14-3-3 proteins are a highly conserved family of phosphoserine binding proteins found in all eukaryotes. The name “14-3-3” comes from the specific ion exchange chromatography elution profiles and starch gel electrophoresis migration patterns of a group of proteins originally isolated from mammalian brain tissue. 14-3-3 proteins play key roles in many diverse physiological processes that involve regulation by phosphorylation. A number of enzymes involved in fundamental processes of plant physiology are regulated in part via interactions with 14-3-3s, including nitrate reductase (NR), sucrose phosphate synthase, starch synthase, glutamate synthase, ATP synthase, and ascorbate peroxidase (Finnie et al., 1999; DeLille et al., 2001). In mammals, 14-3-3s also have been found to be involved in the regulation of cell cycle progression (Liao and Omary, 1996) and cell proliferation and differentiation (Freed et al., 1994) via interactions with regulators including kinases, phosphatases, and transcription factors.
14-3-3 PROTEINS AND THE REGULATION OF TRANSCRIPTION
14-3-3 target binding proteins often are key members of signal transduction cascades or complexes, and they have been found to interact with transcription factors and/or other proteins that affect transcription. For example, in mammals and yeast, interaction with 14-3-3 proteins appears to be critical for the function of Raf-1 kinase, which activates a mitogen-activated protein kinase cascade that mediates the transcription of genes involved in mitogenesis and cell differentiation (Aitken, 1996), and of histone deacetylases that interact with transcription factors to repress gene expression (Grozinger and Schreiber, 2000).
14-3-3 proteins have been found to bind specifically with TATA box binding proteins from plants as well as other eukaryotes and to activate GAL4-dependent β-glucuronidase reporter gene expression when translationally fused with the GAL4 DNA binding domain in a plant transient expression system (Pan et al., 1999). 14-3-3 proteins also have been found to interact with the transcription factors involved in abscisic acid signaling in plants. Schultz et al. (1998) found that 14-3-3 proteins interact with the basic leu-cine zipper factor EmBP1 and with VIVIPAROUS1, both of which activate Em gene expression via binding to the abscisic acid response element Em1a in the Em promoter.
14-3-3 MODE OF ACTION
Crystal structure analysis and binding studies have shown that 14-3-3s are sequence-specific phosphoserine binding proteins that function as homodimers or heterodimers (Xiao et al., 1995; Muslin et al., 1996). The most highly conserved regions of the polypeptides form a large negatively charged channel at the dimerization interface, which provides the binding site for target phosphoproteins. 14-3-3s are not involved in simple phosphorylation/dephosphorylation reactions of target proteins; rather, they appear to interact with their phosphoserine targets in a way that alters the stability, activity, and/or localization of the target protein within the cell.
Binding of 14-3-3 proteins sometimes has been shown to influence the proteolysis or stability of the target protein. For example, binding of 14-3-3 proteins does not appear to activate Raf-1 directly; rather, it may act to stabilize an activatable conformation of Raf-1 (Morrison, 1994). Morrison (1994) and Muslin et al. (1996) proposed that 14-3-3s function as chaperones that stabilize a particular (i.e., activatable) conformation of Raf-1 and other 14-3-3 target proteins. Cotelle et al. (2000) found that selective proteolytic cleavage of a number of 14-3-3 target proteins, including NR, sucrose phosphate synthase, and a calcium-dependent protein kinase, coincided with the loss of 14-3-3 binding in sugar-starved Arabidopsis cells. Extracts from sugar-starved cells were found to contain proteolytic activity against 14-3-3 target proteins, which could be blocked in vitro by their binding to 14-3-3s. Another example from plants is the well-studied interaction of 14-3-3 proteins with the plasma membrane H+-ATPase. This interaction leads to the formation of an activated complex in which the H+-ATPase exhibits enhanced activity (reviewed by Morsomme and Boutry, 2000). Malerba and Bianchetti (2001) found that the interaction with 14-3-3 proteins protects the H+-ATPase from specific proteolysis of the regulatory domain.
