Abstract
Background
In Drosophila, mutations in the gene eyes absent (eya) lead to severe defects in eye development. The functions of its mammalian orthologs Eya1-4 are only partially understood and no mouse model exists for Eya3. Therefore, we characterized the phenotype of a new Eya3 knockout mouse mutant.
Results
Expression analysis of Eya3 by in-situ hybridizations and β-Gal-staining of Eya3 mutant mice revealed abundant expression of the gene throughout development, e.g. in brain, eyes, heart, somites and limbs suggesting pleiotropic effects of the mutated gene. A similar complex expression pattern was observed also in zebrafish embryos.
The phenotype of young adult Eya3 mouse mutants was systematically analyzed within the German Mouse Clinic. There was no obvious defect in the eyes, ears and kidneys of Eya3 mutant mice. Homozygous mutants displayed decreased bone mineral content and shorter body length. In the lung, the tidal volume at rest was decreased, and electrocardiography showed increased JT- and PQ intervals as well as decreased QRS amplitude. Behavioral analysis of the mutants demonstrated a mild increase in exploratory behavior, but decreased locomotor activity and reduced muscle strength. Analysis of differential gene expression revealed 110 regulated genes in heart and brain. Using real-time PCR, we confirmed Nup155 being down regulated in both organs.
Conclusion
The loss of Eya3 in the mouse has no apparent effect on eye development. The wide-spread expression of Eya3 in mouse and zebrafish embryos is in contrast to the restricted expression pattern in Xenopus embryos. The loss of Eya3 in mice leads to a broad spectrum of minor physiological changes. Among them, the mutant mice move less than the wild-type mice and, together with the effects on respiratory, muscle and heart function, the mutation might lead to more severe effects when the mice become older. Therefore, future investigations of Eya3 function should focus on aging mice.
Background
Eya3 is one of four mammalian orthologous genes (Eya1-4) of eyes absent (eya) in Drosophila melanogaster [1,2]. Previous investigations demonstrated that a homozygous knockout of eya function in D. melanogaster results in severe embryonic defects and absence of compound eyes due to eye progenitor cell death [3,4]. Like eyes absent in Drosophila, mammalian Eya genes encode for transcriptional co-activator proteins and are widely co-expressed with Pax6, Six1, Six2 and Dachshund (Dach) genes. It is assumed that Eya genes are part of a hierarchically organized and highly conserved regulatory network during eye development with Pax6 on its top and Six and Dach genes as major players [5].
Eya proteins interact as transcriptional cofactors with Six and Dachshund proteins and do not possess DNA-binding properties. Eya proteins are characterized by a conserved 271-aa C-terminal domain (Eya domain, ED) which is believed to have a dual function [6]: on one hand, it mediates protein-protein interactions with Six and Dachshund proteins [7,8]; it is further required for nuclear translocation of Eya proteins [9]. On the other hand, in-vitro experiments demonstrated a tyrosine-phosphatase activity of the ED suggesting a catalytic function of this domain [10,11]. A second and far less conserved domain is the Eya domain 2 (ED2), which is located in the N-terminal region of the protein and presumably serves as a transactivation domain [12].
The relevance of mutations in the ED for the emergence of diseases has been widely shown for Eya1. In humans, EYA1 haploinsufficiency is responsible for branchio-oto-renal (BOR) syndrome or branchio-oto (BO) syndrome. BOR is an autosomal dominant disorder characterized by branchial cysts, ear malformations, hearing loss and renal abnormalities. Interestingly, the majority of BOR disease-associated missense mutations clusters in the ED and therefore suggest an important role for the ED in the development of these diseases [13]. Mice in which 153 of 271 amino acids of the Eya1-ED have been deleted die at birth and show severe craniofacial and skeletal defects and an absence of ears, kidneys, thymus and parathyroid glands [14]. Eya1+/- mice show a phenotype, which is comparable to humans suffering from BOR syndrome. They display renal abnormalities and conductive hearing loss. Neither Eya1+/- nor Eya1-/- mice reveal ocular defects and only in a few cases could Eya1 mutations be associated with congenital cataracts and ocular anterior segment anomalies in humans [15].
Moreover, Eya genes are associated with the proper development of muscles. Drosophila eya mutants show musculature defects [16]; mouse embryos deficient for Eya1 and Six1 have a complete absence of all hypaxial muscle and a severe reduction of epaxial muscle [17]. While neither in Eya1-/- nor in Eya2-/- mice can a muscle phenotype be observed, Eya1-/-Eya2-/- double knockout embryos display muscle-less limbs [18]. For Eya2 it was shown in tissue culture experiments that it interacts with Six1 and Dach2 to regulate Pax3 and therefore to influence myogenic differentiation [19]. In addition, Eya genes seem to play a role in heart and auditory function in vertebrates. Mutations of the EYA4 gene resulting in truncated EYA4 proteins cause dilated cardiomyopathy and conductive hearing loss in humans [20,21] and Eya4 mutations in mice are responsible for an abnormal morphology of the middle ear [22]. Injection of morpholino oligonucleotides against Eya4 into zebrafish (Danio rerio) embryos indicated an abnormal morphological and physiological phenotype of the heart [20]. Obviously, Eya genes are involved in the proper development of many tissues in vertebrates including the ear, muscles, heart, renal system and possibly the eye.
However, the influence of Eya3 on developmental processes in mice and other mammals is rather unknown. In-vitro experiments showed that Eya3 is important for cell-autonomous proliferation of murine myoblast C2C12 cells [17]. In addition, studies in Xenopus revealed a strong influence of the Eya3 homologous gene Xeya3 on survival and proliferation of neural progenitor cells in the anterior neural plate of Xenopus embryos [23]. There are, however, no indications for a similar expression pattern, neither in fishes nor in mice, and we cannot observe a brain phenotype in our mouse Eya3 mutants. To our knowledge, this Eya3-deficient mutant is the first mouse model for a mutation affecting Eya3 and therefore an excellent model to study its function in mammals. Our systemic analysis within the German Mouse Clinic (GMC) demonstrated a broad spectrum of minor physiological changes in the Eya3-deficient mice. Among them, the reduced movement of the mutant mice and effects on their respiratory, muscle and heart function are the most striking changes.
Results
1. Molecular characterization of Eya3-deficient mice
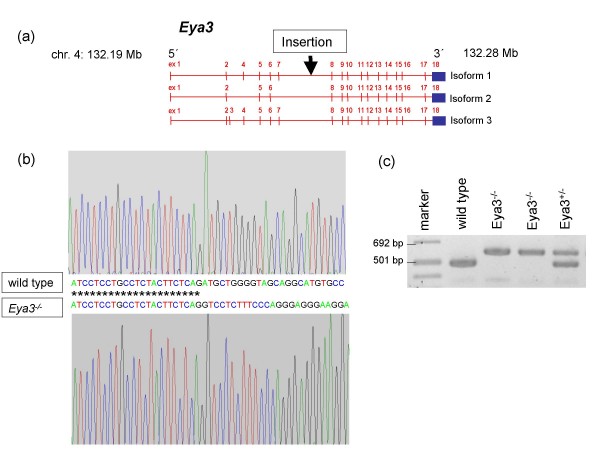
Gene trap vector insertion was detected in a 16.7 kb large intronic region of the Eya3 gene (intron 7) using 5'RACE and PCR experiments and combining different forward primers and a reverse primer in the lacZ cassette of the pT1βgeo-vector. By cloning and sequencing of a 2.1 kb fragment in this region the integration site was determined to 132.23 Mb on chromosome 4 (Ensembl database, release No. 50, 2008) (Fig. 1a, 1b). Genotyping of mice confirmed the vector integration at the determined locus (Fig. 1c).
Figure 1.
Generation of Eya3-deficient mice by insertional mutagenesis. (a) Schematic illustration of the gene trap vector integration into intron 7 of the Eya3 gene resulting in a loss of the following exons. (b) Sequence fragment of the Eya3 gene. The upper chromatogram displays the sequence of wild-type mice, while the lower chromatogram shows the vector integration in mutant mice at 132.23 Mb on chromosome 4 (Ensembl database, release No. 50, 2008). (c) Eya3 mutant mice were genotyped by triplex PCR with a genomic forward primer combined with a genomic reverse primer for identification of wild-type animals and with a reverse primer within the gene trap vector for identification of homozygous mutant animals.
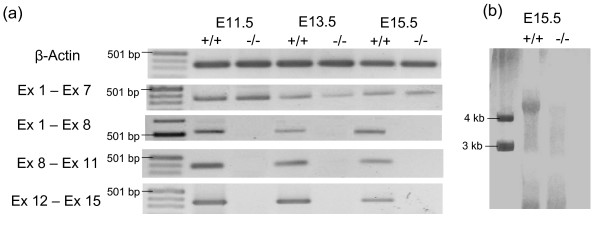
RT-PCR and Northern-Blotting demonstrated the complete loss of exons 8–15 of the Eya3 gene in homozygous mutant animals. Transcripts containing exons 1–8 were detected in wild-type and heterozygous embryos at the age of E11.5 (whole embryo), E13.5 (head) and E15.5 (head). In contrast, no transcripts covering the exons 1–8, 8–11 or 12–15 were detectable in homozygous Eya3-mutants (Fig. 2a). Additionally, using a probe covering exons 8–11, Northern-Blotting (RNA from E15.5) were consistent with the results received from RT-PCR (Fig. 2b).
Figure 2.
Mutation of Eya3 causes a loss of exons 8 – 15 in homozygous mutants. (a) RT-PCR with cDNA of wild type and Eya3-/- embryos at developmental stages E11.5 (whole embryo), E13.5 (head), E15.5 (head) showed the loss of exons 8 – 15 of the Eya3 gene in Eya3-/- mice. (b) Northern-Blotting using RNA of wild-type and Eya3-/- embryos (E15.5) with a riboprobe against exons 8 – 11 confirmed the data received from RT-PCR.
