Abstract
Differential cytosine methylation of genes and transposons is important for maintaining integrity of plant genomes. In Arabidopsis, transposons are heavily methylated at both CG and non-CG sites, whereas the non-CG methylation is rarely found in active genes. Our previous genetic analysis suggested that a jmjC domain-containing protein IBM1 (increase in BONSAI methylation 1) prevents ectopic deposition of non-CG methylation, and this process is necessary for normal Arabidopsis development. Here, we directly determined the genomic targets of IBM1 through high-resolution genome-wide analysis of DNA methylation. The ibm1 mutation induced extensive hyper-methylation in thousands of genes. Transposons were unaffected. Notably, long transcribed genes were most severely affected. Methylation of genes is limited to CG sites in wild type, but CHG sites were also methylated in the ibm1 mutant. The ibm1-induced hyper-methylation did not depend on previously characterized components of the RNAi-based DNA methylation machinery. Our results suggest novel transcription-coupled mechanisms to direct genic methylation not only at CG but also at CHG sites. IBM1 prevents the CHG methylation in genes, but not in transposons.
Keywords: chromatin, IBM1, transposon
Introduction
Transposons and repeats, which are major deleterious constituents in genomes of vertebrates and plants, are epigenetically silenced by DNA methylation (Yoder et al, 1997; Walsh et al, 1998; Miura et al, 2001; Singer et al, 2001; Kato et al, 2003; Bender, 2004; Chan et al, 2005; Gehring and Henikoff, 2007). A central question in epigenetics is how the methylation machinery distinguishes between transposons and cellular genes.
The flowering plant Arabidopsis thaliana serves as a powerful model system to understand control of DNA methylation through genetic and genomic approaches. Genome-wide analyses of DNA methylation in Arabidopsis reveal that transposons are heavily methylated at both CG and non-CG sites, whereas the methylation in genes is at much lower level and limited to CG sites (Lippman et al, 2004; Zhang et al, 2006; Zilberman et al, 2007; Cokus et al, 2008; Lister et al, 2008) (http://neomorph.salk.edu/epigenome.html). The methylation at CG sites is maintained by DNA methyltransferase MET1 (Finnegan et al, 1996; Kankel et al, 2003; Saze et al, 2003). Methylation at non-CG sites depends on DNA methyltransferases CMT3 and DRM2; the former mainly methylates symmetrical CHG sites, whereas the latter can methylate asymmetrical CHH sites (H can be A, C, or T) (Bartee et al, 2001; Lindroth et al, 2001; Cao and Jacobsen, 2002). CMT3-dependent CHG methylation is guided by methylation of histone H3 at lysine 9 (H3K9me) (Jackson et al, 2002; Malagnac et al, 2002). H3K9me is an epigenetic mark of silent ‘heterochromatin' conserved from plants to fungi and animals (Bender, 2004; Chan et al, 2005; Grewal and Elgin, 2007). DNA methylation at specific sequence can be induced by RNA of homologous sequence. This process, called RNA-directed DNA methylation (RdDM), requires de novo methylase DRM2, members of RNAi machinery, such as RDR2, DCL3, AGO4, RNA polymerase IV, as well as a putative chromatin remodeller (DRD1) and an SMC-domain containing protein (DMS3) (Chan et al, 2005; Matzke et al, 2007; Kanno et al, 2008). The RdDM generally affects CG, CHG, and CHH sites, and the CHH methylation is normally associated with small RNA, consistent with the involvement of RNAi machinery.
In addition to these well-investigated positive regulators of DNA methylation, negative regulators of DNA methylation have been recently identified (Agius et al, 2006; Gehring et al, 2006; Saze et al, 2008). Through a genetic screen for mutants with increased methylation of a gene called BONSAI, we have previously identified the jumonji-domain-containing protein IBM1 (increase in BONSAI methylation 1) (Saze and Kakutani, 2007; Saze et al, 2008). IBM1 is a member of the JHDM2/KDM3 family, which is comprised of demethylases of H3K9me conserved from plants to mammals (Klose et al, 2006; Lu et al, 2008; Sun and Zhou, 2008). The ibm1 mutation induces a variety of developmental abnormalities, which are suppressed by mutants of the histone H3K9 methylase KYP/SUVH4 gene and CHG methylase CMT3 gene. The results suggest that ectopic deposition of the chromatin marks, H3K9me and CHG methylation, is responsible for the ibm1-induced developmental abnormalities (Saze et al, 2008). However, actual genomic targets of the IBM1 protein have not been identified, with the exception of the BONSAI gene. The spectrum of genomic targets of IBM1 remained unknown.
