Abstract
The epidermal growth factor receptor (EGFR) is targeted for lysosomal degradation by ubiquitin-mediated interactions with the ESCRTs (endosomal-sorting complexes required for transport) in multivesicular bodies (MVBs). We show that secretory carrier membrane protein, SCAMP3, localizes in part to early endosomes and negatively regulates EGFR degradation through processes that involve its ubiquitylation and interactions with ESCRTs. SCAMP3 is multimonoubiquitylated and is able to associate with Nedd4 HECT ubiquitin ligases and the ESCRT-I subunit Tsg101 via its PY and PSAP motifs, respectively. SCAMP3 also associates with the ESCRT-0 subunit Hrs. Depletion of SCAMP3 in HeLa cells by inhibitory RNA accelerated degradation of EGFR and EGF while inhibiting recycling. Conversely, overexpression enhanced EGFR recycling unless ubiquitylatable lysines, PY or PSAP motifs in SCAMP3 were mutated. Notably, dual depletions of SCAMP3 and ESCRT subunits suggest that SCAMP3 has a distinct function in parallel with the ESCRTs that regulates receptor degradation. This function may affect trafficking of receptors from prelysosomal compartments as SCAMP3 depletion appeared to sustain the incidence of EGFR-containing MVBs detected by immunoelectron microscopy. Together, our results suggest that SCAMP3, its modification with ubiquitin, and its interactions with ESCRTs coordinately regulate endosomal pathways and affect the efficiency of receptor down-regulation.
INTRODUCTION
The internalization of cell surface receptors and transporters coupled to degradation or recycling is critical for nutrient uptake and regulating cell signaling. The epidermal growth factor receptor (EGFR) has been studied extensively as a prototypical receptor that is targeted for lysosomal degradation after ligand-stimulated internalization. Disrupted degradation of the EGFR and other receptors has been linked to the pathogenesis of many diseases including several cancers. An essential element in the down-regulation process is ubiquitin conjugation to EGFR by Cbl, an E3 ubiquitin ligase (reviewed in Marmor and Yarden, 2004). It is thought that the ubiquitin enables successive passage of the receptor to the endosomal-sorting complexes required for transport: ESCRT-0, I, II, and III. These complexes are evolutionarily conserved and specialize in targeting and packaging of membrane proteins into intraluminal vesicles (ILVs) upstream of lysosomal degradation. The ESCRT-0 complex is composed of Hrs, STAM, and Eps15b and is enriched in flat clathrin patches on early endosomes via interactions involving Hrs' FYVE and coiled-coiled domains (Raiborg et al., 2001a, 2002, 2006; Bache et al., 2003b; Roxrud et al., 2008). Ubiquitin-interacting motifs (UIMs) in ESCRT-0 subunits bind ubiquitylated ligands such as EGFR; interaction of Hrs with the UEV domain of ESCRT-I protein Tsg101 is thought to facilitate passage of the receptor to ESCRT-I (Polo et al., 2002; Bache et al., 2003a; Lu et al., 2003). ESCRT-I in turn recruits ESCRT-II, which may function in sequential ligand transfer, although there is currently some uncertainty whether ESCRTs assemble serially and if ESCRT-II is necessary for EGFR down-regulation (Bowers et al., 2006; Malerod et al., 2007; Nickerson et al., 2007). ESCRT-III is thought to mediate packaging of ligands into ILVs, although a more distal involvement in multivesicular body (MVB) biogenesis and/or transport to lysosomes has also been suggested (Bache et al., 2006; Muziol et al., 2006; Shim et al., 2006; Hanson et al., 2008).
Along with the ESCRT complexes, an array of additional proteins is known to contribute to regulating sorting and degradation. Ubiquitin ligases, such as Nedd4 and Tal, have been implicated in ubiquitylating Hrs and Tsg101, respectively, to negatively regulate their ability to bind ubiquitylated cargo (Polo et al., 2002; Amit et al., 2004). Besides the RING finger ubiquitin ligase Cbl, HECT ubiquitin ligases of the Nedd4 family are known to ubiquitylate and down-regulate receptors and channels (e.g., ENaC and CXCR4; Henry et al., 2003; Marchese et al., 2003). Two deubiquitylating enzymes, UBPY and AMSH, have also been implicated in receptor degradation, although currently, their distinct roles in receptor recycling and/or ubiquitin salvage before degradation are still unclear (McCullough et al., 2004; Mizuno et al., 2005; Bowers et al., 2006; Row et al., 2006; Alwan and van Leeuwen, 2007). ESCRT-III also recruits the AAA-ATPase VPS4 and associated proteins that enable ESCRT disassociation, disassembly from clathrin patches, and intraluminal vesicle formation (Sachse et al., 2004; Stuchell-Brereton et al., 2007). Also the sorting nexin, SNX3, and annexin I have emerged as proteins with complementary roles to ESCRTs in facilitating cargo sorting and formation of ILVs (White et al., 2006; Strochlic et al., 2007; Pons et al., 2008).
Within the large spectrum of processes involved in endosomal sorting and down-regulation, most functional insights have featured proteins (e.g., ESCRTs, Rabs, and Rab effectors) that are peripheral components of endosomal membranes, associating mainly through lipid-binding domains (reviewed in Maxfield and McGraw, 2004; Hurley and Emr, 2006; Williams and Urbe, 2007). Information regarding how transmembrane proteins that are not cargoes might contribute to these processes is more limited, although endosomal acidification and turnover of selected cargoes are known to involve transmembrane machinery (Harvey et al., 2002; Hettema et al., 2004; Maxfield and McGraw, 2004; Shearwin-Whyatt et al., 2004). We have been interested in the possibility that secretory carrier membrane proteins, SCAMPs, might contribute to sorting events in endosomes. SCAMPs are tetraspanning, integral membrane proteins that reside in the cell surface recycling system including the trans-Golgi network, early sorting and recycling endosomes, and plasma membrane (Castle and Castle, 2005). In mammalian tissues, the SCAMP family includes four ubiquitous isoforms, SCAMPs 1-4, and a predominantly neuronal isoform SCAMP5 (Fernandez-Chacon and Sudhof, 2000; Hubbard et al., 2000). SCAMP function is presumably conserved through the transmembrane core but is modulated by focal differences in amino acid sequences of the N- and C-termini. This may contribute to subtle differences in localization and more importantly may enable individual SCAMP isoforms to function analogously in distinct membranes and bring unique interactions to complex processes such as regulated exocytosis (Castle and Castle, 2005; Liao et al., 2008).
In the present study, we have focused on SCAMP3 and provide evidence that it acts as a regulator of EGFR trafficking within endosomal membranes. Whereas our previous studies have hinted at a potential functional association of SCAMP3 and EGFR (Wu and Castle, 1998), we now show that this SCAMP is an important determinant of EGFR sorting and rate of receptor degradation. Interestingly, regulation appears to involve ubiquitylation of SCAMP3, its interactions with the ESCRT machinery, and a distinct function in MVB formation or maturation. Other SCAMPs, in particular SCAMP1 and 2, do not appear to participate in these processes.
MATERIALS AND METHODS
Antibodies and Reagents
Monoclonal pan-SCAMP antibody 7C12, SCAMP1 Abs 1ς and 1α, SCAMP2γ Ab, and SCAMP3β have been characterized previously (Singleton et al., 1997; Wu and Castle, 1998; Guo et al., 2002). Mouse mAb, 8E8, to the C-terminus of SCAMP3 ([c]-agvfsnpavrt) was made in the Lymphocyte Culture Center (University of Virginia) according to standard procedures. Other antibodies were obtained as follows: EGFR mAb (F4, having a cytoplasmic epitope) and Flag M2 mAb (Sigma, St. Louis, MO); transferrin (TfR) receptor H68.4 mAb (ATCC, Rockville, MD); Nedd4 mAb and γ-adaptin antibodies (BD Biosciences, San Jose, CA); myc mAb 9E10 hybridoma cells were obtained from J. Thorner (University of California, Berkeley); green fluorescent protein (GFP) mAb (Santa Cruz Biotechnology, Santa Cruz, CA); HA mAb (Covance, Richmond, CA); Hrs and Vps24 polyclonal antibodies were gifts of H. Stenmark (Norwegian Radium Hospital, Norway); Tsg101 mAb (Genetex, San Antonio, TX); and CHMP6 polyclonal antibody was a gift of W. Sundquist (University of Utah). The EGFR mAb 13A9 used for immunostaining and gold conjugation was obtained from Genentech (San Francisco, CA) and has been described previously (Burke et al., 2001).
Protein A Sepharose, N-ethylmaleimide (NEM), protein kinase A, and human holotransferrin (Sigma); glutathione Sepharose and Sephadex G-50 (Amersham Pharmacia Biotech AB, Uppsala, Sweden); HRP-conjugated secondary antibodies (Jackson ImmunoResearch Laboratory, West Grove, PA); Super Signal West Pico ECL Reagent (ThermoScientific, Rockford, IL); Immobilon Western ECL Reagent (Millipore, Billerica, MA); Alexa 488-EGF, Alexa fluor secondary antibodies, and mouse EGF (Invitrogen, Carlsbad, CA); streptavidin agarose and Sulfo-NHS-LC-biotin (Pierce-Endogen, Rockford, IL); cØmplete Roche Protease Inhibitor Cocktail Tablets (Roche, Mannheim, Germany).
Expression Constructs
Glutathione S-transferase (GST) fusion proteins of SCAMP3 N-terminus wild type, Y53A, and L61A were made by PCR amplification from bait clones used in yeast two-hybrid studies (see below) and ligation into pGEX-2TK. Other mutants, P51G, P137A, and L138A, were generated using QuikChange site-directed mutagenesis (Stratagene, La Jolla, CA) to the wild-type SC3 N-terminus in pGEX-2TK. Truncations of the SCAMP3 N-terminus, SC3-NPF (residues 1-89) and SC3-LL (residues 90-164), were created by PCR amplification and ligation into pGEX-2TK. Fusion proteins of GST with fragments of wild-type SCAMP3 (61-73) and its mutant P67L were generated by ligation of the corresponding wild-type or mutant oligonucleotide to human SCAMP3 into BamHI/EcoRI sites in pGEX vector. The SCAMP3 (61-80)–ubiquitin/pGEX chimera was generated by oligonucleotide ligation of the SCAMP3 fragment with BamHI/HindIII sites and PCR amplification of ubiquitin with HindIII/NotI sites. Mouse SCAMP3 mutants Y53A, S67A, and DM (Y53A and S67A), KR6 (K76, 103, 104, 147, 225, and 315R), and W219A were generated using QuikChange site-directed mutagenesis or PCR amplification using nested primers and ligation into pcDNA 3.1.
Other constructs were generous gifts from the following: pEGFP-2xFYVEHrs, H. Stenmark; pKU-HA-Hrs; H. Asao (Tokuku University School of Medicine, Japan); myc-Hrs, myc-Hrs ΔUIM, myc-Hrs ΔVHS/FYVE, myc-Hrs ΔVHS, myc-Hrs Δclathrin-binding domain (ΔCBD) in pcDNA3, M. von Zastrow (University of California, San Francisco) with permission of H. Stenmark; FLAG-Nedd4-2-pcDNA 3.1, Nedd4-pCXN2, mouse Nedd4 WW1-3 and mutant WW1-3 and mouse Nedd4-2 WW1-4 and mutants in pGEX2TK, S. Kumar (Hanson Institute, Adelaide, South Australia); Rab5Q79L in pGreen Lantern, J. Casanova (University of Virginia); UEV domain Tsg101(1-145) in pET 11d, W. Sundquist; HA-Ub-pMT 123, D. Bohmann (University of Rochester, New York); and pEF-HA-Ub KR4, Y. Yarden (Weizmann Institute, Rehovot, Israel).
Two-Hybrid Assay
Bait clones of SCAMP3 N-terminus (aa 1-145) were all constructed in pGBT9 (Clontech, Mountain View, VA) and transformed into yeast strain AH109 (Gietz and Woods, 2002b), whereas prey clones from a 17-d-old mouse embryo cDNA library (Clontech, Mountain View, CA) were constructed in pVP16 (Hollenberg et al., 1995) and transformed into yeast strain Y187. Selection was performed on -leu, -trp, -his, -ade plates according to standard protocols (Gietz and Woods, 2002a).
