Abstract
We report that orotidine 5′-monophosphate decarboxylase (OMPDC) catalyzes exchange of the C-6 proton of uridine 5′-monophosphate (UMP) for deuterium from solvent in D2O at 25 °C and pD 7.0 – 9.3. Kinetic analysis of deuterium exchange gives pKa ≤ 22 for carbon deprotonation of enzyme-bound UMP, which is at least 10 units lower than that for deprotonation of an analog of UMP in water. The observation of enzyme-catalyzed deuterium exchange via a stabilized carbanion provides convincing evidence for the decarboxylation of orotidine 5′-monophosphate (OMP) by OMPDC to give the same carbanion intermediate. The data show that yeast OMPDC stabilizes the bound vinyl carbanion by at least 14 kcal/mol. We conclude that OMPDC also provides substantial stabilization of the late carbanion-like transition state for the decarboxylation of OMP, and that this transition state stabilization constitutes a large fraction, but probably not all, of the enormous 1017-fold enzymatic rate acceleration.
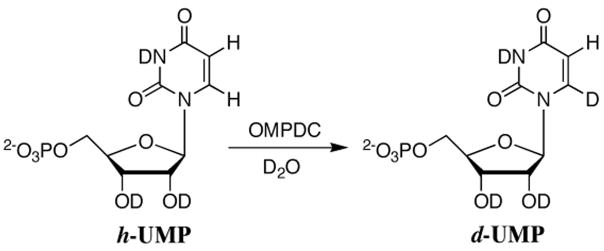
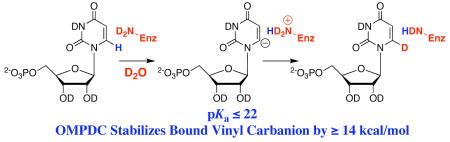
We report that orotidine 5′-monophosphate decarboxylase catalyzes exchange of the C-6 proton of uridine 5′-monophosphate (UMP) for deuterium from solvent in D2O at 25 °C (Scheme 1). Kinetic analysis of deuterium exchange gives pKa ≤ 22 for carbon deprotonation of enzyme-bound UMP, which is at least 10 units lower than that for deprotonation of an analog of UMP in water.
Scheme 1.
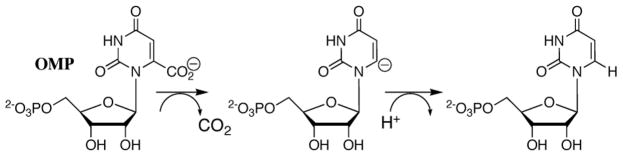
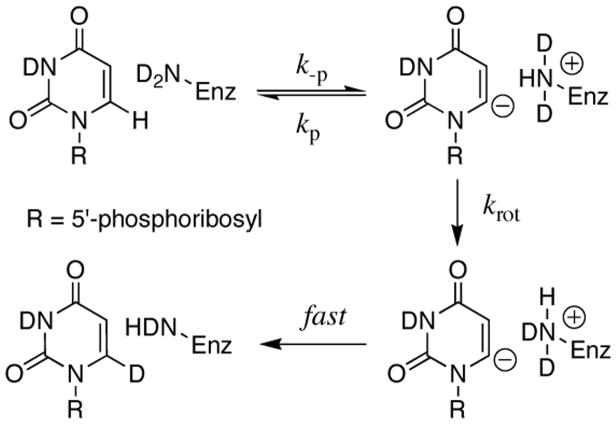
Orotidine 5′-monophosphate decarboxylase (OMPDC) employs no metal ions or other cofactors but yet effects an enormous 1017-fold acceleration of the decarboxylation of orotidine 5′-monophosphate (OMP) to give uridine 5′-monophosphate (UMP).1 The X-ray structure of the yeast enzyme liganded with 6-hydroxyuridine 5′-monophosphate provides strong evidence that the C-6 proton of the product UMP is derived from the terminal NH3+ group of Lys-93.2 The product isotope effect of unity for OMPDC-catalyzed decarboxylation of OMP in 50/50 (v/v) H2O/D2O eliminates a mechanism3 in which proton transfer from Lys-93 to C-6 provides electrophilic push to the loss of CO2 in a concerted reaction.4 This result also provides strong evidence for the formation of a short-lived enzyme-bound carbanion intermediate that shows no discrimination between H and D in the proton transfer step (Scheme 2).4
Scheme 2.
