Abstract
In mammals, X-chromosome dosage compensation is achieved by inactivating one of the two X chromosomes in females. In mice, X inactivation is initially imprinted, with inactivation of the paternal X (Xp) chromosome occurring during preimplantation development. One theory is that the Xp is preinactivated in female embryos, because of its previous silence during meiosis in the male germ line. The extent to which the Xp is active after fertilization and the exact time of onset of X-linked gene silencing have been the subject of debate. We performed a systematic, single-cell transcriptional analysis to examine the activity of the Xp chromosome for a panel of X-linked genes throughout early preimplantation development in the mouse. Rather than being preinactivated, we found the Xp to be fully active at the time of zygotic gene activation, with silencing beginning from the 4-cell stage onward. X-inactivation patterns were, however, surprisingly diverse between genes. Some loci showed early onset (4–8-cell stage) of X inactivation, and some showed extremely late onset (postblastocyst stage), whereas others were never fully inactivated. Thus, we show that silencing of some X-chromosomal regions occurs outside of the usual time window and that escape from X inactivation can be highly lineage specific. These results reveal that imprinted X inactivation in mice is far less concerted than previously thought and highlight the epigenetic diversity underlying the dosage compensation process during early mammalian development.
Keywords: dosage compensation, epigenetics, imprinting, mouse development, X inactivation
In mice, X-chromosome inactivation (XCI) occurs in at least 2 waves during development. The first wave occurs during preimplantation embryogenesis and is subject to imprinting, resulting in preferential inactivation of the paternal X (Xp) chromosome (1). Inactivity of the Xp is maintained in the trophectoderm of the blastocyst but is reversed in the inner cell mass (2, 3), where a second wave of random XCI takes place, affecting either the Xp or the maternal X (Xm) chromosome (4, 5). The initiation of this second wave of XCI is controlled by a key regulatory locus, the X-inactivation center (Xic), which monoallelically upregulates either the paternal or maternal allele of the noncoding Xist transcript, thus triggering XCI in cis during an early time window of differentiation [for review, see Heard and Disteche (6)]. The events underlying the random form of XCI are being unraveled in ES cells. However, the nature of the imprint(s) underlying imprinted XCI and the time window during which silencing actually occurs remain open questions. The imprint could be of maternal or paternal origin, or both. The existence of a maternal resistance to XCI is supported by the fact that maternal disomy for the X chromosome results in early embryonic lethality due to deficient XCI (7, 8). This maternal imprint, which would be laid down during oogenesis (9), may lie at the level of the Xist gene. The maternal Xist allele is repressed during early embryogenesis, and paternally Xist-deficient mice are lethal (10, 11). In the face of the Xm chromosome's resistance to inactivate, the early activity of the paternal genome would result in paternal Xist activity and trigger subsequent Xp inactivation (12). Another hypothesis is that the Xp is transmitted to the zygote in a preinactivated state because of its passage through the male germ line (13). Here, the X and Y undergo meiotic sex chromosome inactivation (MSCI) followed by postmeiotic sex chromatin formation (PMSC) (14, 15). These 2 hypotheses, although not mutually exclusive, remain feasible according to current data [for review, see Reik and Ferguson-Smith (16)]. One way to distinguish between them is to examine the activity of the Xp chromosome after fertilization. To date, X-linked gene activity has mainly been examined from the morula stage, by RT-PCR. At this stage most genes already show some degree of silencing (13). Only a few genes have been examined in 2-cell embryos, and their expression suggests that the Xp is in fact active at this stage (12), with inactivation occurring after paternal Xist RNA coating at the 4-cell stage. This would be consistent with imprinted Xist activity dictating the onset of imprinted Xp inactivation. Nevertheless, it cannot be excluded that in addition to an Xist-mediated process, some degree of paternal X chromosomal silencing is already achieved at the time of fertilization owing to MSCI-dependent preinactivation, or that the MSCI process provides some memory predisposing the Xp to inactivation. A systematic analysis of paternal X-linked gene activity has not, however, been performed at the stages immediately following fertilization to address this. Furthermore, the kinetics and timing of XCI immediately following fertilization have only been examined in very few cases. Although it has been proposed that X-linked genes are inactivated as a gradient across the X chromosome, with genes located closest to Xist being silenced the earliest (13), this was based on morula-stage embryos only. Indeed, we have previously shown that a number of chromatin changes affect the Xp chromosome, from as early as the 4-cell stage. These changes include exclusion of RNA Polymerase II, loss of H3K4me2, and enrichment in H3K27me3 (2) and imply that global silencing may initiate at this stage, but to date only 1 X-linked gene has been examined.
In this study we set out to obtain an in-depth view of the early events in imprinted XCI. The transcriptional activity of a panel of X-linked genes on the Xp was examined during early development, from the time of zygotic gene activity (ZGA) to the blastocyst stage, and beyond in some cases. We show that genes on the Xp are active at the time of ZGA, including those that were silent at the end of spermatogenesis and had been subject to PMSC (14). No correlation could be found between their transcriptional status in the male germ line and the timing and efficiency of XCI in female embryos. Our analysis also revealed highly unusual X-inactivation kinetics for some genes on the Xp. This reveals that different mechanisms of propagation and escape from silencing may be at play in different regions of the X chromosome.
Results
Paternal X-Chromosome Activity at the 2-Cell Stage.