150 offspring from heterozygous matings have been checked: there were 76 males (50.5%) and 74 females (49.5%); 35 animals were wild type (23%), 85 heterozygous (57%) and 30 animals homozygous mutants (20%); the mean litter size was 5.2 offspring. These date indicate no major deviation from the expected numbers according to the Mendelian laws suggesting normal fertility of the heterozygotes and survival of the homozygous mutants.
2. Expression pattern of Eya3 in the mouse and consequences of Eya3 invalidation
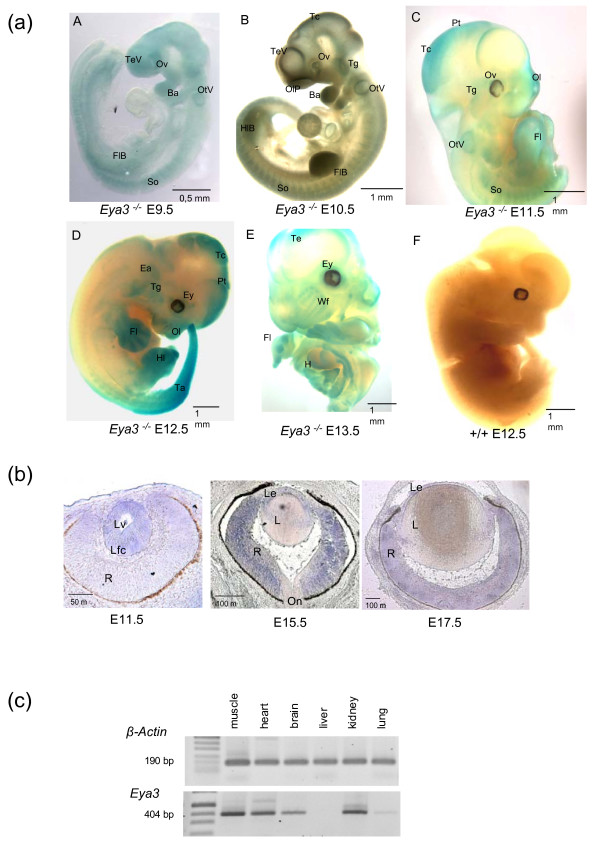
Eya3 shows a complex expression pattern during embryonic development. The expression pattern of Eya3 is identical, if analyzed by in-situ hybridization in wild-type embryos or by β-Gal expression in Eya3-/- mutant mice; data gathered from β-Gal staining of mutant embryos (Fig. 3a) demonstrated an expression of Eya3 at E9.5 in the developing neural tissues predominantly in the neural epithelium of the telencephalic vesicle and the later forebrain. Further on, lacZ staining is found in the somites, in the mandibular and maxillary component of the first branchial arch, in the optic vesicle and in the otic vesicle. In addition, an expression in the forelimb bud is visible. The expression of lacZ continues at the developmental stage E10.5 in the epithelium of the telencephalic vesicle and in tectorial regions of the developing brain. Moreover, Eya3 expression is indicated in the region surrounding the olfactory pit, the somites, fore- and hindlimb bud and in the trigeminal nerve. The first and second branchial arches display Eya3/lacZ transcripts, but the expression in the first branchial arch is mainly restricted to the mandibular component. At E11.5 Eya3 expression is detectable in the olfactory pit, the developing eye, the otic vesicle, tectorial and pretectorial regions of the brain, fore- and hindlimbs and in somites. At E12.5, Eya3 transcripts are most prominent in the limbs, in the tail and in cortical regions of the brain. Later (E13.5), the expression of Eya3 continues in the cerebellar cortex and in the limbs as well as in the ventral and dorsal grey horn of the spinal cord (not shown).
Figure 3.
Expression pattern of Eya3 in mice. (a) Expression analysis of Eya3 by lacZ-staining in homozygous Eya3-/- embryos (A – E, E9.5 – E13.5) revealed an expression of Eya3 in specific regions including the eyes, limbs, somites, branchial arches, tectorial regions, and in different areas of the developing brain. Since the expression pattern of Eya3 is identical, if analyzed by in-situ hybridization in wild-type embryos or by β-Gal expression in Eya3-/- mutant mice, only the mutant data are shown. Wild-type embryos as negative controls did not show lacZ-staining (F). (Ba, branchial arches; Ey, eye; Ea, ear; Fl, forelimb; FlB, forelimb bud; H, heart; Hl, hindlimb; HlB, hindlimb bud; OlP, olfactory pit; Ol, olfactory region; Ov, optic vesicle; OtV, otic vesicle; Pt, pretectorial region; So, somites; Tc, tectorial region; Ta, tail; Te, telencephalon; TeV, telencephalic vesicle; Tg, trigeminal nerve; Wf, follicle of whiskers). (b) Expression analysis of Eya3 in the eye with in-situ hybridization displayed transcripts in the detaching lens at E11.5 and afterwards in the lens epithelium and in the retina (E15.5 – E17.5) (L, lens; Le, lens epithelium; Lfc, lens fiber cells; Lv lens vesicle; R, retina; On, optic nerve). (c) RT-PCR with cDNA of adult mice showed an Eya3 expression in muscle, heart, brain, kidney and lung, however, no Eya3 transcript was found in the liver.
Eya3 expression in the eye (Fig. 3b) is first visible at E10.5 in the detaching lens vesicle. Later on transcripts are restricted to the retina and the lens epithelium (E15.5 – E17.5). In adult mice (Fig. 3c), an expression of Eya3 was traceable by RT-PCR in the brain, skeletal muscles, heart and kidneys. The lung displayed only a very weak Eya3 expression, while in the liver no expression was detected.
3. Eya3 expression in zebrafish development
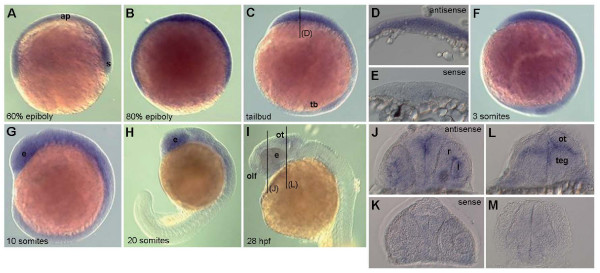
Because of the obvious strong difference in the Eya3 expression pattern between mouse and Xenopus, we investigated Eya3 expression also in zebrafish embryos. Like at early developmental stages in mice, whole mount in-situ hybridization revealed a ubiquitous expression of eya3 in early zebrafish embryos throughout gastrulation and early somitogenesis. At the 10-somite-stage (14 hours post-fertilization, hpf), expression becomes more restricted to the anterior part of the embryo and at the 20-somite-stage (19 hpf) it is most prominent in the brain and developing eye. By the beginning of the second day of development (28 hpf), eya3 expression strongly decreases and becomes only weakly detectable in the eye, optic tectum and olfactory placode (Fig. 4).
Figure 4.
Whole-mount in-situ hybridization for eya3 expression in zebrafish embryos. Whole-mount in-situ hybridization for eya3 expression in zebrafish embryos reveals a ubiquitous expression of eya3 throughout gastrulation and early somitogenesis (A-F). Cross sections at the tailbud stage (level indicated in C) confirm expression in all cell layers (D), compared to a hybridization using the sense probe (E). The expression is more restricted to the anterior part of the embryo at 14 hpf (G). From 19 hpf the expression is most prominent in the developing brain and eye (H). By the beginning of the second day (28 hpf) (I) eya3 expression strongly decreases and is only weakly detectable in the eye, optic tectum, tegmentum and olfactory placode. This is confirmed on cross sections (J, L, at the levels indicated in I) compared to hybridization with the sense probe (K, M). Abbreviations: ap, animal pole; e, eye; l, lens; ot, optic tectum; olf, olfactory placode; r, retina; s, shield; tb, tailbud; teg, tegmentum.
4. General characterization of the Eya3-/- phenotype
In cooperation with the German Mouse Clinic [24] 24 wild-type controls (14 ♂, 10 ♀) and 24 Eya3-/- mice (14 ♂, 10 ♀) have been analyzed in a comprehensive phenotypic screen.
Eya3 mutant animals have been investigated for bone- and weight-related quantitative parameters at the age of 18 weeks. Among these parameters, the homozygous Eya3 mutants showed significant differences in bone mineral content, body weight and body length as compared to the control littermates. At the age of 18 weeks, these three parameters were significantly (p < 0.05) reduced in male mutants. Female mutants also displayed lower values compared to wild-type animals without reaching the level of statistical significance (table 1).
Table 1.
