Abstract
Many flowering plants possess self-incompatibility (SI) systems that prevent inbreeding. In Brassica, SI is controlled by a single polymorphic locus, the S locus. Two highly polymorphic S locus genes, SLG (S locus glycoprotein) and SRK (S receptor kinase), have been identified, both of which are expressed predominantly in the stigmatic papillar cell. We have shown recently that SRK is the determinant of the S haplotype specificity of the stigma. SRK is thought to serve as a receptor for a pollen ligand, which presumably is encoded by another polymorphic gene at the S locus. We previously have identified an S locus gene, SP11 (S locus protein 11), of the S9 haplotype of Brassica campestris and proposed that it potentially encodes the pollen ligand. SP11 is a novel member of the PCP (pollen coat protein) family of proteins, some members of which have been shown to interact with SLG. In this work, we identified the SP11 gene from three additional S haplotypes and further characterized the gene. We found that (i) SP11 showed an S haplotype-specific sequence polymorphism; (ii) SP11 was located in the immediate flanking region of the SRK gene of the four S haplotypes examined; (iii) SP11 was expressed in the tapetum of the anther, a site consistent with sporophytic control of Brassica SI; and (iv) recombinant SP11 of the S9 haplotype applied to papillar cells of S9 stigmas, but not of S8 stigmas, elicited SI response, resulting in inhibition of hydration of cross-pollen. All these results taken together strongly suggest that SP11 is the pollen S determinant in SI.
In Brassica, SI is sporophytically controlled by a highly polymorphic locus, termed the S locus, with more than 100 haplotypes identified so far (1, 2). To date, two stigmatically expressed highly polymorphic genes have been identified at the S locus. One is the S locus glycoprotein (SLG) gene, which encodes a secreted glycoprotein abundantly present in the papillar cell of the stigma surface (3, 4), and the other is the S locus receptor kinase (SRK) gene, which encodes a putative receptor-like serine/threonine protein kinase presumed to span the plasma membrane of the papillar cell (5). The predicted extracellular domain of SRK shares extensive sequence similarity with SLG. Results from our recent gain-of-function experiments have shown that SRK is the sole determinant of the S haplotype specificity of the stigma (6). SRK is thought to function as a receptor for the S determinant of pollen with the same S haplotype. Binding of SRK to the pollen S determinant then would elicit a signaling cascade in the papillar cell, leading to the rejection of self-pollen. Our recent study also has shown that the role of SLG is probably to enhance this recognition process, although how this is accomplished is not yet known (6).
In contrast to the S determinant of the stigma, the pollen S determinant had long remained elusive. Doughty et al. (7, 8) analyzed the pollen coat protein (PCP) of Brassica oleracea and identified a basic 7-kDa protein, termed PCP-A1 (protein 1 of class A PCP), which specifically bound SLGs. However, because PCP-A1 binds SLGs regardless of the S haplotype and its gene is not linked to the S locus, it is unlikely to be the pollen S determinant. Stephenson et al. (9) developed a pollination bioassay system and used it to show that the pollen coating could induce the SI response in the stigma in an S haplotype-specific manner. Fractionation of pollen coating revealed that the molecule (presumably the pollen S determinant) responsible for this activity was likely to be a PCP-A1-like protein with a molecular mass of <10 kDa; however, the true identity of this protein was not determined.
We previously reported sequence analysis of the SLG/SRK genomic region of the S9 haplotype of Brassica campestris and the identification of a potential candidate encoding the pollen S determinant (10). This gene, designated SP11 (S locus protein 11), is specifically expressed in the anther and encodes a small, cysteine-rich basic protein similar to PCP-A1. Very recently, a gene identical to SP11, termed SCR (S locus cysteine-rich), has been identified in the S8 haplotype of B. campestris and shown by loss-of-function and gain-of-function experiments to encode the pollen S determinant of SI (11).
Here we isolated and sequenced the SP11 gene from three additional S haplotypes (S8, S12, S52) of B. campestris and found that it exhibits a very high degree of allelic sequence diversity. We then used in situ hybridization to determine that SP11 has a sporophytic expression pattern, which can explain the sporophytic nature of Brassica SI. Last, we used a pollination bioassay to demonstrate that SP11 has a function expected of the pollen S determinant in the recognition process of SI.