The very large kinetic barrier to the nonenzymatic decarboxylation of OMP (t1/2 = 78 million years)5 arises mainly from the thermodynamic barrier to formation of the highly unstable C-6 vinyl carbanion. This activation barrier may be reduced by interactions with OMPDC that either destabilize bound OMP relative to the bound carbanion intermediate, or by interactions that stabilize the bound carbanion intermediate relative to bound OMP.6 Computational studies support the proposal that binding of OMP to OMPDC induces either electrostatic stress between the protein and the bound substrate in the ground state Michaelis complex,7 or conformational stress in the protein at this complex,8 and that this stress is relieved in the transition state for enzyme-catalyzed decarboxylation. However, other calculations suggest that the enzymatic rate acceleration is due mainly to stabilization of the transition state for decarboxylation.9 These results are difficult to evaluate because there are few experiments that address whether the rate acceleration for OMPDC is due mainly to ground state destabilization,10 to transition state stabilization, or to both effects.
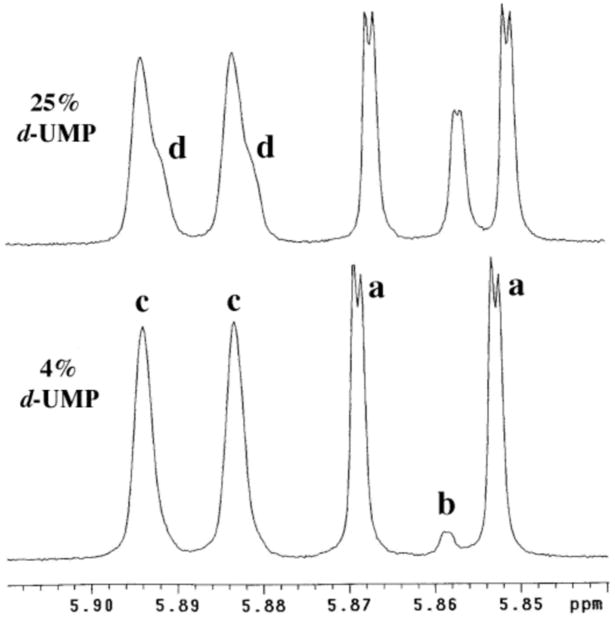
The exchange for deuterium of the C-6 proton of [6-1H]-uridine 5′-monophosphate (h-UMP) to give [6-2H]-uridine 5′-monophosphate (d-UMP) catalyzed by OMPDC from S. cerevisiae (C155S mutant) in D2O (Scheme 1) was monitored by 1H NMR spectroscopy at 500 MHz. Figure 1 shows partial 1H NMR spectra in the region of the anomeric and C-5 protons of recovered UMP ([UMP]total = 2.5 mM) obtained during deuterium exchange at 25 °C catalyzed by OMPDC (0.11 mM, 3.2 mg/mL, monitored for 7 days) in D2O buffered by 100 mM glycylglycine at pD 9.34 (I = 0.1, NaCl). Deuterium exchange results in the disappearance of the double doublet (a) at 5.860 ppm due to the C-5 proton of h-UMP and the appearance of an upfield-shifted broad doublet (b) at 5.857 ppm (Δδ ≈ 0.003 ppm) due to the C-5 proton of d-UMP. The fractional extent of deuterium exchange was obtained from the integrated area of the two downfield peaks of the double doublet due to the C-5 proton of h-UMP (AH) and the combined integrated areas of the upfield peaks of this signal and that of the intervening broad doublet due to the C-5 proton of d-UMP (AD+H), according to eq 1. The observed first-order rate constant for deuterium exchange into UMP, kobsd = 4.90 × 10−7 s−1, was determined from the slope of a semi-logarithmic plot of the fraction of h-UMP remaining, {1 - f(d-UMP)}, against time.