Bone- and weight-related quantitative parameters (18 week old mice)
| Wild type | Eya3-/- | |||
| male | female | male | female | |
| Parameter | n = 9 | n = 9 | n = 10 | n = 9 |
| Bone mineral content [mg] | 557 ± 33 | 467 ± 21 | 422 ± 39* | 447 ± 33 |
| Body length [cm] | 9.06 ± 0.06 | 8.89 ± 0.07 | 8.60 ± 0.15* | 8.67 ± 0.08 |
| Body weight [g] | 27.29 ± 0.63 | 21.05 ± 0.44 | 23.96 ± 1.12* | 20.57 ± 0.82 |
*p < 0.05 (as compared to the corresponding group of wild-type mice); data are presented as mean ± standard error of the mean
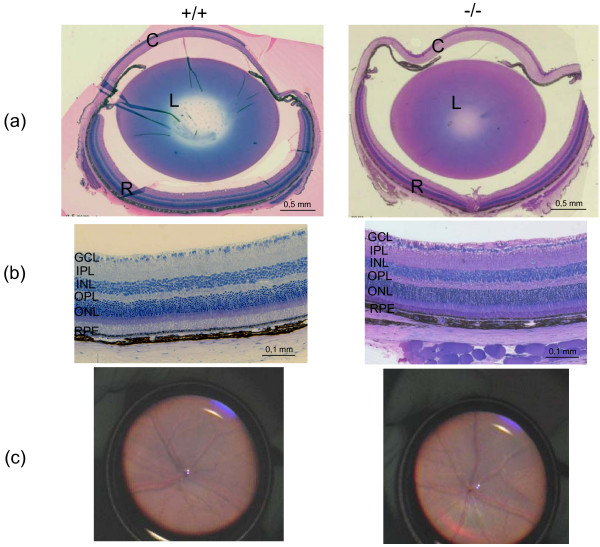
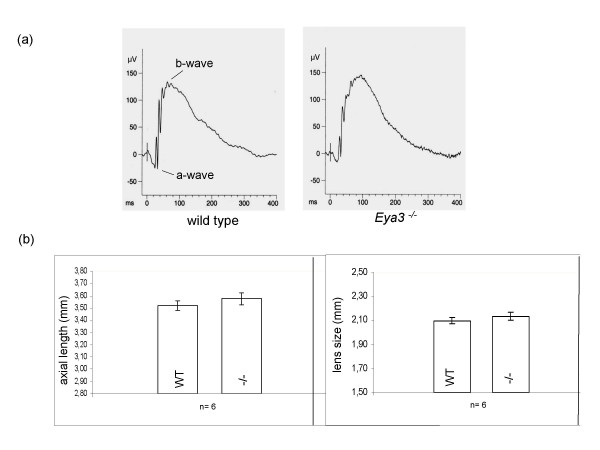
Data from expression analysis demonstrated that Eya3 is expressed in the developing ear and in the adult kidney. However, no significant differences between Eya3 knockout and wild-type animals have been observed in either functional or histological analyses, suggesting that Eya3 is not a major player in these organs. Moreover, expression analysis displayed that Eya3 is expressed in the retina and in the lens epithelium of the eye (Fig. 3b), suggesting that Eya3 may be important for proper lens and retina development. However, histological analysis of eyes taken from adult mice revealed no morphological differences between wild-type controls and homozygous Eya3 mutant mice (Fig. 5a, b). Further on, the investigation of the posterior part of the eye by funduscopy (Fig. 5c) did not show striking phenotypical differences of the retinal structure between wild-type and mutant animals. In addition to the morphological examination of the retina a rough functional analysis was done by electroretinography (ERG) (Fig. 6a). Measurement of retinal and ganglion cell response to the light stimulus displayed no significant (p > 0.05) deviations in mutant animals compared to wild-type controls.
Figure 5.
Histology and funduscopy of the eye in Eya3 mouse mutants. (a) Histology of the eye showed no morphological differences between wild-type mice and homozygous mutants (C, cornea; L, lens; R, retina). In line with these results histological analysis (b) and funduscopy (c) of the retina could not detect structural impairments in mutant animals (GCL, ganglion cell layer; IPL, inner plexiform layer; INL, inner nuclear layer; OPL, outer plexiform layer; ONL, outer nuclear layer; RPE retinal pigment epithelium).
Figure 6.
Electroretinography (ERG) and laser interference biometry. (a) The electroretinography displays no obvious deviations in the shape of the a-wave (response of photoreceptor cells) and b-wave (response of bi-polar cells) between wild-type controls and Eya3 mutant animals. (b) Measurement of the axial length of the entire eye and of the lens size in particular did not reveal significant alterations in Eya3 mutant animals compared to wild-type controls.
An investigation of the eye size by laser interference biometry was performed to identify eye size variation between wild-type controls and Eya3 mutant animals (Fig. 6b). Anterior chamber depth, lens size and axial length were measured revealing no significant (p > 0.05) deviations between wild-type mice and homozygous Eya3 mutant mice.
Alteration in lung, heart and muscle function
The expression analysis (Fig. 3) demonstrated that Eya3 is expressed in heart and muscle, but only weakly in the lung. Nevertheless, systematic analysis of the function of these three organ systems showed significant alterations.
To evaluate the lung function, several parameters have been tested in male Eya3-/- mutants and compared to wild-type littermates (tidal volumes, respiratory rates, minute ventilation, inspiratory and expiratory times as well as peak inspiratory and peak expiratory flow rates). Among these parameters, the tidal volume at rest is significantly reduced (p < 0.05) in the mutants (+/+: 0.25 ± 0.01 ml, n = 5; Eya3-/-: 0.22 ± 0.01 ml, n = 4). This difference is more striking (p < 0.02), if related to the body weight (+/+: 10.1 ± 0.3 μl/g, n = 5; Eya3-/-: 8.7 ± 0.3 μl/g, n = 4). In this context, it might be of interest that the respiratory rate of the mutant mice is enhanced, even if it does not reach the level of statistical significance (+/+: 302.2 ± 9.3/min, n = 5; Eya3-/-: 342.6 ± 19.1/min, n = 4). However, this may suggest that compensatory mechanisms are triggered. The reduced muscle strength – if transferable to respiratory muscles – may limit the potential volume to be inspired and, thus, result in the observed reduced tidal volume. Consequently, the respiratory rate is increased leading to strikingly similar minute ventilations in wild type and Eya3-/- (72.7 ± 3.5 ml/min vs. 71.0 ± 1.6 ml/min).
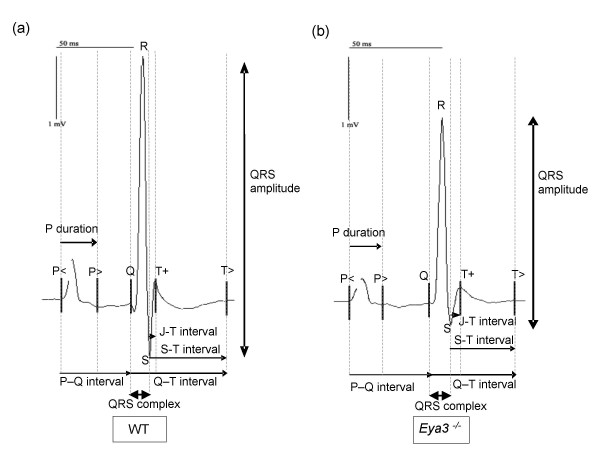
In addition, cardiovascular function was examined by tail-cuff blood pressure and by electrocardiography (ECG). Analysis of blood pressure revealed no genotype-specific differences between wild-type and mutant mice. In contrast, the ECG showed significant genotype-specific differences between controls and mutants of both sexes (Fig. 7). The time from onset of the electrical impulse in the sinu-atrial node until ventricular depolarization (PQ-interval) was increased. Additionally, the time from ventricular depolarization to repolarization (JT-interval) was extended, and the amplitude of the QRS-complex representing ventricular depolarization was highly significantly decreased in mutant animals (table 2).
Figure 7.
Electrocardiography (ECG) of wild-type and Eya3-/- mutant mice. a) Electrocardiogram of a wild-type control. b) Electrocardiogram of an Eya3-/-animal is shown. Eya3 mutant mice of both sexes displayed several alterations compared to the control animals: the PQ-interval and the JT-interval are extended in mutants, while the amplitude of the QRS-complex is reduced in Eya3-/- animals (alterations are indicated by black arrows).
Table 2.
ECG-Parameters (15 week old mice)
| Wild type | Eya3-/- | |||
| male | female | male | female | |
| Parameter | n = 8 | n = 9 | n = 10 | n = 9 |
| PQ interval [ms] | 38.6 ± 0.7 | 40.8 ± 0.7 | 41.3 ± 1.6* | 43.6 ± 0.9* |
| P-wave duration [ms] | 20.4 ± 0.6 | 20.4 ± 0.8 | 20.8 ± 0.6 | 21.4 ± 0.3 |
| QRS-complex duration [ms] | 9.6 ± 0.2 | 9.6 ± 0.2 | 10.0 ± 0.2 | 9.8 ± 0.3 |
| QT interval [ms] | 46.2 ± 1.3 | 41.7 ± 0.6 | 45.6 ± 2.1 | 42.7 ± 0.6 |
| QTcorrected [ms] | 40.6 ± 1.4 | 36.5 ± 0.5 | 40.1 ± 1.5 | 38.2 ± 0.7 |
| RR interval [ms] | 130.6 ± 3.8 | 130.9 ± 4.1 | 130.1 ± 4.9 | 125.2 ± 3.7 |
| Heart rate [bpm] | 463.7 ± 13.5 | 463.4 ± 14.2 | 469.1 ± 18.6 | 482.7 ± 13.3 |
| JT interval [ms] | 4.1 ± 0.6 | 3.7 ± 0.1 | 6.0 ± 1.1* | 5.4 ± 0.4* |
| ST interval [ms] | 36.6 ± 1.5 | 32.1 ± 0.6 | 35.6 ± 2.1 | 32.9 ± 0.6 |
| Q amplitude [mV] | 0.01 ± 0.00 | 0.02 ± 0.01 | 0.04 ± 0.01** | 0.03 ± 0.01** |
| R amplitude [mV] | 3.10 ± 0.18 | 3.16 ± 0.35 | 2.32 ± 0.14** | 2.33 ± 0.29** |
| S amplitude [mV] | -1.26 ± 0.22 | -1.03 ± 0.16 | -0.95 ± 0.14 | -1.01 ± 0.16 |
| QRS amplitude [mV] | 4.35 ± 0.17 | 4.19 ± 0.40 | 3.28 ± 0.13*** | 3.34 ± 0.22*** |
*p < 0.05; **p < 0.01; ***p < 0.001 (as compared to the corresponding group of wild-type mice); data are presented as mean ± standard error of the mean
For evaluation of the behavioral phenotype, the spontaneous activity of mice was measured using a modified hole-board test. The mutants demonstrated a mild increase in exploratory behavior as indicated by a reduced latency to hole exploration. The frequency of rearings in the box was also enhanced in the mutants. However, the difference did not reach the level of statistical significance. In contrast, mutant mice exhibited a reduced forward locomotor activity, measured as reduced distance travel, reduced maximum velocity and reduced frequency of turns; the mean velocity was also reduced, but the difference did not reach the level of significance (table 3). The reduced locomotor activity might be associated with reduced muscle strength in these mice as indicated by the results of the forelimb grip strength test: Eya3 mutant mice of both sexes revealed a significant (p < 0.05) reduction of muscle strength. However, the performance on the Rotarod was only slightly impaired and did not reach the level of statistical significance.