Interactions with 14-3-3 proteins also have been found to alter the intracellular localization of certain target proteins. May and Soll (2000) showed that 14-3-3 proteins are involved in the translocation of nucleus-encoded chloroplast precursor proteins into the chloroplast. Grozinger and Schreiber (2000) found that interactions of the human histone deacetylases HDAC4 and HDAC5 with 14-3-3s leads to the localization of these proteins in the cytosol, whereas the loss of this interaction leads to their translocation into the nucleus. Transcription in eukaryotes is regulated in part by the acetylation and deacetylation of histones. In the nucleus, HDAC4 and HDAC5 interact with HDAC3 and with transcription factors to repress gene expression. Zhao et al. (2001) found that the intracellular localization of HDAC4 is regulated by sequential phosphorylation steps. First, phosphorylation by an unidentified protein kinase promotes 14-3-3 binding to HDAC4, and subsequently the activity of other kinases facilitates nuclear export of the HDAC4–14-3-3 complex. Similarly, the association with 14-3-3s promotes cytoplasmic as opposed to nuclear localization of the protein phosphatase Cdc25, which has a role in the regulation of the cell cycle in Xenopus (Kumagai and Dunphy, 1999).
14-3-3 NUCLEAR SHUTTLE
In this issue of The Plant Cell, Igarashi et al. (pages 2483–2497) show that 14-3-3 proteins regulate the intracellular localization of REPRESSION OF SHOOT GROWTH (RSG), a transcription factor from tobacco that controls the expression of a gibberellin biosynthesis enzyme. RSG is expressed ubiquitously in plant organs, suggesting that some form of post-transcriptional or post-translational modification controls its activity (Fukazawa et al., 2000). A mutant version of RSG that had lost the ability to bind 14-3-3s, as a result of a single amino acid change of serine residue 114 to alanine (RSG-S114A), was localized exclusively in the nucleus (Figure 1) and exhibited higher transcriptional activation capability than wild-type RSG in transient expression assays in tobacco protoplasts. Similar results were observed with wild-type or mutant RSG-S114A constructs fused with green fluorescent protein (GFP) and expressed in transgenic tobacco plants.
Figure 1.
Effects of the S114A Mutation on the Intracellular Localization of RSG.
Tobacco mesophyll protoplast transfected with plasmids encoding the RSG-S114A mutant:GFP fusion protein. GFP fluorescence (green) from the RSG-S114A:GFP fusion protein shows that the mutant protein is localized exclusively in the nucleus. Fluorescence from a wild-type RSG:GFP fusion protein is present throughout the cytoplasm (see Igarashi et al., 2001, in this issue). The S114A mutation disrupts the binding of RSG to 14-3-3 proteins, suggesting that 14-3-3 proteins are involved in controlling the intracellular localization of RSG. (Figure courtesy of Yohsuke Takahashi.)
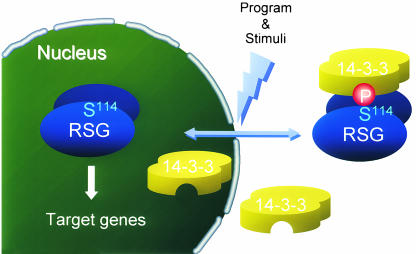
Igarashi et al. further found that wild-type RSG is not localized statically in the cytosol but is capable of shuttling in and out of the nucleus. This was shown in experiments using leptomycin B, an inhibitor of nuclear export that acts by inhibiting Crm1/Exportin1, a receptor that mediates the nuclear export of proteins carrying a nuclear export sequence. Treatment of leaf epidermal cells with leptomycin B caused rapid relocalization of RSG from the cytosol to the nucleus, suggesting that RSG normally continually shuttles between the nucleus and the cytoplasm. The results suggest that 14-3-3 proteins are involved in the regulation of RSG transport into and/or out of the nucleus and that this regulation may play a key role in controlling the activity of RSG as a transcriptional activator and regulator of gibberellic acid (GA) biosynthesis (Figure 2).
