Abstract
Estrogens are critical mediators of breast tumorigenesis. This occurs via the action of estrogens on the estrogen receptor (ER), which regulates the transcriptome of breast cancer cells. Despite the long history of the search for estrogen-regulated genes in breast cancer, knowledge of the E2-regulated transcriptome and its effects is incomplete. We used Affymetrix GeneChips to profile the effects of estradiol on the expression of genes in EFF-3, EFM-19 and MCF-7 cells. In addition to many well-characterized estrogen-regulated genes, this identified a novel group of genes that have roles in vesicle trafficking, including exocytosis. Recent evidence in the literature supports a role for vesicle trafficking in tumorigenesis. We focused on five genes (SYTL5, RAB27B, SNX24, GALNT4 and SLC12A2/NKCC1/BSC2) and confirmed their estrogen-regulation using quantitative real-time PCR (qPCR). qPCR also demonstrated that these five genes were expressed in invasive breast carcinoma tissue. Immunohistochemistry showed expression of SYTL5 in cells of normal breast ductal epithelium, ductal carcinoma in-situ (DCIS) and invasive breast carcinoma. The results suggest that a significant effect of estrogens is to regulate the expression of genes that affect diverse aspects of vesicle trafficking including exocytosis.
Keywords: Breast cancer, vesicle, estrogen, synaptotagmin-like protein, RAB GTPase, exocytosis
Introduction
Estrogens are sex steroid hormones with a plethora of physiological and pathophysiological functions including a central role in the aetiopathogenesis of breast cancer. Estrogens alter gene expression in target cells via the nuclear-localized estrogen receptor (ER), which exists in two forms, ERα and ERβ [1]. 17β-estradiol (E2) has considerably greater biological activity than the other principal types of estrogens (estrone and estratriol) [2]. The estrogen-bound ER complex interacts with estrogen-responsive elements and with other transcription factors to regulate transcription of select genes, which in breast cancer cells, facilitate tumor development and progression [3–5].
One in ten women in Western society will develop breast cancer. There is epidemiologic, clinical and laboratory evidence that estrogens have an important role in breast cancer pathology. Estrogens and estrogen antagonists have proliferative and antiproliferative effects, respectively on breast cancer cells [6, 7]. The successful use of systemic endocrine therapy drives the current interest in the pathophysiology of estrogens in breast cancer. Endocrine treatments include drugs which inhibit the production of estrogens such as the aromatase inhibitors, letrozole and anastrozole, or compete with estrogen for binding to the ER, such as the antiestrogens, tamoxifen and faslodex. Discerning the mechanisms of estrogen signaling, and the development of endocrine resistance will be instrumental in directing the future endocrine management of breast cancer.
Since the advent of DNA microarray technology, a number of studies have been performed to identify E2-regulated genes in breast cancer cell lines building on our previous knowledge [8–16]. These studies have contributed considerably to our knowledge and understanding of E2-regulated gene expression in breast cancer. These studies have tended to focus on three main areas: 1) cataloguing E2-regulated genes, 2) the mechanisms of ER-mediated gene regulation or 3) analysis of E2-regulated gene expression to predict of prognosis. Despite the large body of work performed, the majority of previous studies have been limited to one cell line only (MCF-7). In the present study, in addition to the most studied cell line, MCF-7, we have used two little studied cell lines (EFF-3 and EFM-19) which have been grown in culture for relatively little time [17, 18]. Therefore, EFF-3 and EFM-19 cells are less likely to show a change the E2-regulated transcriptome from that of the original cells from which they are derived. Adding these two cell lines also addresses the issue of heterogeneity of the E2-regulated transcriptome that may exist between different cell lines.
Vesicle trafficking is a constitutive and regulated process in eukaryotic cells, allowing essential cellular processes such as neurotransmission and hormone secretion [19]. One family of vesicle trafficking genes with over 60 members is the RAB GTPases, which have diverse roles including vesicle budding, tethering, fusion and transport, in addition to endocytosis and exocytosis [19]. There is now a growing body of evidence that strongly supports a role for vesicle trafficking and the genes that regulate this, in tumorigenesis [21–23].
In the present study, we tested the hypothesis that novel functional groups of E2-regulated genes exist. We report identification of a group of E2-regulated genes, which are implicated in different aspects of vesicle trafficking. This paper focuses on vesicle trafficking genes, as these represent a previously unrecognized group of E2-regulated genes and this may illuminate our understanding of the actions of E2 in breast cancer.
Materials and Methods
Cell Lines, Culture Conditions and RNA Extraction
E2-responsive ERα+ (EFF-3, EFM-19, MCF-7) and E2-independent ERα- (MDA-MB-231, Hs578T, SK-BR-3) human breast cancer cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum and 1 μg/ml insulin. Cells were grown to approximately 70% confluence in T25 or T75 flasks. Experiments were performed in triplicate for each cell line. Withdrawal from steroids present in the routine culture medium involved culture for 7 days in phenol red-free DMEM containing 10% newborn calf serum (treated with dextran-coated charcoal) and 1 μg/ml insulin. During the first 3 days of steroid withdrawal, cells were washed with PBS twice prior to replenishment with steroid depleted medium. Cells were then cultured in either steroid depleted medium or in depleted medium containing 10−9M E2 for 48 hr. Medium was changed daily.
E2-dose response and time course experiments were performed with MCF-7 cells under the same conditions as described above. E2-dose response experiments were performed in the presence of 0 – 10−7M E2 for 48 hours. Time course experiments were performed under conditions of steroid-withdrawal or in the presence of 10−9M E2 for 0 – 96 hours. Total RNA was extracted using TRIZOL reagent (Invitrogen Life Technologies, Inc., Invitrogen Corp., Carlsbad, CA).
Breast Tumors and RNA Extraction
Ethical permission was obtained from the Local Research Ethics Committee. Invasive breast carcinoma of no special type (NST) samples (n=18) were macro-dissected by a histopathologist from surgical specimens at the Royal Victoria Infirmary, Newcastle upon Tyne. Samples were pulverized at liquid nitrogen temperature using a tungsten carbide ball in a tissue dismembrator and RNA purified following LiCl/urea precipitation [21, 22]. As ERα status was unknown and paraffin blocks were unavailable, owing to the small quantity of sample material, qPCR was used to determine ERα mRNA expression. Tumors were divided into two groups (ERα+ and ERα-) based on ERα expression. The mRNA expression of the five vesicle trafficking genes (described below) was compared between the two groups using the Mann-Whitney test (Minitab software).
RNA Assessment
All RNA samples were assessed for quality by spectrophotometry and agarose gel electrophoresis. Additionally, RNA samples for Affymetrix GeneChip hybridization were purified on Rneasy mini-columns according to the manufacturer's instructions (Qiagen) and were also assessed by Affymetrix test chips (Affymetrix, Inc., Santa Clara, CA).
GeneChip Microarrays
Labeled cRNA was prepared following Affymetrix protocols (www.affymetrix.com). Double-stranded cDNA was synthesized from 5 μg total RNA using a T7-(dT)24 primer (Invitrogen). cRNA was then synthesized by in vitro transcription and biotinylated using the Bioarray High-Yield RNA Transcript Labeling kit (Affymetrix). Labeled targets (20 μg/sample) were fragmented and hybridized to Hu U133 Plus 2.0 GeneChips in a rotating oven at 45°C at the Microarray Facility, University of Newcastle. GeneChips were washed and stained with streptavidin-conjugated phycoerythrin on a fluidics station and scanned twice at 570 nm in a GeneArray Scanner at a resolution of 3 μm. Data was analyzed with MicroArray Suite 5.0 software (Affymetrix). Default settings for absent and present calls were p < 0.04 (present), p 0.04 – 0.06 (marginal) and p>0.06 (absent). Global scaling normalization was used to normalize the expression data to the target value of 150. Gene expression was deemed to be regulated if the signal in the samples differed two-fold (signal log ratio >1.0 or <-1.0; http://www.netaffx.com). It is not possible to know the exact fold-change that represents a significant regulation of gene expression (and this may be different for individual genes). A two-fold cut-off was selected, as this is commonly accepted as appropriate in other microarray studies.