Here, we directly determined genomic targets of IBM1 by high-resolution genome-wide analysis of DNA methylation. The ibm1 mutation induced extensive hyper-methylation in thousands of genes throughout the euchromatic chromosomal arm regions. The hyper-methylation was specific for CHG sites within genes. Unexpectedly, long transcribed genes were most severely affected. The results suggest that not only CG but also CHG methylation in the genes is directed by transcription-coupled mechanisms. We propose that IBM1 protects transcribed genes from the CHG methylation and maintains genome integrity by distinguishing between genes and transposons.
Results
Genome-wide analysis of DNA methylation revealed targets of IBM1 protein
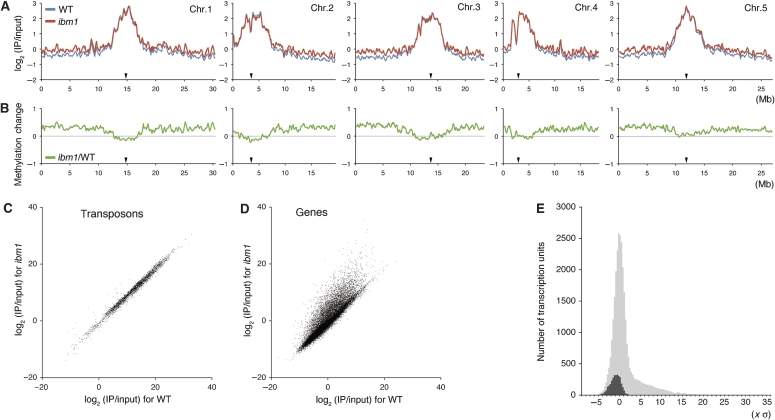
To understand the mode and significance of the control of DNA methylation by the IBM1 protein, we determined DNA methylation across the whole genome in the ibm1 mutant. For that purpose, we adapted a genomic tiling array method combined with immunoprecipitation of the genomic DNA using an anti-methylcytosine antibody. The methylation level was assessed by the hybridization signal ratio of immunoprecipitated DNA to the input DNA. Pericentromeric regions, which are rich in transposons and repeats, show more methylation signal than the gene-rich chromosome arms in the wild-type sample, as has been reported previously (Lippman et al, 2004; Zhang et al, 2006; Zilberman et al, 2007; Cokus et al, 2008; Lister et al, 2008). Compared with the wild type, the ibm1 mutation caused a global increase in the signal intensity over the entire chromosome arms (Figure 1A and B).
Figure 1.
Global view of DNA methylation in the ibm1 mutant and wild-type Columbia plants. (A) Comparison along the chromosomes. Vertical values represent log2 of the immunoprecipitated DNA signal divided by input control signal. Each point reflects sliding window of 300 kb (∼3000 probes). In each chromosome, position of centromere is indicated by black triangle on the x-axis. (B) The difference of the signal between the ibm1 mutant and wild type. (C, D) Methylation of transcription units compared between the ibm1 and wild-type plants. To evaluate significance of the methylation in each transposon or gene, we calculated ‘methylation significance index (MSI)' (see Materials and methods). Most of the transposons were unaffected (C), whereas a subset of genes showed increase in DNA methylation in ibm1 (D). (E) Distribution of difference of the MSI between ibm1 and wild type for genes (grey) and transposons (black). The x value is expressed using standard deviation of differences in the signals between wild type and ibm1 for every probes.
At higher resolution, the comparison revealed that the ibm1-induced hyper-methylation is not uniform within the arms. Interestingly, each region of increased DNA methylation coincides with a transcription unit (Figure 2A and D; Supplementary Figures S2–S13). We therefore calculated the methylation level for each transcription unit based on the recent annotation of the Arabidopsis genome (TAIR7: http://www.arabidopsis.org). When ibm1 and wild type were compared, thousands of genes showed a significant increase in DNA methylation signal (Figure 1D and E). On the other hand, transposons did not show comparable changes (Figure 1C and E). In independent experiments using different plants and different probe amplification methods, the wild-type and ibm1 samples showed consistent results for each gene (Supplementary Figure S1). For further analysis, we selected 3112 genes (651 species of class I genes and 2461 class II genes; with stronger effects in class I genes) that reproducibly showed the most dense increase in methylation signal induced by the ibm1 mutation (Supplementary Figure S1; Supplementary Table S1).