Figure 2.
Scheme of 14-3-3 Regulation of the Intracellular Localization of RSG.
RSG protein continually shuttles back and forth between the nucleus and the cytoplasm. Binding of 14-3-3 proteins to the phosphorylated serine 114 residue of the RSG protein inhibits nuclear import and/or enhances nuclear export to shift the equilibrium of RSG to a predominantly cytoplasmic localization. Inhibition of 14-3-3 binding leads to the rapid redistribution of RSG to the nucleus, where it activates transcription of a GA biosynthetic gene encoding ent-kaurene oxidase. (Figure courtesy of Yohsuke Takahashi.)
This model leads to the hypothesis that inducers of GA biosynthesis should have a direct or indirect effect on the phosphorylation status and 14-3-3 binding capacity of RSG. Thus, the investigation of 14-3-3 binding properties might reveal more details of the regulation of GA levels in plants.
Interestingly, a comparable mode of action was found for the interaction of 14-3-3 proteins with the Cdc25 protein in Xenopus cells. The dual specificity phosphatase Cdc25 dephosphorylates specific sites on Cdc2, which forms part of the maturation-promoting factor complex responsible for triggering the entry into mitosis. Binding of 14-3-3 proteins to Cdc25 suppresses its ability to induce entry into mitosis. Kumagai and Dunphy (1999) found that a wild-type Cdc25:GFP construct expressed in Xenopus cells was localized predominantly in the cytoplasm, whereas mutant Cdc25:GFP, which is unable to bind 14-3-3s, was localized exclusively in the nucleus. As in the experiments of Igarashi et al., leptomycin B treatment elicited a redistribution of wild-type Cdc25:GFP to the nucleus. Thus, it appears that binding of 14-3-3s controls the intracellular localization of Cdc25 in Xenopus in a manner that is very similar, if not identical, to that of RSG localization in plant cells. Kumagai and Dunphy (1999) hypothesized that Cdc25 shuttles continually between the nucleus and the cytoplasm and that binding of 14-3-3 proteins shifts the equilibrium to localization of Cdc25 predominantly in the cytoplasm.
The results of Igarashi et al. and of Kumagai and Dunphy (1999) suggest that the interaction with 14-3-3 proteins is necessary for the modulation of nuclear import and/or nuclear export of RSG and Cdc25. Kumagai and Dunphy (1999) conducted additional experiments to try to distinguish between these two possibilities. Nuclear import of a protein typically requires a nuclear localization sequence and the activity of Importin-α, whereas nuclear export involves the activity of the receptor Crm1/Exportin1, which recognizes one or more nuclear export sequences. Cdc25 has several putative nuclear export sequences, and mutational studies showed that some of these sequences might play a role in localization mediated by 14-3-3s. The authors next asked whether the binding of Importin-α to Cdc25 was affected by the interaction with 14-3-3 proteins. Here they found a clear difference: Importin-α bound to wild-type Cdc25 but not to the mutant version that could not bind 14-3-3 proteins as a result of a single amino acid change of serine residue 287 to alanine. Furthermore, in Xenopus Cdc25, the binding site for 14-3-3 proteins (around serine 287) is immediately adjacent to the putative nuclear localization sequence at residues 298 to 316. These authors suggested that an important function of 14-3-3 proteins is to reversibly hinder the import of Cdc25 into the nucleus (Kumagai and Dunphy, 1999). It will be of interest to determine if this is the case for RSG as well.