Quantitative Real-time PCR (qPCR)
Primers were designed using Primer Express software (Applied Biosystems) so that the amplicon crossed intron–exon boundaries (Table 1.). A nucleotide BLAST search was performed to ensure that primers were specific for the intended transcripts (http://www.ncbi.nlm.nih.gov/blast/). For reverse transcription, 1.0 μg of DNase treated RNA was incubated in 50 mM Tris HCl pH 8.3, 79 mM KCl, 3 mM MgCl2, 10 mM DTT, 2 mM dNTPs, with 0.5 μM oligo dT12–18 (Amersham) and 60 U Moloney murine leukemia virus reverse transcriptase (USB) in 20 μL for 1 hr at 37°C. qPCR reactions contained 120 nM primers in 0.33 μL of the reverse transcription reaction with a final volume of 10 μL using SYBR Green JumpStart Taq (Sigma) as recommended by the manufacturer. Each sample was loaded in triplicate onto an ABI Prism 7900HT Sequence Detection System (Applied Biosystems). Data was analyzed using SDS 2.2 software to assess the level of expression of target genes using the absolute quantification method involving generation of a standard curve for each gene (ABI Prism 7900HT Sequence Detection System and SDS Enterprise Database) (http://www.appliedbiosystems.com). The maximum fluorescence for each gene was normalized for β-actin expression.
Table 1.
Primer sequences and correlation coefficients (R2) for qPCR reactions
| Gene | Forward primer | Reverse Primer | Correlation coefficients (R2) |
|---|---|---|---|
| GALNT4 | GGCCTATATCTTCGTGGAGCTC | CCTGCGGAGGCATGAAAA | 0.9766 |
| RAB27B | GTGCATCTTCAGCTTTGGGAC | GAGACTCCGGAACCGCTCTT | 0.9935 |
| SLC12A2 | ATCAATTTTTCAGTATTCCATGCATC | ACGCCATCCTGGAGATTTTG | 0.9995 |
| SNX24 | TCAAGCAAACTGTCCCACCA | ATATGGATCCCTGAGGAACAGC | 0.9965 |
| SYTL5 | GCCCCAATGGCAGCTG | TTAGCTGCGCGATTTTGTCAC | 0.9866 |
| TFF1 | CCCAGACAGAGACGTGTACAGTG | AACCACAATTCTGTCTTTCACGG | 0.9989 |
| β-actin | GGTCATCACCATTGGCAATG | CCACAGGACTCCATGCCC | 0.9992 |
| RERG | ACCAAACGGTTCATCTGGGA | TCGGTAGGTTGATTCGAGGGT | 0.9990 |
| ER α | GCTCTTGGACAGGAACCAGG | AAGATCTCCACCATGCCCTCT | 0.9843 |
Western Blot Analysis
MCF-7 and Hs-578T cells grown in the presence of 10−9M E2 were lysed using RIPA buffer and proteins prepared for Western blot analysis. 5μg protein samples were subjects to SDS-PAGE and proteins transferred to nitrocellulose membranes. The membranes were incubated with anti-SYTL5 antibodies (raised by PickCell Laboratories) overnight at 4°C followed by horseradish peroxidase conjugated goat anti-rabbit antibody at 1:2000 in PBST/5% non-fat milk for 1 hour at 37°C (Pierce). Filters were visualized using chemiluminescence (Super-Signal West Dura Extended Duration Substrate Kit).
Immunohistochemistry
Ethical approval was obtained from the local research ethics committee. Formalin-fixed paraffin-embedded tissue was used from breast reductions (n=2), ductal carcinoma in situ (DCIS) (n=6) and invasive breast carcinomas (n=13) (invasive ductal carcinoma NST [n=9], mucinous carcinoma [n=3]) ([ERα+: n=6] [ERα: n=7)). ERα-status of invasive breast carcinomas had previously been determined during routine clinical histopathological diagnosis by immunohistochemistry according to national guidelines (The Royal College of Pathologists, UK). 4 μm sections were cut using a microtome and deparaffinized for 10 minutes in xylene, followed by 2 99% IMS rinses and then taken down to water. Sections were subject to a 2 hr pressure cooker treatment (20 minutes at pressure and left to stand for the remaining time) in pH9 retrieval solution (Novocastra). The sections were then loaded onto the Dako Techmate 500 automated stainer. After washing in Buffer (Dako), they are incubated for 10 minute 3% H202 in methanol endogenous peroxidase block. After rinsing the sections were incubated for 10 minutes in Normal Goat Serum block. Then sections were incubated for 1 hour at room temperature with anti-SYTL5 antibody (1:500) (PickCell Laboratories) diluted in PBS. After incubation with primary antibody, sections were rinsed twice and then incubated for 30 minutes with undiluted Envision visualization reagent (Dako). Following further washes the sections are incubated with Dako Envision chromogen at 1:50 dilution for 18 minutes at room temperature. Finally sections are incubated with 5% copper sulphate solution to enhance staining and counterstained with hematoxylin. Then sections were rinsed in water, dehydrated through graded alcohol and xylene, and mounted in DePeX (BDH). Negative controls were performed by omission of the primary antibody and incubation with PBS.
Results
Validation of the Transcriptome Effects of E2 Stimulation and Steroid Withdrawal on Breast Cancer Cell Lines
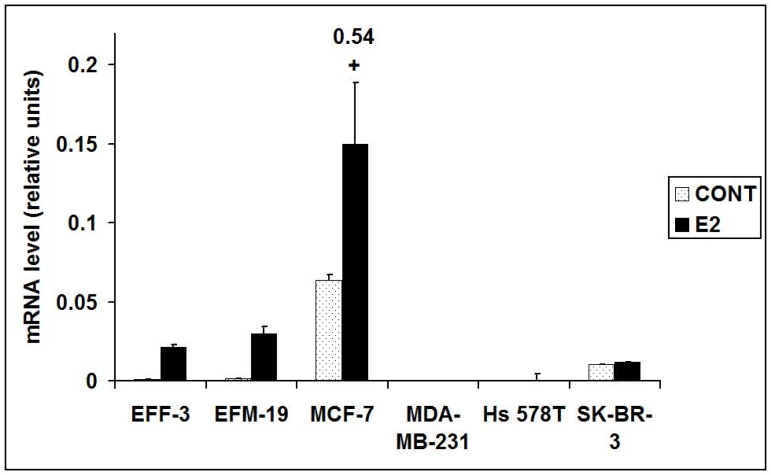
qPCR for the established E2-regulated gene, TFF1, was performed on all cell line RNA samples [16]. This confirmed that E2 stimulated a strong induction of TFF1 mRNA expression in E2-responsive ERα+ cell lines (EFF-3, EFM-19 & MCF-7) at 48hr time points (Figure 1.), and in E2 dose-response and time course experiments (MCF-7). TFF1 mRNA was either not expressed or was expressed at lower levels in E2-independent ERα- cell lines (MDA MB-231, Hs578T & SK-BR-3) and showed no change in expression with E2-stimulation (Figure 1.). The demonstration of these patterns of mRNA expression for the axiomatic E2-regulated gene, TFF1, confirms that experimental conditions including steroid withdrawal and E2-stimulation were appropriate.