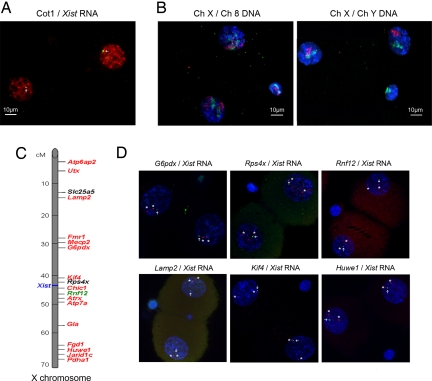
Xist RNA is first detectable from the Xp at the 2-cell stage, around the time of major ZGA in the mouse. At this stage, it appears as a small punctate signal by RNA FISH (Fig. 1A), resembling a primary transcript more than an accumulation over the Xp chromosome. To assess paternal X-chromosome activity at the time of ZGA, we performed Cot 1 RNA FISH, which detects transcribed repeats in the nucleus and has previously been used as an assay for X-chromosome activity (12, 13, 17, 18), although the transcripts detected by such a probe do not necessarily correspond to genic transcripts, as previously shown by our group and others (17, 18). Our analysis on 2-cell female embryos (n = 85) revealed no sign of any particular Cot 1 RNA depletion, or enrichment, in chromatin within or around the Xist RNA signal, compared with the rest of the nucleus (Fig. 1A). Repeat-specific probes, such as B1 or B2 probes of the SINE family, gave a similar pattern of expression (Fig. S1). To assess the size of the Xp-chromosome territory, relative to its maternal homologue, we performed DNA FISH with an X-chromosome paint probe on 2-cell embryos (n = 50). This revealed that both the maternal and paternal X chromosomes occupy a substantial fraction of the total nuclear volume at the 2-cell stage (Fig. 1B). No obvious difference in the intensity and size of FISH signals could be detected for the Xp and Xm chromosome territories, or for the X chromosome compared with an autosome (chromosome 8) (Fig. 1B, Left and Right). Thus, the Xp chromosome does not seem to be particularly condensed or show signs of generalized transcriptional repression at this stage of development. Furthermore, the X-chromosome territory clearly represents a much larger volume than the Xist RNA signal at this stage (compare Fig. 1 A and B). Thus, Xist RNA has not yet coated the Xp chromosome at the 2-cell stage of development.
Fig. 1.
Transcriptional activity of the paternal X chromosome assayed by gene RNA FISH in 2-cell embryos. (A) Deconvolved images of 3D stacks of a representative 2-cell female embryo labeled for RNA FISH with an Xic-specific probe (green) and Cot 1 RNA (red). Cot 1 is present at the site of the Xist RNA. (B) Deconvolved images of 3D stacks of a representative 2-cell embryo labeled for DNA FISH. Left: Female embryo with an Xic-chromosome probe (green) and chromosome 8 probe (red). Right: Male embryo with an Xic-chromosome probe (green) and chromosome Y probe (red). Y chromosome is not present in the second polar body. All of the chromosomes territories are dispersed. (C) X-chromosome map with the 18 studied genes. In red, genes that were shown to be fully or partially repressed during spermatogenesis in round spermatids (14); in green, gene that was shown to be reactivated in round spermatids (14); in black, genes that have been not studied in Namekawa and colleagues' report (14). (D) Representative 2-cell female embryos are shown for Xist transcript (green), gene primary transcripts (red), and DAPI (blue). Two gene signals can be seen in each blastomere of 2-cell embryos.
Although the absence of Cot1 RNA depletion and the lack of Xp-chromosome condensation suggested that the Xp chromosome was transcriptionally active at the 2-cell stage, these criteria could not exclude gene inactivity. We therefore assessed the transcriptional status of the Xp chromosome by studying the expression of 18 X-linked genes using RNA FISH (Fig. 1C). This assay provides a direct read-out of transcription status at the primary transcript level and avoids issues of low mRNA levels and/or maternal and paternal mRNA pools with long half-lives. The genes we examined were selected from across the length of the X chromosome (Fig. 1C). We particularly chose genes that had previously been shown to remain silent in the male germ line after MSCI, because these are the best candidates for possible inactivity (preinactivation) at the 2-cell stage in female embryos after fertilization. Most of the 18 genes tested are thought to remain fully or partially silent in round spermatids after MSCI in the male germ line, with the exception of Rnf12, which becomes reactivated (14). All genes tested were expressed at the 2-cell stage, with the exception of Utx, which could not be efficiently detected by RNA FISH at this stage under our experimental conditions. For genes expressed at the 2-cell stage, 2 primary transcript signals, 1 of which was adjacent to the paternal Xist signal, could be seen in the majority of 2-cell-stage female embryos (Fig. 1D), implying that Xp is indeed active at this stage. The percentage of blastomeres showing a paternal (Xist-linked) or maternal (non–Xist linked) signal was scored for several genes (Fig. S2). No significant difference in the activity of the paternal and maternal alleles of X-linked genes could be found (Fisher's exact test, P > 0.05). For some genes (Atp6ap2, Lamp2, Slc25a5, Mecp2, Fmr1, Fgd1, and Pdha1), the signal of paternal origin was sometimes located at some distance from the paternal Xist transcript (Fig. 1A), making it difficult to define which allele was maternal or paternal. This was consistent with the degree of dispersion of the X-chromosome territories seen at this stage (Fig. 1B, Left). Nevertheless, biallelic expression of these genes could be readily detected at the 2-cell stage (Fig. S2). Thus, we conclude that both the Xp and Xm chromosomes are transcriptionally active at the time of zygotic activation for all genes examined, with no evidence for a continuity of gene silencing on the Xp from the male germ line to the fertilized embryo.