Materials and Methods
Plant Materials.
B. campestris (syn. rapa) homozygous for the S8, S9, and S12 haplotypes have been described previously (4, 12). S52 and S60 haplotypes were derived from B. campestris cv Osome (Takii Seed, Kyoto) and have been characterized previously (13).
Fluorescent Differential Display.
Fluorescent differential display (FDD) was performed according to a previously described method (14). Anthers of S8 and S12 homozygotes of B. campestris were collected from flower buds of 1–10 mm in length that span most developmental stages. To minimize interference from polymorphism of the genetic backgrounds of individual plants unrelated to the S locus, cDNA templates were prepared from anthers collected from 10 S8 and 10 S12 homozygous F2 plants, respectively. These cDNA pools first were compared by FDD to identify S haplotype-specific bands, whose segregation with the S locus was confirmed further by a second FDD analysis by using 10 additional cDNA templates prepared from other F2 progeny. The FDD analyses using 240 independent primer combinations yielded 15 S8 haplotype-specific bands and 11 S12 haplotype-specific bands. Among them, the SP11–8 gene (the S8 haplotype of SP11) was identified as an S8 haplotype-specific band (180 bp) that had been amplified with anchored primer 5′-XRITC-GT15VA-3′ (XRITC: Rhodamine X isothiocyanate, V = mixture of A, C, and G) and an arbitrary primer 5′-CGTGGCTTTC-3′. Because the nucleotide sequence of this DNA fragment did not contain the initiation codon or the polyadenylation signal consensus sequence (AATAAA), the 5′ and 3′ rapid amplification of cDNA ends (RACE) cloning strategy was performed to obtain a full-length cDNA of SP11–8. The genomic structure of SP11–8 also was determined by long and accurate–PCR analysis with primers designed to the extreme 5′ and 3′ ends of the coding region.
Amplification of SP11–12 by PCR.
Total RNA was extracted from anthers (collected from buds at 0–7 days before anthesis) by using Isogen (Nippon Gene, Toyama, Japan). Approximately 20 μg of RNA was subjected to first-strand cDNA synthesis by using Superscript II (GIBCO) and an EcoRI-NotI oligo(dT) primer (15). An SP11–12 (the S12 haplotype of SP11) cDNA fragment was obtained by PCR using a primer (5′-ATGAAATCTGCTGTTTATGTTTC-3′) designed based on the conserved signal peptide sequence and an EcoRI-NotI primer. The 5′ end of the SP11–12 gene was determined by 5′ RACE.
Construction and Screening of cDNA Library.
Anthers of flower buds 5.0–6.5 mm in length were collected from B. campestris S52 haplotype. Poly(A) RNA was isolated from the anthers with a Micro-FastTrack mRNA isolation kit (Invitrogen). cDNA synthesized from the poly(A) RNA using a cDNA synthesis kit (Pharmacia LKB) was used for cDNA library construction in λ gt10 vector (Stratagene). The library was screened by plaque hybridization with digoxigenin-labeled SP11–9 (the S9 haplotype of SP11) probe. The probe was prepared by random-primed DNA labeling by using the digoxigenin DNA-labeling kit (Roche Diagnostics). Hybridization and detection were carried out as described previously (16).
Pulse-Field Gel Electrophoresis Gel Blot Analysis.
Megabase DNA embedded in agarose plugs was prepared from young leaf tissue of S52 haplotype by a rapid method described previously (16). The DNA was digested with infrequently cutting restriction enzymes and electrophoresed on a 1% SeaKem GTG agarose gel (FMC) with a CHEF-DR II apparatus (Bio-Rad) at 170 V and 14°C, with switching times ramped from 30 to 50 sec for 24 hr. Transfer, hybridization, and detection were carried out as described previously (16).
Molecular Cloning of BcPCP-A1.