Figure 1.
qPCR demonstrating the mRNA expression of TFF1 in EFF-3, EFM-19, MCF-7, MDA-MB-231, Hs578T and SK-BR-3 cells after 48 hours E2-stimulation (E2) compared with steroid withdrawn (CONT) samples. (Error bars are standard deviation of three independent replicates.)
GeneChip Microarray Analysis of EFF-3, EFM-19 and MCF-7 Cell Lines
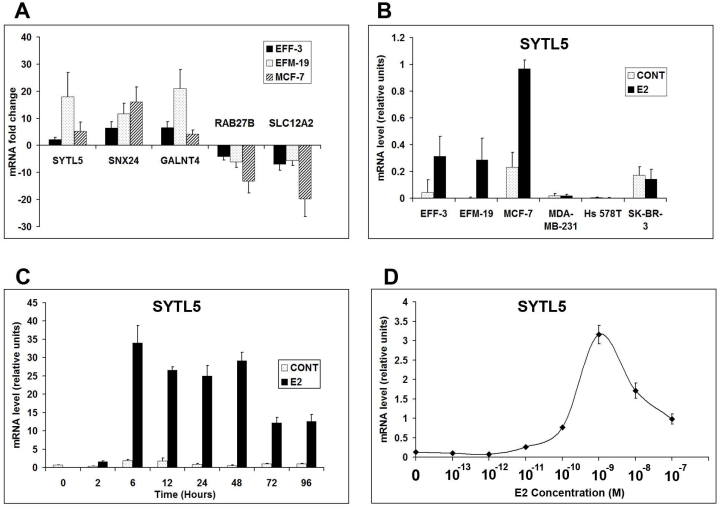
qPCR was performed on RNA samples used for microarray studies to determine mRNA expression of TFF1, RERG, SYTL5, RAB27B, SLC12A2, SNX24 and GALNT4. This demonstrated a strong correlation between microarray data and qPCR, including the direction of regulation (induction or repression) and the degree of regulation (Supplemental Table 1; Figures 1, 2A and 2B).
Figure 2.
qPCR demonstrating E2-regulation of SYTL5, GALNT4, SNX24, RAB27B and SLC12A2. A. E2-regulation of all 5 genes in EFF-3, EFM-19 and MCF-7 cells showing mRNA fold change after 48 hours E2-treatment. B. The effects of E2-stimulation for 48 hours on SYTL5 mRNA expression on 6 breast cancer cell lines comparing steroid withdrawn (CONT) with E2-stimulated (E2) samples. C. SYTL5 mRNA expression in MCF-7 cells after E2-stimulation for 0 to 96 hours, comparing steroid withdrawn (CONT) with E2-stimulated (E2) samples. D. SYTL5 mRNA expression in MCF-7 cells after 48 hours in the presence of 0 – 10−7M E2. (Error bars are standard deviation of three independent replicates.)
255, 748 and 2077 transcripts in EFF-3, EFM-19 and MCF-7 cells respectively, showed =2-fold regulation of expression by E2. Numerous established E2-regulated genes were identified, including PgR, TFF1, PDZK1, GREB1, IRS1, IGF1R, CDC2, CDC6, IGFBP4, RERG, STC2, CXCL12 and BLNK. The identification of known E2-regulated genes further validates the experimental conditions and the microarray data.
We believed that while existing gene ontology systems have significant value to help elucidate the functional significance of microarray data, that they also have limitations. Therefore, in order to avoid classifying genes in a pre-defined manner or focusing on gene networks, a literature search was performed for all characterized genes detected by the arrays (www.pubmed.com). The most notable finding was the unexpected identification of 147 genes implicated in various aspects of membrane, protein and vesicle trafficking, including exocytosis (Supplemental Table 1). Instead of cataloging all E2-regulated genes or studying mechanistic aspects of ER function, we wished to focus on a novel group of yet unrecognized E2-regulated genes: vesicle trafficking genes.
Varying patterns of induction and repression of expression of vesicle trafficking genes were seen in EFF-3, EFM-19 and MCF-7 cells with the majority being regulated in one or two out of the three cell lines. Based on a 2-fold cutoff, only eight vesicle trafficking genes showed regulation in all three cell lines (GALNT4, PDZK1, PPFIA4, RAB27B, RERG, SLC12A2, SNX24 and SYTL5). All of these genes were up-regulated except SLC12A2 and RAB27B, which were down-regulated. RERG and PDZK1 are known E2-regulated genes that have already been studied in breast cancer [13]. One striking finding was the identification of a large number of RAB GTPases, RABlike/related genes and Rho-related genes. Many of the genes identified are implicated or known to have roles in exocytosis, including RAB GTPases, SNAP23, SLC12A2, STX16, RIMS3, SYTL4, SYTL5, VAMP2 and VAMP8 (Supplemental Table 1).
E2 Regulation and Expression of 5 Vesicle Trafficking Genes in Breast Cancer Cell Lines
We selected five vesicle trafficking genes (SYTL5, RAB27B, SNX24, GALNT4 and SLC12A2), which were consistently E2-regulated in EFF-3 EFM-19, and MCF-7 cells for further investigation. qPCR was performed with all 6 cell lines used in the study with steroid withdrawn and E2-stimulated samples at 48 hour time points. This confirmed that all five genes showed a strong pattern of E2-regulation of mRNA expression in EFF-3, EFM-19 and MCF-7 cells, which was ≥2-fold in all cases with the same direction of regulation (induction or repression) as predicted by microarray data (Figure 2A). SNX24 and RAB27B showed a similar degree of relative regulation with microarray analysis and qPCR. The other genes showed more variation between cell lines. However, despite the accuracy of Affymetrix GeneChips, in the current setting, their principal role is as a genome-wide screening method, rather than as a tool for precise quantification of expression of individual genes. Therefore, exact correlation of the degree of regulation between the two methods cannot be expected.
Whereas all five genes showed a consistent pattern of E2-regulation in all three ERα+ cell lines, these genes showed no evidence of regulation in the three ERα- cell lines. Furthermore, all five genes were generally either not expressed or expressed at much lower levels in the ERα- cell lines. These findings are concordant with what is expected and support their existence as E2-regulated genes. Data is further illustrated for one gene, SYTL5 in Figure 2B recapitulating what has been described but showing the relative level of mRNA in control and E2-treated samples. Figure 2B shows that EFF-3, EFM-19 and MCF-7 cells demonstrated a strong induction of expression after 48 hours of E2-stimulation. It also confirms the lack of regulation in MDAMB-231, Hs-578T and SK-BR-3 cells. SYTL5 was expressed at very low or insignificant levels in MDA-MB-231 and Hs-578T cells. However, it was expressed at comparable but slightly lower levels in SK-BR-3 cells than in the E2-treated samples of EFF-3 and EFM-19 cells. EFM-19 cells showed the greatest degree of E2-regulation (17.96-fold), compared with EFF-3 and MCF-7 cells (Figure 2A). Despite this, EFM-19 cells had the lowest levels of SYTL5 expression after E2-stimulation compared with EFF-3 and MCF-7 cells (Figure 2B). MCF-7 cells showed the highest level of SYTL5 expression after E2-stimulation despite having a lower fold change in expression (Figures 2A and 2B).