Small Interfering RNAs
All SCAMP siRNAs were made against human isoforms and obtained from Dharmacon (Lafayette, CO) or Invitrogen. Sense sequences for small interfering RNAs (siRNAs) were as follows: for SCAMP3, (1) 5′-CAGCTACTCGACAGAACAA-3′ and (2) 5′-AACGGATCCACTCCTTATA-3′; for SCAMP1, 5′-TCATCTCACTAGTTAATGTT-3′; for SCAMP2, 5′-CACTGTAGCCAACTTGCAT-3′. siRNA sequences for Hrs, Tsg101, and Vps24 have been described previously (Bache et al., 2003b, 2006; Razi and Futter, 2006). For controls, a nonspecific siRNA no. 2 (Invitrogen) or a scrambled siRNA to SCAMP3 siRNA-1 (5′-GAAGCGGCGUAAUGACCAAdTdT-3′) was used.
Cell Culture and Transfections
HeLa and HEK-293T cells were grown in DMEM (Invitrogen), supplemented with 10% fetal bovine serum and antibiotics. DNA plasmids were transfected with Lipofectamine 2000 or Lipofectamine PLUS Reagent (Invitrogen) according to manufacturer's directions. For siRNA transfections using Oligofectamine (Invitrogen), 75 nM siRNA was used to transfect HeLa cells. For siRNA transfections using Lipofectamine RNAiMax (Invitrogen), HeLa cells were transfected on sequential days with 8.3 nM siRNA for each transfection. Cells were analyzed 48 h after the second transfection.
GST Pulldowns and Coimmunoprecipitation Assays
For GST pulldowns, 20–25 μg of fusion protein conjugated to glutathione Sepharose was used. For each pulldown 100 μl of postnuclear supernatant prepared from rat parotid gland was diluted with 2× TGE (20 mM Tris-HCl, 300 mM K-glutamate, 2 mM EDTA, pH 7.5) with protease inhibitors, and Triton X-100 was added to 0.5%. Pulldowns were incubated overnight at 4°C and then washed three times with TGE and 0.5% Triton X-100. For immunoprecipitations to detect ubiquitylated SCAMP3 and interactions with Nedd4 and Nedd4-2, HEK-293T cells were lysed in RIPA buffer (1% Triton X-100, 1% sodium deoxycholate [DOC], 10 mM Tris, 150 mM NaCl, protease inhibitors, pH 7.4) on ice for 20 min. To the lysate, 0.1% SDS was added before incubation with protein A-Sepharose and 2 μg SCAMP3β antibody or 10 μg rabbit IgG for 1 h at RT or overnight at 4°C. IPs were washed three times with wash buffer (1% Triton X-100, 150 mM NaCl, 50 mM HEPES, 5 mM EDTA, 0.05% SDS, protease inhibitors, pH 7.4) and one time with PBS. For immunoprecipitations with Hrs and EGFP-2xFYVEHrs, HEK-293T cells were lysed in Triton lysis buffer (1% Triton X-100, 10 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH 7.4) containing protease inhibitors and with or without 10 mM NEM and then incubated with antibody overnight at 4°C. Protein A-Sepharose was added the next day for 0.5–2 h. The immunoprecipitations were washed three times with lysis buffer and one time with PBS. All pulldowns and immunoprecipitations were resuspended in Laemmli buffer with 4% SDS and 50 mM DTT, boiled, and run on SDS-PAGE.
Far Western Analysis of SCAMP3 N-Terminus Interaction with Nedd4 WW Domains
Purified WW domains of mouse Nedd4 and Nedd4-2 were labeled with [γ-32P]ATP using protein kinase A according to the manufacturer's instructions. The reaction was quenched with 25 mm HEPES, pH 7.4, 12.5 mM MgCl2, 20% wt/vol glycerol, 100 mM KCl, 1 mg/ml BSA, and 1 mM DTT and fractionated on a 3-ml Sephadex G-50 column. One microgram of GST or fusion proteins was run on SDS-PAGE and transferred to nitrocellulose or stained with Coomassie Blue. The nitrocellulose was blocked with 5% milk in Hyb-75 (20 mM HEPES, pH 7.7, 75 mM KCl, 0.1 mM EDTA, 2.5 mM MgCl2, 0.05% NP-40, 1 mM DTT) for 4 h and then incubated in 5.0 × 105 cpm/ml radiolabeled WW domains and 30 μg unlabeled GST in 1% milk in Hyb-75 overnight at 4°C. The membranes were washed three times with 1% milk in Hyb-75 and two times in Hyb-75, dried, and exposed to a phosphoimager.
Binding Analysis by Surface Plasmon Resonance
All experiments were performed at 25°C. Purification of the UEV domain and immobilization of GST antibody onto a research grade CM5 chip were done as previously described (Garrus et al., 2001). Bacterial lysates expressing GST proteins were diluted ten times in HBS-EP (0.01 M HEPES, pH 7.4, 0.15 M NaCl, 3 mM EDTA, 0.005% vol/vol Surfactant P20) + 1 mg/ml BSA and captured on antibody surfaces to densities of 1–1.5 kilo response units. UEV domain (25 μl) was injected in triplicate at a flow rate of 50 μl/min. Binding-specific responses were obtained by subtracting responses in the absence of GST protein from responses in the presences of GST protein. The equilibrium dissociation constants were obtained from 1:1 interaction binding isotherms using Biaevaluation (Biacore AB, Uppsala, Sweden) and Origin 6.1 (OriginLab Corp., Northampton, MA) (Myszka, 1999).
Immunofluorescence Microscopy and Quantitation
Immunofluorescent labeling was performed according to standard procedures (Liu et al., 2002). For staining of Hrs, cells were permeabilized briefly (2 min) with 0.05% saponin, 80 mM K-PIPES, 5 mM EGTA, and 1 mM MgCl2, pH 6.8, before fixation as described in Simonsen et al. (1998a). Images were captured using either a Zeiss Axiovert 100 wide-field microscope (63× oil objective, 1.4 NA; Thornwood, NY) or a Nikon TE2000E confocal microscope (100× oil objective, 1.45 NA, Melville, NY). For quantitation of the fluorescent signal, Z-stack images (0.25 μm) were captured using a 63× oil objective and deconvolved using volume deconvolution, and the fluorescent intensity was measured using Openlab software (Improvision, Lexington, MA). More than 25 cells were analyzed for each sample to obtain average fluorescence intensity per cell. The Student's t test was used to determine statistical significance between control and experimental samples.
EGF internalization was measured in HeLa cells that were starved 1–2 h in DMEM and 0.1% BSA, labeled on ice for 1 h with 100 ng/ml Alexa 488-EGF, chased for various times in the presence of 100 ng/ml unlabeled EGF, washed, and processed for immunofluorescence microscopy. To visualize internalized EGFR, cells were labeled with 1 μg/ml EGFR mAb 13A9 for 1 h on ice, washed, stimulated with 100 ng/ml EGF at 37°C, chilled quickly, and stripped with 200 mM glycine and 150 mM NaCl, pH 2.5 on ice for 5 min. The cells were then processed for immunofluorescence microscopy. To quantitate recycled EGFR, cells were labeled with 1 μg/ml EGFR mAb 13A9 for 1 h on ice, washed, and then stimulated with 100 ng/ml EGF for 2 h. Cells were then fixed and labeled with an anti-mouse Alexa 594 secondary antibody to detect cell surface–bound EGFR before permeabilization and labeling with SCAMP3β antibody and anti-rabbit Alexa 488 secondary antibody. Cells were scored for the presence or absence of surface-associated fluorescent EGFR signal compared with nontransfected cells.
125I-EGF and 125I-TfR Degradation, Recycling, and Uptake Assays
Mouse EGF and human holotransferrin were iodinated using IODO-GEN Precoated Iodination Tubes (Pierce) according to manufacturer's instructions. All incubations were performed at 37°C. Cells were incubated with DMEM and 0.1% BSA containing 0.5–1 μCi/ml 125I-EGF or 125I-TfR for 1–2 h or 30 min, respectively. The cells were washed and chased in DMEM and 0.1% BSA for indicated times. An excess of unlabeled EGF was added to the chase media for EGF assays. For TfR, the cells were stripped briefly with 50 mM glycine, 100 mM NaCl, and 1 mg/ml PVP, pH 3.0, before incubation with chase medium. At each time point, the medium was removed and replaced with fresh medium. After incubation, cells were lysed in 1 M NaOH and 0.1–1% Triton X-100. For EGF assays, the chase media were precipitated with 5% trichloroacetic acid (TCA)/1% phosphotungstic acid (PTA) to separate degraded (TCA/PTA soluble) from recycled (TCA/PTA insoluble) material (Sorkin et al., 1988). For TfR assays, all the 125I-Tf in the medium was considered recycled. All values were normalized to the total 125I-EGF or 125I-Tf present in media and cells. To measure uptake of EGF, cells were incubated in the presence of 0.5–1 μCi/ml 125I-EGF on ice for 45 min, washed to remove unbound ligand, and chased at 37°C for indicated times. Cells were rapidly chilled, the medium was removed, and cells were acid stripped with 0.2 M acetic acid and 0.5 M NaCl (pH 3.0) and lysed. 125I-EGF in the lysates was considered internalized, and the values were normalized to total 125I-EGF in the acid strip medium and lysate.
EGFR and TfR Degradation Assays
To monitor EGFR degradation, cells were starved 1–2 h in DMEM, 0.1% BSA, stimulated with 100 ng/ml EGF for specified times, and lysed on ice in Triton lysis buffer with protease inhibitors. Cleared lysates were subjected to SDS-PAGE and immunoblotting. Chemiluminescent signal was detected using a FujiFilm LAS-3000 imager (FujiFilm Life Sciences, Stamford, CT) and analyzed with MultiGauge 3.0 (FujiFilm Life Sciences). Protein levels were normalized using an antibody to a γ-adaptin or other nonaffected protein. To measure TfR degradation, HeLa cells were starved 2 h in DMEM, 0.1% BSA and then chilled on ice for 5 min. The cell surface was labeled with 0.5 mg/ml Sulfo-NHS-LC-biotin at 4°C, washed, and incubated with complete media for 0–48 h at 37°C. Cells were scraped and lysed in Triton lysis buffer with protease inhibitors. The lysates were incubated with streptavidin-agarose for 1–2 h at room temperature, washed, and subjected to SDS-PAGE. Undegraded, biotinylated TfR remaining at each time point was detected by enhanced chemiluminescence (ECL) using an antibody to TfR and quantitated as the fraction of total biotinylated TfR present initially.
Electron Microscopy
EGFR mAb 13A9 antibody was conjugated to 10-nm colloidal gold (Ted Pella, Redding, CA), harvested, and washed by centrifugation using a previously described procedure (Slot and Geuze, 1981). EGF-stimulated uptake of the colloidal gold conjugate was documented by immunofluorescence microscopy (data not shown). HeLa cells, transfected with nonspecific control siRNA or SCAMP3 siRNA-2, were serum starved 1 h in DMEM, 0.1% BSA. The cells were then chilled on ice, labeled 1 h with 13A9-gold antibody, washed, and stimulated at 37°C with 100 ng/ml EGF for 30 or 60 min. At the end of the incubation, the samples were washed briefly with prewarmed PBS and fixed 45 min at room temperature by addition of warmed 2.5% glutaraldehyde, 0.1 M cacodylate, pH 7.5. After fixation, the samples were washed in 5% sucrose, 0.1 M cacodylate, postfixed 45 min in 0.5% OsO4, 0.1 M cacodylate containing 0.1% potassium ferrocyanide, washed in 0.1 M NaCl, and stained 40min in 0.5% uranyl acetate, and 0.1 M acetate, pH 6.1. Cells were scraped from the culture dishes, embedded in 1.75% LMP-agarose, dehydrated, and embedded according to standard procedures. Sections were examined unstained and photographed at 12,000× using a Jeol 1230 TEM microscope equipped with a digital camera (Tokyo, Japan). Grids were systematically scanned, and all gold visualized was photographed until at least 30 images per experiment of each sample, including >1500 μm2 cytoplasm over three separate experiments, had been obtained. EGFR-gold was evaluated for location (MVB, late endosome/lysosome, or tubule/vesicle/other), and when possible, the number of ILVs was manually counted and the area of EGFR-labeled MVBs and cytoplasm was calculated using the measurements module in ImageJ (http://rsb.info.nih.gov/ij/).