The BcPCP-A1 protein was isolated first as a major SLG-binding protein from pollen coat extracts of B. campestris S9 homozygotes by the combination of reversed-phase HPLC and cation-exchange HPLC. During purification, binding activity was monitored by using BIAcore (Pharmacia) with a sensor chip immobilized with SLG9, which had been purified from stigmas as described previously (12). A partial amino acid sequence (KGKPCYSEEPD) of BcPCP-A1 was obtained after deblocking the N-terminal pyroglutamic acid by pyroglutamate aminopeptidase digest. A partial cDNA fragment of BcPCP-A1 was obtained by PCR by using a degenerate primer [5′-AA(A/G)GG(A/T/G/C)AA(A/G)CCATG(C/T)TA (C/T)TCAGA-3′], designed based on the amino acid sequence shown above, and an EcoRI-NotI oligo(dT) primer. Subsequent PCR was conducted on this fragment by using another degenerate primer [5′-TG(C/T)TA(C/T)TCAGA(A/G)GA (A/G)CCAGA-3′] and an EcoRI-NotI primer. The 5′ end of the BcPCP-A1 cDNA was amplified by 5′ RACE. BcPCP-A1 shows 95% sequence identity with PCP-A1. The GenBank accession number for the BcPCP-A1 cDNA is AB035564.
RNA Gel Blot Analysis.
Total RNA was isolated from several tissues of B. campestris S8 homozygotes by using Isogen. The probes (SP11–8 coding region and BcPCP-A1 coding region) were 32P-labeled by random priming (Roche Diagnostics). The RNA (20 μg) was size-fractionated on a 1.2% agarose-formaldehyde gel, transferred to a Gene-screen Plus (DuPont) membrane, hybridized overnight (in 5× SSC/5× Denhardt's/1% SDS at 60°C), and washed at high stringency [in 0.1× SSPE (standard saline phosphate/EDTA)/2% SDS at 60°C].
In Situ Hybridization.
Anthers at developmental stage 5 (bud length: 4–5 mm) and stage 7 (7–10 mm) were collected from B. campestris S8 homozygotes. Digoxigenin-labeled sense and antisense RNA probes of SP11–8 (coding region) and BcPCP-A1 (coding region) were prepared by using the SP6/T7 digoxigenin RNA labeling kit (Roche Diagnostics). In situ hybridization to 10-μm-thick Paraplast (Sigma) sections of formaldehyde-fixed anthers was performed as described previously (8).
Recombinant SP11 Protein.
The SP11–9 protein was expressed in E. coli (BL21) as a glutathione S-transferase (GST)-fusion protein by using pGEX-5X-3 vector (Amersham Pharmacia). The amino acid at the N-terminal end of mature SP11–9 cannot be determined unambiguously; however, the asparagine residue at +1 is a likely candidate (Fig. 2). In this experiment, we made the expression construct so that after the fusion protein was cleaved to remove the GST portion, the recombinant SP11–9 protein would begin with a glutamine residue, which is indicated with an asterisk in Fig. 2. The GST-fusion protein was extracted by using B-PER (Pierce) and was affinity-purified with a glutathione-Sepharose 4B column (Amersham Pharmacia). After cleavage with factor Xa, the recombinant SP11–9 protein was purified by reversed-phase HPLC by using a Capcell Pak C18 UG300 column (Shiseido, Tokyo).
Figure 2.
Alignment of predicted amino acid sequences of four allelic variants of SP11 and of PCP-A1. Gaps (hyphens) were introduced to optimize the alignment. The amino acids of the putative signal peptide for each protein are shown in italics; amino acid residues conserved in all four allelic variants of SP11 are shown in bold; and the conserved cysteine residues are boxed. The asterisk (*) indicates the glutamine residue at the N terminus of the recombinant SP11–9 protein used in pollination bioassays. GenBank accession nos. for SP11–12 and SP11–52 are AB035503 and AB035505, respectively.
Pollination Bioassay.
Before carrying out the assay, 4–10 papillar cells in one region of each stigma were selected under microscope, and self- or cross-pollen was attached to each papillar cell by using a micromanipulator. Only those pollen sources and stigmas that were found to behave normally in SI were selected for the pollination bioassay performed immediately after this test. The assay was performed essentially according to a previously described method (9). Briefly, pollen coat (PC) was isolated from fully dehiscent anthers of 50 flowers of either S8 or S9 haplotype. PC was mixed with 1 μl of a solution of the recombinant SP11–9 protein (0.06 μg/μl in water), and the mixture was applied, using a micromanipulator, to papillar cells in a separate region of the stigma that had been selected for bioassay. PC mixed with water was used as a negative control.