Time course and E2 dose-response experiments with MCF-7 cells demonstrated that all five genes showed a strong pattern of induction (GALNT4, SNX24 and SYTL5) or repression (RAB27B and SLC12A2) of mRNA expression by E2. Corresponding control samples showed relatively lower (GALNT4, SNX24 and SYTL5) or higher (RAB27B and SLC12A2) expression levels than E2-treated samples. All five genes showed rapid E2-regulation of expression within 6 hours, which was maintained up to 72 hours (RAB27B and GALNT4) or 96 hours (SYTL5, SNX24 and SLC12A2). The time to reach the maximum (SYTL5, SNX24 and GALNT4) or minimum (RAB27B and SLC12A2) expression level was 6 hours (SYTL5), 12 hours (SNX24 and RAB27B), 48 hours (GALNT4) and 72 hours (SLC12A2). All five genes showed a pattern of expression reflecting a pharmacological E2 dose-response effect with maximum regulation between 10−10M and 10−7M. Figures 2C and 2D illustrate time course and E2 dose-response data respectively for SYTL5. SYTL5 showed a rapid and maximal induction at 6 hours, which was persistent over time but showed a lower level of induction at 72 and 96 hours. Figure 2D shows E2-regulation of SYTL5 expression with a characteristic sinusoidal dose-response curve with maximal regulation at 10−9M, a dose that is considered to be near physiological [6, 14, 15]. Together these data support the mRNA expression of all five genes being under the regulation of E2. The early induction of these genes and the E2 dose-response patterns suggest that these genes are regulated directed by E2 rather than via secondary effects of other E2-regulated translated proteins.
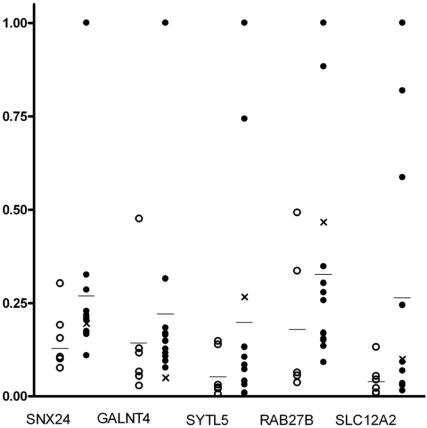
mRNA Expression of 5 Vesicle Trafficking Genes in Invasive Breast Carcinoma
Concentrations of ERα mRNA varied over 700 fold. Six tumour samples had a low expression of ERα, similar to the levels in the ERα- breast cancer cell lines, and were designated ERα-. The remaining tumours expressed ERα at levels equivalent to or greater than those in the ERα+ breast cancer cell lines and were designated ERα+. Figure 3 illustrates the mRNA expression levels in relative units for the five vesicle trafficking genes (GALNT4, SNX24, SYTL5, RAB27B and SLC12A2) in 18 breast tumours. All five genes were expressed in breast tumours, each gene showing varying patterns of expression. The mean level of expression was higher for each gene in the ERα+ tumours than the ERα- tumours. However, this did not reach statistical significance with SNX24 having the lowest p-value (p=0.0593). As association of E2-regulated genes with ERα-status was not the focus of this study, this was not further investigated.
Figure 3.
mRNA expression of SYTL5, GALNT4, SNX24, RAB27B and SLC12A2 in invasive breast carcinoma tissue samples and MCF-7 cells. (• ERα+ tumors, ○ ERα- tumors, – mean mRNA expression, X MCF-7 cells. Y-axis: mRNA expression in relative units.)
Protein Expression of SYTL5 in Breast Tumors
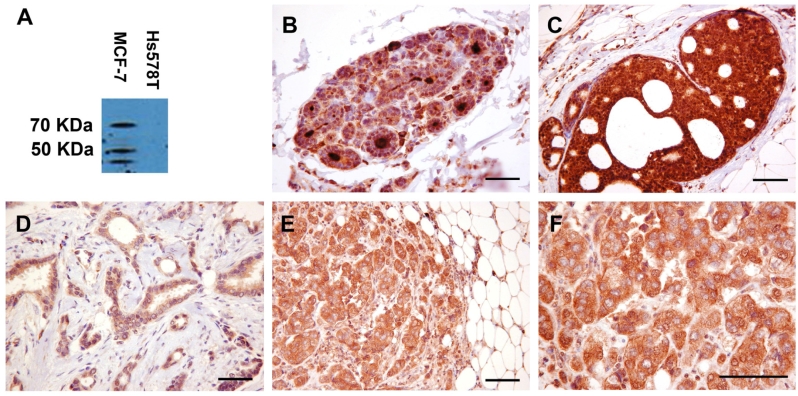
One vesicle trafficking gene, SYTL5, was selected for investigation of protein expression in breast tumours. Anti-SYTL5 polyclonal rabbit antibody was validated by performing Western blotting with protein extracts from breast cancer cell lines. Strong immunoreactive bands were identified in MCF-7 corresponding to proteins at 70KDa, 50KDa and 45KDa after 20 minutes exposure (Figure 4A). Although SYTL5 is predicted to encode an 81.52KDa protein, this is the first study of SYTL5 protein in human tissues. Exact correlations between predicted and actual molecular weights are not unexpected. No bands were identified with the Hs-578T protein. SYTL5 protein and mRNA expression correlate strongly, both being expressed at high levels in MCF-7 cells but absent or at very low levels in Hs-578T cells (Figures 2B and 4A).
Figure 4.
SYTL5 protein expression in breast cancer. A. SYTL5 protein expression determined by Western blotting in MCF-7 and Hs578T cells. B–F. Immunohistochemistry showing SYTL5 protein expression in normal breast tissue (strong staining) (B), DCIS (strong staining) (C), ER- invasive breast carcinoma NST (moderate staining) (D) and ER+ invasive breast carcinoma NST (strong staining) (E and F). (Scale bar = 100μm)
Immunohistochemistry controls with no primary antibody showed no staining in all tissue samples used including normal breast tissue and invasive breast carcinoma specimens. There was moderate to strong immunostaining for SYTL5 within the ductal epithelial cells and within the lumina of breast ducts of normal breast tissue (Figure 4B).
There was moderate SYTL5 immunostaining in three DCIS samples and strong staining in the remaining three DCIS samples (Figure 4C). The majority or all DCIS cells stained in five samples and moderate numbers of DCIS cells stained in one sample. All 13 invasive breast carcinomas showed SYTL5 immunostaining of carcinoma cells with strong staining in eight tumours and moderate staining in five tumours (Figures 4D-F). No difference in intensity of immunostaining was detected between ERα+ and ERα- tumours. Ten tumours showed staining of the majority or all carcinoma cells and three tumours showed staining of moderate numbers of carcinoma cells. Staining was present in a diffuse fine granular cytoplasmic pattern in carcinoma cells (Figure 4F). Nuclear or cell membrane staining was not apparent. The extracellular matrix did not stain. With the exception of occasional cells, stromal fibroblasts did not stain. Adipocytes and vascular smooth muscle cells showed no immunostaining. The pattern of immunostaining in these samples including the lack of staining in the negative control and in non-cancer cells suggests that the staining was not non-specific, and that SYTL5 protein is expressed in breast cancer.
