Abstract
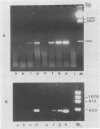
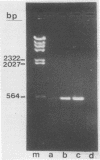
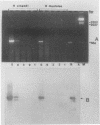
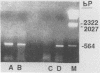
Sequence data on Helicobacter pylori 16S rRNA were used to select two 22-base oligonucleotide primers for use in a polymerase chain reaction (PCR) for detection of H. pylori. H. pylori cells were treated with lysis buffer, boiled, and chloroform extracted. Reverse transcription of rRNA was followed by PCR amplification (RT-PCR) of the synthesized cDNA and 16S rRNA gene. The amplified PCR products were analyzed by agarose gel electrophoresis and Southern blotting. Using ethidium bromide-stained agarose gels, we were able to detect the expected 500-bp DNA fragment from as few as two H. pylori organisms per reaction. The specificity of the RT-PCR assay was tested with 27 clinical isolates and related reference strains; although the number of bacterial cells used per reaction was 10(5)-fold greater than the number of H. pylori organisms used, amplification was detected only with bacteria in the same genus, H. cinaedi and H. mustelae. Ten H. pylori organisms per biopsy specimen were detected on agarose gels when organisms were added to samples prepared from a processed colon biopsy sample. RT-PCR results were consistent with urea breath test and culture results in 14 of 15 gastric biopsy specimens; the specificity was 100%. RT-PCR of rRNA from H. pylori increased the sensitivity of pathogen detection at least 25- to 50-fold compared with that of previous PCR assays. This low level of detection by RT-PCR assay may prove to be well suited for verifying eradication following therapy.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barthel J. S., Everett E. D. Diagnosis of Campylobacter pylori infections: the "gold standard" and the alternatives. Rev Infect Dis. 1990 Jan-Feb;12 (Suppl 1):S107–S114. doi: 10.1093/clinids/12.supplement_1.s107. [DOI] [PubMed] [Google Scholar]
- Clayton C. L., Kleanthous H., Coates P. J., Morgan D. D., Tabaqchali S. Sensitive detection of Helicobacter pylori by using polymerase chain reaction. J Clin Microbiol. 1992 Jan;30(1):192–200. doi: 10.1128/jcm.30.1.192-200.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D. J., Jr, Evans D. G., Graham D. Y., Klein P. D. A sensitive and specific serologic test for detection of Campylobacter pylori infection. Gastroenterology. 1989 Apr;96(4):1004–1008. doi: 10.1016/0016-5085(89)91616-8. [DOI] [PubMed] [Google Scholar]
- Fox J. G., Paster B. J., Dewhirst F. E., Taylor N. S., Yan L. L., Macuch P. J., Chmura L. M. Helicobacter mustelae isolation from feces of ferrets: evidence to support fecal-oral transmission of a gastric Helicobacter. Infect Immun. 1992 Feb;60(2):606–611. doi: 10.1128/iai.60.2.606-611.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girvitz S. C., Bacchetti S., Rainbow A. J., Graham F. L. A rapid and efficient procedure for the purification of DNA from agarose gels. Anal Biochem. 1980 Aug;106(2):492–496. doi: 10.1016/0003-2697(80)90553-9. [DOI] [PubMed] [Google Scholar]
- Graham D. Y. Helicobacter pylori: its epidemiology and its role in duodenal ulcer disease. J Gastroenterol Hepatol. 1991 Mar-Apr;6(2):105–113. doi: 10.1111/j.1440-1746.1991.tb01448.x. [DOI] [PubMed] [Google Scholar]