RESULTS
Interactions with Ubiquitin-dependent Sorting Machinery
Putative Binding Motifs in SCAMP3.
Along the ESCRT pathway, sorting is accomplished by a number of intermolecular interactions involving binding of ubiquitin and sequence motifs such as PY and P(S/T)AP. These motifs have been shown to interact with Nedd4 ubiquitin ligases and the ESCRT-I subunit Tsg101 (reviewed in Morita and Sundquist, 2004). A sequence alignment across various SCAMP3 orthologues revealed the presence of conserved PY and PSAP motifs and a less conserved PTEP sequence within the cytoplasmic domain (Figure 1A). Interestingly, these motifs are unique to SCAMP3 and are not found in SCAMPs 1 and 2.
Figure 1.
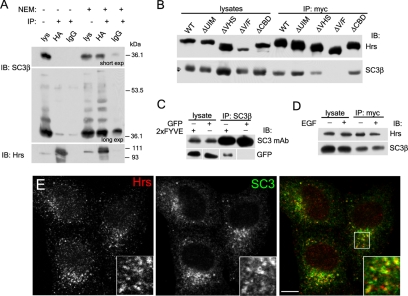
SCAMP3 contains conserved PY and P(S/T)AP motifs and is multimonoubiquitylated. (A) Organization of SCAMP3 featuring its proline-rich segment containing PY, PSAP, and PTEP motifs. (B) Interaction of SCAMP3 N-terminus with WW domain-containing proteins in a yeast two-hybrid screen; two Nedd4 ubiquitin ligases, Nedd4 and WWP2, and a third protein, YAP65, were identified as interaction partners. Mutation of SCAMP3's PY motif to PPAA (SC3Y53A) but not mutation of a distal PLPP (PAPP, SC3L61A) abrogated the interaction. (C) Pulldowns of SCAMP3 from lysates of rat parotid gland using GST fusions of mouse Nedd4 WW domains (WW1-3), corresponding mutant WW domains, and fusion protein made from Nedd4 cDNA retrieved in yeast two-hybrid screen (N4-Y2H). (D) Coimmunoprecipitation of untagged Nedd4 and FLAG-tagged Nedd4-2 expressed in HEK-293T cells using anti-SCAMP3 antibody or control IgG followed by immunoblotting with Nedd4 or FLAG antibody. (E) Ubiquitylation of SCAMP3. Lysates of HA-ubiquitin–transfected cells were immunoprecipitated with isoform-specific SCAMP1, 2, or 3 antibodies and immunoblotted with HA antibody (top), pan-SCAMP mAb (middle), or SCAMP3 mAb (bottom). Asterisk indicates ubiquitylated SCAMP3 species. (F) SCAMP3 is multimonoubiquitylated. Samples were immunoprecipitated as in E from lysates expressing wild-type or polymerization-defective HA-Ub KR4. The identical pattern of bands for Ub and UbKR4 demonstrates multimonoubiquitylation rather than addition of ubiquitin polymers of different length. Closed and open arrowheads denote the positions of SCAMP3 monomer and dimer, respectively, in C, E, and F.
Interaction with Nedd4 HECT Ubiquitin Ligases.
The PY motif is known to bind to WW domains of proteins including those of the Nedd4 HECT ubiquitin ligase family (Staub et al., 1996). Using the intracellular N-terminal domain of human SCAMP3 as bait in a yeast two-hybrid screen, two members of the Nedd4 family, Nedd4 and WWP2, and Yes-associated protein 1 YAP65 were identified as binding partners (Figure 1B). We suspected that the interaction occurred via the PY motif on SCAMP3 because all three binding partners contained WW domains. The prey cDNA of Nedd4 that was retrieved contained the entire second WW domain (WW2) and part of the third one (WW3). To confirm that SCAMP3 could bind WW domains, we used GST fusion proteins of individual wild-type or interaction-deficient WW domains from mouse Nedd4 to pull down SCAMP3 from lysates and also to detect binding on far Western blots (Figure 1C, Supplemental Figure S1). In both cases we detected strong binding of the WW2 domain and much weaker binding of the WW3 domain; no binding was detected with the WW1 domain. Furthermore, mutation of the SCAMP3 PY motif (Y53A) abrogated binding in both the yeast two-hybrid and far Western assays, whereas a mutation in a PPLP motif (L61A) did not, confirming that interactions with Nedd4 ubiquitin ligases are mediated through SCAMP3's PY motif (Figure 1B, Supplemental Figure S1). We were able to coimmunoprecipitate SCAMP3 from lysates of cells expressing Nedd4. In addition, we found that SCAMP3 also coimmunoprecipitated with another Nedd4 ubiquitin ligase, Nedd4-2 (Figure 1D). The PY motif is redundant as an interaction site for WW domains in Nedd4 ubiquitin ligases. Although these results demonstrate that SCAMP3 may have the potential to bind several Nedd4 ligases in vivo, they do not distinguish which ligase(s) is a physiological binding partner.
SCAMP3 Ubiquitylation.
Because SCAMP3 can interact with Nedd4 ubiquitin ligases, we were curious whether SCAMP3 could itself be ubiquitylated. To evaluate this possibility, we transfected cells with hemagglutinin (HA)-tagged ubiquitin, immunoprecipitated SCAMP3, and analyzed the presence of ubiquitin by Western blotting of HA. We observed a discrete ladder of bands, ∼7–9 kDa apart, extending upwards from ∼43 kDa (∼7 kDa above the bulk of SCAMP3 antigen, Figure 1E), consistent with attachment of multiple ubiquitin moeities to the SCAMP. We also analyzed the ability of SCAMP1 and 2, which do not contain a PY motif, to be ubiquitylated. SCAMP1 appears to be ubiquitylated to a small degree, whereas ubiquitylation of SCAMP2 was not detected (Figure 1E). Thus, ubiquitylation is selective for SCAMP3.
Previous studies have shown that receptors undergoing down-regulation are derivatized at multiple sites by monoubiquitin (multimonoubiquitylation) although addition of ubiquitin polymers (polyubiquitylation) has been reported as well and may facilitate sorting for degradation (Hicke, 2001; Huang et al., 2006; Barriere et al., 2007; Umebayashi et al., 2008). Furthermore, the ubiquitin adaptors Eps15 and epsin as well as members of the ESCRT complexes are themselves regulated by multimonoubiquitylation (Mosesson and Yarden, 2006). To determine if the ubiquitylated SCAMP3 species reflect multimonoubiquitylation or polyubiquitylation, we used an ubiquitin mutant (KR4) in which four of its major lysines were mutated to arginine (K11, 29, 48, and 63R), thus suppressing polyubiquitylation (Amit et al., 2004). We observed identical banding patterns for immunoprecipitations from cells expressing wild-type or KR4 ubiquitin, indicating that SCAMP3 is most likely multimonoubiquitylated (Figure 1F).
Interaction with Tsg101.
P(S/T)AP motifs have been shown to bind the UEV domain of Tsg101 in a region that is distinct from the ubiquitin-binding site (Pornillos et al., 2002). We used surface plasmon resonance to evaluate the potential for the PSAP motif of SCAMP3 to associate with the UEV domain of Tsg101. A GST fusion protein containing a small peptide from SCAMP3, which includes the PSAP motif, was able to bind the UEV domain of Tsg101 with significant, albeit low affinity (Figure 2). In contrast, a peptide containing a mutation in the PSAP motif (P67L) did not detectably bind to the UEV domain. Previous studies have shown that covalent attachment of ubiquitin to P(S/T)AP-containing peptides enhances binding synergistically due to the proximity of the P(S/T)AP motif and ubiquitin-binding sites (Garrus et al., 2001). Because SCAMP3 can be ubiquitylated, we hypothesized that ubiquitylated SCAMP3 may bind the UEV domain more efficiently. We generated a SCAMP3 peptide–ubiquitin chimera by substituting a single ubiquitin in place of a naturally occurring lysine in the SCAMP3 (Figure 2). When we tested its affinity, we saw a 20-fold increase in binding to the UEV domain (Figure 2), comparable to that observed for another ubiquitin chimera (Garrus et al., 2001). We attempted to coimmunoprecipitate the Tsg101-SCAMP3 complex in cell lysates using overexpression of Tsg101 and/or SCAMP3 but were not successful. This may be a reflection of the low binding affinity of even the SCAMP3–ubiquitin chimera, suggesting that the interaction is weak and perhaps transient or may involve only a small fraction of the SCAMP3 and Tsg101.
Figure 2.
Interaction of the PSAP motif in SCAMP3 with the UEV domain of Tsg101. (A) GST fusion proteins of SCAMP3 used in this study and calculated Kd from a representative binding experiment shown in B. n.d., not determined.
Interaction with Hrs.
Although our results above suggest a possible interaction with Tsg101, the localization of SCAMP3 on early endosomes (see below; Castle and Castle, 2005) may allow it to have additional binding partners that function in ubiquitin-mediated sorting. We thus considered whether the ESCRT-0 protein, Hrs, is a putative binding partner for SCAMP3. To test this we coimmunoprecipitated SCAMP3 from HA-Hrs–expressing cell lysates. Immunoprecipitations were carried out in parallel on lysates with or without NEM, a sulfhydryl-blocking agent that has been used to inhibit deubiquitylating enzymes and AAA-ATPases. As seen in Figure 3A, SCAMP3 coimmunoprecipitated with HA-Hrs but only in the presence of NEM. The predominant form recovered exhibited a mobility corresponding to underivatized SCAMP3 (see panel A, short exp); however, upon longer exposure, we were able to detect the presence of additional higher molecular weight bands. It is likely that these bands correspond to ubiquitylated SCAMP3.
Figure 3.
SCAMP3 interacts with Hrs. (A) HA-Hrs–transfected HEK-293T cells were lysed in the absence or presence of 10 mM N-ethylmaleimide (NEM), immunoprecipitated with anti-HA or control IgG, and immunoblotted with anti-SCAMP3 (top and middle panel) or anti-Hrs antibodies (bottom panel). The top panel is a short exposure, and the middle panel is a longer exposure of the same immunoblot to show ubiquitylated SCAMP3 species. (B). Mapping of the site on Hrs involved in SCAMP3 interaction. Lysates from cells expressing myc-tagged wild-type Hrs or indicated deletion mutants (ΔUIM, ΔVHS, ΔV/F, and ΔCBD) were immunoprecipitated with anti-myc antibody in the presence of 10 mM NEM and immunoblotted with anti-Hrs (top) or anti-SCAMP3 (bottom). (C) Interaction of the FYVE domain with SCAMP3. Lysates expressing GFP or GFP-2xFYVEHrs were immunoprecipitated with SCAMP3 antibody and immunoblotted with SCAMP3 mAb (top panels) or GFP (bottom panels). (D) Cells expressing myc-tagged Hrs were incubated for 30 min in the presence or absence of 100 ng/ml EGF, lysed, immunoprecipitated as in B, and immunoblotted with Hrs (top) or SCAMP3 antibody (bottom). (E) Endogenously expressed Hrs and SCAMP3 are partially colocalized. HeLa cells were permeabilized before fixation and immunostained for Hrs and SCAMP3. Bar, 10 μm.