Table 3.
Results of behavioural observations (8–10 week old mice).
| Wild type | Eya3-/- | |||
| male | female | male | female | |
| Parameter | n = 15 | n = 9 | n = 15 | n = 9 |
| Forward locomotor activity (total distance travelled) [cm] | 3387.2 ± 124.9 | 3534.3 ± 115.5 | 3086.8 ± 126.5* | 3248.6 ± 71.4* |
| Speed of movement (maximum velocity) [cm/sec] | 62.9 ± 1.9 | 59.8 ± 3.2 | 55.0 ± 1.9* | 58.2 ± 1.9* |
| Turns [frequency] | 1714.9 ± 46.1 | 1776.8 ± 41.7 | 1610.3 ± 41.6* | 1688.8 ± 39.8* |
| Exploratory behaviour (hole exploration) [latency] | 14.6 ± 3.7 | 13.9 ± 3.1 | 8.1 ± 1.5* | 8.5 ± 1.6* |
| Grip strength (force) [p] | 121.7 ± 2.8 | 99.3 ± 3.7 | 102.8 ± 4,9** | 89.9 ± 3.9** |
*p < 0.05; **p < 0.01 (as compared to the corresponding group of wild-type mice); data are presented as mean ± standard error of the mean
Analysis of differentially expressed genes in Eya3 mutant mice
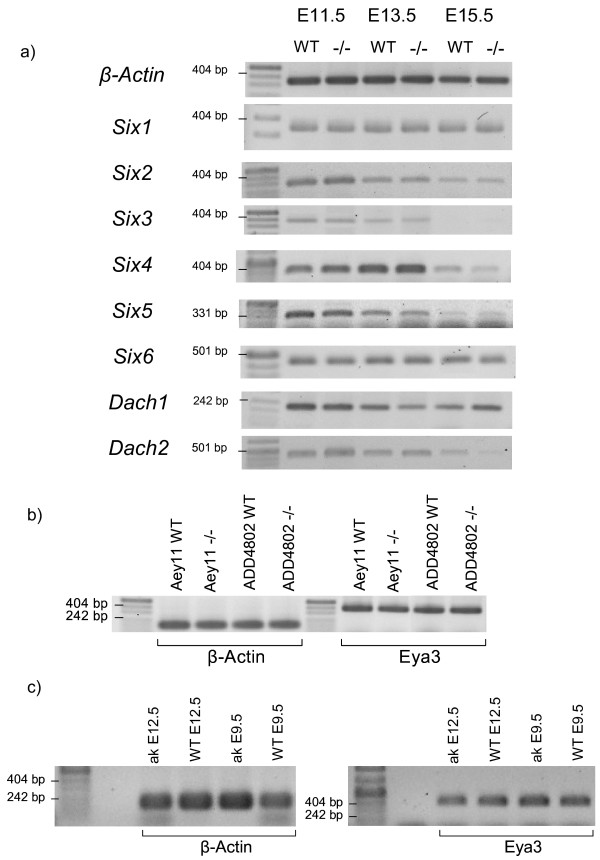
Since the members of the Eya gene family are discussed to be members of a conserved regulatory network during eye development including also Six and Dach genes and Pax6 on its top [5], we tested the influence of the loss of Eya3 activity on their expression. Fig. 8a demonstrates that all six Six genes as well as the two Dach genes are not changed in their overall expression at least within the critical phase of early eye development between E11.5 and E15.5. On the other side, we checked also the Eya3 expression in two Pax6 mutant alleles (Fig. 8b; allele symbols Aey11, ADD4802; 39); the overall Eya3 expression is obviously not altered even in homozygous Pax6 mutant embryos. The same is true in a Pitx3 mutant aphakia (allele symbol ak; Fig. 8c; [25]). These results indicate that Eya3 is no major player in the regulatory network of eye development.
Figure 8.
Eya3 and the retinal determination network. a) The expression of all six Six genes (Six1 – Six6) and the two Dach (Dach1, Dach2) genes have been investigated in wild-type and homozygous mutant embryos by RT-PCR using total mRNA; β-actin was used as loading control. b) Eya3 expression was tested in embryos of two homozygous Pax6 mutant alleles (Pax6Aey11 and Pax6ADD4802); β-actin was used as loading control. b) Eya3 expression was tested in embryos of a homozygous Pitx3 mutant allele (Pitx3ak); β-actin was used as loading control.
To obtain a systematic overview on the expression of downstream target genes, we performed an expression profiling analysis using a genome-wide cDNA microarray approach. Gene expression patterns of skeletal muscle, heart, and brain of adult, homozygous Eya3 mutants were compared to wild-type littermates. In skeletal muscles of Eya3 mutant mice no significantly regulated genes were detected between mutant and wild-type tissue. In contrast, 93 differentially expressed genes were identified in brain (13 up- and 80 down-regulated genes; Fig. 9a). In heart tissue all 17 differentially regulated genes showed decreased expression levels in the Eya3 mutant mice (Fig. 9b). Due to the limited number of regulated genes, no over-representation of Gene Ontology (GO) terms could be identified in these 17 genes. However, among the differentially regulated genes in the brain, the macromolecule biosynthetic process was over-represented for biological processes, and the structural constituent of cytoskeleton and structural constituent of ribosome were over-represented for molecular functions.
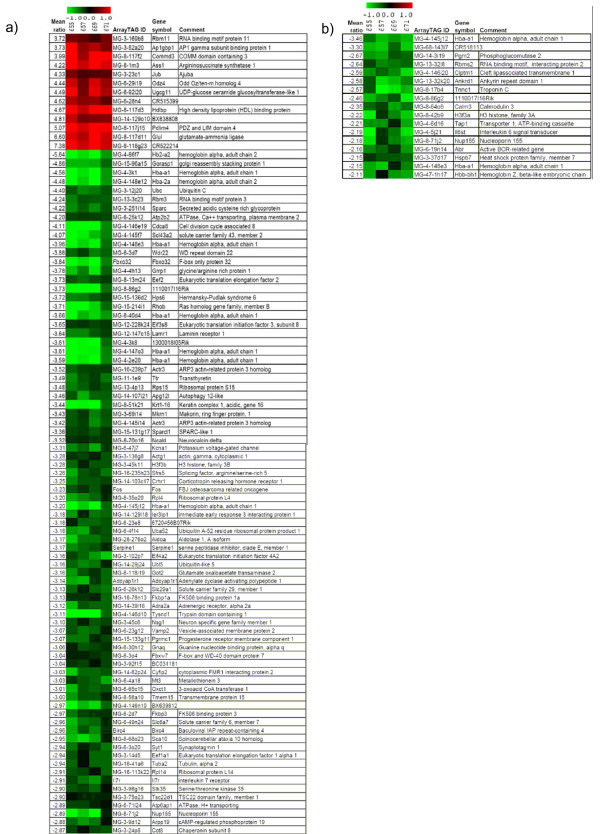
Figure 9.
Heat plots of gene expression profiles from 8 DNA microarray experiments of Eya3 mutants versus control mice. Differential gene expression was analyzed in the brain (a) and in the heart (b). One dye-flip pair represents two experimental replicates of each analyzed mouse (A-D). One ArrayTAG ID is the unique probe identifier from the LION Bioscience clone set. Official gene symbols are given. The scale bar encodes the ratio of the fold induction; 0.7% (a) or 1.3% (b) of the elements are above the upper limit of the color range selection (red is up-regulated and green down-regulated in the Eya3 mutant mice).
Among these differentially regulated genes, Nup155 (Nucleoporin 155) was significantly down-regulated both, in heart (2.1-fold) and brain (2.8-fold). Validation of Nup155 expression level with real-time PCR confirmed the differential regulation in both organs of the Eya3-/- mutant mice (heart: 1.6-fold, p < 0.05; brain: 2.2-fold, p < 0.01). Since the Nup155 protein is discussed to be involved in the transport of proteins and mRNA, its reduced expression in brain and heart might also contribute to the alterations observed in the behavioral phenotype of the Eya3 mutant mice and of their slightly altered cardiac function.
Discussion
The gene eyes absent was shown to have a prominent function during eye development of Drosophila melanogaster [3]. In contrast, the mammalian orthologs seem to play a far less important role in eye development.
In contrast to Xenopus embryos, where the expression of Xeya3 is restricted exclusively to the anterior neural plate and cells of the prospective brain and eye within neurulation (Kriebel et al., 2007), our results from mice and zebrafish display a ubiquitous expression of Eya3 in early embryos during neural plate stage. It is only by the end of neurulation that a restriction of Eya3 expression to distinct regions like forebrain and eye in mice or optic tectum and eye in zebrafish is observable. Thus, our expression analysis for Eya3 in mouse and zebrafish demonstrates striking differences between these organisms and Xenopus, suggesting the possibility of different functions for Eya3 among vertebrate species.
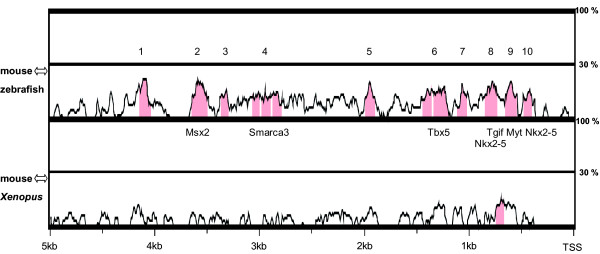
This might be due to differences in regulatory mechanisms of Eya3 between these organisms. Comparative sequence analysis of the Eya3 upstream region using mVISTA (Fig. 10) revealed a significantly higher number of conserved non-coding sequences (CNS) between mouse and zebrafish than between mouse and Xenopus (mouse ↔ zebrafish: 13 CNS, 1325 bp at 52,9%; mouse ↔ Xenopus: 1 CNS, 78 bp, 50%), indicating the possibility that Eya3 genes gained individual expression patterns and functions among vertebrate species due to evolutionary determined diversity in regulatory mechanisms.