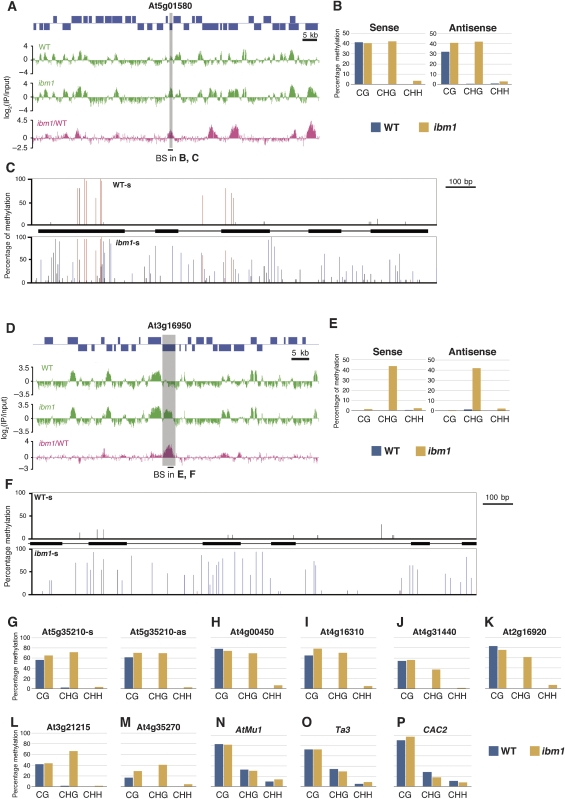
Figure 2.
DNA methylation status of the ibm1 mutant and wild-type Columbia plants in 12 representative loci, which are analysed by bisulphite sequencing. For all the 12 loci, the array results with biological replica and full bisulphite sequencing results are shown in Supplementary Figures S2–S13. (A, D) DNA methylation analysed by tiling array for two loci (A, position 172 500–277 500; D, position 5 745 000–5 835 000). Blue boxes indicate genes, which oriented 5′–3′ on the top and the reverse in the bottom. Each vertical green bar represents the log2 signal of the immunoprecipitated DNA divided by input control. Magenta bars represent ibm1/WT ratios of signals. (C, F) Results of bisulphite sequencing in wild type and ibm1 mutants within two genes shown in (A, D). Results of sense strands are shown. Results for both sense and antisense strands are shown in Supplementary Figures S2–S4. The percentage of methylated cytosines at each site is indicated by vertical bar (red, CG; blue, CHG; black, asymmetric). Exons and introns are shown by black boxes and lines between the results of wild type and ibm1 mutant. (B, E, G–P) The percentage of methylated cytosines found in different contexts at genic sequences (B, E, G–M) and transposon sequences (N–P) in wild type and ibm1.
To validate the tiling array results, DNA methylation was determined at the nucleotide level by the bisulphite sequencing method for 12 loci (6 class I genes, 3 class II genes, and 3 transposons). For all the examined class I and II genes, the bisulphite sequencing confirmed the increase in DNA methylation detected by the tiling array (Figure 2; Supplementary Figures S2–S10); extensive hyper-methylation was noted. The bisulphite sequencing results also revealed that the increase in DNA methylation was mainly at CHG sites (Figure 2). This sequence specificity contrasts with genic methylation in wild type, which is almost exclusively at CG sites (Cokus et al, 2008; Lister et al, 2008) (http://neomorph.salk.edu/epigenome.html) (Figure 2B, G–M). Both sense and antisense strands of the genes were affected similarly (Figure 2B, E and G; Supplementary Figures S2–S4). On the other hand, transposons were methylated at CG, CHG, and CHH sites already in wild type, and this methylation was not affected by the ibm1 mutation (Figure 2N–P; Supplementary Figures S11–S13). The bisulphite sequencing data confirmed that the targets of the IBM1 function were not transposons but genes, as was suggested from the tiling array results (Figure 1).
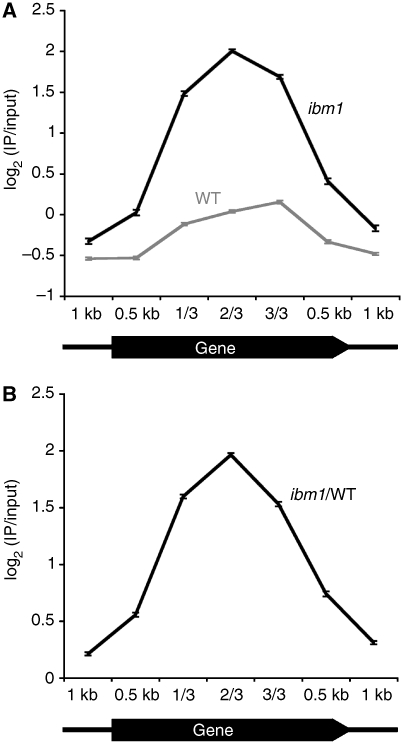
In Arabidopsis, another class of negative regulators of DNA methylation has been identified; DEMETER, ROS1, DML1, and DML2 are structurally related DNA demethylases (Agius et al, 2006; Gehring et al, 2006). Triple mutation of ros1, dml1, and dml2 (rdd) causes increased methylation in ∼200 genes (Penterman et al, 2007; Lister et al, 2008). The rdd triple mutation can induce hyper-methylation in the 5′ and 3′ regions of the gene. In contrast, the ibm1 mutation most severely affected the central region of transcription units, whereas the 5′ and 3′ terminal regions were least affected (Figure 3). This bias is similar to the distribution of genic CG methylation in wild type (Zhang et al, 2006; Zilberman et al, 2007). In the ibm1 mutant, the bias was enhanced by including extensive CHG methylation.