Hrs is a multidomain protein that includes an N-terminal VHS (Vps27/Hrs/STAM) domain, an FYVE (Fab1/YOTB/Vac1/EEA1) domain, a UIM, a coiled-coiled domain, and a C-terminal clathrin-binding box (Raiborg et al., 2001b). We used deletion mutants of myc-tagged Hrs to determine which domain was responsible for the interaction with SCAMP3. Deletion of the clathrin-binding domain did not affect binding. Deletion of the UIM also did not affect binding, suggesting that ubiquitylation of SCAMP3 is not necessary for interaction (Figure 3B). This is consistent with the finding that coimmunoprecipitation primarily brings down nonubiquitylated SCAMP3 (Figure 3A). Interestingly, we found a significant decrease in binding when the VHS domain is deleted and complete abrogation of the interaction when both the VHS and FYVE domains are deleted (Figure 3B). Binding of Hrs' VHS domain has been implicated previously in several interactions, but interaction with the FYVE domain was unexpected (Nielsen et al., 2001; Puertollano et al., 2001; Zhu et al., 2001). To test further that the FYVE domain can contribute to association of Hrs with SCAMP3, we attempted a coimmunoprecipitation from HEK-293T cells expressing the GFP-2xFYVEHrs chimera. We found that SCAMP3 could specifically coimmunoprecipitate the dual FYVEHrs domain alone (Figure 3C). Finally, we inquired whether the interaction between Hrs and SCAMP3 was affected by addition of EGF; however, we found that a 30-min stimulation had no effect, suggesting that the association is constitutive (Figure 3D). To date our efforts to detect coimmunoprecipitation of endogenous Hrs and SCAMP3 have been unsuccessful. Comparative examination of their distributions by immunofluorescence revealed a pattern of partial overlap (Figure 3E), thus supporting a potential interaction of endogenous proteins. The inability to detect it by coimmunoprecipitation could reflect any of several factors including its low affinity, transient nature, or mediation by other proteins.
SCAMP3 Localizes to Early Endosomes That Traffic EGFR
SCAMP3 has been shown to colocalize with endocytosed TfR in early sorting endosomes (Castle and Castle, 2005). Also, electrostatic associations with polyanionic phospholipids, especially phosphoinositides, have been implicated as a conserved property of SCAMPs (Ellena et al., 2004; Liao et al., 2007). Consequently, we sought to determine if SCAMP3 localized to sorting portions of early endosomes, which are known to be enriched in phosphatidylinositol-3-phosphate (PI3P). For this purpose we transfected cells with constitutively active GFP-Rab5 (Rab5Q79L), GFP-2xFYVEHrs, or myc-Hrs, each of which causes early endosomes to enlarge and accumulate PI3P (Stenmark et al., 1994; Komada et al., 1997; Gillooly et al., 2000). We observed localization of endogenous SCAMP3 in enlarged endosomes in Rab5Q79L- and 2xFYVEHrs-expressing cells, but SCAMP3 was unevenly concentrated along and protruding from the more evenly stained profiles of the two GFP constructs (Figure 4, A and B). Expressed SCAMP3 also localized with enlarged endosomes of Hrs expressing cells (Figure 4C).
Figure 4.
Localization of SCAMP3 in relation to enlarged early endosomes. HeLa cells were transfected with GFP-Rab5Q79L (A) or GFP-2xFYVEHrs (B) or cotransfected with myc-Hrs and untagged mouse SCAMP3 (C). All samples were immunostained for SCAMP3; sample in C was also stained for myc. Enlargements of indicated areas are shown below the respective panels and illustrate focal localization of SCAMP3 along the membranes of expanded early endosomes stained by GFP (A and B) and substantial overlap with Hrs (C). Bar, 10 μm.
Previous studies from our lab using EGFR-overexpressing cells indicated the possibility that the receptor traffics through SCAMP3-containing compartments (Wu and Castle, 1998). To revisit this issue for endogenous EGFR, we labeled the surface of HeLa cells with an antibody to the EGFR, stimulated the cells with EGF, and tracked antibody appearance in SCAMP3 compartments. As seen in Figure 5, there is moderate colocalization of internalized EGFR and SCAMP3 at 15 min, mainly in smaller peripheral puncta. At later times, 30 and 60 min, colocalization increases in larger perinuclear puncta (Figure 5). The timing of colocalization corresponds well with the presence of SCAMP3 in early endosomes as well as endosomally derived carrier vesicles. Notably, SCAMP3 is not found in late endosomes and lysosomes marked by BMP/LBPA and Lamp1 (data not shown; Brand et al., 1991).
Figure 5.
EGFR traffics to SCAMP3-rich compartments. HeLa cells were labeled with EGFR Ab 13A9, washed, and stimulated with 100 ng/ml EGF for 15, 30, or 60 min. Cells were then acid-stripped with low pH glycine to remove surface-associated EGFR antibody, fixed, permeabilized, and immunostained to detect internalized EGFR and SCAMP3. Bar, 10 μm.
Knockdown of SCAMP3 Accelerates EGFR Degradation
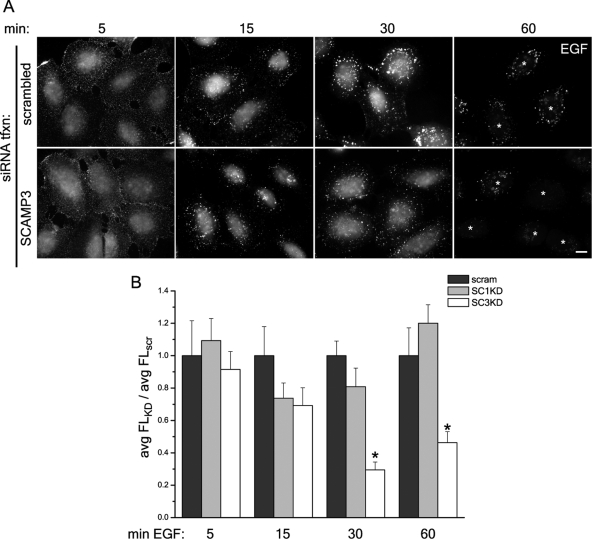
We next set out to determine whether SCAMP3 has a role in regulating EGFR degradation by evaluating the trafficking of EGF and EGFR in HeLa cells that had been transfected with siRNAs designed specifically to human SCAMP3. Two different siRNAs each reduced the level of SCAMP3 expression >90% compared with control transfected cells (see example in Figure 7C). Initially, we followed trafficking of fluorescently labeled EGF (Figure 6). Labeling in control and SCAMP3 knockdown cells was similar at early time points, 5 and 15 min, and was characterized by fine punctate staining dispersed throughout the cytoplasm. At 30 min, EGF concentrated into bright perinuclear accumulations in control cells as observed in early studies (e.g., Shearwin-Whyatt et al., 2004). However, in cells depleted of SCAMP3, the fluorescent ligand was found in smaller more widely distributed puncta. At 60 min, EGF fluorescence was notably decreased, especially in SCAMP3 knockdown cells, where puncta were barely visible (Figure 6A). To compare ligand levels quantitatively, the average fluorescent EGF signal per cell was measured and compared at each time point between control and knockdown samples. This analysis revealed a significantly lower signal at 30 and 60 min in SCAMP3-depleted cells (Figure 6B). Accelerated loss of fluorescent EGF was observed with two independent siRNAs for SCAMP3 but was not observed for SCAMP1 knockdown, which produced profiles similar to the control (Figure 6B and data not shown).
Figure 7.
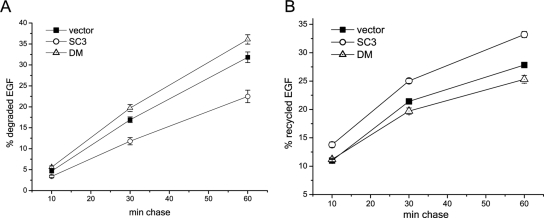
Knockdown of SCAMP3 accelerates EGF and EGFR degradation. Degradation (A) and recycling (B) of 125I-EGF in HeLa cells depleted of SCAMP3 (siRNA no. 2). Results from four independent experiments show that EGF degradation is accelerated and recycling inhibited compared with control cells. (C and D) Degradation of EGFR in HeLa cells transfected with control or SCAMP3-specific siRNA (no. 2). Cells were stimulated with 100 ng/ml EGF for indicated times, and the lysates were immunoblotted with specified antibodies. (C) A representative experiment is shown. (D) Quantitation of EGFR remaining after each time point from three independent experiments. EGFR levels were normalized for protein loading and quantitated as fraction of EGFR at time 0. Error bars, SEM.
Figure 6.
RNAi-mediated knockdown of SCAMP3 enhances disappearance of fluorescent EGF. HeLa cells were transfected with scrambled, SCAMP1-, or SCAMP3-specific siRNA (no. 1), labeled with 100 ng/ml Alexa-488 EGF at 4°C, washed, and chased with 100 ng/ml EGF for indicated times at 37°C. (A) Representative images of cells transfected with scrambled or SCAMP3-specific siRNAs are shown; bar, 10 μm; asterisks at 60 min identify positions of cells in the images. (B) EGF fluorescence/cell was quantitated from Z-stacks of deconvolved images as described in Materials and Methods and was expressed as the average fluorescence ratio (knockdown/control) for each time point. The ratio for cells transfected with scrambled siRNA was set to 1.0. Error bars, SEM; * p < 0.01.
The disappearance of fluorescently labeled EGF in SCAMP3-depleted cells could indicate an acceleration of the rate of recycling or degradation. To distinguish between these two possibilities, we used two complementary assays: analysis of the fate of internalized 125I-labeled EGF and analysis of EGFR levels by quantitative Western blotting. Knockdown of SCAMP3 did not affect uptake of 125I-EGF into the cells (Supplemental Figure S2). As shown in Figure 7, A and B, knockdown of SCAMP3 accelerated degradation while concurrently decreasing the rate of 125I-EGF recycling compared with the control. Consistent with this outcome, Western blotting of the receptor in lysates confirmed that degradation was accelerated in the absence of SCAMP3. The same results were obtained using two different siRNAs in both assays, confirming the specificity of SCAMP3 knockdown on EGFR degradation (Figure 7, C and D, and data not shown). The rate of receptor degradation was not affected in cells depleted of either SCAMP1 or 2 (data not shown).
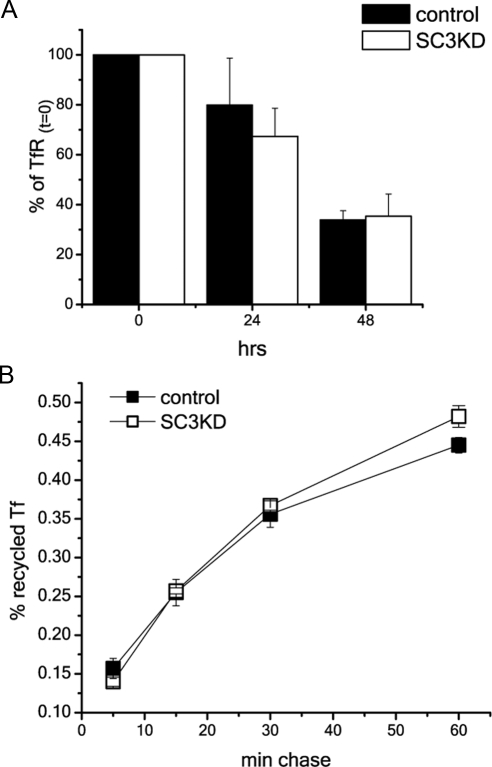
Potentially, acceleration of EGFR degradation and inhibition of recycling may reflect a general shift such that more cargo is directed toward lysosomal degradation. We therefore looked at whether degradation or recycling of the TfR receptor, which normally recycles with high efficiency, was similarly affected by knockdown of SCAMP3. To measure degradation of TfR, we followed the disappearance of biotinylated surface TfR over 48 h. There was no significant difference in the amount of receptor recovered between control and SCAMP3-depleted cells at any time point (Figure 8A). To complement this finding, we examined the fate of 125I-TfR in cells preloaded to steady state. The results show that knockdown of SCAMP3 did not affect recycling (Figure 8B). Therefore, SCAMP3 depletion does not affect constitutive recycling.
Figure 8.