Diverse Kinetics of XCI During Preimplantation Mouse Development.
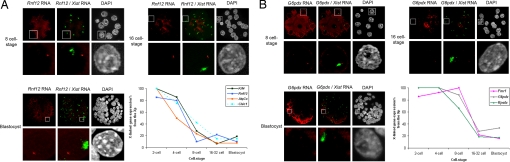
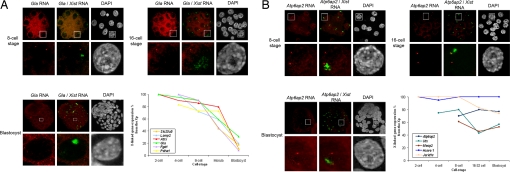
To determine the exact timing of XCI for different genes after the 2-cell stage, we performed RNA FISH at different stages of in vivo preimplantation mouse development (4-cell, 8-cell, 16–32-cell, and blastocyst stages). All experiments were performed on embryos in vivo. Embryos were generated from superovulated females for 2-cell to 32-cell stages and from natural matings for blastocysts and postimplantation stages. Differences between superovulated females and natural matings were not evaluated. No in vitro culture was used because this can perturb the kinetics of XCI (I.O. and C.P., unpublished data). For each gene, we normalized the primary transcript detection frequency obtained for the paternal (Xist RNA-associated) allele in female embryos against that obtained for the maternal allele in male embryos at the same stage (Figs. S3 and S4). This normalization step is essential to distinguish between gene silencing due to X inactivation vs. stage-specific changes in gene expression during early embryogenesis. We found different X-inactivation profiles for the genes tested, in terms of time of onset, kinetics, and efficiency. We divided the profiles into 4 broad categories. The first corresponded to genes such as Rnf12, Kif4, Chic1, and Atp7a, which were rapidly silenced from as early as the 4-cell stage and maintained in this silent state through to the blastocyst stage (Fig. 2A). For these genes, Xp expression dropped from >96% detection at the 2-cell stage to just 25% by the 8-cell stage. The second category corresponded to genes (G6pdx, Fmr1, and Rps4x) that initiated silencing later but that were efficiently silenced by the morula stage (Fig. 2B). Biallelic expression could be detected at the 2-cell, 4-cell, and 8-cell stages, but Xp expression had disappeared in more than 80% of blastomeres by the 16–32-cell stage, and this level of silencing was maintained to the blastocyst stage. A third category of genes (Lamp2, Slc25a5, Atrx, Gla, Fgd1, and Pdha1) exhibited a slower rate of silencing on the Xp, initiating after the 8-cell stage and only reaching maximum levels of silencing by the blastocyst stage (Fig. 3A). For the above categories a certain degree of escape from XCI could be detected for some genes by the blastocyst stage in up to 20% of cells. However, for a fourth category of genes (Atp6ap2, Utx, Mecp2, Jarid1c, and Huwe1), very little or no XCI could be detected throughout preimplantation development. These genes were still active on the Xp in the majority of trophectoderm cells by the blastocyst stage (Fig. 3B). Two of these genes have previously been shown to escape from XCI (Utx and Jarid1c). However, the partial inactivation of Atp6ap2 and Mecp2, as well as the complete lack of inactivation observed in the case of Huwe1, were unexpected.
Fig. 2.
Representative genes of a specific profile of transcriptional activity during X-imprinted inactivation assayed by RNA FISH in female embryos. (A) Representative gene silenced from the 8-cell stage. Rnf12 primary transcript signals (red) evidenced by specific RNA FISH at 8-cell (Upper Left), 16-cell (Upper Right), and blastocyst (Bottom Left) stages. One signal, distant from the Xist domain (green), is present in the majority of the cells from 8-cell to blastocyst stages. For each cell stage, a representative blastomere is shown. Bottom Right: Kinetics of expression of the 4 genes Rnf12, Atp7a, Chic1, and Kif4 silenced from the 8-cell stage. (B) Representative gene silenced from the morula stage. G6pdx primary transcripts (red) shown by specific RNA FISH at 8-cell (Upper Left), 16-cell (Upper Right), and blastocyst (Bottom Left) stages. Two G6pdx signals, with 1 within the Xist domain (green), could be seen in the majority of the blastomeres at the 8-cell stage, whereas only 1 signal could be observed in the majority of the blastomeres at the 16-cell and blastocyst stages. For each cell stage, a representative blastomere is shown. Bottom Right: Kinetics of expression of the 3 genes G6pdx, Rps4x, and Fmr1 silenced from the 16-cell stage.
Fig. 3.
Representative genes of a specific profile of transcriptional activity during X-imprinted inactivation assayed by RNA FISH in female embryos. (A) Representative gene silenced at the blastocyst stage. Gla primary transcripts (red) shown by specific RNA FISH at 8-cell (Upper Left), 16-cell (Upper Right), and blastocyst (Bottom Left) stages. Two Gla signals, with 1 within the Xist domain (green), could be seen in the majority of the blastomeres at the 8-cell and 16-cell stages, whereas only 1 signal could be observed in the majority of the blastomeres at the blastocyst stage. For each cell stage, a representative blastomere is shown. Bottom Right: Kinetics of expression of the 6 genes Slc25a5, Lamp2, Atrx, Gla, Fgd1, and Pdha1 silenced from the blastocyst stage. (B) Representative gene escaping to silence. Atp6ap2 primary transcripts (red) shown by specific RNA FISH at 8-cell (Upper Left), 16-cell (Upper Right), and blastocyst (Bottom Left) stages. Two Atp6ap2 signals, with 1 within the Xist domain (green), could be seen in the majority of the blastomeres throughout the preimplantation stages. For each cell stage, a representative blastomere is shown. Bottom Right: Kinetics of expression of the 5 genes Atp6ap2, Utx, Mecp2, Huwe1, and Jarid1c escaping from silencing.