Figure 3.
Distribution of DNA methylation. Each gene was divided into five regions (5′ and 3′ 500-bp segments, and three internal regions with equal lengths) with 1000-bp flanking segments. (A) Average of signals in probes in each segment was plotted for ibm1 and wild type. In total, 862 genes hyper-methylated in ibm1 and longer than 2 kb were analysed. (B) Difference of ibm1 and wild type.
Long transcribed genes were most severely affected by the ibm1 mutation
Each transcription unit seems to be affected in a coordinated manner; some transcription units are affected over the entire gene, whereas others are not affected at all (Figures 1, 2A and D; Supplementary Figures S2–S13). This possibility was examined quantitatively by comparing the effects of the ibm1 mutation between the 5′ and 3′ halves of a transcription unit. This analysis revealed that they indeed behave in a coordinated manner (Supplementary Figure S14). The correlation between the 5′ and 3′ halves of a transcription unit was significant (r=0.67). In other words, when the ibm1 mutation affected the 5′ half of one gene, then the 3′ half of that gene tended to be affected as well. This behaviour was not simply due to the spread of hyper-methylation to nearby sequences, because the correlation with flanking regions outside the gene was much lower; r=0.29 for the 5′ regions and r=0.36 for the 3′ regions (Supplementary Figure S14).
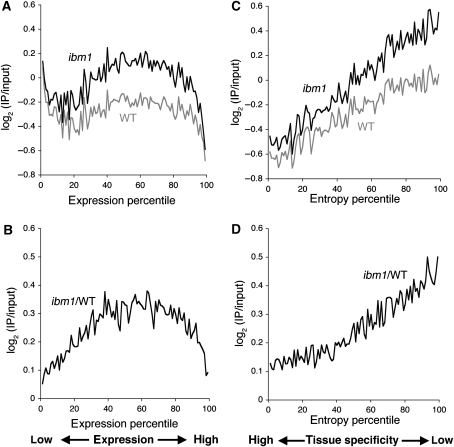
In wild-type plants, moderately transcribed genes are most likely to be methylated, whereas genes at either extreme with regard to the expression level are less likely (Zhang et al, 2006; Zilberman et al, 2007). Notably, the ibm1 mutation affected moderately transcribed genes and constitutively expressed genes most severely (Figure 4A–D). The spectrum of affected genes was consistent with that seen for genic CG methylation in wild type, and the methylation was also found at CHG sites in the ibm1 mutant. In addition, some genes unmethylated in wild type also showed hyper-methylation in the ibm1 mutant (Figures 1D and 2D–F). Interestingly, genes unmethylated in wild type and hyper-methylated in ibm1 were also transcribed in wild type (Supplementary Figure S15). Overall, the methylation of transcribed genes was much enhanced by the ibm1 mutation (Figure 4).
Figure 4.
Transcription and effect of ibm1 on DNA methylation. (A, B) Bias to moderately transcribed genes. All genes were rank-ordered and binned based on the sum of expression in all of Arabidopsis tissues (see Supplementary data). The vertical value shows mean methylation signal for genes in each percentile. (C, D) Bias to constitutively expressed genes. Same as (A, B), except that the genes were rank-ordered based on tissue specificity of expression measured by entropy level (see Supplementary data).