Knockdown of SCAMP3 does not affect turnover and trafficking of transferrin receptor (TfR). (A) Surface-biotinylated cells were incubated for indicated times, lysed, adsorbed with streptavidin-agarose, and immunoblotted for TfR. The amount of TfR was normalized to the total biotinylated TfR (time 0). Results are averaged from three independent experiments and show no significant difference between control transfected or SCAMP3-depleted cells. Error bars, SEM. (B) Recycling of transferrin (125I-TfR) is the same in control transfected and SCAMP1- or 3-depleted HeLa cells. Recycling was measured in triplicate in three experiments as described in Materials and Methods. A representative experiment is shown. Error bars, SEM.
Overexpression of Normal but Not Mutant SCAMP3 Increases EGFR Recycling
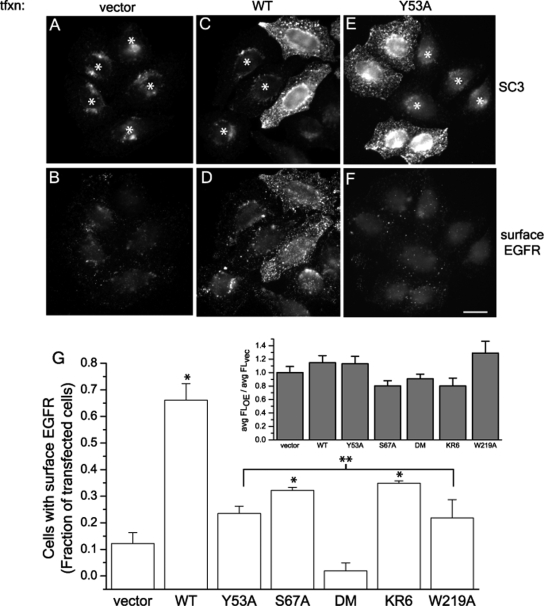
Because knockdown of SCAMP3 enhances EGFR degradation and inhibits receptor recycling, we hypothesized that overexpression of SCAMP3 may have the opposite effect of increasing receptor recycling. To test this possibility, we developed an assay to monitor recycled EGFR using fluorescent secondary antibody to detect the appearance of receptor-antibody complexes at the cell surface after 2 h of EGF stimulation as described in Materials and Methods. Because internalization of EGF and EGFR was not affected (Figure 9G, inset, and Supplemental Figure S3), we could reasonably assume that the presence of surface-associated receptors at 2 h is indicative of recycling. Moreover, we confirmed in separate controls 1) that cell surfaces were largely cleared of anti-EGFR at 30 min (Supplemental Figure S3) and 2) that intracellular antigen EEA1 was not detected without permeabilization, thereby documenting surface exposure of recycled complexes. We found that 66% of SCAMP3-transfected cells had enhanced surface-associated EGFR compared with 12% in vector-transfected cells (Figure 9, B–F and G). Thus, overexpression of SCAMP3 increases receptor recycling.
Figure 9.
Recycling of EGFR is induced by SCAMP3 overexpression and is dependent on an intact PSAP motif and ubiquitylation. HeLa cells were transfected with vector (A and B), wild-type (WT) mouse SCAMP3 (C and D), SCAMP3 PY mutant Y53A (E and F), or other mutants not shown: PSAP mutant (S67A), double mutant (DM), ubiquitylation-deficient KR mutant (KR6), or W219A. Cells were surface labeled with EGFR Ab 13A9 and then stimulated with 100 ng/ml EGF for 2 h. After fixation, surface-associated EGFR was labeled with Alexa-594 secondary antibody (B, D, and F), and the cells were then permeabilized and immunostained for SCAMP3 using a low concentration of antibody to distinguish transfected from nontransfected cells (A, C, and E). Asterisk indicates nontransfected/low-expressing cells; bar, 10 μm. (G) Transfected cells were scored for the presence of surface-exposed EGFR relative to neighboring nontransfected cells. The average of 3–4 experiments is shown in the bar graph. * p < 0.05 compared with vector alone, ** p < 0.05 compared with wild-type (WT) transfected. Inset, internalization of EGF is not affected in overexpressing cells. Cells were labeled with 100 ng/ml Alexa-488 EGF at 4°C, washed, incubated with 100 ng/ml unlabeled EGF for 15 min at 37°C, and processed for immunostaining with a low concentration of SCAMP3 antibody. EGF fluorescence of SCAMP3 WT or mutant-expressing cells was quantitated as described in Materials and Methods, and results are expressed as average fluorescence ratio: SCAMP3 constructs overexpressed (OE):vector transfected (vec). Error bars, SEM.
Our biochemical data suggest that SCAMP3 function may be closely tied with its ability to be ubiquitylated and to interact with the ESCRT-I protein Tsg101. Using this recycling assay, we then tested whether ubiquitylation sites and putative interaction motifs on SCAMP3 were necessary to observe recycling. We made the following mutations in mouse SCAMP3: 1) Y53A, in which the ubiquitin ligase binding site PPxY was abrogated; 2) S67A, in which the PSAP motif was altered to inhibit possible binding to the ESCRT-I protein Tsg101; 3) a double mutation Y53A, S67A (DM); 4) KR6, in which six of the nine cytoplasmic lysines were mutated to arginine, rendering SCAMP3 incapable of being ubiquitylated (K76,103,104,147, 225, and 315R; data not shown); and 5) W219A, a change of a highly conserved residue that in SCAMP2 creates a dominant inhibitor of exocytosis (Liu et al., 2002). Expression of each of these mutants yields immunostaining that closely approximates that of endogenous and exogenous wild-type SCAMP3, although the efficiency of transfection, particularly for DM and W219A, was lower than for wild-type SCAMP3 (Supplemental Figure S4, data not shown). Also, each of these mutants did not affect internalization of EGF (Figure 9G, inset). Notably, all the mutations substantially reduced the ability of overexpressed SCAMP to elicit increased EGFR recycling. DM (Y53A, S67A) was especially potent in this regard (Figure 9G).
As a complementary approach to analyzing the effect of overexpression of SCAMP3 and select mutants on EGFR recycling, we examined the recycling versus degradation of 125I-EGF. As shown in Figure 10, overexpression of SCAMP3 decreased degradation and increased recycling as compared with cells transfected with vector alone. Quite interestingly, overexpression of the DM mutant slightly enhanced degradation and decreased recycling as compared with vector-transfected cells. The effect of this mutant resembles the effect of SCAMP3 knockdown and suggests that the mutant acts in a dominant-inhibitory manner. In a separate set of analyses, overexpression of mutant KR6 decreased recycling as compared with wild-type SCAMP3, but the effect was not as strong as for the mutant DM (data not shown). In these experiments, the levels of overexpression of mutants KR6 and DM were about one-third and the same as wild-type SCAMP3, respectively. Taken together, these results of the EGFR and 125I-EGF recycling assays provide strong support that SCAMP3 interaction with ESCRT machinery and its ubiquitylation promote EGFR recycling.
Figure 10.
Overexpression of wild-type SCAMP3 but not a double mutant inhibits EGF degradation and increases recycling. HeLa were transfected with vector (pcDNA 3.1), wild-type mouse SCAMP3, or DM-SCAMP3. (A) Degradation of 125I-EGF. (B) Recycling of 125I-EGF. Error bars, SEM, determined from three separate experiments.
SCAMP3 Functions Coincidentally with ESCRT Complexes
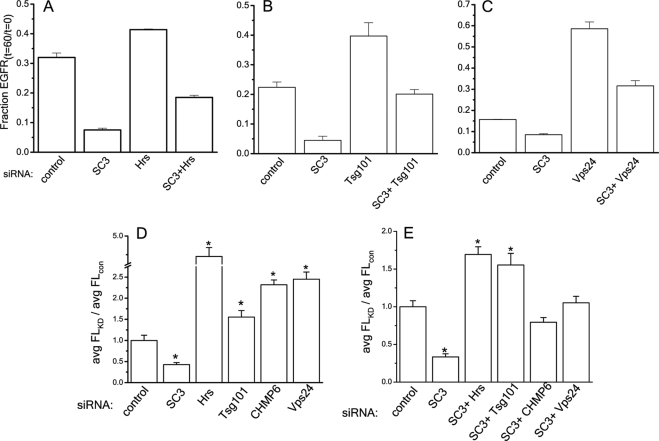
One current model of ESCRT-mediated degradation suggests that ubiquitylated receptors are passed serially through ESCRT complexes to facilitate packaging into internal vesicles (Babst, 2005). A corollary of this model is that knockdown of subunits of ESCRT complexes inhibits EGFR degradation. Indeed, previous studies have shown that knockdowns of the ESCRT proteins Hrs, Tsg101, and Vps24/CHMP3, individually severely inhibit EGFR degradation (Bache et al., 2006; Raiborg et al., 2008). Given that the distribution of SCAMP3 encompasses at least the early portion of the degradative pathway where the ESCRTs function and that SCAMP3 is thought to interact with Hrs and Tsg101, we were curious whether SCAMP3 functions in series with the ESCRTs. To this end, we used siRNAs to deplete the ESCRT proteins Hrs, Tsg101, and Vps24 individually and in paired combination with SCAMP3 (Supplemental Figure S5) and followed degradation of EGFR. As in the earlier studies, we found that single knockdowns of Hrs, Tsg101, and Vps24 inhibited EGFR degradation compared with control transfected cells (Figure 11, A–C). We anticipated that if SCAMP3 functions solely in series with the ESCRT complexes then inhibition of receptor degradation in paired SCAMP3/ESCRT knockdowns would be comparable to ESCRT knockdown alone. However, we consistently found that all paired knockdowns of SCAMP3/ESCRT subunits resulted in a rate of receptor degradation that was faster than for knockdown of the ESCRT subunits alone but slower than for knockdown of SCAMP3 alone (Figure 11, A–C). These outcomes would seem to rule out a strictly serial relationship between SCAMP3 and the ESCRTs and raise the possibility that they may have parallel roles or that SCAMP3 has compound functions acting both in series and in parallel with ESCRTs.
Figure 11.
Accelerated degradation of EGFR in SCAMP3-depleted cells is not blocked by simultaneous knockdown of ESCRT subunits. HeLa cells were transfected with control siRNA, SCAMP3 siRNA, and Hrs siRNA or Hrs and SCAMP3 siRNAs (A), Tsg101 siRNA or Tsg101 and SCAMP3 siRNAs (B), and Vps24 siRNA or Vps24 and SCAMP3 siRNAs (C). The cells were stimulated with 100 ng/ml EGF for 60 min, lysed, and immunoblotted for EGFR using ECL for quantitation. EGFR levels were normalized for protein levels and calculated as the fraction of total from time 0 (fraction EGFRt=60/t=0) in three independent experiments. Error bars, SEM. (D and E) EGF degradation in HeLa cells after knockdown of SCAMP3 and ESCRT subunits alone (D) and in paired combination (E). Cells were labeled with Alexa488-EGF and chased with 100 ng/ml EGF for 60 min. EGF fluorescence was quantitated as described in Materials and Methods, and results are expressed as average fluorescence ratio: knockdown:control transfected. Error bars, SEM; * p < 0.01.
To complement these findings we carried out the same knockdowns of ESCRTs, alone and in combination with SCAMP3, and measured cellular fluorescence of Alexa488-EGF–labeled cells 1 h after stimulation. We included an additional set where the ESCRT-III subunit CHMP6 was knocked down. As expected, we found that cells with knockdowns of individual ESCRT subunits had significantly more Alexa488-EGF than control cells, ∼1.5–4.5-fold increase, indicating that degradation was inhibited (Figure 11D). When we tested the effect of paired knockdowns with SCAMP3, there was, in most cases, an increase in fluorescence compared with control, ∼0.8–1.75-fold; however, it was less than for knockdowns of individual ESCRT subunits alone but greater than for knockdown of SCAMP3 alone (Figure 11E). Except in the case of Tsg101, these results do not appear to be a consequence of defects in internalization (Supplemental Figure S6) and support the possibility that SCAMP3 function may coincide with and complement ESCRT function.