These unexpectedly diverse patterns of X-inactivation that we found during preimplantation embryogenesis are summarized in Fig. S5. No apparent correlation could be found between gene inactivity in the male germ line and kinetics of XCI. For example, Rnf12, which reactivates after MSCI (14), is rapidly and efficiently silenced, whereas genes such as Pdha1, which shows slow XCI, and Huwe1, which shows no silencing in preimplantation stages, are both subject to MSCI and PMSC (14). Furthermore, no clear-cut correlation between X-linked gene location and the rate or efficiency of XCI could be made. However, we did find a significant correlation between proximity to the Xist locus and the time of onset of gene silencing (P < 0.02; linear regression test). This is particularly true for the Kif4–Atp7a cluster spanning the Xic. Our data are therefore supportive of a role for Xic-mediated initiation of XCI, although the kinetics of gene silencing are too diverse to be considered as a simple gradient from the Xic.
Early Imprinted Inactivation of Atrx Is Followed by Its Lineage-Specific Escape.
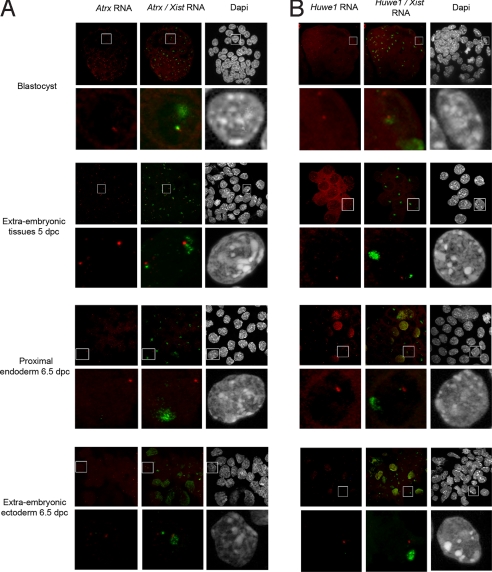
Previous reports had shown that Atrx escapes XCI in extraembryonic tissues, particularly in trophectoderm-derived tissues (19). We were therefore surprised to find that Atrx is fully silenced in the mural trophectoderm by the blastocyst stage (Fig. 4 and Fig. S6), as well as in trophoblast stem (TS) cells, which are derived from polar trophectoderm. In the ICM, Atrx is found to be active and then undergoes random inactivation in the epiblast lineage (I.O., unpublished data). We therefore examined early postimplantation-stage embryos for Atrx gene silencing. Consistent with previous reports, escape from X inactivation was detected for Atrx in extraembryonic tissues at 5 days postcoitum (dpc) and 6.5 dpc (Fig. 4), with 93% of extraembryonic ectoderm showing biallelic expression (Table S1). On the other hand, complete Atrx silencing (100%) was observed in proximal endoderm, because only 1 signal, not associated with the Xist RNA–coated chromosome, was detected in all these cells (Fig. 4). Thus, Atrx is subject to complete XCI in embryonic tissues, but there is tissue-specific release of Atrx gene silencing in trophectoderm-derived cells at postimplantation stages. This contrasts with the escape patterns observed for genes in our fourth category (Fig. 3B), in which a high proportion of blastomeres show a lack of silencing from the outset.
Fig. 4.
Specific profile of Atrx and Huwe1 transcriptional activity during X-imprinted inactivation assayed by RNA FISH in pre- and postimplantation stages. (A) Atrx primary transcript signals (red) using specific RNA FISH at blastocyst stage (Top), in extraembryonic tissues at 5 dpc (Upper Middle), proximal endoderm (Bottom Middle), and extraembryonic ectoderm at 6.5 dpc (Bottom). A loss of Atrx signal from the paternal allele was observed in the trophectoderm at the blastocyst stage. Activity from the paternal allele was observed in extraembryonic tissue as early as 5 dpc, as assessed by the presence of 2 Atrx signals. This pattern was confirmed in extraembryonic tissue at 6.5 dpc. Only 1 signal, distant from the Xist RNA domain, was observed in proximal endodermic cells at 6.5 dpc. (B) Huwe1 primary transcript signals (red) using specific RNA FISH at blastocyst stage (Top), in extraembryonic tissues at 5 dpc (Upper Middle), proximal endoderm (Bottom Middle), and extraembryon or embryon at 6.5 dpc (Bottom). Two signals are present, 1 within the Xist domain (green), at the blastocyst stage. The paternal allele is silenced in the extraembryonic tissues as early as 5 dpc because only 1 signal, near the maternal Xist pinpoint, persists. The same pattern was observed in extraembryonic tissues at 6.5 dpc.
Huwe1 Shows Delayed Imprinted XCI.