Results
Identification of the SP11 Gene of S8, S12, and S52 Haplotypes.
SP11 was identified previously in the S9 haplotype of B. campestris from sequencing analysis of the SLG9/SRK9 region of the S locus (10): SP11 is located at the S locus, expressed exclusively in the anther, and encodes a small, cysteine-rich basic protein similar to PCP-A1, which has been shown to interact with SLG (8). Genomic DNA gel blot analyses using the SP11–9 cDNA as a probe revealed that SP11–9 hybridized either very weakly or not at all to the genomic DNA of other S haplotypes. These results suggested the possibility that SP11 is a polymorphic gene (10), as would be expected for the gene encoding the pollen S determinant.
In this work, we used the technique of FDD to search for the gene encoding the pollen S determinant and identified SP11–8. Briefly, we used FDD to compare anther cDNAs from S8 and S12 homozygotes to identify genes that were expressed specifically in the anther of either S haplotype. SP11–8 was identified first as an S8 haplotype-specific 180-bp band, and the full-length cDNA was obtained by the 5′ and 3′ RACE cloning strategy (Fig. 1). The SP11–8 cDNA sequence contained an ORF of 222 bp, encoding a 74-aa protein similar to that encoded by SP11–9 (Fig. 2). SP11–8 contains a putative signal peptide, and its most likely cleavage site is indicated in Fig. 2.
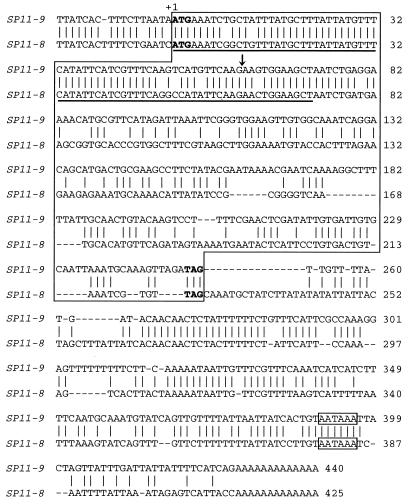
Figure 1.
Alignment of nucleotide sequences of SP11–9 and SP11–8 (GenBank accession nos. AB022078 and AB035504, respectively). Gaps (hyphens) were introduced to optimize the alignment. The coding region and the polyadenylation signal consensus sequence for each gene are boxed. The sequence for the putative signal peptide is underlined. Conserved nucleotides are indicated by vertical lines. The arrow indicates the conserved location of the intron.
Alignment of the nucleotide sequences of SP11–8 and SP11–9 revealed a striking similarity in the region encoding the signal peptide (Fig. 1). The nucleotide sequence identity in the coding region for the signal peptide is 92%, whereas that in the coding region for the mature protein is only 48%. Therefore, we used a primer designed based on the conserved signal peptide region and a 3′ end primer for PCR to obtain SP11–12. The high degree of sequence conservation in the signal peptide observed here appears to be a common feature of SP11, because using the primer designed based on this conserved signal peptide sequence and an oligo(dT) primer, we succeeded in using PCR to obtain SP11 cDNA from all of the class I S haplotypes of B. campestris examined (M.W., unpublished results). The deduced amino acid sequence of SP11–12 is shown in Fig. 2. In the linkage analysis using an F2 progeny segregating for S8 and S12 haplotypes, SP11–8 and SP11–12 were found to cosegregate, respectively, with SLG8 and SLG12 without exception, suggesting their genetic linkage to the S locus.
We also found from DNA gel blot analysis that the S52 haplotype contained an SP11-related sequence (results not shown), and we isolated a full-length SP11–52 (the S52 haplotype of SP11) cDNA clone from an S52 anther cDNA library by using the SP11–9 cDNA as a probe.
Alignment of the predicted amino acid sequences of the four allelic forms of SP11 revealed that all of their mature proteins contain eight conserved cysteine residues, one of the hallmarks of the PCP family proteins (8). However, the position of the fourth conserved cysteine residue from the N-terminal end is different from that of class A proteins of the PCP family (8). Thus, allelic variants of SP11 form a novel class of the PCP family. The amino acid sequences of the predicted mature proteins of the four SP11s are extremely divergent, with the amino acid sequence identity ranging from 26% to 40%. Only 2 aa, other than the eight conserved cysteine residues, are conserved among these four SP11s (Fig. 2). This high degree of allelic sequence diversity is consistent with the potential role of SP11 as the pollen S determinant in SI.