Figure 10.
Conserved elements in Eya3 promoter/enhancer region. Sequence analysis of 5 kb upstream from Eya3 transcriptional start site for mouse, zebrafish and Xenopus using mVISTA (parameters: 50% sequence identity, 80 bp window length). The analysis revealed 10 conserved non-coding regions within the mouse-zebrafish alignment (sequence identity: 1325 bp at 52.9%, highlighted in red) and only 1 conserved non-coding region for the mouse-Xenopus alignment (sequence identity: 78 bp at 50%). Transcription factor binding sites of a matrix similarity of 1.000 are mentioned. TSS, transcription start site.
The results of our phenotypic study on Eya3 are in line with earlier findings in other mammalian Eya genes indicating that their loss does not have an effect on eye development and ocular functions. Eye disorders have not been observed in Eya1, Eya2 or Eya4 mutant mice [14,18,22]. Additionally, histological analysis and detailed investigation of ocular morphology and function by tests such as electroretinography and the optokinetic reflex in the Eya3 knockout mutants presented here did not show any differences between the eyes of wild-type controls and hetero- or homozygous Eya3 mutant animals, although expression of Eya3 was detected in the retina and in the lens epithelium during eye development. In addition to the striking differences between the function of Eya genes in Drosophila and mice, there are also important differences between the phenotypes of Eya mutants among vertebrates. In contrast to morpholino-injected Xenopus embryos [23], Eya3 mutant mice showed a normal development of neural tissues.
Nevertheless, further investigations revealed clear differences in bone mineral content and body length between wild-type animals and Eya3 mutant mice, suggesting an impact of Eya3 on physical constitution in general, leading to a reduction of growth. Moreover, Eya3 is expressed in the developing muscle, and concomitantly, the forelimb grip strength is reduced in the mutant mice. This might be a reason for the observed decreased locomotor activity of the Eya3 mutant mice. Muscle deficiencies have been reported also for Eya1-/-Eya2-/- mutant mice [18], and also for Six1, encoding a potential interaction partner of Eya proteins, which is also required for proper muscle development [26].
Moreover, the results from additional physiological investigations indicate that Eya3 mutant mice suffer also from some heart and lung problems as indicated by some ECG parameters (increased PQ and JT intervals, decreased QRS amplitude) and a decreased tidal volume of the lung. Indeed, Eya4 is also involved in cardiac processes as demonstrated previously in human and zebrafish [20]. Taking the lung, heart and muscle alterations together, the significantly reduced locomotion might be understood as a consequence of all these features. Since we have tested the mice only at a young age, these deficits might accumulate during ageing and therefore be responsible for more severe deficits when the mice become older.
To understand the molecular network involving Eya3, we analyzed differentially expressed genes in the mutants. We identified several regulated genes in heart and brain indicating regulatory effects of the Eya3 mutation or secondary affects based upon missing or changed cell types. Among them, Nup155 was confirmed by real-time PCR to be significantly down-regulated in both organs. Nup155 is involved in nuclear envelope formation during cell division and is important for the export of macromolecules like mRNAs from the nucleus to the cytoplasm. Depletion of Nup155 leads to a failure of nuclear lamina formation and defects in chromosome segregation at anaphase [27] and therefore has an important impact on cell proliferation. Nup155 encodes a protein containing nucleoporin, a non-repetitive/WGA-negative domain. Gene ontology (GO) terms indicate that Nup155 protein is located in a nuclear pore and in a membrane.
Among the 17 down-regulated genes in the heart of Eya3 mutant mice, some are associated with specific cardiac functions, e.g. Tnnc1 is involved in cardiac muscle contraction, Calm3 plays a role in calcium signaling, and Ankrd1 is a transcriptional co-factor being required during cardiac myogenesis [28,29].
Furthermore, for several regulated genes in the brain an association with neurodegeneration, particularly with Alzheimer disease has been found. Among the up-regulated genes, ASS1 expression levels were significantly higher in glial cells of Alzheimer's disease brains [30]. Among the genes being down-regulated in the brains of Eya3 mutants, several are reported to be down-regulated also in Alzheimer's disease, e.g. ARPP19 [31], BIRC4 [32], MT3 [33], SYT1 [34] or TTR [35]. Moreover, the SCA10 gene, which is down-regulated in the brain of the Eya3 mutants, is also down-regulated in patients suffering from Parkinson's disease [36]. This short comparison between the differentially expressed genes in the Eya3 mutant mice with corresponding features in neurodegenerative disorders in human patients points to an interesting putative role of EYA3 in Alzheimer's and Parkinson's disease. This function might be attributed by putative interaction of Eya3 with either Six1 (Six1 binding sites were reported in the Birc4 promoter) or with Pax6 (Pax6 binding sites have been identified in the Ass1 promoter) (data accumulated by Bibliosphere). These functional considerations are based upon associative interpretations of available literature and comparison with the data obtained during the basic characterization of the Eya3 mutant opening several interesting avenues for further detailed experimental studies.
Conclusion
Our study demonstrates that Eya3 affects particularly heart, lung and muscle functions. Even if the exploratory behavior is increased, the mutant mice move less than the wild-type mice. Together with the effects on respiratory, muscle and heart function, it might lead to more severe effects, when the mice become older. Therefore, future investigations of Eya3 function should focus on effects on aging mice and consider also effects related to neurodegenerative disorders like Alzheimer's or Parkinson's disease.
Methods
Generation and molecular characterization of Eya3 mutant mice
The Eya3 deficient mouse line was generated by insertional mutagenesis in a large-scale gene trap screen (German Gene Trapping Consortium, GGTC). A promoterless reporter gene (lacZ) was inserted into the mouse genome by random integration of a gene trap vector (pT1βgeo) [37]. ES cells were derived from strain Sv129S2 (formerly known as Sv129Pasteur). Mutant mice were generated through blastocyst injection and resulting chimeras were bred to C57BL/6 females and crossed to C57BL/6 for four generations. Thus, mice were predominantly C57BL/6. All experiments and housing of the animals were performed according to the German Law on the Protection of Animals (regulation 209.1/211-2531-112/02 by the Government of Upper Bavaria [Regierung von Oberbayern]).
For detection of the gene trap vector integration in mutant mice a genomic forward primer was combined with a reverse primer in the lacZ cassette of the gene trap vector. Genotyping of mice was performed by Triplex-PCR. A genomic forward primer was either combined with a genomic reverse primer for wild-type amplification or with a reverse primer within the gene trap vector for mutant amplification (table 4). PCR was carried out using the following cycles: 1 × 3 min 94°C; 35 × 94°C 30 sec, 62°C 45 sec, 72°C 60 sec; 1 × 72°C 5 min (PCR-cycler: MJ Research PTC-225, Bio-Rad Laboratories, Munich, Germany).
Table 4.
PCR primer for characterization of the insertion of the gene trap vector
| Primer | Sequence (5' → 3') |
| gene trap forward primer (genomic) | TGGGATGCAAACTCGGGTCCTA |
| gene trap reverse primer (lacZ cassette) | CAAGGCGATTAAGTTGGGTAACG |
| genotyping forward primer (wild type/mutant) | CAGGAACAGACTGAAACCATGC |
| genotyping reverse primer (wild type) | AGGTTGTCAGACTGGCACTTCA |
| genotyping reverse primer (mutant) | CACGCCATACAGTCCTCTTCAC |
| ex 1_forward primer | CAGAGTGGGTCCGTAATCGT |
| ex 7_reverse primer | TGCCCAGGATAAGTGGTTTC |
| ex 8_reverse primer | ATTGCAGATGAAGTGGGTATCAG |
| ex 8_forward primer | CCAGCCTGATACCCACTTCA |
| ex 11_reverse primer | ATACCCGTTCCAATTCACTGTC |
| ex 12_forward primer | CCAACAGTAGTAATTGGCTCAGGT |
| ex 15_reverse primer | ACCTCGATCTCTGCTCTGAGTCT |
RNA isolation, RT-PCR and Northern blotting
Total RNA was prepared with RNA-Bee (Tel-Test Inc., Friendswood, TX, USA) according to the suppliers protocol. Tissue (50 μg each) was taken from wild-type animals and homozygous mutants from 3 different embryonic stages: E11.5 (whole embryo), E13.5 (head) and E15.5 (head). cDNA was generated by using Superscript II RT (Invitrogen, Karlsruhe, Germany).
Northern Blotting was done with RNA from E15.5 embryos. Samples were initially denatured for 10 min at 68°C and loaded onto an agarose-gel (1,2%, 1 × MOPS-buffer) and separated under denaturing conditions for 4 h (60 V). RNA samples were blotted to Hybond-N+ membrane (Amersham Biosciences, GE Healthcare-Buchler, Braunschweig, Germany) overnight and UV-fixed. Afterwards, the membrane was prehybridized for 2 h with CHURCH-buffer (7% SDS, 50% formamide, 5 × SSC, 2% Roche blocking reagent, 50 mM Na2HPO4, 0,1% laurylsarcosyl) and then hybridized overnight at 52°C with a digoxigenin-labeled riboprobe (Roche Diagnostics, Mannheim, Germany) against exons 8 – 11 of Eya3. Hybridization was followed by washing in 2 × SSC (30 mM Na-Citrat pH 7.0; 300 mM NaCl), 0.1% SDS for 5 min and in 0.1 × SSC, 0.1% SDS for 40 min at 52°C. After incubation with blocking reagent in 100 mM Tris-HCl pH 9.5 and 150 mM NaCl for 30 min at 52°C the membrane was washed in 100 mM Tris-HCl pH 9.5, 150 mM NaCl and incubated with anti-digoxygenin antibody (Roche, diluted 1:5000 in 100 mM Tris-HCl pH 9.5, 150 mM NaCl) for 30 min at room temperature. Finally, the membrane was stained with NBT/BCIP in 100 mM Tris-HCl pH 9.5, 100 mM NaCl, 50 mM MgCl2 for 1 h in the dark.