The Huwe1 gene showed a highly unusual X-inactivation profile. Given that completely biallelic expression was observed in female preimplantation embryos (Fig. 4 and Fig. S6), we expected this gene to be a new example of an escapee. However, we found that Huwe1 is actually monoallelically expressed and silent on the Xist RNA–coated chromosome in TS cells (Fig. S7). We therefore examined the activity of this gene at early postimplantation stages (5.0–6.5 dpc) and found it to be silent on the inactive X in almost all cells of extraembryonic tissues by 5.0 dpc and fully silent by 6.5 dpc (Fig. 4 and Table S1). It also showed complete XCI in the embryonic ectoderm (Table S1). In summary, Huwe1 shows a novel and unexpected pattern of XCI. It is fully active on the Xp chromosome throughout preimplantation development, during the time window in which all other X-linked genes examined initiate and complete XCI, but then shows initiation and completion of imprinted and random XCI immediately after implantation. Thus, for some genes, the time window in which X-linked gene silencing can occur is much later than previously thought.
Discussion
In this study we have directly assessed the transcriptional activity of the paternal X chromosome using primary transcript analysis, throughout murine preimplantation development, from the time of ZGA. We found no sign of reduced transcription from the Xp chromosome at the 2-cell stage, either by Cot1 RNA FISH or at the level of X-linked genes. We also show that the paternal X chromosome at the 2-cell stage does not seem to be in a particularly condensed state at the time of major ZGA, despite its previous heterochromatinization in the male germ line. Collectively our data show that imprinted inactivation of the Xp chromosome only begins after fertilization. Thus, even though paternal X-linked genes, along with the rest of the genome, are silent in sperm, this is rapidly reversed after fertilization, perhaps by an immediate egg cytoplasmic effect. This still leaves open the possibility that the paternal X is predisposed to XCI after fertilization, owing to an imprint or memory that it carries from the male germ line and that is only read after ZGA. Such a memory mark cannot be MSCI dependent, because autosomal Xic transgenes that do not undergo MSCI in the germ line can still trigger imprinted inactivation when paternally transmitted (16). Rather, this predisposition may be a general characteristic of the male genome. In particular, it could be linked to the extensive chromatin remodeling and enhanced transcription activation of the paternal genome that occurs immediately after fertilization, when protamines are replaced by histones (20). This would result in paternal Xist activation at the 2-cell stage, which would then trigger XCI from the 4-cell stage onward.
In this study we also found that genes located closest to the Xic show the earliest onset of XCI. This is consistent with a previous report proposing that XCI occurs as a gradient from the Xic (13), although earlier cleavage stages had not been examined. Here we demonstrate that genes closest to the Xic clearly tend to be silenced most rapidly, from the 4–8-cell stage. Nevertheless, there are exceptions, with some genes close to the Xic showing late onset of silencing (e.g., Atrx), and for genes that are located further from the Xic more variable times of onset are observed (e.g., compare Gla and Fmr1). This could be due to the 3D organization of the X chromosome and the regions coated by Xist RNA, or else other processes may be at play. Furthermore, although time of onset of XCI correlates well with the proximity to the Xic, no correlation between Xic location and the speed or efficiency of gene silencing could be made. A cluster of genes in XqF3 (Fgd1–Pdha1) provides a particularly striking example of such diversity within a single 9-Mb region. Fgd1 and Pdha1 both show rapid and efficient XCI. Huwe1 shows no XCI at all during preimplantation development and efficient XCI after implantation, whereas the nearby Jarid1c gene shows partial XCI and continued escape throughout development. Imprinted XCI is thus far more piecemeal and less concerted than previously thought. Our findings also have important implications for the time window in which XCI is believed to occur. Studies in mouse ES cells, using inducible Xist cDNA transgenes, have indicated that XCI can only be induced during an early differentiation time window (21) and that silencing is achieved in just 1 or 2 cell cycles. For the majority of genes we examined a similar timing of inactivation relative to Xist coating was found during preimplantation development. However, Huwe1 is fully active on the Xp throughout preimplantation development and becomes inactivated only after implantation. This pattern is difficult to reconcile with current models of XCI. Our results suggest that different regions of the X chromosome may initiate silencing during different time windows of development. Indeed, this could potentially be due to alternative mechanisms and actors that only come into play at later stages. Interesting parallels can be drawn between our findings and those of imprinted clusters in which sequential and lineage-specific gene silencing linked to noncoding RNAs, such as Kcnq1ot1, have been reported. In the Kcnq1-imprinted cluster the Kcnq1ot1 RNA is paternally expressed at the 2-cell stage and is believed to participate in silencing of flanking genes on the paternal chromosome (22). It has been suggested that Kcnq1ot1 RNA represses its most proximal genes in all cells (both embryonic and extraembryonic) but that silencing spreads to more distant genes only in the trophoblast, after implantation. The epigenetic basis for this late onset of silencing and lineage specificity is not understood but may share common features with the differential X-inactivation kinetics reported here.