14-3-3S AS FINE-TUNERS OF COMPLEX REGULATORY PATHWAYS
The interaction of 14-3-3 proteins with plant NR is well characterized and provides a good example of what may be a general characteristic of 14-3-3 interactions: a primary role in the tight control of complex regulatory pathways. Plant NR is subject to rapid changes in activity in response to a wide variety of environmental factors that involve Ca2+, phosphorylation, interactions with 14-3-3 proteins, regulation of proteolysis, and coordination with photosynthetic rate (reviewed by MacKintosh and Meek, 2001). NR cycles between inactive and active forms, and inactivation occurs via phosphorylation by a specific NR kinase. However, phosphorylation alone does not alter NR activity; rather, it creates a binding site for 14-3-3 proteins, and it is 14-3-3 binding to NR that inhibits activity. Coordination of NR activity with photosynthesis is regulated in part via the interaction with 14-3-3 proteins; extracellular sugars influence the phosphorylation status and the binding of 14-3-3 proteins to NR, and 14-3-3 binding also may influence the proteolysis of NR.
The complex and extremely rapid regulation of NR activity is believed to minimize the major costs and possible hazards associated with the synthesis of amino acids from nitrogenous minerals and sugars. The conversion of nitrate to amino acids requires the reducing power of a considerable amount of NADH and NADPH. Furthermore, nitrite generated in the cytosol via NR activity is highly toxic. Photosynthesis normally produces enough reductant to eliminate nitrite, but nitrite poisoning might pose a threat to the cell if photosynthesis were inhibited. 14-3-3 interactions often form part of such tightly regulated processes.
14-3-3 proteins usually are members of reasonably large gene families. For example, there are 15 family members in the Arabidopsis genome, and at least 12 of these are expressed (Rosenquist et al., 2001). Given 12 different isoforms, a large number of combinations of homodimers and heterodimers is possible. Rosenquist et al. (2000) observed large differences in binding affinity between nine Arabidopsis 14-3-3 isoforms and the 14-3-3 binding domain of the Arabidopsis H+-ATPase, and they suggested that these differences might be related to functional specificity. The association of specific isoforms or specific combinations of isoforms with specific roles in the cell is a possibility that needs to be investigated more fully.
Despite the high degree of conservation of the phosphoserine site and evidence that 14-3-3 proteins function as dimers, there is some evidence that not all 14-3-3 proteins require phosphoserine binding or dimerization. Pan et al. (1999) found that the highly specific interactions of 14-3-3 proteins with the TATA box binding protein and with transcription factor IIB required residues in the 14-3-3 box 1 domain and were not dependent on dimerization or on known 14-3-3 recognition motifs containing phosphoserine. Thus, 14-3-3 proteins may have more than one standard mode of action.
As with the regulation of NR activity, hormonal regulation of biochemical pathways usually is found to be highly complex and tightly regulated and it may involve rapid shifts in enzyme activities and/or regulation of gene expression. 14-3-3 protein interactions appear to be a universal fine-tuner of biochemical regulation, perhaps offering a highly flexible and somewhat variable mode of action that can be applied to a variety of metabolic processes that require the tight control of numerous inputs and outputs.
References
- Aitken, A. (1996). 14-3-3 and its possible role in co-ordinating multiple signalling pathways. Trends Cell Biol. 6 341–347. [DOI] [PubMed] [Google Scholar]
- Cotelle, V., Meek, S.E., Provan, F., Milne, F.C., Morrice, N., and MacKintosh, C. (2000). 14-3-3s regulate global cleavage of their diverse binding partners in sugar-starved Arabidopsis cells. EMBO J. 19 2869–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLille, J.M., Sehnke, P.C., and Ferl, R.J. (2001). The Arabidopsis 14-3-3 family of signaling regulators. Plant Physiol. 126 35–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnie, C., Borch, J., and Collinge, D.B. (1999). 14-3-3 proteins: Eukaryotic regulatory proteins with many functions. Plant Mol. Biol. 40 545–554. [DOI] [PubMed] [Google Scholar]