Location of SP11 in the S Locus Region.
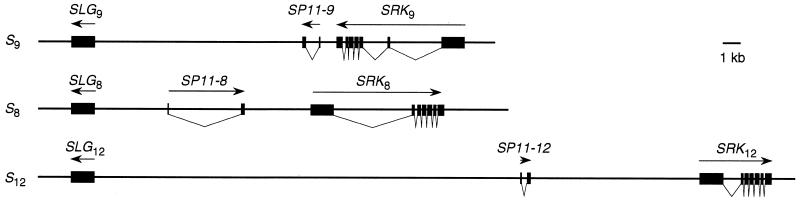
To confirm the allelic relationship of SP11–8, SP11–9, and SP11–12, we determined the location of each of these genes at the S locus (Fig. 3). We have shown previously that SP11–9 is located between SRK9 and SLG9 based on the sequencing analysis of a PAC clone (E89) obtained from MluI-digested genomic DNA of the S9 haplotype (10). For the S8 haplotype, we used long and accurate–PCR to amplify the region between SP11–8 and SLG8/SRK8 and sequenced the amplified fragments to determine the position and direction of transcription of SP11–8 (Fig. 3). The S locus of the S12 haplotype also was analyzed by long and accurate–PCR to determine the relative location of SLG12, SRK12, and SP11–12, which was confirmed further by the sequencing analysis of a bacterial artificial chromosome clone containing these three genes (H.S., unpublished results). Although the precise location and the direction of transcription of SLG52, SRK52, and SP11–52 were not determined in the S52 haplotype, the pulse-field gel electrophoresis gel blot analysis suggested that SP11–52 is located downstream of the SRK52 gene and within a distance of 30 kb (G.S., unpublished results).
Figure 3.
Genomic organization of the SLG/SRK region of the S locus of B. campestris. The arrows indicate the direction of transcription of each gene. Exons are indicated by solid boxes and introns are indicated by dips.
Although the S8, S9, and S12 alleles of the SP11 gene are all located between SRK and SLG, their location relative to SLG/SRK and their direction of transcription are divergent (Fig. 3). Further, although all three alleles of the SP11 gene contain a single intron that is located close to the 3′ end of the signal peptide-coding region (Fig. 1), the size of the intron is divergent (SP11–8, ≈4 kb; SP11–9, ≈0.7 kb; SP11–12, ≈0.3 kb; SP11–52, ≈0.3 kb). This divergent genomic organization of the SLG/SRK region might be one of the reasons why recombination is suppressed at the S locus. Also, the very short distance between the SRK gene and the SP11 gene can explain why recombination has not been detected between the genes encoding the female and male S determinants.
Expression of SP11 Gene.
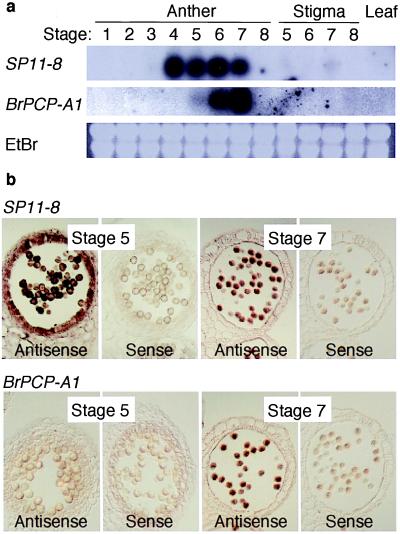
Because of the sporophytic nature of SI in Brassica, the gene encoding the pollen S determinant is likely to be expressed sporophytically in the anther (17). Because many proteins of pollen coating or tryphine are derived from the sporophytic tapetal cells that line the locule, the pollen S determinant might be one of these tapetum-derived proteins (18). However, the PCP family proteins examined thus far all show a strictly gametophytic expression pattern (8). We analyzed the temporal and spatial expression pattern of SP11 along with BcPCP-A1, a putative B. campestris ortholog of the PCP-A1 gene of B. oleracea (see Materials and Methods). As was true for PCP-A1, RNA gel blot analysis revealed that expression of BcPCP-A1 was maximal at late developmental stages, when the tapetal degradation was virtually complete (Fig. 4a, stages 6 and 7). However, expression of SP11–8 started at very early stages, when the tapetal cells were fully intact (Fig. 4a, stages 4 and 5). In situ hybridization analysis also showed that BcPCP-A1 expression was not detectable in early stages of anthers (Fig. 4b, stage 5) and was detectable only in the microspores of late developmental stages (Fig. 4b, stage 7), confirming a strictly gametophytic mode of expression. In contrast, expression of SP11–8 was detectable in the tapetal cells of early stages as well as in the microspores at late stages, suggesting both sporophytic and gametophytic expression of SP11–8. A similar expression pattern also was observed for SP11–9 and SP11–52 (data not shown). The tapetal expression pattern can explain the sporophytic nature of Brassica SI.