Our study also demonstrates that different mechanisms probably underlie the capacity of genes to escape from XCI. Constitutive escape was found for some loci, such as Jarid1c, whereas lineage-specific escape was found in others, such as Atrx. Recently Jarid1c has been shown to have an intrinsic capacity to escape, whatever its location (23). Understanding the sequence determinants and epigenetic basis for escape from X inactivation is an important question for the future. Our findings for the Atrx gene highlight the fact that there may be very diverse requirements for dosage compensation of X-linked genes during early development. Genes that show lineage-specific escape, such as Atrx, may need to be silenced in certain tissues (epiblast) but not in others (trophoblast). Indeed, given that Atrx is involved in chromatin remodeling and heterochromatin formation (24, 25) and is enriched on the inactive X chromosome in extraembryonic tissues (26), one can speculate that escape from XCI and increased dosage may be useful for the X-inactivation process in specific lineages during development. The fact that several genes show only partial silencing during early development also raises the issue of whether gene dosage regulation is critical for all X-linked loci or whether some genes are simply bystanders to this chromosome-wide process that evolved to regulate dose for a critical subset of loci. The very late but efficient silencing of a gene such as Huwe1 indicates that adequate dosage compensation can be achieved within rather different time frames, depending on the region of the X chromosome.
In summary, we provide definitive evidence, based on X-linked gene expression patterns, that imprinted XCI in mice is not dependent on MSCI but is probably dependent on the early activity of the paternal genome and the paternal Xist locus in particular. This implies that paternal XCI may have evolved more than once, in different mammals. In marsupials it may well be an MSCI-dependent process, whereas in eutherian mammals, such as rodents, it seems to depend on Xist imprinting. Our study also reveals the diverse kinetics of gene silencing and the existence of specific silencing processes affecting different domains of the X chromosome. The extent to which such piecemeal silencing is dependent on Xist RNA, nuclear organization, and epigenetic context of different regions remain important questions for the future.
Materials and Methods
Recovery of Embryos.
Procedures for handling and experimentation followed ethical guidelines established by the Federation of European Laboratory Animal Science Associations.
Mice were exposed to light daily between 7:00 AM and 7:00 PM. The midpoint of the dark period (midnight on the day of the plug is 0.5 dpc) was taken as the mating time. All preimplantation embryos were obtained from F1 (C57BL/6 × DBA/2J) female mice. Preimplantation embryos at 2-cell, 4-cell, 8-cell, 16-cell, and 32-cell stages were recovered by flushing the oviducts of superovulated females in M2 medium at 45–47 h, 52–53 h, 62–63 h, or 73–75h after human chorionic gonadotropin (hCG) injection, respectively (2). Females were superovulated by i.p. injection of 8 IU pregnant mare serum gonadotropin (Chronogest; Intervet), followed 47 h later by an i.p. injection of 5 IU of hCG (Chorulon; Intervet), and then individually mated with F1 males. Embryos at the blastocyst stage were obtained from naturally ovulating F1 females individually mated with F1 males, by flushing the uterus at 3.5 dpc in M2 medium. Embryos at 5.0 and 6.5 dpc were obtained by natural matings and dissected from decidua in PBS. Concepti were dissected out from decidua and then Reichert's membrane was removed with fine forceps. Thereafter, extraembryonic and embryonic regions were separated by a fine glass needle according to the morphologic landmarks (27). The 6.5-dpc visceral endoderm was removed from embryonic and extraembryonic regions with fine glass needles. A part of each tissue was divided into subfragments using a fine glass needle. The subfragments were incubated for 10 min in PBS at room temperature and subsequently used as specimens for RNA FISH.
Preparation of Embryos for RNA FISH.
Embryos were rinsed in PBS with 6 mg/mL BSA then transferred onto a Denhardt's solution–coated coverslip and dried down for 30 min at room temperature, as previously described (2). A supplementary step of decompaction by incubating embryos in PBS with 0.02% EDTA and 0.02% trypsin at 37 °C for 10–30 min was added for 16–32-cell embryos, after zona pellucida removal. Embryos were then fixed in 3% paraformaldehyde for 10 min at room temperature. Permeabilization of embryos was performed on ice in PBS with 0.5% Triton X-100 and 2 mM vanadyl ribonucleoside complex (New England Biolabs) for 3–10 min, depending on the embryo cell stage, then progressively dehydrated in ethanol. The coverslips were kept in 70% ethanol at −20 °C before RNA FISH.
Preparation of 2-Cell-Stage Embryos for Chromosome Painting.
Embryos were dried down onto a Denhardt's solution–coated coverslip for 20 sec at room temperature, as previously described (2). A 0.25-μL drop of 0.01 N HCl/0.1% Tween 20 solution (28) was applied onto the embryos coverslip. The embryo coverslips were left to dry for 15 min and thereafter washed in PBS for 5 min and dehydrated through an ethanol series.
RNA FISH on Mouse Embryos.
Probes used for RNA FISH were a 19-kb genomic fragment derived from a λ clone that covers most of Xist gene for Xist; overlapping λ clones for Chic1. For X-linked loci, BAC probes were used (Table S2) after verification of their location and single-copy nature by DNA FISH together with yeast artificial chromosome PA-2 (29) on male fibroblast metaphase spread. Probes were labeled with either SpectrumGreen-dUTP or with SpectrumRed-dUTP (Vysis) by nick translation using manufactory conditions. Probes for the B1 family of SINE elements were made by PCR amplification of the element from genomic DNA (B1-forward: 5′-GAGTGGTGGCGCACGCCTTTA-3′; B1-reverse: 5′CTGGCTGTCCTGGAACTCACT-3′) and labeled by random priming (Roche).