LacZ-staining and in-situ hybridization of Eya3
LacZ-staining was performed on whole mouse embryos of developmental stages E11.5 – E13.5. Mouse embryos were dissected and fixed in 4% paraformaldehyde in PBS. For staining embryos were washed three times in PBS containing 0.01% Na-deoxicholate, 0.02% Non-idet P40, 5 mM EGTA (pH 8.0) and 2 mM MgCl2 after fixation. Afterwards, embryos were stained overnight at 37°C in PBS containing 0.5 mg/ml X-gal, 2 mM MgCl2, 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, 0.01% Na desoxycholate, and 0.02% NP-40 in the dark. Before postfixation in 4% paraformaldehyde in PBS at 4°C, several wash steps were carried out in PBS. Embryos were stored in 70% ethanol at 4°C.
In-situ hybridizations with paraffin sections were performed to evaluate Eya3 expression in the eye. Embryos were fixed in 4% paraformaldehyde in PBS at 4°C, dehydrated and embedded in Jung Histowax (Cambridge Instruments, Nussloch, Germany). Sections (8 μm) were cut with the RM 2065-microtome (Leica, Wetzlar, Germany), mounted onto slices and treated as previously described [38].
Analysis of zebrafish embryos
Embryos were produced by paired matings of zebrafish (Danio rerio, AB strain), and staged in hours post-fertilization (hpf), as described previously [39,40]. In-situ hybridization for zebrafish embryos was performed as previously described [41]. For sectioning at the tailbud stage, the stained embryos were cryoprotected in 15% sucrose in phosphate buffer, embedded in 7.5% gelatin, 15% sucrose in PBS, frozen and cryostat-sectioned at 15 μm. For sectioning at the 28 hpf stage, the stained embryos were embedded in 3% agarose and vibratome-sectioned at 70 μm. As probe, we used an eya3 cDNA clone generously provided by G. Nica (LMU Munich).
Phenotyping of mice
A broad phenotypical analysis of 24 wild-type controls (14 ♂, 10 ♀) and 24 Eya3-/- mice (14 ♂, 10 ♀) was done in collaboration with the German Mouse Clinic [24]. For histological analysis of the eye 11 week old mice were sacrificed and eyes were removed immediately. Fixation and staining with hematoxylin and eosin were performed using standard-protocols. Funduscopy (age: 11 weeks) and electroretinography (age: 11 weeks, 9 months) were performed as described before [42]. Laser interference biomicroscopy (age: 13 weeks; n = 6) was carried out as previously described [43]. Data from controls and mutant animals were compared using the Student's t-test. Tables show mean value ± standard error of the mean (s.e.m.).
Behavioral phenotyping was done at the age of 8 weeks by using the modified hole board test [44] and carried out as previously described [45]. Data were statistically analyzed using SPSS software (SPSS Inc, Chicago, USA). The forelimb grip strength was tested at the age of 10 weeks with the grip strength meter system, which determines the muscle strength of a mouse. The device exploits the tendency of a mouse to grasp a horizontal metal bar while being pulled by its tail. During the measurement the mouse grasps a special adjustable grip (2 mm) mounted on a force sensor. Five trials were undertaken for each mouse within one minute. Values were presented as means ± standard error of the mean (SEM) and analyzed for the effects of genotype, sex and weight by fitting linear mixed effect models.
To examine the cardiovascular phenotype a surface limb ECG was obtained at the age of 12 – 15 weeks in anesthetized mice. The complete setup was located in a faraday cage. A shape analysis of the ECG traces was performed using the software ECG-auto (EMKA Technologies, Paris, France). For each animal, intervals and amplitudes were measured from five different sets of averaged beats. In addition, the recordings were screened for arrhythmias, including supraventricular and ventricular extrasystoles and conduction blockages. Analysis of variance (ANOVA) was used for multi-factorial analysis of quantitative traits with respect to sex and genotype.
Lung function was determined by whole body plethysmography according to a previously described principle [46]. For measurement a system from Buxco® Electronics (Sharon, Connecticut, USA) was used to assess breathing patterns in unrestrained animals by measuring pressure changes which arise from inspiratory and expiratory temperature and humidity fluctuations during breathing. The system allows transforming pressure swings into flow and volume signals, so that automated data analysis provides tidal volumes, respiratory rates, minute ventilation, inspiratory and expiratory times as well as peak inspiratory and peak expiratory flow rates. Measurements were always performed between 8 a.m. and 11 a.m. to account for potential diurnal variations in breathing.
Gene expression of putative downstream genes were analyzed using Glass cDNA-chips containing the fully sequenced 20K cDNA mouse arrayTAG library (Lion Bioscience, Heidelberg, Germany) [47-50]. A full description of the probes on the microarray is available in the GEO database under GPL4937 [51]. The expression data of Eya3 analysis of heart, muscle and brain have been submitted to the GEO database (GSE9999). Total RNA from brain, heart and muscle of 12 – 13 week old male animals (5 controls, 4 mutants) was used for cDNA chip hybridization according to a modified TIGR protocol [52]. Two chip hybridizations including a dye-swap experiment were performed from all selected organs of each individual mutant mouse against the identical pool of the same organ of reference RNAs. Slides were scanned with a GenePix 4000A microarray scanner (GenePix Pro 6.1 image processing software, Axon Instruments, USA). TIGR software package for Microarray analysis [53] including MIDAS (Microarray Data Analysis System) was used for normalization and SAM (Significance Analysis of Microarrays) [54] for the identification of significantly differentially expressed genes. Genes were ranked according to their relative difference value d(i) and selected as significantly regulated with d(i) > 0.5. To estimate the false discovery rate, false positive genes were identified by calculating 1000 permutations of the measurements. The selection of the top differentially expressed genes with reproducible up- or down-regulation includes less than 10% false positives.
The commercial software tool Bibliosphere Pathway Edition (Genomatix Software GmbH, Germany) was used to identify pathways and gene functions that were significantly abundant among the regulated genes. Regulated genes were filtered for two categories of Gene Ontology terms: biological processes and molecular functions. A statistical analysis was performed based on the relative number of observed versus expected annotations of each term. The z-score (> 5.0) indicates whether particular GO annotations are over-represented in a dataset of genes.
Results from Microarray analysis were controlled by real-time PCR for differentially expressed genes in heart and brain with the ABI Prism 7000 Detection System (Applied Biosystems, Darmstadt, Germany). Therefore, the identical total RNA as for the microarray experiments was used. Afterwards, cDNA was prepared with the Ready-to-go T-primed First-strand Kit (Amersham Biosciences, Braunschweig, Germany) according to the manufacturers protocols. Results from real-time PCR were analyzed with the ABI Prism 7000 SDS software and REST (Relative Expression Software Tool [55]).
Sequence Comparison
For sequence comparison of the 5 kb upstream of the Eya3 transcription start sites we used the program mVISTA (http://genome.lbl.gov/vista/mvista/about.shtml[56])
Abbreviations
ANOVA: analysis of variance; BO: branchio-oto syndrome; BOR: branchio-oto-renal syndrome; ECG: electrocardiography; ED: Eya domain; GGTC: German Gene Trap Consortium; GMC: German Mouse Clinic; GO: gene ontology; PBS: phosphate-buffered saline, RACE: rapid amplification of cDNA ends; RT-PCR: reverse transcription – polymerase chain reaction; SEM: standard error of the mean.
Authors' contributions
TS, performed the genetic analysis and breeding of the mice, the expression analysis and wrote the manuscript; CD, performed most of the studies of the eyes, analyzed the data and wrote the manuscript; OP, performed some of the studies of the eyes and analyzed the data; TF, created the mouse mutant; LB, performed the neurological screen and analyzed the data; IB, performed the lung function screen and analyzed the data; JF, performed some of the studies at the eyes, analyzed the data and wrote the manuscript; WH, performed the dysmorphology screen and analyzed the data; SMH, performed the behavioral screen, analyzed the data and wrote the manuscript; MH, designed the expression profiling studies and performed the statistical and functional analysis; MK, performed the behavioral screen and analyzed the data; EK, performed the neurological screen and analyzed the data; CM, performed the clinical-chemical screen and analyzed the data; AS, performed the cardio-vascular screen and analyzed the data; CS, designed the experiments on zebrafish and analyzed the data; ST, performed some of the expression studies on zebrafish; VGD, design and coordination of primary workflow and wrote the manuscript; BN, coordination primary workflow; JB, designed the expression profiling and wrote the manuscript; HF, design and coordination of primary workflow, performed the dysmorphology screen, and wrote the manuscript; BI, designed the cardiovascular screen, validated and discussed its results; TK, designed the neurology screen; HS, designed the lung function screen and wrote the manuscript; EW, designed the clinical-chemical screen; WW, designed the gene-trap approach for mouse mutant generation and designed the behavioral screen; LBC, designed the zebrafish experiments, analyzed the data and wrote the manuscript; MHdA, design of concept GMC, primary workflow; study design; JG, designed the experiments, discussed the results at all stages with various partners and wrote the manuscript. All authors read and improved the final manuscript.