Figure 4.
Expression of SP11. (a) Northern blot analysis. Total RNA from anther, pistil, and leaf tissues was hybridized with an SP11–8 (coding region) probe and a BcPCP-A1 (coding region) probe. The stage numbers correspond to different bud sizes, with 1 = 0–1 mm, 2 = 1–2 mm, 3 = 2–3 mm, 4 = 3–4 mm, 5 = 4–5 mm, 6 = 5–7 mm, 7 = 7–10 mm, and 8 = open flower. EtBr, ethidium bromide staining of the gel before blotting. (b) In situ hybridization. Anther sections derived from flower buds of stages 5 and 7 were hybridized with SP11–8 and BcPCP-A1 antisense riboprobes and their sense riboprobes (negative control).
Biological Activity of Recombinant SP11 Protein.
Results from a pollination bioassay reported recently have provided evidence that the pollen coating contains the pollen S determinant that can modulate pollen hydration in an S-specific manner (9). This determinant was expected to be a protein with a molecular mass of <10 kDa and copurified with PCP-A class protein by gel filtration and reversed-phase HPLC. These characteristics of the pollen S determinant are consistent with those predicted for the SP11 protein. To confirm that SP11 itself is the pollen S determinant, we prepared the recombinant SP11–9 protein and tested its biological activity by using the pollination bioassay system. The results are summarized in Table 1. B. campestris S8 and S9 homozygotes exhibited a strong self-incompatibility phenotype, and the hydration of self-pollen was inhibited completely on the papillar cell. Pretreatment of the papillar cell of a stigma (e.g., S9 stigma) with cross-pollen coat (e.g., S8 PC) did not affect the hydration of cross-pollen (e.g., S8 pollen), as reported previously (9). Therefore, we used the cross-PC as a carrier and tested the biological effect of SP11–9. When the recombinant SP11–9 protein was applied to the S9 papillar cells before pollination, the hydration of S8 (cross) pollen was retarded significantly. Because the recombinant SP11–9 protein did not affect the hydration of cross-pollen when applied on the S8 papillar cells, this inhibitory effect of SP11–9 was not due to toxicity of this protein, but rather was due to an S haplotype-specific induction of SI response.
Table 1.
The effect of recombinant SP11-9 protein on pollen hydration
|
S9 stigma N = 16*
|
S8 stigma N = 14†
|
|||||||
|---|---|---|---|---|---|---|---|---|
| S9 pollen | S8 pollen | S8 PC + H2O + S8 pollen | S8 PC + SP11-9 + S8 pollen | S8 pollen | S9 pollen | S9 PC + H2O + S9 pollen | S9 PC + SP11-9 + S9 pollen | |
| No hydration | 51 (98) | 28 (28) | 7 (29) | 74 (89) | 36 (90) | 17 (29) | 12 (39) | 14 (39) |
| Hydration | 1 (2) | 73 (72) | 17 (71) | 9 (11) | 4 (10) | 42 (71) | 19 (61) | 22 (61) |
The data represent the number (percentage in brackets) of pollen grains that hydrated in the pollination bioassay. In the experiment, the recombinant SP11-9 protein was added to supplement isolated PC and applied on papillar cells before pollination. In the control, water was added to PC.