For dual RNA FISH experiments on mouse embryos, DNA probes to be used for RNA (Xist and X-linked genes primary transcripts) were prepared as previously described (30). Before hybridization, 0.1 μg of probe is ethanol precipitated together with 10 μg of salmon sperm with or without Cot 1 (1–10 μg, depending on the probe), washed twice in 70% ethanol and resuspended in formamide. The probe is denaturated at 75 °C for 7 min, mixed with an equal quantity of 2× hybridization buffer, and kept on ice before use. Embryos were hybridized with labeled probes overnight at 37 °C in a dark and humid chamber. After 3 washes in 50% formamide/2× SSC, 3 washes in 2× SSC at 42 °C, and DAPI (1 μg/mL) counterstaining, coverslips were mounted on slides in 90% glycerol, 0.1× PBS, and 0.1% p-phenylenediamine (Aldrich) and visualized using a fluorescence microscope.
DNA FISH on 2-Cell Embryos.
The DNA FISH procedure as previously described (29) was used for 2-cell embryos. Briefly, the nuclei were digested with 0.01% pepsin in 0.01 N HCl for 10 min at 37 °C. After washing in PBS, they were fixed 1% paraformaldehyde in PBS at room temperature and rinsed in PBS and then dehydrated through an ethanol series. Dual DNA FISH was performed with combination of biotin-labeled mouse X and Cy3-labeled mouse Y chromosome probes or biotin-labeled mouse X and FITC-labeled mouse 8 chromosome probes (Cambio). The probe mixture was denatured for 10 min at 65 °C and preannealed at 37 °C for 1 h. Embryo coverslips were denatured through immersion in 70% formamide/2× SSC (pH 7.0) at 80 °C for 10 min in an oven, dehydrated through an ice-cold ethanol series, and air-dried. The 2.5 μL of probe mixture was applied on the embryos directly. The coverslips were placed in a moist chamber, and hybridization was allowed to take place for 72 h at 37 °C. Posthybridization washes were carried out 3 times for 7 min at 42 °C with 50% formamide/2× SSC (pH 7.3), then twice for 5 min at 42 °C with 2× SSC. The biotin-labeled probe was detected by 2-h incubation with streptavidin-Alexa Fluor 488 or 568 (Molecular Probes). After incubation, coverslips were washed 3 times in 4× SSC/0.05% Nonidet P-40 for 5 min at 42 °C, then twice in 2× SSC at room temperature. DAPI (3 μg/mL) counterstaining coverslips were mounted as described above.
Fluorescence Microscopy.
A 200M Axiovert fluorescence microscope (Zeiss) equipped with an ApoTome system was used for image acquisition and the generation of optical sections in 3D. Sequential z axis images were collected in 0.3-μm steps. At the blastocyst stage, only cells corresponding to the trophoblast were taken into account. At each stage of development, counts were performed on ≥10 embryos at the 2-cell stage and ≥5 embryos for each of the 4-cell to blastocyst stages. A mean ± SD (range) of 20.7 ± 1.3 (10–33), 5.4 ± 0.3 (5–7), 8.1 ± 0.8 (5–15), 8.3 ± 0.7 (5–15), and 6.0 ± 0.4 (6–11) were examined for each gene at the 2-cell, 4-cell, 8-cell, morula, and blastocyst stage, respectively. Embryos were obtained from at least 5 different females.
Statistical Analysis.
When possible, Fisher's exact tests were used to compare Xm and Xp expression rates at the 2-cell stage for several genes. In other respects, polynomial regression models were used to study the relationship between Xp expression rate for a gene (regarded as a continuous dependant variable) and the distance between the gene and Xist on the chromosome. Powers up to 2 were included to account for potential nonlinear effects of the distance; P < 0.05 was considered to be statistically significant.
Supplementary Material
Acknowledgments.
The authors thank S. Augui, J. Chaumeil, C. Ciaudo, V. Colot, P. Fauque, P. Jouannet, and C. Corbel for discussions and critical reading of the manuscript; and M. Blanche and I. Grandjean for help with mice. This work was supported by the Centre National de la Recherche Scientifique, the French Ministry of Research, the Institut Curie, the Agence pour la Biomedicine, the Agence National de la Recherche, and the European Union Network of Excellence Epigenome.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810683106/DCSupplemental.