Figure 1.
Partial 1H NMR spectra (500 MHz) of recovered UMP obtained during exchange for deuterium of the C-6 proton catalyzed by OMPDC (0.11 mM) in D2O at pD 9.34 and 25 °C (I = 0.1, NaCl). Disappearance of the double doublet (a) due to the C-5 proton of h-UMP at 5.860 ppm (J = 8.1, 0.5 Hz, coupled to the C-6 and anomeric protons) is accompanied by the appearance of a upfield-shifted broad doublet (b) due to the C-5 proton of d-UMP at 5.857 ppm (J ≈ 0.5 Hz, coupled to the anomeric proton). The broad doublet (c) due to the anomeric proton of h-UMP at 5.889 ppm (J = 5.3 Hz, coupled to the C-2′ proton) exhibits “shoulders” (d) due to the slightly upfield-shifted doublet for the anomeric proton of d-UMP.
| (1) |
The values of kobsd (s−1) determined for enzyme-catalyzed deuterium exchange in D2O at pD 9.34 with [UMP]total = 2.5 – 10 mM show a good fit to eq 2 that was derived for Scheme 3 (see Supporting Information), with Kd ≪ [UMP]total. The data give the first-order rate constant for deuterium exchange into saturating enzyme-bound UMP at pD 9.34 as kex = 1.15 × 10−5 s−1. Similar experiments using ca. 0.3 mM OMPDC (9 mg/mL) gave values of kex (s−1) for the turnover of saturating UMP (2.5 – 5 mM) at pD 8.13 (100 mM glycylglycine buffer), and at pD 7.58 and 7.03 (48 mM imidazole buffer), at 25 °C and I = 0.1 (NaCl).11
Scheme 3.
| (2) |
Figure 2 shows the pD-rate profile of the values of kex (s−1) for turnover of enzyme-bound h-UMP to give d-UMP by yeast OMPDC in D2O at 25 °C and I = 0.1 (NaCl). The large increase in kex (s−1) with increasing pD and the leveling off at pD > 8 shows that deuterium exchange is promoted by the basic form of an amino acid side chain at the active site of OMPDC.12 We suggest that the catalytic base is the neutral form of Lys-93,13 so that deuterium exchange arises from the reverse of the proton transfer “half reaction” that occurs in the active site during the physiological decarboxylation of OMP to give UMP (Scheme 2). Analysis of the data in Figure 2 gives p(Ka)Lys = 8.0 for the catalytic base in D2O at I = 0.1, and (kex)max = 1.24 × 10−5 s−1 for proton transfer from bound h-UMP to the neutral side chain of Lys-93 to give the enzyme-bound vinyl carbanion (Scheme 4).
Figure 2.