*SP11-9 supplemented S8 PC experimental vs. S8 PC control: χ2 = 36.42; df = 1; P < 0.001.
†SP11-9 supplemented S9 PC experimental vs. S9 PC control: χ2 = 0.0002; df = 1; P > 0.98.
Discussion
SP11 is the first PCP-like gene identified at the S locus of Brassica and has been proposed to be a good candidate for encoding the pollen ligand of the SI system possessed by Brassica (10). In this communication, we present several lines of evidence that strongly suggest that SP11 encodes the pollen S determinant of SI. The pollen S determinant is expected to exhibit the following properties: (i) it is encoded by a gene at the S locus; (ii) it exhibits S haplotype-specific sequence polymorphism; (iii) it is a pollen coat protein expressed in the anther; and (iv) it can interact with SRK to elicit an S haplotype-specific SI response in the stigma, leading to pollen rejection. Although the interaction between SP11 and SRK remains to be demonstrated, we have shown that SP11 has all the properties expected of the pollen S determinant of SI.
In this study, we determined the allelic relationship of the SP11 gene identified from three S haplotypes, S8, S9, and S12, by analyzing the location of this gene at the S locus. The S locus regions of these three S haplotypes show divergent structural organization, as reported previously (19, 20). However, the SP11 gene is invariably located between SLG and SRK, being upstream of SLG and closer to SRK than to SLG. The conservation of the location of SP11 relative to the SLG/SRK gene pair supports the allelic relationship of the SP11 gene identified from these three S haplotypes. This organization is also consistent with the previous observation that the gene organization downstream of SLG is highly colinear (20), supporting the possibility that the SLG downstream region could be one border of the S locus.
Recently, Schopfer et al. (11) reported the identification of a cysteine-rich protein gene, termed SCR, at the S locus, and the demonstration by loss-of-function and gain-of-function experiments that SCR encodes the pollen S determinant. In this work, they identified the SCR gene from the sequence analysis of the SLG/SRK genomic region of B. campestris S8 haplotype. SCR8 is completely identical in sequence to SP11–8 identified in our present study, suggesting that the SCR gene is the same as the SP11 gene. The results of this transformation experiment provide another line of evidence that SP11 encodes the pollen S determinant of SI. In our independent gain-of-function experiment, we also have obtained similar results: i.e., pollen from B. campestris S52S60 heterozygotes transformed with an SP11–8 transgene has acquired S8 haplotype specificity (H.S., unpublished work).
In the present study, we ascertained the function of SP11 by a pollination bioassay by using a recombinant SP11–9 protein produced in Escherichia coli. The recombinant protein applied on the papillar cells of S9 stigmas before pollination inhibited the hydration of cross-pollen (S8 haplotype). The same recombinant protein when applied on the papillar cells of S8 stigmas did not inhibit the hydration of cross-pollen (S9 haplotype). These results suggest that SP11–9 itself is sufficient to induce an SI response in the papillar cell of the same S haplotype, but not of a different S haplotype. This S haplotype-specific SI response of the stigma induced by SP11 demonstrates that SP11 is the pollen S determinant.
Because of the sporophytic nature of SI in Brassica, the pollen S determinant has been postulated to be a protein expressed sporophytically in the anther (17). In contrast to class A PCP proteins, which show a strictly gametophytic expression pattern, SP11 shows both sporophytic and gametophytic expression patterns. Although the possibility remains that gametophytically produced SP11 molecules might be secreted to the anther locule and redistributed to the pollen coat, as suggested by Doughty et al. (8), the sporophytic expression of the SP11 gene alone can explain the sporophytic control of Brassica SI on the male side. It will be of interest to determine whether SP11 indeed is transported from the tapetum to the anther locule and becomes incorporated into the pollen coat.
Acknowledgments
We thank the technical help of T. Nakanishi, A. Arai, and K. Fujii. We thank Dr. S. Kawasaki (National Institute of Agrobiological Resources) for providing a bacterial artificial chromosome vector and technical advice. This work was supported in part by Grants-in-Aid for Special Research on Priority Areas (A, 07281103; and B, 11238205) and for Scientific Research (B, 09480195 and 11460056) from the Ministry of Education, Science, Sports and Culture, Japan.