Acknowledgments
Acknowledgements
The excellent technical assistance of Miriam Backs, Erika Bürkle, Elfi Holupirek, Regina Kneuttinger, Maria Kugler, Sandra Schädler, Reinhard Seeliger, Monika Stadler, Susanne Wittich and Claudia Zeller (Helmholtz Center Munich) is gratefully acknowledged. This work was supported at least in part by the German National Genome Research Network (NGFN; Grants 01GR0430, 01GR0434, 01GR0438, 01KW0106, 01KW9948 and R0313435B) and by the European Union (FP6, EUMODIC, LSHG-CT-2006-037188).
Contributor Information
Torben Söker, Email: tsoeker@web.de.
Claudia Dalke, Email: dalke@helmholtz-muenchen.de.
Oliver Puk, Email: oliver.puk@helmholtz-muenchen.de.
Thomas Floss, Email: tfloss@helmholtz-muenchen.de.
Lore Becker, Email: lore.becker@helmholtz-muenchen.de.
Ines Bolle, Email: ines.bolle@freenet.de.
Jack Favor, Email: favor@helmholtz-muenchen.de.
Wolfgang Hans, Email: wolfgang.hans@helmholtz-muenchen.de.
Sabine M Hölter, Email: hoelter@helmholtz-muenchen.de.
Marion Horsch, Email: horsch@helmholtz-muenchen.de.
Magdalena Kallnik, Email: magdalena.kallnik@web.de.
Eva Kling, Email: eva2308@gmx.de.
Corinna Moerth, Email: moerth@biochem.mpg.de.
Anja Schrewe, Email: anja.schrewe@helmholtz-muenchen.de.
Christian Stigloher, Email: stiglohe@biologie.ens.fr.
Stefanie Topp, Email: ToppStefanie@aol.com.
Valerie Gailus-Durner, Email: gailus@helmholtz-muenchen.de.
Beatrix Naton, Email: beatrix.naton@helmholtz-muenchen.de.
Johannes Beckers, Email: beckers@helmholtz-muenchen.de.
Helmut Fuchs, Email: hfuchs@helmholtz-muenchen.de.
Boris Ivandic, Email: boris_ivandic@med.uni-heidelberg.de.
Thomas Klopstock, Email: Thomas.Klopstock@med.uni-muenchen.de.
Holger Schulz, Email: Schulz@helmholtz-muenchen.de.
Eckhard Wolf, Email: ewolf@lmb.uni-muenchen.de.
Wolfgang Wurst, Email: wurst@helmholtz-muenchen.de.
Laure Bally-Cuif, Email: bally@helmholtz-muenchen.de.
Martin Hrabé de Angelis, Email: hrabe@helmholtz-muenchen.de.
Jochen Graw, Email: graw@helmholtz-muenchen.de.
References
- Xu PX, Woo IW, Her H, Beier DR, Maas RL. Mouse Eya homologues of the Drosophila eyes absent gene require Pax6 for expression in lens and nasal placode. Development. 1997;124:219–231. doi: 10.1242/dev.124.1.219. [DOI] [PubMed] [Google Scholar]
- Borsani G, DeGrandi A, Ballabio A, Bulfone A, Bernard L, Banfi S, Gattuso C, Mariani M, Dixon M, Donnai D, Metcalfe K, Winter R, Robertson M, Axton R, Brown A, van Heyningen V, Hanson I. Eya4, a novel vertebrate gene related to Drosophila eyes absent. Hum Mol Genet. 1999;8:11–23. doi: 10.1093/hmg/8.1.11. [DOI] [PubMed] [Google Scholar]
- Bonini NM, Leiserson WM, Benzer S. The eyes absent gene: genetic control of cell survival and differentiation in the developing Drosophila eye. Cell. 1993;72:379–395. doi: 10.1016/0092-8674(93)90115-7. [DOI] [PubMed] [Google Scholar]
- Bonini NM, Leiserson WM, Benzer S. Multiple roles of the eyes absent gene in Drosophila. Dev Biol. 1998;196:42–57. doi: 10.1006/dbio.1997.8845. [DOI] [PubMed] [Google Scholar]
- Rebay I, Silver SJ, Tootle TL. New vision from Eyes absent: transcription factors as enzymes. Trends Genet. 2005;21:163–171. doi: 10.1016/j.tig.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Jemc J, Rebay I. The eyes absent family of phosphotyrosine phosphatases: properties and roles in developmental regulation of transcription. Annu Rev Biochem. 2007;76:513–538. doi: 10.1146/annurev.biochem.76.052705.164916. [DOI] [PubMed] [Google Scholar]
- Pignoni F, Hu B, Zavitz KH, Xiao J, Garrity PA, Zipursky SL. The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell. 1997;91:881–891. doi: 10.1016/S0092-8674(00)80480-8. [DOI] [PubMed] [Google Scholar]
- Chen R, Amoui M, Zhang Z, Mardon G. Dachshund and Eyes absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila. Cell. 1997;81:893–904. doi: 10.1016/S0092-8674(00)80481-X. [DOI] [PubMed] [Google Scholar]
- Ohto H, Kamada S, Tago K, Tominaga SI, Ozaki H, Sato S, Kawakami K. Cooperation of Six and Eya in activation of their target genes through nuclear translocation of Eya. Mol Cell Biol. 1999;19:6815–6824. doi: 10.1128/mcb.19.10.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tootle TL, Silver SJ, Davies EL, Newman V, Latek RR, Mills IA, Selengut JD, Parlikar BE, Rebay I. The transcription factor Eyes absent is a protein tyrosine phosphatase. Nature. 2003;426:299–302. doi: 10.1038/nature02097. [DOI] [PubMed] [Google Scholar]
- Rayapureddi JP, Kattamuri C, Steinmetz BD, Frankfort BJ, Ostrin EJ, Mardon G, Hegde RS. Eyes absent represents a class of protein tyrosine phosphatases. Nature. 2003;426:295–298. doi: 10.1038/nature02093. [DOI] [PubMed] [Google Scholar]
- Silver SJ, Davies EL, Doyon L, Rebay I. Functional dissection of Eyes absent reveals new modes of regulation within the Retinal Determination Gene Network. Mol Cell Biol. 2003;23:5989–5999. doi: 10.1128/MCB.23.17.5989-5999.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelhak S, Kalatzis V, Heilig R, Compain S, Samson D, Vincent C, Levi-Acobas F, Cruaud C, Le Merrer M, Mathieu M, König R, Vigneron J, Weissenbach J, Petit C, Weil D. Clustering of mutations responsible for branchio-oto-renal (BOR) syndrome in the eyes absent homologous region (eyaHR) of EYA1. Hum Mol Genet. 1997;6:2247–2255. doi: 10.1093/hmg/6.13.2247. [DOI] [PubMed] [Google Scholar]
- Xu PX, Adams J, Peters H, Brown MC, Heaney S, Maas R. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat Genet. 1999;23:113–117. doi: 10.1038/12722. [DOI] [PubMed] [Google Scholar]
- Azuma N, Hirakiyama A, Inoue T, Asaka A, Yamada M. Mutations of a human homologue of the Drosophila eyes absent gene (EYA1) detected in patients with congenital cataracts and ocular anterior segment anomalies. Hum Mol Genet. 2000;9:363–366. doi: 10.1093/hmg/9.3.363. [DOI] [PubMed] [Google Scholar]
- Clark IBN, Boyd J, Hamilton G, Finnegan DJ, Jarman AP. D-six4 plays a key role in patterning cell identities deriving from the Drosophila mesoderm. Dev Biol. 2006;294:220–231. doi: 10.1016/j.ydbio.2006.02.044. [DOI] [PubMed] [Google Scholar]
- Li X, Oghi KA, Zhang J, Krones A, Bush KT, Glass CK, Nigam SK, Aggarwal AK, Maas R, Rose DW, Rosenfeld MG. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature. 2003;426:247–265. doi: 10.1038/nature02083. [DOI] [PubMed] [Google Scholar]
- Grifone R, Demignon J, Giordani J, Niro C, Souil E, Bertin F, Laclef C, Xu PX, Maire P. Eya1 and Eya2 proteins are required for hypaxial somitic myogenesis in the mouse embryo. Dev Biol. 2006;302:602–616. doi: 10.1016/j.ydbio.2006.08.059. [DOI] [PubMed] [Google Scholar]
- Heanue TA, Reshef R, Davis RJ, Mardon G, Oliver G, Tomarev S, Lassar AB, Tabin CJ. Synergistic regulation of vertebrate muscle development by Dach2, Eya2 and Six1, homologs of genes required for Drosophila eye formation. Genes Dev. 1999;13:3231–3243. doi: 10.1101/gad.13.24.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönberger J, Wang L, Shin JT, Kim SD, Depreux F, Zhu H, Zon L, Pizard A, Kim JB, MacRae CA, Mungall AJ, Seidman JG, Seidman CE. Mutation in the transcriptional coactivator Eya4 causes dilated cardiomyopathy and sensorineural hearing loss. Nat Genet. 2005;37:418–422. doi: 10.1038/ng1527. [DOI] [PubMed] [Google Scholar]
- Makishima T, Madeo AC, Brewer CC, Zalewski CK, Butman JA, Sachdev V, Arai AE, Holbrook BM, Rosing DR, Griffith AJ. Nonsyndromic hearing loss DFNA10 and a novel mutation of EYA4. Am J Med Gen. 2007;143A:1592–1598. doi: 10.1002/ajmg.a.31793. [DOI] [PubMed] [Google Scholar]
- Depreux FF, Darrow K, Conner DA, Eavey RD, Liberman MC, Seidman CE, Seidman JG. Eya4-deficient mice are a model for heritable otitis media. J Clin Invest. 2008;118:651–658. doi: 10.1172/JCI32899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriebel M, Müller F, Hollemann T. Xeya3 regulates survival and proliferation of neural progenitor cells within the anterior neural plate of Xenopus embryos. Dev Dyn. 2007;236:1526–1534. doi: 10.1002/dvdy.21170. [DOI] [PubMed] [Google Scholar]
