osteogenic cells from murine bone marrow by two-color cell sorting using anti-Sca-1 monoclonal antibody and wheat germ agglutinin. Blood 84:753–763 [PubMed]
- Werntz JR, Lane JM, Burstein AH et al (1996) Qualitative and quantitative analysis of orthotopic bone regeneration by marrow. J Orthop Res 14:85–93. doi:10.1002/jor.1100140115 [DOI] [PubMed]
- Yoshikawa T, Ohgushi H, Nakajima H et al (2000) In vivo osteogenic durability of cultured bone in porous ceramics: a novel method for autogenous bone graft substitution. Transplantation 69:128–134. doi:10.1097/00007890-200001150-00022 [DOI] [PubMed]