Abbreviations
- FDD
fluorescent differential display
- SI
self-incompatibility
- SLG
S locus glycoprotein
- SRK
S receptor kinase
- PCP
pollen coat protein
- SP11
S locus protein 11
- SP11–8 (SP11–9
SP11–12, or SP11–52), SP11 of the S8 (S9, S12, or S52) haplotype
- RACE
rapid amplification of cDNA ends
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AB035503–AB035505 and AB035564).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.040556397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.040556397
References
- 1.Bateman A J. Heredity. 1955;9:53–68. [Google Scholar]
- 2.Nou I S, Watanabe M, Isogai A, Hinata K. Sex Plant Reprod. 1993;6:79–86. [Google Scholar]
- 3.Nasrallah J B, Kao T-H, Goldberg M L, Nasrallah M E. Nature (London) 1985;318:263–267. [Google Scholar]
- 4.Takayama S, Isogai A, Tsukamoto C, Ueda Y, Hinata K, Okazaki K, Suzuki A. Nature (London) 1987;326:102–104. [Google Scholar]
- 5.Stein J C, Howlett B, Boyes D C, Nasrallah M E, Nasrallah J B. Proc Natl Acad Sci USA. 1991;88:8816–8820. doi: 10.1073/pnas.88.19.8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takasaki, T., Hatakeyama, K., Suzuki, G., Watanabe, M., Isogai, A. & Hinata, K. (2000) Nature (London), in press. [DOI] [PubMed]
- 7.Doughty J, Hedderson F, McCubbin A, Dickinson H. Proc Natl Acad Sci USA. 1993;90:467–471. doi: 10.1073/pnas.90.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doughty J, Dixon S, Hiscock S J, Willis A C, Parkin I A P, Dickinson H G. Plant Cell. 1998;10:1333–1347. doi: 10.1105/tpc.10.8.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stephenson A G, Doughty J, Dixon S, Elleman C, Hiscock S, Dickinson H G. Plant J. 1997;12:1351–1359. [Google Scholar]
- 10.Suzuki G, Kai N, Hirose T, Fukui K, Nishio T, Takayama S, Isogai A, Watanabe M, Hinata K. Genetics. 1999;153:391–400. doi: 10.1093/genetics/153.1.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schopfer C R, Nasrallah M E, Nasrallah J B. Science. 1999;286:1697–1700. doi: 10.1126/science.286.5445.1697. [DOI] [PubMed] [Google Scholar]
- 12.Isogai A, Takayama S, Tsukamoto C, Ueda Y, Shiozawa H, Hinata K, Okazaki K, Suzuki A. Plant Cell Physiol. 1987;28:1279–1291. [Google Scholar]
- 13.Takasaki T, Hatakeyama K, Watanabe M, Toriyama K, Isogai A, Hinata K. Plant Mol Biol. 1999;40:659–668. doi: 10.1023/a:1006274525421. [DOI] [PubMed] [Google Scholar]
- 14.Ito T, Kito K, Adati N, Mitsui Y, Hagiwara H, Sakaki Y. FEBS Lett. 1994;351:231–236. doi: 10.1016/0014-5793(94)00867-1. [DOI] [PubMed] [Google Scholar]
- 15.Phillips A L, Huttly A K. Cell. 1994;12:1133–1142. [Google Scholar]
- 16.Suzuki G, Watanabe M, Toriyama K, Isogai A, Hinata K. Gene. 1997;199:133–137. doi: 10.1016/s0378-1119(97)00358-2. [DOI] [PubMed] [Google Scholar]
- 17.de Nettancourt D. Incompatibility in Angiosperms. Berlin: Springer; 1997. [Google Scholar]
- 18.Dickinson H G, Lewis D. Proc R Soc London Ser B. 1973;184:148–165. [Google Scholar]
- 19.Boyes D C, Nasrallah M E, Vrebalov J, Nasrallah J B. Plant Cell. 1997;9:237–247. doi: 10.1105/tpc.9.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui Y, Brugiere N, Jackman L, Bi Y-M, Rothstein S J. Plant Cell. 1999;11:2217–2231. doi: 10.1105/tpc.11.11.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]