Discussion
Identification of E2-regulated Vesicle Trafficking Genes in Breast Cancer
In the present study we report 147 vesicle trafficking genes that showed E2-regulation of mRNA expression in breast cancer cells. Five of these (SYTL5, RAB27B, SNX24, GALNT4 and SLC12A2) have been validated in detail using qPCR and showed a consistent pattern of E2-regulation in EFF-3, EFM-19 and MCF-7 cells. These five genes were also expressed as mRNA in invasive breast carcinoma tissues. SYTL5 protein was expressed in MCF-7 cells but not in Hs578T cells. SYTL5 protein was also expressed in benign breast ductal epithelial cells and in breast carcinoma cells (DCIS and invasive carcinoma). SYTL5 was expressed with a fine granular cytoplasmic pattern suggesting that it may be localized to vesicles on the micron or submicron scale. Although there was a pattern of higher SYTL5 mRNA expression in ERα+ than ERα- cell lines, there was no statistically significant association with ERα status of primary tumors. SYTL5 protein expression also showed no clear association with ERα status. This may be explained in part by sample size. Also, it should be considered that this finding is not necessarily unexpected as E2-regulated genes and ERα-associated genes are not synonymous [13].
Varying patterns of E2-regulation of vesicle trafficking genes were detected between cell lines. The majority of genes only showed E2-regulation in one or two of the three cell lines and a few, for example RAB31, showed bi-directional E2-regulation between cell lines. Only eight genes were consistently E2-regulated in all three cell lines, five of which we have studied in detail.
These findings are in keeping with the current knowledge of E2-regulated genes in breast cancer [13]. It is incorrect to assume that the E2-regulated transcriptome of MCF-7 cells is identical to that of other breast cancer cells, both in culture and in vivo. It is well known that there is considerable heterogeneity of gene expression between different tumors and between different cancer cell lines [24]. Previous work has shown that only a minority of E2-regulated genes are common to different cell lines. Rae et al only reported three such genes out of 811 that were common to three breast cancer cell lines [10]. Soulez and Parker identified cytochrome P450 IIB as the most up-regulated (>100 fold) gene by E2 in ZR75-1 cells, and also reported that it was neither expressed nor E2-regulated in MCF-7 cells [9]. The reader should also consider that ERα itself is an E2-regulated gene that is bi-directionally regulated between different cell lines [25]. All of this points towards there being considerable heterogeneity of the E2-regulated transcriptome in breast cancer.
There are a number of possible reasons why previous authors have not given full recognition to vesicle trafficking genes. These include the tendency to limit studies to one cell line, gene ontology classification systems, a focus on genes with the greatest degree of regulation or a focus on the predictive value of gene signatures rather than gene function [11–13]. There has also often been a focus on E2-regulation of genes that regulate the cell cycle with less recognition being given to other E2-regulated genes [4].
Although the present study describes a novel group of E2-regulated genes in breast cancer, these findings are not entirely unexpected when the wider literature is considered. E2 has been shown to regulate the expression of six proteins involved in exocytosis in the rat pituitary gland (synaptotagmin 1, syntaxin 1, RAB3A, SNAP25, synaptobrevin 2 and cellubrevin) [26]. E2 also regulates the expression of RAB11 in endometrial cells, which also correlates with our findings [27]. RAB31 and RAB5C-like expression has previously been reported to be E2-regulated in MCF-7 and ZR75-1 cells respectively [8, 9]. However, little recognition has been given to the significance of their function. In 1982, Vic et al studied the effects of E2 on the ultrastructure of MCF-7 cells and reported that E2 lead to formation of small intracellular vesicles, which were released into the extracellular fluid by an exocytosis-like process [28]. However, the significance of these electron microscopy results has been given very little subsequent recognition or investigated further. There are significant parallels between the morphology described by Vic et al and the gene expression findings of the present study.
SYTL5, RAB27B, SNX24, GALNT4 and SLC12A2
Synaptotagmin-like 5 protein (SYTL5) is a novel 730 amino acid E2–regulated protein [29]. It belongs to a group of proteins thought to be involved in vesicle trafficking and exocytosis. Synaptotagmin-like proteins are mediators of neurotransmitter release and have a role in hormone release from endocrine cells. SYTL5 and SYTL4 (which was also identified as E2-regulated) are effector molecules that specifically bind and regulate the GTP-bound form of the GTPase RAB27A/B [29, 30]. In this regard, the concurrent identification of SYTL5 and its hypothetical partner molecule RAB27B may be significant. Interestingly, SYTL1 and RAB27A are known vesicle trafficking partner molecules in androgen-regulated secretion of prostate-specific antigen (PSA) and prostatic-specific acid phosphatase (PSAP) in prostate cancer cells [31].
Ras-related protein RAB27B (RAB27B) is a 218 amino acid membrane-bound member of the Ras oncogene family of GTPases. RAB27A and RAB27B are closely related GTPases involved in vesicle trafficking and exocytosis [30, 32]. RAB27B is involved in melanosome trafficking, and exocytosis of hormones, enzymes and platelet dense core granules. RAB27A is implicated in lymph node metastasis in murine xenograft breast cancer models [33].
Sorting nexin 24 (SNX24) is a little studied PX domain-containing vesicle trafficking protein of 199 amino acids. Sorting nexins form a diverse family, some of which are known to be regulators of endosomal sorting [34]. This includes protein and membrane trafficking between the sorting endosome compartment and the cell membrane, the Golgi body and lysosomes. Although there is evidence that SNX1, SNX2, SNX10 and SNX16 are involved in tumorigenesis, SNX24 remains uninvestigated [33, 35–38].
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N acetylgalactosaminyltransferase 4 (GALNT4) is a 578 amino acid type II membrane protein that catalyzes the initial reaction in O-linked oligosaccharide biosynthesis, the transfer of an N-acetyl-Dgalactosamine residue to a serine or threonine residue on the protein [39]. It is located in the Golgi body where it forms part of the protein and vesicle trafficking pathway.
Solute carrier family 12 (sodium/ potassium/chloride transporters) member 2 (SLC12A2/NKCC1/BSC2) is a bumetanide-sensitive 1,191 amino acid integral membrane protein that is expressed ubiquitously, in particular, in the basolateral membrane of epithelial cells and in the nervous system. It is an electroneutral cation-chloride cotransporter that mediates sodium and chloride reabsorption and plays a vital role in the regulation of ionic balance and cell volume [40]. It mediates exocytosis of catecholamines from chromaffin cells and is involved in neuroregulation and breast morphogenesis [41, 42].
The Significance of E2-regulated Vesicle Trafficking Genes in Breast Tumorigenesis
In 2002, Palmer et al reported that a chimeric transcription factor (EWS-WT1), a fusion product of chromosomal translocation in desmoplastic small round cell tumor, led to a large induction of BAIAP3, a protein involved in exocytosis. This introduced what appeared to be a new paradigm of exocytosis-mediated tumorigenesis [21, 22].
There is now a large body of evidence that vesicle trafficking and exocytosis are important in tumorigenesis [21–23]. A growing number of vesicle trafficking genes have been shown to have roles in tumourigenesis or are over-expressed in diverse types of cancer [23]. This includes at least 21 RAB GTPases, synaptotagmin I, syntaxins, septins, TPD52, SNAP25, sorting nexins, LIMK1, RAC1, RAP1A and REBL1 [23, 35–38, 43–47].
Cheng et al showed that of the genes in the region of the 1q22 amplification in ovarian and breast cancers, RAB25 was over-expressed [48, 49]. Increased RAB25 expression increased tumourigenesis in vitro and in vivo. In murine xenograft breast and ovarian cancer models, RAB25 over-expression increased tumour cell growth and worsened prognosis, whereas reduced RAB25 expression decreased tumour proliferation. More recently, Hou et al showed that RAB23 is an amplified over-expressed gene in gastric cancer that has an important role in invasion [50].