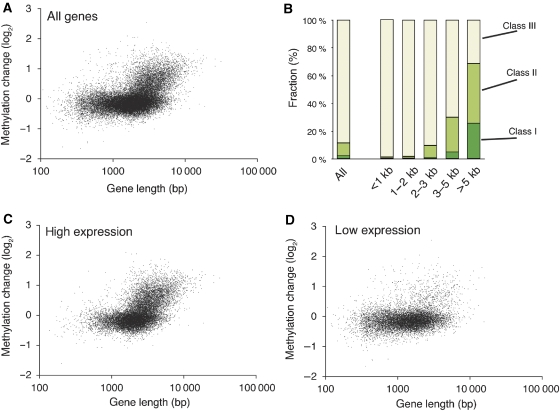
Another notable feature of the effect of the ibm1 mutation is that longer genes showed a much stronger response (Figure 5A and B). For genes longer than 5 kb, about two-thirds belong to ibm1-affected genes in class I or II, and the class I genes were enriched more than 10-fold compared with the total population of genes (Figure 5B). As short genes tend to have fewer introns, we examined whether the intron number mediates the effect of the gene length on ibm1-induced hyper-methylation. However, even within the genes of the same intron number, longer genes tend to be affected severely by the ibm1 mutation (Supplementary Figure S16).
Figure 5.
Long and expressed genes were affected by ibm1. (A) Relationship between the gene length and increase in methylation in ibm1. The methylation change shows mean of ibm1/WT signals in each gene. (B) ibm1 affects long genes. Genes were divided into five groups based on length. The proportions of class I–III genes are shown by different colours. These three classes were categorized on the basis of the mean of log2(ibm1/WT) signals in each gene, with class I being the most severely affected and class III the least affected by ibm1 (Supplementary Figure S1). (C, D) Genes shown in (A) were divided into high expressers (15 000 genes in (C)) and low expressers (10 620 genes in (D)) based on the transcript level in leaves. The effect of the length is less evident in the latter population.
The severe response of long genes was less evident in genes with low levels of expression (Figure 5D), suggesting that the effect of the gene length on the response to the ibm1 mutation also depends on the transcription.
The ibm1-induced CHG methylation did not depend on previously characterized RNAi-based components for DNA methylation
The results presented above suggest that the genic methylation of both CG and CHG sites can be induced by transcription-coupled mechanisms, and that the latter is masked by the function of IBM1 protein, which contributes to the differential methylation of genes and transposons. A question would be how transcription could affect genic methylation. A change in the chromatin state might be induced by passage of the transcription machinery itself, or by the action of the resultant transcript.
For possible effects of the RNA on chromatin states, the most extensively studied system in plants is RdDM, which is connected to small RNA. We therefore tested whether known components of the RdDM process, namely, DRM2, RDR2, RDR6, AGO4, and NRPD1a, are necessary for the ibm1-induced hyper-methylation or not. Methylation in the double mutants was examined for two target genes of IBM1, BONSAI and ERL2 (ERECTA-LIKE 2) (Figure 6; Supplementary Figure S17). Interestingly, in all the double mutants, the ibm1-induced hyper-methylation was detected. The results suggest that the ibm1-induced CHG methylation does not depend on any of these previously characterized RNAi-based components of the DNA methylation machinery. Taken together, these results suggest that novel transcription-coupled mechanisms direct genic methylation not only at CG but also at CHG sites.
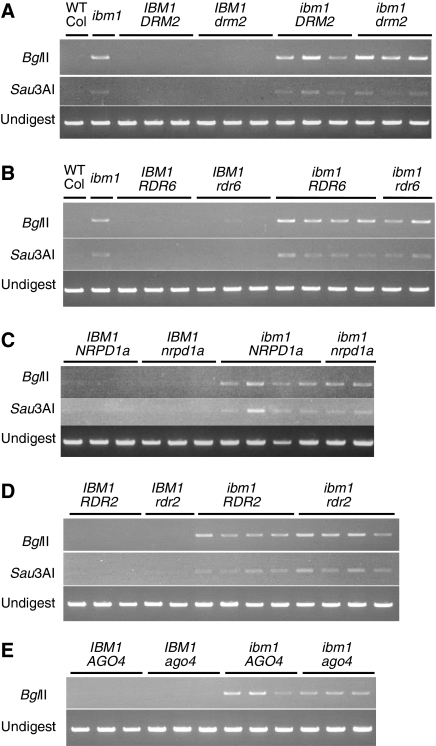
Figure 6.
Known components of RNAi-based DNA methylation are dispensable for the ibm1-induced genic hyper-methylation. DNA methylation status of the BONSAI gene was examined by methylation-sensitive restriction digestion and subsequent PCR. Detailed conditions were as described previously (Saze and Kakutani, 2007). The band can be detected when the restriction site is methylated and undigestible. Essentially the same results were obtained for another target gene, ERL2 (ERECTA-LIKE 2, At5g07180) (Supplementary Figure S17). (A) Four types of homozygotes segregating in self-pollinated progeny of a double heterozygote IBM/ibm1, DRM2/drm2. The double mutant plants also showed hyper-methylation. (B–E) Methylation status was examined in double mutants with RDR6, NRPD1a, RDR2, or AGO4 gene. Plants in F3 generation were examined in (C).