References
- 1.Takagi N, Sasaki M. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature. 1975;256:640–642. doi: 10.1038/256640a0. [DOI] [PubMed] [Google Scholar]
- 2.Okamoto I, Otto AP, Allis CD, Reinberg D, Heard E. Epigenetic dynamics of imprinted XCI during early mouse development. Science. 2004;303:644–649. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- 3.Mak W, et al. Reactivation of the paternal X chromosome in early mouse embryos. Science. 2004;303:666–669. doi: 10.1126/science.1092674. [DOI] [PubMed] [Google Scholar]
- 4.Rastan Timing of X-chromosome inactivation in postimplantation mouse embryos. J Embryol Exp Morphol. 1982;71:11–24. [PubMed] [Google Scholar]
- 5.Takagi N, Sugawara O, Sasaki M. Regional and temporal changes in the pattern of X chromosome replication during the early post-implantation development of the female mouse. Chromosoma. 1982;85:275–286. doi: 10.1007/BF00294971. [DOI] [PubMed] [Google Scholar]
- 6.Heard E, Disteche CM. Dosage compensation in mammals: Fine-tuning the expression of the X chromosome. Genes Dev. 2006;20:1848–1867. doi: 10.1101/gad.1422906. [DOI] [PubMed] [Google Scholar]
- 7.Lyon M, Rastan S. Parental source of chromosome imprinting and its relevance for X chromosome inactivation. Differentiation. 1984;26:63–67. doi: 10.1111/j.1432-0436.1984.tb01375.x. [DOI] [PubMed] [Google Scholar]
- 8.Shao C, Takagi N. An extra maternally derived X chromosome is deleterious to early mouse development. Development. 1990;110:969–975. doi: 10.1242/dev.110.3.969. [DOI] [PubMed] [Google Scholar]
- 9.Tada T, et al. Imprint switching for non-random X-chromosome inactivation during mouse oocyte growth. Development. 2000;127:3101–3103. doi: 10.1242/dev.127.14.3101. [DOI] [PubMed] [Google Scholar]
- 10.Kay GF, Barton SC, Surani MA, Rastan S. Imprinting and X chromosome counting mechanisms determine Xist expression in early mouse development. Cell. 1994;77:639–650. doi: 10.1016/0092-8674(94)90049-3. [DOI] [PubMed] [Google Scholar]
- 11.Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- 12.Okamoto I, et al. Evidence for de novo imprinted X-chromosome inactivation independent of meiotic inactivation in mice. Nature. 2005;438:369–373. doi: 10.1038/nature04155. [DOI] [PubMed] [Google Scholar]
- 13.Huynh KD, Lee JT. Inheritance of a pre-inactivated paternal X chromosome in early mouse embryos. Nature. 2003;426:857–862. doi: 10.1038/nature02222. [DOI] [PubMed] [Google Scholar]
- 14.Namekawa SH, et al. Postmeiotic sex chromatin in the male germline of mice. Curr Biol. 2006;16:660–667. doi: 10.1016/j.cub.2006.01.066. [DOI] [PubMed] [Google Scholar]
- 15.Turner JM, Mahadevaiah SK, Ellis PJ, Mitchell MJ, Burgoyne PS. Pachytene asynapsis drives meiotic sex chromosome inactivation and leads to substantial postmeiotic repression in spermatids. Dev Cell. 2006;10:521–529. doi: 10.1016/j.devcel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 16.Reik W, Ferguson-Smith AC. The X-inactivation yo-yo. Nature. 2005;438:297–298. doi: 10.1038/438297a. [DOI] [PubMed] [Google Scholar]
- 17.Chaumeil J, Le Baccon P, Wutz A, Heard E. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 2006;20:2223–2237. doi: 10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clemson CM, Hall LL, Byron M, McNeil J, Lawrence JB. The X chromosome is organized into a gene-rich outer rim and an internal core containing silenced nongenic sequences. Proc Natl Acad Sci USA. 2006;103:7688–7693. doi: 10.1073/pnas.0601069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garrick D, et al. Loss of Atrx affects trophoblast development and the pattern of X-inactivation in extraembryonic tissues. PLoS Genet. 2006;2:438–450. doi: 10.1371/journal.pgen.0020058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aoki F, Worrad D, Schultz R. Regulation of transcriptional activity during the first and second cycles in the pre-implantation embryo. Dev Biol. 1997;181:296–307. doi: 10.1006/dbio.1996.8466. [DOI] [PubMed] [Google Scholar]
- 21.Wutz A, Jaenisch R. A shift from reversible to irreversible X-inactivation is triggered during ES cell differentiation. Mol Cell. 2000;5:695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- 22.Lewis A, et al. Epigenetic dynamics of the Kcnq1 imprinted domain in the early embryo. Development. 2006;133:42303–44210. doi: 10.1242/dev.02612. [DOI] [PubMed] [Google Scholar]
- 23.Lin N, Carrel L. Escape from X chromosome inactivation is an intrinsic property of the Jarid1c locus. Proc Natl Acad Sci USA. 2008;105:17055–17060. doi: 10.1073/pnas.0807765105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibbons RJ, et al. Mutations in transcriptional regulator ATRX establish the functional significance of a PHD-like domain. Nat Genet. 1997;17:146–148. doi: 10.1038/ng1097-146. [DOI] [PubMed] [Google Scholar]
- 25.Gibbons RJ, et al. Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nat Genet. 2000;24:368–371. doi: 10.1038/74191. [DOI] [PubMed] [Google Scholar]
- 26.Baumann C, De La Fuente R. ATRX marks the inactive X chromosome (Xi) in somatic cells and during imprinted X chromosome inactivation in trophoblast stem cells. Chromosoma. 2008 doi: 10.1007/s00412-008-0189-x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Downs KM, Davies T. Staging of gastrulating mouse embryos by morphological landmarks in the dissecting microscope. Development. 1993;118:1255–1266. doi: 10.1242/dev.118.4.1255. [DOI] [PubMed] [Google Scholar]
- 28.Coonen E, Dumoulin JC, Ramaekers FC, Hopman AH. Optimal preparation of preimplantation embryo interphase nuclei for analysis by fluorescence in-situ hybridization. Hum Reprod. 1994;9:533–537. doi: 10.1093/oxfordjournals.humrep.a138540. [DOI] [PubMed] [Google Scholar]
- 29.Heard E, Mongelard F, Arnaud D, Avner P. Xist yeast artificial chromosome transgenes function as X-inactivation centres only in multicopy arrays and not as single copies. Mol Cell Biol. 1999;9:3156–3166. doi: 10.1128/mcb.19.4.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaumeil J, Okamoto I, Guggiari M, Heard E. Integrated kinetics of X chromosome inactivation in differentiating embryonic stem cells. Cytogenet Genome Res. 2004;99:75–84. doi: 10.1159/000071577. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.