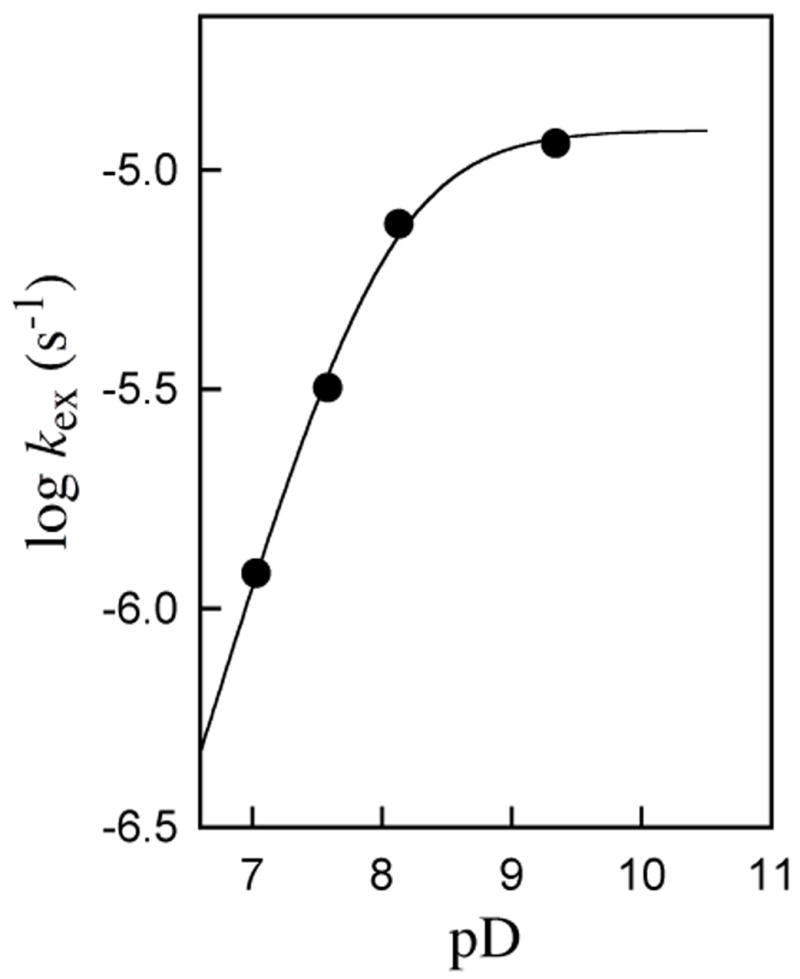
pD-Rate profile of kex (s−1) for turnover of enzyme-bound h-UMP to give d-UMP by OMPDC from Saccharomcyes cerevisiae (C155S mutant) in D2O at 25 °C and I = 0.1 (NaCl). The solid line shows the calculated profile for a catalytic base of pKa = 8.0.
Scheme 4.
We have shown that proton transfer from protonated Lys-93 to the vinyl carbanion must be faster than any molecular motion that exchanges the positions of the N-L+ hydrons of Lys-93, so that kp ≫ krot (Scheme 4).4 Therefore, the observed deuterium exchange reaction consists of the pre-equilibrium reversible chemical step of proton transfer from UMP to Lys-93, followed by the occasional rotation of the terminal CH2-ND2H+ group of Lys-93 into a position to deliver a deuteron to the vinyl carbanion (krot, Scheme 4). The CH2-NH3+ group of Lys-93 is hydrogen-bonded to the carboxylate groups of Asp-91 and Asp-96,2 and the barrier to CH2-ND2H+ bond rotation and hydron exchange is expected to be at least 5 kcal/mol, so that krot ≤ 109 s−1.14 The limit of krot ≤ 109 s−1 can be substituted into eq 3, derived for the mechanism shown in Scheme 4, with (kex)max = 1.24 × 10−5 s−1 and (Ka)Lys = 10−8 M to give p(Ka)UMP ≤ 22 for the C-6 proton of enzyme-bound UMP.
| (3) |
The observation of enzyme-catalyzed deuterium exchange via formation of a stabilized carbanion provides convincing evidence for decarboxylation of OMP by yeast OMPDC to give the same carbanion.15 The value of pKa ≤ 22 for the C-6 proton of enzyme-bound UMP determined here is at least 10 units lower than the estimated values of pKa = 30 – 34 for the C-6 proton of 1,3-dimethyluracil in water.16–18 Therefore, yeast OMPDC stabilizes the bound vinyl carbanion by at least 14 kcal/mol. We conclude that OMPDC also provides substantial stabilization of the late carbanion-like transition state for the decarboxylation of OMP, and that this transition state stabilization constitutes a large fraction, but probably not the entire, enzymatic rate acceleration. Further experimental studies directed at elucidating the origin of the transition state stabilization for OMPDC will provide insight into its so far unexplained extraordinary catalytic power.