Knockdown of SCAMP3 Perturbs EGFR-containing MVBs
To search for changes in endosomal morphology accompanying SCAMP3 knockdown more quantitatively and at higher resolution, we examined compartments containing internalized EGFR by immunoelectron microscopy. HeLa cells were labeled with EGFR mAb 13A9-gold, stimulated with EGF, and processed for observation at 30 and 60 min. Three separate experiments resulted in very similar outcomes. We found that gold-labeled MVBs from SCAMP3-depleted cells and control cells were similar in size and morphology (Figure 12, Table 1). Also, the number of internal vesicles/μm2 MVB cross-sectional area was about the same, indicating that ILV formation is not affected (Table 1). Together, these results indicated that depletion of SCAMP3 did not detectably affect the structure of individual MVBs. However we found an interesting difference in the density (number/μm2 cytoplasm) of EGFR-labeled MVBs between SCAMP3 knockdown and control cells. Although both types of cells exhibited the same density at 30 min after initiating uptake, this density decreased by one-third in control cells but was unchanged at 60 min in knockdown cells. Notably, the density of gold-labeled late endosomes/lysosomes was essentially the same in both types of cells at 30 and 60 min (Table 2). Therefore it appears that depletion of SCAMP3 does not affect the kinetics of trafficking to lysosomes but rather the number of gold-labeled MVBs enroute to the lysosome. When we examined the EGFR-gold containing MVBs from both control and SCAMP3-depleted cells more closely, we often found small buds or tubules extending from and occasionally into them, suggesting that these MVBs are actively sorting cargo and have not fully matured (Figure 12, B, D, F, and G; Woodman and Futter, 2008). We also examined the sections for the presence of EGFR-labeled tubulovesicular compartments, which may represent an intermediate for recycling cargo between MVBs and the ERC or TGN. We were able to identify some compartments that appeared tubulovesicular in morphology (data not shown); however, we were not able to sufficiently resolve these compartments to quantify them. Occasionally we also found gold residing on the plasma membrane and on extracellular material. Nevertheless, these results suggest that depletion of SCAMP3 affects trafficking of EGFR upstream of degradation in the lysosome, possibly during maturation or formation of MVBs (see Discussion).
Figure 12.
EGFR-containing MVBs from SCAMP3-depleted cells have normal morphology. HeLa cells were transfected with control (A, B, E, F, and G) or SCAMP3 siRNA (C, D, H, I, and J), labeled with EGFR mAb 13A9 conjugated to gold, stimulated with 100 ng/ml EGF for 30 (A–D) or 60 min (E–J), and then processed for immunoelectron microscopy. Arrowheads indicate buds/tubules extending from the MVB. N, nucleus; LE, late endosome; bar, 0.25 μm.
Table 1.
Characteristics of EGFR-gold labeled MVBs
| EGFR-gold labeled MVBs area (μm2)a | No. of ILVs/EGFR-gold labeled MVB | No. of ILVs/μm2 EGFR-gold labeled MVB area | |
|---|---|---|---|
| control, 30min | 0.091 ± 0.005 | 9.5 ± 0.6 | 116.1 ± 5.3 |
| SC3KD, 30min | 0.085 ± 0.006 | 8.5 ± 0.7 | 106.3 ± 5.2 |
| control, 60min | 0.107 ± 0.007 | 11.7 ± 0.8 | 114.6 ± 6.1 |
| SC3KD, 60min | 0.100 ± 0.006 | 10.5 ± 0.7 | 111.8 ± 4.4 |
a MVBs were identified by presence of internal vesicles and the absence of lamellae.
Table 2.
Cytoplasmic densities of EGFR-gold labeled MVBs and late endosomes/lysosomes
| μm2 Cytoplasm | No. of EGFR-gold labeled MVBs | No. of EGFR-gold labeled MVBs/μm2 cytoplasm | No. of EGFR-gold labeled LE/LYa | No. of EGFR-gold labeled LE or LY/μm2 cytoplasm | |
|---|---|---|---|---|---|
| Control, 30 min | 1892 | 118 | 0.062 | 20 | 0.011 |
| SC3KD, 30 min | 1936 | 121 | 0.062 | 18 | 0.009 |
| Control, 60 min | 1765 | 70 | 0.040 | 37 | 0.021 |
| SC3KD, 60 min | 1855 | 120 | 0.065 | 39 | 0.021 |
a LE/LY, late endosome/lysosome, were identified by increased density, lamellae, and may or may not contain internal vesicles.
DISCUSSION
We have shown that SCAMP3 contributes to the regulation of EGFR trafficking and degradation and have identified novel interactions between SCAMP3 and the endocytic machinery involved in ubiquitin-mediated receptor degradation. SCAMP3 can be ubiquitylated at multiple sites, probably via association of its PY motif with one or more Nedd4 ubiquitin ligase(s); it contains a PSAP motif that can bind Tsg101, and it can associate with Hrs, by an interaction involving Hrs' VHS and FYVE domain. As the FYVE domain binds PI3P and contributes to Hrs localization to early endosomes, the association with SCAMP3 may either enhance targeting or otherwise influence Hrs function in sorting (Raiborg et al., 2001a). It will be interesting to determine the reciprocal interaction module on SCAMP3 and whether SCAMP3-FYVE domain association is involved in other endocytic processes, such as Rab5 binding to EEA1 (Simonsen et al., 1998b).
Ubiquitylation of SCAMP3 and its interactions with ESCRTs are reminiscent of several adaptors that function in endosomal sorting. In particular, Ndfip1/2 and the yeast adaptors Bsd2, Ear1p, and Ssh4p are all membrane proteins that contain PY motifs and are ubiquitylated by Nedd4/Rsp5p (Harvey et al., 2002; Konstas et al., 2002; Hettema et al., 2004; Shearwin-Whyatt et al., 2004; Oberst et al., 2007; Leon et al., 2008). Additionally, Nedd4 ubiquitylates GGA3 to promote transport of the membrane protein LAPTM5 to lysosomes (Pak et al., 2006). The PSAP motif of Hrs recruits Tsg101 to facilitate cargo transfer from ESCRT-0 to ESCRT-I (Garrus et al., 2001; Katzmann et al., 2003; Pornillos et al., 2003; Yanagida-Ishizaki et al., 2008), and similar to SCAMP3, PY and PSAP motifs on the membrane protein SIMPLE/LITAF bind Nedd4 and Tsg101, probably to support lysosomal sorting from the Golgi and plasma membrane (Shirk et al., 2005).
Given the similarities with these adaptors and SCAMP3's localization to membranes, including PI3P-rich endosomes (present studies; Castle and Castle, 2005), we sought to determine whether knockdown or overexpression of SCAMP3 affected trafficking of EGFR. Surprisingly, depletion of SCAMP3 accelerated receptor degradation, whereas overexpression enhanced recycling. Moreover, the ability to enhance receptor recycling depended on SCAMP3's PY and PSAP motifs and its capability to be ubiquitylated. Together, these observations imply that unlike the adaptors described above, which promote degradation, SCAMP3 inhibits lysosomal degradation of EGFR. One mode of SCAMP3 function may involve inhibition of ESCRT-mediated sorting through competitive binding of Hrs and Tsg101, a process that may be enhanced by SCAMP3 ubiquitylation. This would be consistent with various studies that show Hrs and Tsg101 promote EGFR degradation and reduce recycling (Razi and Futter, 2006; Raiborg et al., 2008). Therefore, SCAMP3 may oppose Hrs and Tsg101 function at this level to support recycling.
However, our results suggest SCAMP3 has additional function(s), in parallel to ESCRTs that contribute to EGFR sorting. First, simultaneous knockdown of SCAMP3 with Hrs or Tsg101 did not inhibit EGF/EGFR degradation to the same extent as knockdown of Hrs and Tsg101 alone. Second, SCAMP3 knockdown in combination with the downstream proteins, Vps24 or CHMP6, also reduced the rates of EGF/EGFR degradation compared with knockdown of ESCRT-III subunits alone. Third, recycling of EGFR in SCAMP3-overexpressing cells was suppressed when a mutation of a highly conserved residue (W219A) was introduced. This residue is found in a cytoplasmic domain that likely contributes to the conserved function of all SCAMPs (Guo et al., 2002; Liu et al., 2002, 2005; Liao et al., 2007), but is not involved in ESCRT binding or ubiquitylation. Together, these results clearly conflict with the idea that SCAMP3 merely antagonizes ESCRT-EGFR interaction.
To further understand SCAMP3's role in regulating EGFR sorting, we examined whether MVB structure, formation, and incidence were also affected by SCAMP3-depletion. Our analysis by electron microscopy (EM) indicated that the structure of EGFR-labeled MVBs (size and incidence of ILVs), was not affected in SCAMP3-depleted cells. And the equal densities of labeled MVBs (number/cytoplasmic area) in control and knockdown cells at 30 min suggested to a first approximation that the rate of EGF-induced MVB formation (White et al., 2006) was not altered. However, we observed a striking disparity in the incidence of EGFR-labeled MVBs at 60 min. In control cells, the density of MVBs decreased one-third, but in SCAMP3-depleted cells the density remained unchanged. This could not be accounted for by an increase in transport to lysosomes because appearance of EGFR-labeled late endosomes/lysosomes in control and SCAMP3-depleted cells at both time points was similar. Possibly a portion of EGFR relocated to the cell surface or to other endocytic organelles (non-MVB/late endosomal/lysosomal), and although we attempted to assess this quantitatively, no consistent picture has emerged. Nevertheless, the decrease of EGFR-labeled MVBs in control cells implies the presence of a previously unrealized process by which SCAMP3 normally facilitates loss of EGFR from this compartment.
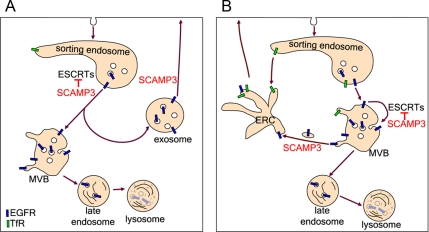
On the basis of our results, we propose that SCAMP3 regulates EGFR trafficking by inhibiting ubiquitin-dependent sorting through competitive binding of ESCRTs and by promoting recycling. We have thought of two mechanisms of SCAMP3 action that could account for the loss of EGFR-labeled MVBs (Figure 13). First, SCAMP3 may promote formation of MVBs that function in recycling rather than degradation. There is already precedence for heterogeneity among MVBs (White et al., 2006) and including those that might be specialized for exocytosis (Trajkovic et al., 2008). Also EGFRs and Ndfip1, which is structurally similar to SCAMP3, have been identified in exosomes, probably resulting from exocytosis of MVBs (Putz et al., 2008; Sanderson et al., 2008). The formation of specific MVBs from PI3P-containing endosomes may be supported by SCAMP3 because SCAMPs are thought to contribute to the formation and function of polyphosphoinositide-containing domains in other contexts (e.g., exocytosis; Liu et al., 2005; Liao et al., 2007, 2008). In the second possibility, SCAMP3 may facilitate removal of receptors from maturing, lysosomally directed MVBs to a recycling pathway largely distal to those used by TfR or induced by low EGF concentration (Sigismund et al., 2008). In this model, SCAMP3 antagonizes ESCRT function by preventing (or reversing secondarily through retrofusion; van der Goot and Gruenberg, 2006) sorting of receptors into ILVs and redirects them into tubules (seen in many of our EMs) that facilitate recycling. Notably, SCAMP3-regulated recycling may apply to receptors that are not polyubiquitylated and are subjected to deubiquitylation (Clague and Urbe, 2006; Umebayashi et al., 2008). Moreover, the two mechanisms suggested are not mutually exclusive and may even occur in tandem.
Figure 13.
Model of SCAMP3 function in inhibiting EGFR degradation and promoting recycling. After internalization, a fraction of EGFRs is sorted for lysosomal degradation in an ubiquitin and ESCRT-dependent manner. This sorting is negatively regulated by SCAMP3 through at least two possible mechanisms. (A) SCAMP3 inhibits ESCRT-mediated sorting of receptors into lysosomally directed MVBs and also promotes formation of a distinct population of MVBs destined for recycling. (B) Receptors are sorted into MVBs that mature en route to the lysosome. SCAMP3 inhibits ESCRT-mediated sorting of receptors into ILVs and facilitates their removal from maturing MVBs and trafficking through the endocytic recycling compartment (ERC).