- Gray M. W., Sankoff D., Cedergren R. J. On the evolutionary descent of organisms and organelles: a global phylogeny based on a highly conserved structural core in small subunit ribosomal RNA. Nucleic Acids Res. 1984 Jul 25;12(14):5837–5852. doi: 10.1093/nar/12.14.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammar M., Tyszkiewicz T., Wadström T., O'Toole P. W. Rapid detection of Helicobacter pylori in gastric biopsy material by polymerase chain reaction. J Clin Microbiol. 1992 Jan;30(1):54–58. doi: 10.1128/jcm.30.1.54-58.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshina S., Kahn S. M., Jiang W., Green P. H., Neu H. C., Chin N., Morotomi M., LoGerfo P., Weinstein I. B. Direct detection and amplification of Helicobacter pylori ribosomal 16S gene segments from gastric endoscopic biopsies. Diagn Microbiol Infect Dis. 1990 Nov-Dec;13(6):473–479. doi: 10.1016/0732-8893(90)90079-b. [DOI] [PubMed] [Google Scholar]
- Morotomi M., Hoshina S., Green P., Neu H. C., LoGerfo P., Watanabe I., Mutai M., Weinstein I. B. Oligonucleotide probe for detection and identification of Campylobacter pylori. J Clin Microbiol. 1989 Dec;27(12):2652–2655. doi: 10.1128/jcm.27.12.2652-2655.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedenskov-Sørensen P., Aase S., Bjørneklett A., Fausa O., Bukholm G. Sampling efficiency in the diagnosis of Helicobacter pylori infection and chronic active gastritis. J Clin Microbiol. 1991 Apr;29(4):672–675. doi: 10.1128/jcm.29.4.672-675.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster B. J., Lee A., Fox J. G., Dewhirst F. E., Tordoff L. A., Fraser G. J., O'Rourke J. L., Taylor N. S., Ferrero R. Phylogeny of Helicobacter felis sp. nov., Helicobacter mustelae, and related bacteria. Int J Syst Bacteriol. 1991 Jan;41(1):31–38. doi: 10.1099/00207713-41-1-31. [DOI] [PubMed] [Google Scholar]
- Reed K. C., Mann D. A. Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res. 1985 Oct 25;13(20):7207–7221. doi: 10.1093/nar/13.20.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romaniuk P. J., Zoltowska B., Trust T. J., Lane D. J., Olsen G. J., Pace N. R., Stahl D. A. Campylobacter pylori, the spiral bacterium associated with human gastritis, is not a true Campylobacter sp. J Bacteriol. 1987 May;169(5):2137–2141. doi: 10.1128/jb.169.5.2137-2141.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Totten P. A., Fennell C. L., Tenover F. C., Wezenberg J. M., Perine P. L., Stamm W. E., Holmes K. K. Campylobacter cinaedi (sp. nov.) and Campylobacter fennelliae (sp. nov.): two new Campylobacter species associated with enteric disease in homosexual men. J Infect Dis. 1985 Jan;151(1):131–139. doi: 10.1093/infdis/151.1.131. [DOI] [PubMed] [Google Scholar]
- Valentine J. L., Arthur R. R., Mobley H. L., Dick J. D. Detection of Helicobacter pylori by using the polymerase chain reaction. J Clin Microbiol. 1991 Apr;29(4):689–695. doi: 10.1128/jcm.29.4.689-695.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Berg F. M., Zijlmans H., Langenberg W., Rauws E., Schipper M. Detection of Campylobacter pylori in stomach tissue by DNA in situ hybridisation. J Clin Pathol. 1989 Sep;42(9):995–1000. doi: 10.1136/jcp.42.9.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandamme P., Falsen E., Rossau R., Hoste B., Segers P., Tytgat R., De Ley J. Revision of Campylobacter, Helicobacter, and Wolinella taxonomy: emendation of generic descriptions and proposal of Arcobacter gen. nov. Int J Syst Bacteriol. 1991 Jan;41(1):88–103. doi: 10.1099/00207713-41-1-88. [DOI] [PubMed] [Google Scholar]
- Waters A. P., McCuthan T. F. Ribosomal RNA: nature's own polymerase-amplified target for diagnosis. Parasitol Today. 1990 Feb;6(2):56–59. doi: 10.1016/0169-4758(90)90071-b. [DOI] [PubMed] [Google Scholar]
- Wetherall B. L., McDonald P. J., Johnson A. M. Detection of Campylobacter pylori DNA by hybridisation with non-radioactive probes in comparison with a 32P-labelled probe. J Med Microbiol. 1988 Aug;26(4):257–263. doi: 10.1099/00222615-26-4-257. [DOI] [PubMed] [Google Scholar]