Supplementary Material
Experimental procedures, kinetic analysis, semi-logarithmic plots, table of values of kobsd and kex.
Acknowledgments
We acknowledge the National Institutes of Health (Grant GM39754 to J.P.R. and Grant GM65155 to J.A.G.) for generous support of this work.
References
- 1.Miller BG, Wolfenden R. Annu Rev Biochem. 2002;71:847–885. doi: 10.1146/annurev.biochem.71.110601.135446. [DOI] [PubMed] [Google Scholar]
- 2.Miller BG, Hassell AM, Wolfenden R, Milburn MV, Short SA. Proc Natl Acad Sci USA. 2000;97:2011–2016. doi: 10.1073/pnas.030409797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Appleby TC, Kinsland C, Begley TP, Ealick SE. Proc Natl Acad Sci USA. 2000;97:2005–2010. doi: 10.1073/pnas.259441296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toth K, Amyes TL, Wood BM, Chan K, Gerlt JA, Richard JP. J Am Chem Soc. 2007;129:12946–12947. doi: 10.1021/ja076222f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Radzicka A, Wolfenden R. Science. 1995;267:90–93. doi: 10.1126/science.7809611. [DOI] [PubMed] [Google Scholar]
- 6.Jencks WP. Adv Enzymol Relat Areas Mol Biol. 1975;43:219–410. doi: 10.1002/9780470122884.ch4. [DOI] [PubMed] [Google Scholar]
- 7.Wu N, Mo Y, Gao J, Pai EF. Proc Natl Acad Sci USA. 2000;97:2017–2022. doi: 10.1073/pnas.050417797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao J. Curr Opin Struct Biol. 2003;13:184–192. doi: 10.1016/s0959-440x(03)00041-1. [DOI] [PubMed] [Google Scholar]
- 9.Warshel A, Strajbl M, Villa J, Florian J. Biochemistry. 2000;39:14728–14738. doi: 10.1021/bi000987h. [DOI] [PubMed] [Google Scholar]
- 10.Callahan BP, Wolfenden R. J Am Chem Soc. 2004;126:14698–14699. doi: 10.1021/ja0450049. [DOI] [PubMed] [Google Scholar]
- 11.The reported failure to detect deuterium exchange in an earlier study is likely due to the use of only a small amount of enzyme and a relatively short reaction time [ Smiley JA, DelFraino BJ, Simpson BA. Arch Biochem Biophys. 2003;412:267–271. doi: 10.1016/s0003-9861(03)00062-6.
- 12.The exchange reaction does not occur in the presence of a saturating amount of the competitive inhibitor 6-aza-uridine 5′-monophosphate.
- 13.The pH-dependence of kcat for turnover of OMP indicates the involvement of the acidic form of a catalytic residue of pKa = 8.8 [ Porter DJT, Short SA. Biochemistry. 2000;39:11788–11800. doi: 10.1021/bi001199v.
- 14.krot = 1011 s−1 for unhindered rotation within an ion pair in water [ Richard JP, Tsuji Y. J Am Chem Soc. 2000;122:3963–3964.
- 15.We have observed that deuterium exchange into UMP is also catalyzed by the OMPDCs from Methanobacterium thermoautotrophicum and Escherichia coli.
- 16.Sievers A, Wolfenden R. J Am Chem Soc. 2002;124:13986–13987. doi: 10.1021/ja021073g. [DOI] [PubMed] [Google Scholar]
- 17.Wong FM, Capule CC, Wu W. Org Lett. 2006;8:6019–6022. doi: 10.1021/ol0624981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeoh FY, Cuasito RR, Capule CC, Wong FM, Wu W. Bioorg Chem. 2007;35:338–343. doi: 10.1016/j.bioorg.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Experimental procedures, kinetic analysis, semi-logarithmic plots, table of values of kobsd and kex.