Discussion
In this study, we determined genomic targets of the jmjC domain protein IBM1 by genome-wide analysis of DNA methylation. The ibm1 loss-of-function mutation induced extensive hyper-methylation in thousands of genes. Long transcribed genes were most severely affected. Methylation of transcribed genes is limited to CG sites in wild type, but those genes were also methylated at CHG sites in the ibm1 mutant. Thus, the IBM1 protein prevents CHG methylation of transcribed genes. These observations broaden our understanding of the control of genic methylation, but important questions remain, as illustrated below.
Why are long and transcribed genes body-methylated?
It has been shown previously that transcribed genes tend to be body-methylated at CG sites (Zhang et al, 2006; Zilberman et al, 2007). Here, we report that, without the IBM1 function, transcribed genes are also body-methylated at CHG sites. For the body methylation of transcribed genes, Zilberman et al (2007) proposed that disruption of chromatin during the passage of transcription machinery allows activation of cryptic promoters, resulting in aberrant transcript formation, and subsequent DNA methylation. With regard to the ibm1-induced hyper-methylation of gene body, the entire transcription unit behaved in a coordinated manner (Supplementary Figure S14). If sporadic events, such as activation of cryptic promoters, affect the entire gene, a long gene would have more chance to initiate such an event. This prediction is consistent with our observation that long genes tend to be affected most severely by the ibm1 mutation (Figure 5).
On the other hand, our genetic analyses suggest that the ibm1-induced hyper-methylation does not depend on previously characterized components of RdDM, which include RNAi machinery and the de novo DNA methylase DRM2 (Figure 6; Supplementary Figure S17). Even if aberrant RNA is involved in genic methylation, the latter might use an as yet uncharacterized pathway.
Alternatively, the passage of transcription machinery itself, rather than the RNA produced, might mediate a change in the chromatin state and facilitate gene body methylation. In yeast, transcription machinery can be associated with histone methylases, such as SET1 and SET2 (Krogan et al, 2003; Ng et al, 2003). SET2-induced H3K36 methylation recruits histone deacetylase complex, which negatively regulates the transcription (Keogh et al, 2005). Similar mechanisms might also be used for generating repressive histone marks such as H3K9me directly or indirectly. In addition, passage of transcription machinery can enhance histone replacement, which may also affect chromatin structure (Martin and Zhang, 2007).
The ibm1 mutation induces genic CHG methylation, which is rarely found in wild type. Interestingly, the genic CHG methylation is also induced in the mutant background of CG methylase gene MET1 (Cokus et al, 2008; Lister et al, 2008). The CHG methylation in the met1 mutant might function to rescue the deleterious effects of the loss of the CG methylation (Mathieu et al, 2007). We compared the spectrum of genes showing CHG methylation between the met1 and ibm1 mutants. The correlation was found to be significant between genes showing CHG hyper-methylation in ibm1 and genes with CG hypo-methylation and CHG hyper-methylation in the met1 mutant (Supplementary Figure S18), suggesting that IBM1 might be linked to a compensatory methylation pathway induced by CG hypo-methylation.
Is the CHG hyper-methylation sufficient for silencing the gene?
Considering the large impact of the ibm1 mutation on gene body methylation shown in this study, it may be surprising that effects of the ibm1 mutation on developmental phenotypes are not catastrophic (Saze et al, 2008). Indeed, in our preliminary results on gene expression, the CHG hyper-methylation of gene body induced by the ibm1 mutation is not always associated with transcriptional silencing. Within the genes hyper-methylated by ibm1 mutation, some showed a decrease in the transcript level, whereas others were unaffected or even showed an increased expression (unpublished). On the other hand, in the wild-type background, non-CG methylation is not found in active genes, but found only in silent transposons and repeats. Of the transposon-specific non-CG methylation, CHG methylation is connected to H3K9me (Jackson et al, 2002; Malagnac et al, 2002), whereas CHH methylation is associated with small RNA (Matzke et al, 2007). If CHG methylation is not sufficient for the gene silencing, interaction and/or collaboration with other layers of epigenetic modifications might be important for generating silent heterochromatin.