The work of Montel et al has highlighted that a significant number of metastasis-associated genes in their study had vesicle trafficking roles [33, 51]. These included RAB27A, RAB38, SNX10, syntaxin 7, SNAP23 and VAMP-A, which were differentially expressed in murine xenograft models of breast cancer metastasis.
Extracellular microvesicles are <1μm diameter vesicles that are released from normal and cancer cells into the extracellular fluid [52, 53, 54]. Microvesicles (MV) are considered to be ectoorganelles with cell to cell communication roles including via growth factors, and horizontal transfer of receptors (e.g. CXCR4) and mRNA (e.g. VEGF, MMPs) between cells [55, 56]. They are formed within and released from cells by intracellular vesicle trafficking, although these pathways are largely unstudied. MV have been purified from cancer cell lines and malignant effusions [57]. They are well studied and there is strong evidence that they have important roles in diverse areas of tumourigenesis. This includes tumor invasion, growth factor release, angiogenesis, the p53 response, tumor immune privilege, neoplastic coagulopathy and chemotherapy resistance [58–64]. MV have been shown to contain proteins (e.g. VEGF, MMPs, FGF, tissue factor, CXCR4 & HER2), DNA, mRNA, microRNAs and even mitochondria [23, 52, 53, 55, 57].
The exact roles and interactions of the large number of E2-regulated vesicle trafficking genes in breast cancer remain unclear. However, by identifying this novel group of genes, it is hoped that this will broaden our perspective and understanding of breast cancer. Previous authors have already shown that some of these genes or other members of the same gene family are involved in tumourigenesis. Future work is now required to elucidate the roles of vesicle trafficking in breast cancer and to integrate it with our current knowledge. Furthermore, the function of individual genes requires characterization in relation to the exocytosis-like process described by Vic et al, which has now been “re-discovered” over a quarter of a century later [28].
Supplementary Material
Acknowledgments
This work was supported by grants from The Royal College of Surgeons of Edinburgh, The Cancer and Polio Research Fund, The Newcastle upon Tyne NHS Hospitals Charity, Breast Cancer Campaign, Cancer Research UK and The Wellcome Trust. PKW was funded by the Robertson Research Training Fellowship from The Royal College of Surgeons of Edinburgh.
References
- 1.Nilsson S, Makela S, Treuter E, Tujaque M, Thomsen J, Andersson G, Enmark E, Pettersson K, Warner M, Gustafsson JA. Mechanisms of estrogen action. Physiol Rev. 2001;81:1535–1565. doi: 10.1152/physrev.2001.81.4.1535. [DOI] [PubMed] [Google Scholar]
- 2.Ackermann GE, Carr BR. Estrogens. Rev Endocr Metab Disord. 2002;3:225–230. doi: 10.1023/a:1020024409888. [DOI] [PubMed] [Google Scholar]
- 3.Osborne CK, Schiff R. Estrogen-receptor biology: continuing progress and therapeutic implications. J Clin Oncol. 2005;23:1616–1622. doi: 10.1200/JCO.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 4.Doisneau-Sixou SF, Sergio CM, Carroll JS, Hui R, Musgrove EA, Sutherland RL. Estrogen and antiestrogen regulation of cell cycle progression in breast cancer cells. Endocr Relat Cancer. 2003;10:179–186. doi: 10.1677/erc.0.0100179. [DOI] [PubMed] [Google Scholar]
- 5.Cunliffe HE, Ringner M, Bilke S, Walker RL, Cheung JM, Chen Y, Meltzer PS. The gene expression response of breast cancer to growth regulators: patterns and correlation with tumor expression profiles. Cancer Res. 2003;63:7158–7166. [PubMed] [Google Scholar]
- 6.Johnson MD, Westley, May FEB. Oestrogenic activity of tamoxifen and its metabolites on gene regulation and cell proliferation in MCF-7 breast cancer cells. Br J Cancer. 1989;59:727–738. doi: 10.1038/bjc.1989.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Platet N, Cathiard AM, Gleizes M, Garcia M. Estrogens and their receptors in breast cancer progression: a dual role in cancer proliferation and invasion. Crit Rev Oncol Hematol. 2004;51:55–67. doi: 10.1016/j.critrevonc.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 8.Frasor J, Danes JM, Komm B, Chang KC, Lyttle CR, Katzenellenbogen BS. Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: Insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology. 2003;144:4562–4574. doi: 10.1210/en.2003-0567. [DOI] [PubMed] [Google Scholar]
- 9.Soulez M, Parker MG. Identification of novel oestrogen receptor target genes in human ZR75-1 breast cancer cells by expression profiling. J Mol Endocrinol. 2001;27:259–274. doi: 10.1677/jme.0.0270259. [DOI] [PubMed] [Google Scholar]
- 10.Rae JM, Johnson MD, Scheys JO, Cordero KE, Larios JM, Lippman ME. GREB1 is a critical regulator of hormone dependent breast cancer growth. Breast Cancer Res Treat. 2005;92:141–149. doi: 10.1007/s10549-005-1483-4. [DOI] [PubMed] [Google Scholar]
- 11.Kininis M, Chen BS, Diehl AG, Isaacs GD, Zhang T, Siepel AC, Clark AG, Kraus WL. Genomic analysis of transcription factor binding, histone acetylation and gene expression reveal mechanistically distinct classes of estrogen-regulated genes. Mol Cell Biol. 2007;27:5090–5104. doi: 10.1128/MCB.00083-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu J, Yu J, Cordero KE, Johnson MD, Ghosh D, Rae JM, Chinnaiyan AM, Lippman ME. A transcriptional fingerprint of estrogen in human breast cancer predicts patient survival. Neoplasia. 2008;10:79–88. doi: 10.1593/neo.07859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Westley BR, May FEB. Identification of steroid hormone-regulated genes in breast cancer. Methods Mol Med. 2006;120:363–388. doi: 10.1385/1-59259-969-9:363. [DOI] [PubMed] [Google Scholar]
- 14.May FEB, Westley BR. Cloning of estrogen-regulated messenger sequences from human breast cancer cells. Cancer Res. 1986;46:6034–6040. [PubMed] [Google Scholar]
- 15.Henry JA, Nicholson S, Hennessy C, Lennard TWJ, May FEB, Westley BR. Expression of the oestrogen regulated pNR-2 mRNA in human breast cancer: relation to oestrogen receptor mRNA levels and response to tamoxifen therapy. Br J Cancer. 1990;61:32–38. doi: 10.1038/bjc.1990.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prest SJ, May FE, Westley BR. The estrogen-regulated protein, TFF1, stimulates migration of human breast cancer cells. FASEB J. 2002;16:592–594. doi: 10.1096/fj.01-0498fje. [DOI] [PubMed] [Google Scholar]