A final point comes from realizing the extensive loss of fluorescent EGF by 60 min in SCAMP3 knockdown cells (Figure 6), the same time point when most gold-labeled EGFR is upstream of lysosomes. This loss of fluorescent EGF may indicate that depletion of SCAMP3 leads to prelysosomal digestion in MVBs, perhaps because of increased acidification. Indeed, SCAMPs have been proposed to control the trafficking of NHE7 and the Na+/H+ exchanger that may restrict endosomal acidification (Lin et al., 2005; Orlowski and Grinstein, 2007). Depletion of SCAMP3 may prevent association of NHEs or other pH regulatory machinery with MVBs.
In conclusion, by studying the effects of SCAMP3 knockdown and overexpression, we have been able to show that SCAMP3 can regulate the degradative versus recycling fate of the EGFR. This regulation putatively involves SCAMP3 ubiquitylation and interactions with ESCRTs and may entail formation of recycling MVBs or recycling from lysosomally directed MVBs. In the future, a major challenge will be to clarify SCAMP3's specific contributions to receptor sorting and recycling, how its function is regulated by ubiquitylation/deubiquitylation and tyrosine phosphorylation, and how these events influence EGFR's growth-promoting actions.
Supplementary Material
ACKNOWLEDGMENTS
We thank numerous colleagues identified in the text as well as Genentech for their generous gifts of reagents. We also thank John Shannon (Biomolecular Research Facility, University of Virginia) for help in surface plasmon resonance experiments. We are grateful to Jim Casanova and Judy White for their insightful comments on the manuscript. Studies were supported in part by National Institutes of Health Grant DE09655 (D.C.) and by Predoctoral Training Grant T32 GM008136 (Q.A.).
Glossary
Abbreviations used:
- ILV
intraluminal vesicle
- MVB
multivesicular body
- SCAMP
secretory carrier membrane protein.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-09-0894) on January 21, 2009.
REFERENCES
- Alwan H. A., van Leeuwen J. E. UBPY-mediated epidermal growth factor receptor (EGFR) de-ubiquitination promotes EGFR degradation. J. Biol. Chem. 2007;282:1658–1669. doi: 10.1074/jbc.M604711200. [DOI] [PubMed] [Google Scholar]
- Amit I., et al. Tal, a Tsg101-specific E3 ubiquitin ligase, regulates receptor endocytosis and retrovirus budding. Genes Dev. 2004;18:1737–1752. doi: 10.1101/gad.294904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M. A protein's final ESCRT. Traffic. 2005;6:2–9. doi: 10.1111/j.1600-0854.2004.00246.x. [DOI] [PubMed] [Google Scholar]
- Bache K. G., Brech A., Mehlum A., Stenmark H. Hrs regulates multivesicular body formation via ESCRT recruitment to endosomes. J. Cell Biol. 2003a;162:435–442. doi: 10.1083/jcb.200302131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bache K. G., Raiborg C., Mehlum A., Stenmark H. STAM and Hrs are subunits of a multivalent ubiquitin-binding complex on early endosomes. J. Biol. Chem. 2003b;278:12513–12521. doi: 10.1074/jbc.M210843200. [DOI] [PubMed] [Google Scholar]
- Bache K. G., Stuffers S., Malerod L., Slagsvold T., Raiborg C., Lechardeur D., Walchli S., Lukacs G. L., Brech A., Stenmark H. The ESCRT-III subunit hVps24 is required for degradation but not silencing of the epidermal growth factor receptor. Mol. Biol. Cell. 2006;17:2513–2523. doi: 10.1091/mbc.E05-10-0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barriere H., Nemes C., Du K., Lukacs G. L. Plasticity of polyubiquitin recognition as lysosomal targeting signals by the endosomal sorting machinery. Mol. Biol. Cell. 2007;18:3952–3965. doi: 10.1091/mbc.E07-07-0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers K., Piper S. C., Edeling M. A., Gray S. R., Owen D. J., Lehner P. J., Luzio J. P. Degradation of endocytosed epidermal growth factor and virally ubiquitinated major histocompatibility complex class I is independent of mammalian ESCRTII. J. Biol. Chem. 2006;281:5094–5105. doi: 10.1074/jbc.M508632200. [DOI] [PubMed] [Google Scholar]
- Brand S. H., Laurie S. M., Mixon M. B., Castle J. D. Secretory carrier membrane proteins 31-35 define a common protein composition among secretory carrier membranes. J. Biol. Chem. 1991;266:18949–18957. [PubMed] [Google Scholar]
- Burke P., Schooler K., Wiley H. S. Regulation of epidermal growth factor receptor signaling by endocytosis and intracellular trafficking. Mol. Biol. Cell. 2001;12:1897–1910. doi: 10.1091/mbc.12.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle A., Castle D. Ubiquitously expressed secretory carrier membrane proteins (SCAMPs) 1-4 mark different pathways and exhibit limited constitutive trafficking to and from the cell surface. J. Cell Sci. 2005;118:3769–3780. doi: 10.1242/jcs.02503. [DOI] [PubMed] [Google Scholar]
- Clague M. J., Urbe S. Endocytosis: the DUB version. Trends Cell Biol. 2006;16:551–559. doi: 10.1016/j.tcb.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Ellena J. F., Moulthrop J., Wu J., Rauch M., Jaysinghne S., Castle J. D., Cafiso D. S. Membrane position of a basic aromatic peptide that sequesters phosphatidylinositol 4,5 bisphosphate determined by site-directed spin labeling and high-resolution NMR. Biophys. J. 2004;87:3221–3233. doi: 10.1529/biophysj.104.046748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Chacon R., Sudhof T. C. Novel SCAMPs lacking NPF repeats: ubiquitous and synaptic vesicle-specific forms implicate SCAMPs in multiple membrane-trafficking functions. J. Neurosci. 2000;20:7941–7950. doi: 10.1523/JNEUROSCI.20-21-07941.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrus J. E., et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- Gietz R. D., Woods R. A. Screening for protein-protein interactions in the yeast two-hybrid system. Methods Mol. Biol. 2002a;185:471–486. doi: 10.1385/1-59259-241-4:471. [DOI] [PubMed] [Google Scholar]
- Gietz R. D., Woods R. A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002b;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- Gillooly D. J., Morrow I. C., Lindsay M., Gould R., Bryant N. J., Gaullier J. M., Parton R. G., Stenmark H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO. J. 2000;19:4577–4588. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Liu L., Cafiso D., Castle D. Perturbation of a very late step of regulated exocytosis by a secretory carrier membrane protein (SCAMP2)-derived peptide. J. Biol. Chem. 2002;277:35357–35363. doi: 10.1074/jbc.M202259200. [DOI] [PubMed] [Google Scholar]
- Hanson P. I., Roth R., Lin Y., Heuser J. E. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J. Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey K. F., Shearwin-Whyatt L. M., Fotia A., Parton R. G., Kumar S. N4WBP5, a potential target for ubiquitination by the Nedd4 family of proteins, is a novel Golgi-associated protein. J. Biol. Chem. 2002;277:9307–9317. doi: 10.1074/jbc.M110443200. [DOI] [PubMed] [Google Scholar]
- Henry P. C., Kanelis V., O'Brien M. C., Kim B., Gautschi I., Forman-Kay J., Schild L., Rotin D. Affinity and specificity of interactions between Nedd4 isoforms and the epithelial Na+ channel. J. Biol. Chem. 2003;278:20019–20028. doi: 10.1074/jbc.M211153200. [DOI] [PubMed] [Google Scholar]
- Hettema E. H., Valdez-Taubas J., Pelham H. R. Bsd2 binds the ubiquitin ligase Rsp5 and mediates the ubiquitination of transmembrane proteins. EMBO J. 2004;23:1279–1288. doi: 10.1038/sj.emboj.7600137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L. Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2001;2:195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- Hollenberg S. M., Sternglanz R., Cheng P. F., Weintraub H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol. Cell. Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F., Kirkpatrick D., Jiang X., Gygi S., Sorkin A. Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol. Cell. 2006;21:737–748. doi: 10.1016/j.molcel.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Hubbard C., Singleton D., Rauch M., Jayasinghe S., Cafiso D., Castle D. The secretory carrier membrane protein family: structure and membrane topology. Mol. Biol. Cell. 2000;11:2933–2947. doi: 10.1091/mbc.11.9.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J. H., Emr S. D. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 2006;35:277–298. doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann D. J., Stefan C. J., Babst M., Emr S. D. Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J. Cell Biol. 2003;162:413–423. doi: 10.1083/jcb.200302136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komada M., Masaki R., Yamamoto A., Kitamura N. Hrs, a tyrosine kinase substrate with a conserved double zinc finger domain, is localized to the cytoplasmic surface of early endosomes. J. Biol. Chem. 1997;272:20538–20544. doi: 10.1074/jbc.272.33.20538. [DOI] [PubMed] [Google Scholar]
- Konstas A. A., Shearwin-Whyatt L. M., Fotia A. B., Degger B., Riccardi D., Cook D. I., Korbmacher C., Kumar S. Regulation of the epithelial sodium channel by N4WBP5A, a novel Nedd4/Nedd4-2-interacting protein. J. Biol. Chem. 2002;277:29406–29416. doi: 10.1074/jbc.M203018200. [DOI] [PubMed] [Google Scholar]
- Leon S., Erpapazoglou Z., Haguenauer-Tsapis R. Ear1p and ssh4p are new adaptors of the ubiquitin ligase rsp5p for cargo ubiquitylation and sorting at multivesicular bodies. Mol. Biol. Cell. 2008;19:2379–2388. doi: 10.1091/mbc.E08-01-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H., Ellena J., Liu L., Szabo G., Cafiso D., Castle D. Secretory carrier membrane protein SCAMP2 and phosphatidylinositol 4,5-bisphosphate interactions in the regulation of dense core vesicle exocytosis. Biochemistry. 2007;46:10909–10920. doi: 10.1021/bi701121j. [DOI] [PubMed] [Google Scholar]
- Liao H., Zhang J., Shestopal S., Szabo G., Castle A., Castle D. Nonredundant function of secretory carrier membrane protein isoforms in dense core vesicle exocytosis. Am J. Physiol. Cell Physiol. 2008;294:C797–C809. doi: 10.1152/ajpcell.00493.2007. [DOI] [PubMed] [Google Scholar]
- Lin P. J., Williams W. P., Luu Y., Molday R. S., Orlowski J., Numata M. Secretory carrier membrane proteins interact and regulate trafficking of the organellar (Na+,K+)/H+ exchanger NHE7. J. Cell Sci. 2005;118:1885–1897. doi: 10.1242/jcs.02315. [DOI] [PubMed] [Google Scholar]
- Liu L., Guo Z., Tieu Q., Castle A., Castle D. Role of secretory carrier membrane protein SCAMP2 in granule exocytosis. Mol. Biol. Cell. 2002;13:4266–4278. doi: 10.1091/mbc.E02-03-0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Liao H., Castle A., Zhang J., Casanova J., Szabo G., Castle D. SCAMP2 interacts with Arf6 and phospholipase D1 and links their function to exocytotic fusion pore formation in PC12 cells. Mol. Biol. Cell. 2005;16:4463–4472. doi: 10.1091/mbc.E05-03-0231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Hope L. W., Brasch M., Reinhard C., Cohen S. N. TSG101 interaction with HRS mediates endosomal trafficking and receptor down-regulation. Proc. Natl. Acad. Sci. USA. 2003;100:7626–7631. doi: 10.1073/pnas.0932599100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malerod L., Stuffers S., Brech A., Stenmark H. Vps22/EAP30 in ESCRT-II mediates endosomal sorting of growth factor and chemokine receptors destined for lysosomal degradation. Traffic. 2007;8:1617–1629. doi: 10.1111/j.1600-0854.2007.00630.x. [DOI] [PubMed] [Google Scholar]