In this context, it may be interesting that double mutant of the IBM1 gene and a chromatin remodelling gene DDM1 (decrease in DNA methylation 1) shows much more severe developmental phenotypes than either single mutant (Saze et al, 2008). As is the case for the ibm1 mutation, the ddm1 mutation induces methylation of the BONSAI gene. Unlike ibm1, however, ddm1 mutation causes BONSAI hyper-methylation in all the three contexts: CG, CHG, and CHH (Saze and Kakutani, 2007). In addition, although the ddm1-induced BONSAI methylation depends on the presence of a LINE retrotransposon nearby (Saze and Kakutani, 2007), our genome-wide analysis revealed that genes affected by the ibm1 mutation are not necessarily located near transposons (Supplementary Table S2). Targets of IBM1 are low copy genes, and repetitive sequences were unaffected (Figures 1 and 2). On the other hand, the direct targets of the DDM1 gene appears to be repeats (Vongs et al, 1993; Lippman et al, 2004), and the effect of ddm1 mutations on low copy sequences, such as BONSAI, might be indirect (Saze and Kakutani, 2007). Even if ibm1 and ddm1 mutations trigger the hyper-methylation by different mechanisms, these pathways may converge on overlapping targets, with possible interactions of different layers of epigenetic marks. The possible interactions may be clarified at the genome level by combination of genomics and genetics using these and other Arabidopsis mutations.
Materials and methods
Plant materials
Isolation and initial characterization of the ibm1 mutants were described previously (Saze et al, 2008). A presumed null allele ibm1-4, which has T-DNA insertion in the coding region, was used throughout. Alleles or origins of the other mutants are rdr6-11, ago4-1, SALK059661 (RDR2), SALK128426 (NRPD1a), and cs6366 (DRM2).
Array design and data analysis
We used NimbleGen 3 × 385K array (Zilberman et al, 2007), which covers the entire sequenced Arabidopsis genome with an interval of ∼100 bp. Details of the experimental procedures are described in the Supplementary data. The methylation signal for each probe was represented as log2 signal ratio of immunoprecipitated DNA to input DNA. Methylation of a gene (Figures 3, 4 and 5; Supplementary Figures S1 and S15) is represented by the average of the log2 signal for all probes covered by the gene. In the results shown in Figure 1C–E, we used ‘methylation significance index' of a gene, which is the sum of methylation signal for the probes covered by the gene divided by root of the probe number, because fluctuation of that value would be independent of the probe number per gene, assuming that the signal for each probe fluctuates independently.
Bisulphite sequencing
Bisulphite sequencing was performed as described previously (Saze et al, 2008). In total, 38 amplicons were sequenced for the 12 loci examined. At least 12 clones were sequenced for each amplicon. Restriction enzymes and primer sequences used for the bisulphite sequencing are shown in Supplementary Table S3.
Supplementary Material
Supplementary Information
Supplementary Table S1
Acknowledgments
We thank Akiko Terui for technical assistance; Steve Henikoff for advice; Ryan Lister and Joe Ecker for list of the genes CHG hyper-methylated in the met1 mutant; and Susumu Hirose, Eric Richards, Hiroyuki Sasaki, and Daniel Zilberman for critical comments on the paper. This study was supported by Takeda Science Foundation and Grant-in-Aid for Scientific Research (19207002 and 19060014).
References
- Agius F, Kapoor A, Zhu JK (2006) Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc Natl Acad Sci USA 103: 11796–11801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartee L, Malagnac F, Bender J (2001) Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev 15: 1753–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender J (2004) DNA methylation and epigenetics. Ann Rev Plant Biol 55: 41–68 [DOI] [PubMed] [Google Scholar]
- Cao X, Jacobsen SE (2002) Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc Natl Acad Sci USA 99: 16491–16498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SW, Henderson IR, Jacobsen SE (2005) Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet 6: 351–360 [DOI] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE (2008) Shotgun bisulfite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452: 215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan E, Peacock J, Dennis E (1996) Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc Natl Acad Sci USA 93: 8449–8454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Henikoff S (2007) DNA methylation dynamics in plant genomes. Biochim Biophys Acta 1769: 276–286 [DOI] [PubMed] [Google Scholar]
- Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, Goldberg RB, Fischer RL (2006) DEMETER DNA glycosylase establishes MEDEA polycombe gene self-imprinting by allele-specific demethylation. Cell 124: 495–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Elgin SC (2007) Transcription and RNA interference in the formation of heterochromatin. Nature 447: 399–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JP, Lindroth AM, Cao X, Jacobsen SE (2002) Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416: 556–560 [DOI] [PubMed] [Google Scholar]