- 17.Simon WE, Albrecht M, Trams G, Dietel M, Holzel F. In vitro growth promotion of human mammary carcinoma cells by steroid hormones, tamoxifen, and prolactin. J Natl Cancer Inst. 1984;73:313–321. doi: 10.1093/jnci/73.2.313. [DOI] [PubMed] [Google Scholar]
- 18.Healicon RM, Westley BR, May FEB. Isolation and characterization of an oestrogen-responsive breast cancer cell line, EFF-3. Int J Cancer. 1993;53:388–394. doi: 10.1002/ijc.2910530308. [DOI] [PubMed] [Google Scholar]
- 19.Burgoyne RD, Morgan A. Secretory granule exocytosis. Physiol Rev. 2003;83:581–632. doi: 10.1152/physrev.00031.2002. [DOI] [PubMed] [Google Scholar]
- 20.Jordens I, Marsman M, Kuijl C, Neefjes J. Rab proteins, connecting transport and vesicle fusion. Traffic. 2005;6:1070–1077. doi: 10.1111/j.1600-0854.2005.00336.x. [DOI] [PubMed] [Google Scholar]
- 21.Chan AM, Weber T. A putative link between exocytosis and tumor development. Cancer Cell. 2002;2:427–428. doi: 10.1016/s1535-6108(02)00210-6. [DOI] [PubMed] [Google Scholar]
- 22.Palmer RE, Bong Lee S, Wong JC, Reynolds PA, Zhang H, Truong V, Oliner JD, Gerald WL, Haber DA. Induction of BAIAP3 by EWS-WT1 chimeric fusion implicates regulated exocytosis in tumorigenesis. Cancer Cell. 2002;2:497–505. doi: 10.1016/s1535-6108(02)00205-2. [DOI] [PubMed] [Google Scholar]
- 23.Wright PK. Targeting vesicle trafficking: An important approach to cancer chemotherapy. Recent Patents Anti-Cancer Drug Discov. 2008;3:137–147. doi: 10.2174/157489208784638730. [DOI] [PubMed] [Google Scholar]
- 24.Bertucci F, Viens P, Tagett R, Nguyen C, Houlgatte R, Birnbaum D. DNA arrays in clinical oncology: promises and challenges. Lab Invest. 2003;83:305–316. doi: 10.1097/01.lab.0000059936.28369.19. [DOI] [PubMed] [Google Scholar]
- 25.Westley BR, May FEB. Oestrogen regulates oestrogen receptor mRNA levels in an oestrogen-responsive human breast cancer cell line. Biochem Biophys Res Commun. 1988;155:1113–1118. doi: 10.1016/s0006-291x(88)81255-5. [DOI] [PubMed] [Google Scholar]
- 26.Majo G, Lorenzo MJ, Blasi J, Aguado F. Exocytotic protein components in rat pituitary gland after long-term estrogen administration. J Endocrinol. 1999;161:323–331. doi: 10.1677/joe.0.1610323. [DOI] [PubMed] [Google Scholar]
- 27.Chen D, Ganpathy P, Zhu L, Xu X, Li Q, Bagchi IC, Bagchi MK. Potential regulation of membrane trafficking by estrogen receptor α via induction of rab11 in uterine glands during implantation. Mol Endocrinol. 1999;13:993–1004. doi: 10.1210/mend.13.6.0287. [DOI] [PubMed] [Google Scholar]
- 28.Vic P, Vignon F, Derocq D, Rochefort H. Effect of estradiol on the ultrastructure of the MCF-7 human breast cancer cells in culture. Cancer Res. 1982;42:667–673. [PubMed] [Google Scholar]
- 29.Kuroda TS, Fukuda M, Ariga H, Mikoshiba K. Synaptotagmin-like protein 5: a novel Rab27A effector with C-terminal tandem C2 domains. Biochem Biophys Res Commun. 2002;293:899–906. doi: 10.1016/S0006-291X(02)00320-0. [DOI] [PubMed] [Google Scholar]
- 30.Fukuda M. Versatile role of Rab27 in membrane trafficking: focus on the Rab27 effector families. J Biochem (Tokyo) 2005;137:9–16. doi: 10.1093/jb/mvi002. [DOI] [PubMed] [Google Scholar]
- 31.Johnson JL, Ellis BA, Noack D, Seabra MC, Catz SD. The Rab27a binding protein, JFC1, regulates the androgen-dependent secretion of prostate-specific antigen and prostate-specific acid phosphatase. Biochem J. 2005;391:699–710. doi: 10.1042/BJ20050380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramalho JS, Tolmachova T, Hume AN, McGuigan A, Gregory-Evans CY, Huxley C, Seabra MC. Chromosomal mapping, gene structure and characterization of the human and murine RAB27B gene. BMC Genet. 2001;2:2e. doi: 10.1186/1471-2156-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Montrel V, Huang T, Mose E, Pesonjamasp K, Tarin D. Expression profiling of primary tumours and matched lymphatic and lung metastases in a xenogeneic breast cancer model. Am J Path. 2005;166:1565–1579. doi: 10.1016/S0002-9440(10)62372-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Worby CA, Dixon JE. Sorting out the cellular functions of sorting nexins. Nat Rev Mol Cell Biol. 2002;3:919–931. doi: 10.1038/nrm974. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen LN, Holdren MS, Nguyen AP, Furuya MH, Bianchini M, Levy E, Mordoh J, Liu A, Guncay GD, Campbell JS, Parks WT. Sorting Nexin 1 Down-Regulation Promotes Colon Tumourigenesis. Clin Cancer Res. 2006;12:6952–6959. doi: 10.1158/1078-0432.CCR-06-0317. [DOI] [PubMed] [Google Scholar]
- 36.Fuchs U, Rehkamp G, Hass OA, Slany R, Konig M, Bojesen S, Bohle RM, Damm-Welk C, Ludwig WD, Harbott J, Borkhardt A. The human formin-binding protein 17 (FBP17) interacts with sorting nexin, SNX2, and is an MLL-fusion partner in acute myelogenous leukaemia. Proc Natl Acad Sci USA. 2001;98:8756–8761. doi: 10.1073/pnas.121433898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watahiki A, Waki K, Hayatsu N, Shiraki T, Kondo S, Nakamura M, Sasaki D, Arakawa T, Kawai J, Harbers M, Hayashizaki Y, Carninci P. Libraries enriched for alternatively spliced exons reveal splicing patterns in melanocytes and melanomas. Nature Methods. 2004;1:233–239. doi: 10.1038/nmeth719. [DOI] [PubMed] [Google Scholar]
- 38.Osman I, Bajorin DF, Sun TT, Zhong H, Douglas D, Scattergood J, Zheng R, Han M, Marshall KW, Liew CC. Novel blood biomarkers of human urinary bladder cancer. Clin Cancer Res. 2006;12:3374–3380. doi: 10.1158/1078-0432.CCR-05-2081. [DOI] [PubMed] [Google Scholar]
- 39.Bennett EP, Hassan H, Mandel U, Mirgorodskaya E, roepstorff P, Burchell J, Taylor-Papadimitriou J, Hollingsworth MA, Merkx G, van Kessel AG, Eiberg H, Steffensen R, Clausen H. Cloning of a human UDP-Nacetyl-α-D-galactosamine:polypeptide Nacetylgalactosaminyltransferase that complements other GalNAc-transferases in complete O-glycosylation of the MUC1 tandem repeat. J Biol Chem. 1998;27:30472–30481. doi: 10.1074/jbc.273.46.30472. [DOI] [PubMed] [Google Scholar]
- 40.Hebert SC, Mount DB, Gamba G. Molecular physiology of cation-coupled Cl- cotransport: the SLC12 family. Pflugers Arch - Eur J Physiol. 2004;447:580–593. doi: 10.1007/s00424-003-1066-3. [DOI] [PubMed] [Google Scholar]
- 41.Xie Z, Currie KPM, Cahill AL, Fox AP. Role of Cl- Co-transporters in excitation produced by GABAA receptors in juvenile bovine adrenal chromaffin cells. J Neurophysiol. 2003;90:3828–3837. doi: 10.1152/jn.00617.2003. [DOI] [PubMed] [Google Scholar]