- Gailus-Durner V, Fuchs H, Becker L, Becker L, Bolle I, Brielmeier M, Calzada-Wack J, Elvert R, Ehrhardt N, Dalke C, Franz TJ, Grundner-Culemann E, Hammelbacher S, Hölter SM, Hölzlwimmer G, Horsch M, Javaheri A, Kalaydjiev SV, Klempt M, Kling E, Kunder S, Lengger C, Lisse T, Mijalski T, Naton B, Pedersen V, Prehn C, Przemeck G, Racz I, Reinhard C, Reitmeir P, Schneider I, Schrewe A, Steinkamp R, Zybill C, Adamski J, Beckers J, Behrendt H, Favor J, Graw J, Heldmaier G, Höfler H, Ivandic B, Katus H, Kirchhof P, Klingenspor M, Klopstock T, Lengeling A, Müller W, Ohl F, Ollert M, Quintanilla-Martinez L, Schmidt J, Schulz H, Wolf E, Wurst W, Zimmer A, Busch DH, Hrabé de Angelis M. Introducing the German Mouse Clinic: open access platform for standardized phenotyping. Nat Meth. 2005;2:404–404. doi: 10.1038/nmeth0605-403. [DOI] [PubMed] [Google Scholar]
- Grimm C, Chatterjee B, Favor J, Immervoll T, Löster J, Klopp N, Sandulache R, Graw J. Aphakia (ak), a mouse mutation affecting early eye development: fine mapping, consideration of candidate genes and altered Pax6 and Six3 gene expression pattern. Dev Genet. 1998;23:299–316. doi: 10.1002/(SICI)1520-6408(1998)23:4<299::AID-DVG5>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Laclef C, Souil E, Demignon J, Maire P. Thymus, kidney and craniofacial abnormalities in Six1 deficient mice. Mech Dev. 2003;120:669–679. doi: 10.1016/S0925-4773(03)00065-0. [DOI] [PubMed] [Google Scholar]
- Franz C, Askjaer P, Antonin W, Iglesias CL, Haselmann U, Schelder M, de Marco A, Wilm M, Antony C, Mattaj IW. Nup155 regulates nuclear envelope and nuclear pore complex formation in nematodes and vertebrates. EMBO J. 2005;24:3519–3531. doi: 10.1038/sj.emboj.7600825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaborit N, Le Bouter S, Szuts V, Varro A, Escande D, Nattel S, Demolombe S. Regional and tissue specific transcript signatures of ion channel genes in the non-diseased human heart. J Physiol. 2007;582:675–693. doi: 10.1113/jphysiol.2006.126714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Evans S, Chen J, Kuo HC, Harvey RP, Chien KR. CARP, a cardiac ankyrin repeat protein, is downstream in the Nkx2-5 homeobox gene pathway. Development. 1997;124:793–804. doi: 10.1242/dev.124.4.793. [DOI] [PubMed] [Google Scholar]
- Heneka MT, Wiesinger H, Dumitrescu-Ozimek L, Riederer P, Feinstein DL, Klockgether T. Neuronal and glial coexpression of argininosuccinate synthetase and inducible nitric oxide synthase in Alzheimer disease. J Neuropathol Exp Neurol. 2001;60:906–916. doi: 10.1093/jnen/60.9.906. [DOI] [PubMed] [Google Scholar]
- Kim SH, Nairn AC, Cairns N, Lubec G. Decreased levels of ARPP-19 and PKA in brains of Down syndrome and Alzheimer's disease. J Neural Transm Suppl. 2001:263–272. doi: 10.1007/978-3-7091-6262-0_21. [DOI] [PubMed] [Google Scholar]
- Christie LA, Su JH, Tu CH, Dick MC, Zhou J, Cotman CW. Differential regulation of inhibitors of apoptosis proteins in Alzheimer's disease brains. Neurobiol Dis. 2007;26:165–173. doi: 10.1016/j.nbd.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Isart N, Jensen LT, Zerbe O, Winge DR, Vasak M. Engineering of metallothionein-3 neuroinhibitory activity into the inactive isoform metallothionein-1. J Biol Chem. 2002;277:37023–37028. doi: 10.1074/jbc.M205730200. [DOI] [PubMed] [Google Scholar]
- Yoo BC, Cairns N, Fountoulakis M, Lubec G. Synaptosomal proteins, beta-soluble N-ethylmaleimide-sensitive factor attachment protein (beta-SNAP), gamma-SNAP and synaptotagmin I in brain of patients with Down syndrome and Alzheimer's disease. Dement Geriatr Cogn Disord. 2001;12:219–225. doi: 10.1159/000051261. [DOI] [PubMed] [Google Scholar]
- Gloeckner SF, Meyne F, Wagner F, Heinemann U, Krasnianski A, Meissner B, Zerr I. Quantitative analysis of transthyretin, tau and amyloid-beta in patients with dementia. J Alzheimers Dis. 2008;14:17–25. doi: 10.3233/jad-2008-14102. [DOI] [PubMed] [Google Scholar]
- März P, Probst A, Lang S, Schwager M, Rose-John S, Otten U, Ozbek S. Ataxin-10, the spinocerebellar ataxia type 10 neurodegenerative disorder protein, is essential for survival of cerebellar neurons. J Biol Chem. 2004;279:35542–35550. doi: 10.1074/jbc.M405865200. [DOI] [PubMed] [Google Scholar]
- Wiles MV, Vauti F, Otte J, Füchtbauer EM, Ruiz P, Füchtbauer A, Arnold HH, Lehrach H, Metz T, von Melchner H, Wurst W. Establishment of a gene-trap sequence tag library to generate mutant mice from embryonic stem cells. Nat Genet. 2000;24:13–14. doi: 10.1038/71622. [DOI] [PubMed] [Google Scholar]
- Grimm C, Chatterjee B, Favor J, Immervoll T, Löster J, Klopp N, Sandulache R, Graw J. Aphakia (ak), a mouse mutation affecting early eye developmentfine mapping, consideration of candidate genes and altered Pax6 and Six3 gene expression pattern. Dev Genet. 1998;23:299–316. doi: 10.1002/(SICI)1520-6408(1998)23:4<299::AID-DVG5>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio) 3. Eugene, OR: University of Oregon Press; 1995. [Google Scholar]
- Hauptmann G, Gerster T. Two-color whole-mount in-situ hybridization to vertebrate and Drosophila embryos. Trends Genet. 1994;10:266. doi: 10.1016/0168-9525(90)90008-T. [DOI] [PubMed] [Google Scholar]
- Dalke C, Löster J, Fuchs H, Gailus-Durner V, Soewarto D, Favor J, Neuhäuser-Klaus A, Pretsch W, Gekeler F, Shinoda K, Zrenner E, Meitinger T, Hrabé de Angelis M, Graw J. Electroretinography as a screening method for mutations causing retinal dysfunction in mice. Invest Ophthalmol Vis Sci. 2004;45:601–609. doi: 10.1167/iovs.03-0561. [DOI] [PubMed] [Google Scholar]
- Puk O, Dalke C, Favor J, Hrabé de Angelis M, Graw J. Variations of eye size parameters among different strains of mice. Mamm Genome. 2006;17:851–857. doi: 10.1007/s00335-006-0019-5. [DOI] [PubMed] [Google Scholar]
- Ohl F, Sillaber I, Binder E, Keck ME, Holsboer F. Differential analysis of behavior and diazepam-induced alterations in C57BL/6N and BALB/c mice using the modified hole board test. J Psychiatr Res. 2001;35:147–154. doi: 10.1016/S0022-3956(01)00017-6. [DOI] [PubMed] [Google Scholar]
- Kallnik M, Elvert R, Ehrhardt N, Kissling D, Mahabir E, Welzl G, Faus-Kessler T, Hrabé de Angelis M, Wurst W, Schmidt J, Hölter SM. Impact of IVC housing on emotionality and fear learning in male C3HeB/FeJ and C57BL/6J mice. Mamm Genome. 2007;18:173–186. doi: 10.1007/s00335-007-9002-z. [DOI] [PubMed] [Google Scholar]
- Drorbaugh JE, Fenn WO. A barometric method for measuring ventilation in newborn infants. Pediatrics. 1955;16:81–87. [PubMed] [Google Scholar]
- Greenwood AD, Horsch M, Stengel A, Vorberg I, Leib-Mösch C. Cell line dependent RNA expression profiles of prion-infected mouse neuronal cells. J Mol Biol. 2005;349:487–500. doi: 10.1016/j.jmb.2005.03.076. [DOI] [PubMed] [Google Scholar]
- Seltmann M, Horsch M, Drobyshev A, Chen Y, Beckers J. Assessment of a systematic expression profiling approach in ENU-induced mouse mutant lines. Mamm Genome. 2005;16:1–10. doi: 10.1007/s00335-004-3012-x. [DOI] [PubMed] [Google Scholar]
- Beckers J, Herrmann F, Rieger S, Drobyshev AL, Seliger B. Identification and validation of novel ERBB2 (HER2, NEU) targets including genes involved in angiogenesis. Int J Cancer. 2005;114:590–597. doi: 10.1002/ijc.20798. [DOI] [PubMed] [Google Scholar]
- Horsch M, Schädler S, Gailus-Durner V, Fuchs H, Meyer H, Hrabé de Angelis M, Beckers J. Systematic gene expression profiling of mouse model series reveals coexpressed genes. Proteomics. 2008;8:1248–1256. doi: 10.1002/pmic.200700725. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucl Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde P, Qi R, Abernathy K, Gay C, Dharap S, Gaspard R, Hughes JE, Snesrud E, Lee N, Quackenbush J. A concise guide to cDNA microarray analysis. Biotechniques. 2000;29:548–556. doi: 10.2144/00293bi01. [DOI] [PubMed] [Google Scholar]
- Saeed A, Sharov V, White J, Li J, Lioang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucl Acids Res. 2002;30:36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA computational tools for comparative genomics. Nucl Acids Res. 2004;32:W273–279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]