- Kankel MW, Ramsey DE, Stokes TL, Flowers SK, Haag JR, Jeddeloh JA, Riddle NC, Verbsky ML, Richards EJ (2003) Arabidopsis MET1 cytosine methyltransferase mutants. Genetics 163: 1109–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T, Bucher E, Daxinger L, Huettel B, Boemdorfer G, Gregor W, Kreil DP, Matzke M, Matzke AJ (2008) A structural-maintenance-of-chromosomes hinge domain-containing protein is required for RNA-directed DNA methylation. Nat Genet 40: 670–675 [DOI] [PubMed] [Google Scholar]
- Kato M, Miura A, Bender J, Jacobsen SE, Kakutani T (2003) Role of CG and non-CG methylation in immobilization of transposons in Arabidopsis. Curr Biol 13: 421. [DOI] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, Boone C, Emili A, Weissman JS, Hughes TR, Strahl BD, Grunstein M, Greenblatt JF, Buratowski S, Krogan NJ (2005) Cotranslational set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123: 593–605 [DOI] [PubMed] [Google Scholar]
- Klose RJ, Kallin EM, Zhang Y (2006) JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet 7: 715–727 [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, Shilatifard A, Buratowski S, Greenblatt J (2003) Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol 23: 4207–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth AM, Cao X, Jackson JP, Zilberman D, McCallum CM, Henikoff S, Jacobsen SE (2001) Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292: 2077–2080 [DOI] [PubMed] [Google Scholar]
- Lippman Z, Gendrel AV, Black M, Vaughn MW, Dedhia N, McCombie WR, Lavine K, Mittal V, May B, Kasschau KD, Carrington JC, Doerge RW, Colot V, Martienssen R (2004) Role of transposable elements in heterochromatin and epigenetic control. Nature 430: 471–476 [DOI] [PubMed] [Google Scholar]
- Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR (2008) Highly integrated single-base resolution maps of epigenome in Arabidopsis. Cell 133: 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Li G, Cui X, Liu C, Wang XJ, Cao X (2008) Comparative analysis of jmjC-domain-containing proteins reveals the potential histone demethylases in Arabidopsis and rice. J Integr Plant Biol 50: 886–896 [DOI] [PubMed] [Google Scholar]
- Malagnac F, Bartee L, Bender J (2002) An Arabidopsis SET domain protein is required for maintenance but not establishment of DNA methylation. EMBO J 21: 6842–6852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Zhang Y (2007) Mechanisms of epigenetic inheritance. Curr Opin Cell Biol 19: 266–272 [DOI] [PubMed] [Google Scholar]
- Mathieu O, Reinders J, Caikovski M, Smathajitt C, Paszkowski J (2007) Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell 130: 851–862 [DOI] [PubMed] [Google Scholar]
- Matzke M, Kanno T, Huettel B, Daxinger L, Matzke AJ (2007) Targets of RNA-directed DNA methylation. Curr Opin Plant Biol 10: 512–519 [DOI] [PubMed] [Google Scholar]
- Miura A, Yonebayashi S, Watanabe K, Toyama T, Shimada H, Kakutani T (2001) Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 411: 212–214 [DOI] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K (2003) Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell 11: 709–719 [DOI] [PubMed] [Google Scholar]
- Penterman J, Zilberman D, Huh JH, Ballinger T, Henikoff S, Fischer RL (2007) DNA demethylation in the Arabidopsis genome. Proc Natl Acad Sci USA 104: 6752–6757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze H, Kakutani T (2007) Heritable epigenetic mutation of a transposon-flanked Arabidopsis gene due to lack of the chromatin-remodeling factor DDM1. EMBO J 26: 38–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze H, Mittelsten-Scheid O, Paszkowski J (2003) Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat Genet 34: 65–69 [DOI] [PubMed] [Google Scholar]
- Saze H, Shiraishi A, Miura A, Kakutani T (2008) Control of genic DNA methylation by a jmjC domain-containing protein in Arabidopsis thaliana. Science 319: 462–465 [DOI] [PubMed] [Google Scholar]
- Singer T, Yordan C, Martienssen RA (2001) Robertson's mutator transposons in A. thaliana are regulated by the chromatin-remodeling gene decrease in DNA methylation (DDM1). Genes Dev 15: 591–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Zhou DX (2008) Rice jmjC domain-containing gene JMJ706 encodes H3K9 demethylase required for floral organ development. Proc Natl Acad Sci USA 105: 13679–13684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vongs A, Kakutani T, Martienssen RA, Richards EJ (1993) Arabidopsis thaliana DNA methylation mutants. Science 260: 1926–1928 [DOI] [PubMed] [Google Scholar]
- Walsh CP, Chaillet JR, Bestor TH (1998) Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet 20: 116–117 [DOI] [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, Bestor TH (1997) Cytosine methylation and the ecology of intragenomic parasites. Trends Genet 13: 335–340 [DOI] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sudaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, Ecker JR (2006) Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126: 1189–1201 [DOI] [PubMed] [Google Scholar]
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S (2007) Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet 39: 61–69 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Table S1