- 42.Schillingford JM, Miyoshi K, Flagella M, Shull GE, Hennighasuen L. Mouse mammary epithelial cells express the Na-K-Cl cotransporter, NKCC1: Characterization, localization and involvement in ductal development and morphogenesis. Mol Endocrinol. 2002;16:1309–1321. doi: 10.1210/mend.16.6.0857. [DOI] [PubMed] [Google Scholar]
- 43.Montalbano J, Jin W, Sheikh MS, Huang Y. RBEL1 Is a novel gene that encodes a nucleocytoplasmic Ras superfamily GTP-binding protein and Is overexpressed in breast cancer. J Biol Chem. 2007;282:37640–37649. doi: 10.1074/jbc.M704760200. [DOI] [PubMed] [Google Scholar]
- 44.Nishimura Y, Yoshiokab K, Bernardc O, Himenoa M, Itohb K. LIM kinase 1: evidence for a role in the regulation of intracellular vesicle trafficking of lysosomes and endosomes in human breast cancer cells. Eur J Cell Biol. 2004;83:369–380. doi: 10.1078/0171-9335-00382. [DOI] [PubMed] [Google Scholar]
- 45.Kominsky SL, Argani P, Korz D, Evron E, Raman V, Garrett E, Rein A, Sauter G, Kallioniemi OP, Sukumar S. Loss of the tight junction protein claudin-7 correlates with histological grade in both ductal carcinoma in situ and invasive ductal carcinoma of the breast. Oncogene. 2003;22:2021–2033. doi: 10.1038/sj.onc.1206199. [DOI] [PubMed] [Google Scholar]
- 46.Bravo-Cordero JJ, Marrero-Diaz R, Megi D, Genis L, García-Grande A, Garcia MA, Arroyo AG, Montoya MC. MT1-MMP proinvasive activity is regulated by a novel Rab8-dependent exocytic pathway. EMBO J. 2007;26:1499–1510. doi: 10.1038/sj.emboj.7601606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jayachandran G, Sazaki J, Nishizaki M, Xu K, Girard L, Minna JD, Roth JA, Ji L. Fragile histidine triad-mediated tumor suppression of lung cancer by targeting multiple components of the Ras/Rho GTPase molecular switch. Cancer Res. 2007;67:10379–10388. doi: 10.1158/0008-5472.CAN-07-0677. [DOI] [PubMed] [Google Scholar]
- 48.Cheng KW, Lahad JP, Kuo WL, Lapuk A, Yamada K, Auersperg N, Liu J, Smith-McCune K, Lu KH, Fishman D, Gray JW, Mills GB. The RAB25 small GTPase determines aggressiveness of ovarian and breast cancers. Nat Med. 2004;10:1251–1256. doi: 10.1038/nm1125. [DOI] [PubMed] [Google Scholar]
- 49.Cheng KW, Lahad JP, Gray JW, Mills GB. Emerging role of RAB GTPases in cancer and human disease. Cancer Res. 2005;65:2516–2519. doi: 10.1158/0008-5472.CAN-05-0573. [DOI] [PubMed] [Google Scholar]
- 50.Hou Q., Wu YH, Grabsch H, Zhu Y, Leong SH, Ganesan K, Cross D, Tan LK, Tao J, Gopalakrishnan V, Tanq BL, Kon OL, Tan P. Integrative genomics identifies RAB23 as an invasion mediator gene in diffuse-type gastric cancer. Cancer Res. 2008;68:4623–4630. doi: 10.1158/0008-5472.CAN-07-5870. [DOI] [PubMed] [Google Scholar]
- 51.Steeg PS. New insights into the tumor metastatic process revealed by gene expression profiling. Am J Path. 2005;166:1291–1294. doi: 10.1016/S0002-9440(10)62348-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hugel B, Martinez MC, Freyssinet J. Membrane microparticles: Two sides of the coin. Physiology. 2005;20:22–27. doi: 10.1152/physiol.00029.2004. [DOI] [PubMed] [Google Scholar]
- 53.Fevrier B, Raposo G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr Opin Cell Biol. 2004;16:415–421. doi: 10.1016/j.ceb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 54.Taylor DD, Black PH. Shedding of plasma membrane fragments. Neoplastic and developmental importance. Dev Biol. 1985;3:33–57. doi: 10.1007/978-1-4684-5050-7_3. [DOI] [PubMed] [Google Scholar]
- 55.Ratajczak J, Miekus K, Kucia M, Zhang J, Reca R, Dvorak P, Ratajczak MZ. Embryonic stem cell-derived microvesicles reprogram hematopoetic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukaemia. 2006;20:847–856. doi: 10.1038/sj.leu.2404132. [DOI] [PubMed] [Google Scholar]
- 56.Baj-Krzyworzeka M, Szatanek R, Weglarczyk K, Baran J, Urbanowicz B, Branski P, Ratajczak MZ, Zembala M. Tumour-derived microvesicles carry several surface determinants and mRNA of tumour cells and transfer some of these determinants to monocytes. Cancer Immunol Immunother. 2006;55:808–818. doi: 10.1007/s00262-005-0075-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Andre F, Schartz NE, Movassagh M, Flament C, Pautier P, Morice P, Pomel C, Lhomme C, Escudier B, Le Chevalier T, Tursz T, Amigorena S, Raposo G, Anqevin E, Zitvogel L. Malignant effusions and immunogenic tumour-derived exosomes. Lancet. 2002;360:295–305. doi: 10.1016/S0140-6736(02)09552-1. [DOI] [PubMed] [Google Scholar]
- 58.Graves LE, Ariztia EV, Navari JR, Matzel HJ, Stack MS, Fishman DA. Proinvasive properties of ovarian cancer ascites-derived membrane vesicles. Cancer Res. 2004;64:7045–7049. doi: 10.1158/0008-5472.CAN-04-1800. [DOI] [PubMed] [Google Scholar]
- 59.Kim CW, Lee HM, Lee TH, Kang C, Kleinman HK, Gho YS. Extracellular membrane vesicles from tumor cells promote angiogenesis via sphingomyelin. Cancer Res. 2002;62:6312–6317. [PubMed] [Google Scholar]
- 60.Kim JW, Wieckowski E, Taylor DD, Reichert TE, Watkins S, Whiteside TL. Fas ligand-positive membranous vesicles isolated from sera of patients with oral cancer induce apoptosis of activated T lymphocytes. Clin Cancer Res. 2005;11:1010–1020. [PubMed] [Google Scholar]
- 61.Schnaeker EM, Ossig R, Ludwig T, Dreier R, Oberleithner H, Wilhelmi M, Schneider SW. Microtubule-dependent matrix metalloproteinase-2/matrix metalloproteinase-9 exocytosis: prerequisite in human melanoma cell invasion. Cancer Res. 2004;64:8924–8931. doi: 10.1158/0008-5472.CAN-04-0324. [DOI] [PubMed] [Google Scholar]
- 62.Valenti R, Huber V, Iero M, Filipazzi P, Parmiani G, Rivoltini L. Tumor-released microvesicles as vehicles of immunosuppression. Cancer Res. 2007;67:2912–2915. doi: 10.1158/0008-5472.CAN-07-0520. [DOI] [PubMed] [Google Scholar]
- 63.Yu JL, May L, Lhotak V, Shahrzad S, Shirasawa S, Weitz JI, Coomber BL, Mackman N, Rak JW. Oncogenic events regulate tissue factor expression in colorectal cancer cells: implications for tumor progression and angiogenesis. Blood. 2005;105:1734–1741. doi: 10.1182/blood-2004-05-2042. [DOI] [PubMed] [Google Scholar]
- 64.Yu X, Harris SL, Levine AL. The regulation of exosome secretion: a novel function of the p53 protein. Cancer Res. 2006;66:4795–4801. doi: 10.1158/0008-5472.CAN-05-4579. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.