- Freed, E., Symons, M., Macdonald, S.G., McCormick, F., and Ruggieri, R. (1994). Binding of 14-3-3 proteins to the protein kinase Raf and effects on its activation. Science 265 1713–1716. [DOI] [PubMed] [Google Scholar]
- Fukazawa, J., Sakai, T., Ishida, S., Yamaguchi, I., Kamiya, Y., and Takahashi, Y. (2000). REPRESSION OF SHOOT GROWTH, a bZIP transcriptional activator, regulates cell elongation by controlling the level of gibberellins. Plant Cell 12 901–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozinger, C.M., and Schreiber, S.L. (2000). Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc. Natl. Acad. Sci. USA 97 7835–7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi, D., Ishida, S., Fukazawa, J., and Takahashi, Y. (2001). 14-3-3 proteins regulate intracellular localization of the bZIP transcriptional activator RSG. Plant Cell 13 2483–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai, A., and Dunphy, W.G. (1999). Binding of 14-3-3 proteins and nuclear export control the intracellular localization of the mitotic inducer Cdc25. Genes Dev. 13 1067–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, J., and Omary, M.B. (1996). 14-3-3 proteins associate with phosphorylated simple epithelial keratins during cell cycle progression and act as a solubility factor. J. Cell Biol. 133 345–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKintosh, C., and Meek, S.E.M. (2001). Regulation of plant NR activity by reversible phosphorylation, 14-3-3 proteins and proteolysis. Cell. Mol. Life Sci. 58 205–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malerba, M., and Bianchetti, R. (2001). 14-3-3 protein-activated and autoinhibited forms of plasma membrane H+-ATPase. Biochem. Biophys. Res. Commun. 286 984–990. [DOI] [PubMed] [Google Scholar]
- May, T., and Soll, J. (2000). 14-3-3 proteins form a guidance complex with chloroplast precursor proteins in plants. Plant Cell 12 53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison, D. (1994). 14-3-3: Modulators of signaling proteins? Science 266 56–57. [DOI] [PubMed] [Google Scholar]
- Morsomme, P., and Boutry, M. (2000). The plant plasma membrane H+-ATPase: Structure, function and regulation. Biochim. Biophys. Acta 1465 1–16. [DOI] [PubMed] [Google Scholar]
- Muslin, A.J., Tanner, J.W., Allen, P.M., and Shaw, A.S. (1996). Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 84 889–897. [DOI] [PubMed] [Google Scholar]
- Pan, S., Sehnke, P.C., Ferl, R.J., and Gurley, W.B. (1999). Specific interactions with TBP and TFIIB in vitro suggest that 14-3-3 proteins may participate in the regulation of transcription when part of a DNA binding complex. Plant Cell 11 1591–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenquist, M., Sehnke, P., Ferl, R.J., Sommarin, M., and Larsson, C. (2000). Evolution of the 14-3-3 protein family: Does the large number of isoforms in multicellular organisms reflect functional specificity? J. Mol. Evol. 51 446–458. [DOI] [PubMed] [Google Scholar]
- Rosenquist, M., Alsterfjord, M., Larsson, C., and Sommarin, M. (2001). Data mining the Arabidopsis genome reveals fifteen 14-3-3 genes: Expression is demonstrated for two out of five novel genes. Plant Physiol. 127 142–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, T.F., Medina, J., Hill, A., and Quatrano, R.S. (1998). 14-3-3 proteins are part of an abscisic acid–VIVIPAROUS1 (VP1) response complex in the Em promoter and interact with VP1 and EmBP1. Plant Cell 10 837–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, B., Smerdon, S.J., Jones, D.H., Dodson, G.G., Soneji, Y., Aitken, A., and Gamblin, S.J. (1995). Structure of a 14-3-3 protein and implications for coordination of multiple signalling pathways. Nature 376 188–194. [DOI] [PubMed] [Google Scholar]
- Zhao, X., Ito, A., Kane, C.D., Liao, T.S., Bolger, T.A., Lemrow, S.M., Means, A.R., and Yao, T.P. (2001). The modular nature of histone deacetylase HDAC4 confers phosphorylation-dependent intracellular trafficking. J. Biol. Chem. 276 35042–35048. [DOI] [PubMed] [Google Scholar]