- Marchese A., Raiborg C., Santini F., Keen J. H., Stenmark H., Benovic J. L. The E3 ubiquitin ligase AIP4 mediates ubiquitination and sorting of the G protein-coupled receptor CXCR4. Dev. Cell. 2003;5:709–722. doi: 10.1016/s1534-5807(03)00321-6. [DOI] [PubMed] [Google Scholar]
- Marmor M. D., Yarden Y. Role of protein ubiquitylation in regulating endocytosis of receptor tyrosine kinases. Oncogene. 2004;23:2057–2070. doi: 10.1038/sj.onc.1207390. [DOI] [PubMed] [Google Scholar]
- Maxfield F. R., McGraw T. E. Endocytic recycling. Nat. Rev. Mol. Cell Biol. 2004;5:121–132. doi: 10.1038/nrm1315. [DOI] [PubMed] [Google Scholar]
- McCullough J., Clague M. J., Urbe S. AMSH is an endosome-associated ubiquitin isopeptidase. J. Cell Biol. 2004;166:487–492. doi: 10.1083/jcb.200401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno E., Iura T., Mukai A., Yoshimori T., Kitamura N., Komada M. Regulation of epidermal growth factor receptor down-regulation by UBPY-mediated deubiquitination at endosomes. Mol. Biol. Cell. 2005;16:5163–5174. doi: 10.1091/mbc.E05-06-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E., Sundquist W. I. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- Mosesson Y., Yarden Y. Monoubiquitylation: a recurrent theme in membrane protein transport. Isr. Med. Assoc. J. 2006;8:233–237. [PubMed] [Google Scholar]
- Muziol T., Pineda-Molina E., Ravelli R. B., Zamborlini A., Usami Y., Gottlinger H., Weissenhorn W. Structural basis for budding by the ESCRT-III factor CHMP3. Dev. Cell. 2006;10:821–830. doi: 10.1016/j.devcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Myszka D. G. Improving biosensor analysis. J. Mol. Recognit. 1999;12:279–284. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Nickerson D. P., Russell M. R., Odorizzi G. A concentric circle model of multivesicular body cargo sorting. EMBO Rep. 2007;8:644–650. doi: 10.1038/sj.embor.7401004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M. S., Madsen P., Christensen E. I., Nykjaer A., Gliemann J., Kasper D., Pohlmann R., Petersen C. M. The sortilin cytoplasmic tail conveys Golgi-endosome transport and binds the VHS domain of the GGA2 sorting protein. EMBO J. 2001;20:2180–2190. doi: 10.1093/emboj/20.9.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberst A., et al. The Nedd4-binding partner 1 (N4BP1) protein is an inhibitor of the E3 ligase Itch. Proc. Natl. Acad. Sci. USA. 2007;104:11280–11285. doi: 10.1073/pnas.0701773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlowski J., Grinstein S. Emerging roles of alkali cation/proton exchangers in organellar homeostasis. Curr. Opin. Cell Biol. 2007;19:483–492. doi: 10.1016/j.ceb.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak Y., Glowacka W. K., Bruce M. C., Pham N., Rotin D. Transport of LAPTM5 to lysosomes requires association with the ubiquitin ligase Nedd4, but not LAPTM5 ubiquitination. J. Cell Biol. 2006;175:631–645. doi: 10.1083/jcb.200603001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo S., Sigismund S., Faretta M., Guidi M., Capua M. R., Bossi G., Chen H., De Camilli P., Di Fiore P. P. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature. 2002;416:451–455. doi: 10.1038/416451a. [DOI] [PubMed] [Google Scholar]
- Pons V., Luyet P. P., Morel E., Abrami L., van der Goot F. G., Parton R. G., Gruenberg J. Hrs and SNX3 functions in sorting and membrane invagination within multivesicular bodies. PLoS Biol. 2008;6:e214. doi: 10.1371/journal.pbio.0060214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pornillos O., Alam S. L., Rich R. L., Myszka D. G., Davis D. R., Sundquist W. I. Structure and functional interactions of the Tsg101 UEV domain. EMBO J. 2002;21:2397–2406. doi: 10.1093/emboj/21.10.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pornillos O., Higginson D. S., Stray K. M., Fisher R. D., Garrus J. E., Payne M., He G. P., Wang H. E., Morham S. G., Sundquist W. I. HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein. J. Cell Biol. 2003;162:425–434. doi: 10.1083/jcb.200302138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puertollano R., Aguilar R. C., Gorshkova I., Crouch R. J., Bonifacino J. S. Sorting of mannose 6-phosphate receptors mediated by the GGAs. Science. 2001;292:1712–1716. doi: 10.1126/science.1060750. [DOI] [PubMed] [Google Scholar]
- Putz U., Howitt J., Lackovic J., Foot N., Kumar S., Silke J., Tan S. S. Nedd4 family-interacting protein 1 (Ndfip1) is required for the exosomal secretion of Nedd4 family proteins. J. Biol. Chem. 2008;283:32621–32627. doi: 10.1074/jbc.M804120200. [DOI] [PubMed] [Google Scholar]
- Raiborg C., Bache K. G., Gillooly D. J., Madshus I. H., Stang E., Stenmark H. Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat. Cell Biol. 2002;4:394–398. doi: 10.1038/ncb791. [DOI] [PubMed] [Google Scholar]
- Raiborg C., Bache K. G., Mehlum A., Stang E., Stenmark H. Hrs recruits clathrin to early endosomes. EMBO J. 2001a;20:5008–5021. doi: 10.1093/emboj/20.17.5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C., Bache K. G., Mehlum A., Stenmark H. Function of Hrs in endocytic trafficking and signalling. Biochem. Soc. Trans. 2001b;29:472–475. doi: 10.1042/bst0290472. [DOI] [PubMed] [Google Scholar]
- Raiborg C., Malerod L., Pedersen N. M., Stenmark H. Differential functions of Hrs and ESCRT proteins in endocytic membrane trafficking. Exp. Cell Res. 2008;314:801–813. doi: 10.1016/j.yexcr.2007.10.014. [DOI] [PubMed] [Google Scholar]
- Raiborg C., Wesche J., Malerod L., Stenmark H. Flat clathrin coats on endosomes mediate degradative protein sorting by scaffolding Hrs in dynamic microdomains. J. Cell Sci. 2006;119:2414–2424. doi: 10.1242/jcs.02978. [DOI] [PubMed] [Google Scholar]
- Razi M., Futter C. E. Distinct roles for Tsg101 and Hrs in multivesicular body formation and inward vesiculation. Mol. Biol. Cell. 2006;17:3469–3483. doi: 10.1091/mbc.E05-11-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Row P. E., Prior I. A., McCullough J., Clague M. J., Urbe S. The ubiquitin isopeptidase UBPY regulates endosomal ubiquitin dynamics and is essential for receptor down-regulation. J. Biol. Chem. 2006;281:12618–12624. doi: 10.1074/jbc.M512615200. [DOI] [PubMed] [Google Scholar]
- Roxrud I., Raiborg C., Pedersen N. M., Stang E., Stenmark H. An endosomally localized isoform of Eps15 interacts with Hrs to mediate degradation of epidermal growth factor receptor. J. Cell Biol. 2008;180:1205–1218. doi: 10.1083/jcb.200708115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachse M., Strous G. J., Klumperman J. ATPase-deficient hVPS4 impairs formation of internal endosomal vesicles and stabilizes bilayered clathrin coats on endosomal vacuoles. J. Cell Sci. 2004;117:1699–1708. doi: 10.1242/jcs.00998. [DOI] [PubMed] [Google Scholar]
- Sanderson M. P., Keller S., Alonso A., Riedle S., Dempsey P. J., Altevogt P. Generation of novel, secreted epidermal growth factor receptor (EGFR/ErbB1) isoforms via metalloprotease-dependent ectodomain shedding and exosome secretion. J. Cell. Biochem. 2008;103:1783–1797. doi: 10.1002/jcb.21569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearwin-Whyatt L. M., Brown D. L., Wylie F. G., Stow J. L., Kumar S. N4WBP5A (Ndfip2), a Nedd4-interacting protein, localizes to multivesicular bodies and the Golgi, and has a potential role in protein trafficking. J. Cell Sci. 2004;117:3679–3689. doi: 10.1242/jcs.01212. [DOI] [PubMed] [Google Scholar]
- Shim J. H., et al. CHMP5 is essential for late endosome function and down-regulation of receptor signaling during mouse embryogenesis. J. Cell Biol. 2006;172:1045–1056. doi: 10.1083/jcb.200509041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirk A. J., Anderson S. K., Hashemi S. H., Chance P. F., Bennett C. L. SIMPLE interacts with NEDD4 and TSG 101, evidence for a role in lysosomal sorting and implications for Charcot-Marie-Tooth disease. J. Neurosci. Res. 2005;82:43–50. doi: 10.1002/jnr.20628. [DOI] [PubMed] [Google Scholar]
- Sigismund S., Argenzio E., Tosoni D., Cavallaro E., Polo S., Di Fiore P. P. Clathrin-mediated internalization is essential for sustained EGFR signaling but dispensable for degradation. Dev. Cell. 2008;15:209–219. doi: 10.1016/j.devcel.2008.06.012. [DOI] [PubMed] [Google Scholar]
- Simonsen A., Bremnes B., Ronning E., Aasland R., Stenmark H. Syntaxin-16, a putative Golgi t-SNARE. Eur. J. Cell Biol. 1998a;75:223–231. doi: 10.1016/S0171-9335(98)80116-7. [DOI] [PubMed] [Google Scholar]
- Simonsen A., Lippe R., Christoforidis S., Gaullier J. M., Brech A., Callaghan J., Toh B. H., Murphy C., Zerial M., Stenmark H. EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature. 1998b;394:494–498. doi: 10.1038/28879. [DOI] [PubMed] [Google Scholar]
- Singleton D. R., Wu T. T., Castle J. D. Three mammalian SCAMPs (secretory carrier membrane proteins) are highly related products of distinct genes having similar subcellular distributions. J. Cell Sci. 1997;110(Pt 17):2099–2107. doi: 10.1242/jcs.110.17.2099. [DOI] [PubMed] [Google Scholar]
- Slot J. W., Geuze H. J. Sizing of protein A-colloidal gold probes for immunoelectron microscopy. J. Cell Biol. 1981;90:533–536. doi: 10.1083/jcb.90.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin A. D., Teslenko L. V., Nikolsky N. N. The endocytosis of epidermal growth factor in A431 cells: a pH of microenvironment and the dynamics of receptor complex dissociation. Exp. Cell Res. 1988;175:192–205. doi: 10.1016/0014-4827(88)90266-2. [DOI] [PubMed] [Google Scholar]
- Staub O., Dho S., Henry P., Correa J., Ishikawa T., McGlade J., Rotin D. WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle's syndrome. EMBO J. 1996;15:2371–2380. [PMC free article] [PubMed] [Google Scholar]
- Stenmark H., Parton R. G., Steele-Mortimer O., Lutcke A., Gruenberg J., Zerial M. Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J. 1994;13:1287–1296. doi: 10.1002/j.1460-2075.1994.tb06381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strochlic T. I., Setty T. G., Sitaram A., Burd C. G. Grd19/Snx3p functions as a cargo-specific adapter for retromer-dependent endocytic recycling. J. Cell Biol. 2007;177:115–125. doi: 10.1083/jcb.200609161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuchell-Brereton M. D., Skalicky J. J., Kieffer C., Karren M. A., Ghaffarian S., Sundquist W. I. ESCRT-III recognition by VPS4 ATPases. Nature. 2007;449:740–744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- Trajkovic K., Hsu C., Chiantia S., Rajendran L., Wenzel D., Wieland F., Schwille P., Brugger B., Simons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- Umebayashi K., Stenmark H., Yoshimori T. Ubc4/5 and c-Cbl continue to ubiquitinate EGF receptor after internalization to facilitate polyubiquitination and degradation. Mol. Biol. Cell. 2008;19:3454–3462. doi: 10.1091/mbc.E07-10-0988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Goot F. G., Gruenberg J. Intra-endosomal membrane traffic. Trends Cell Biol. 2006;16:514–521. doi: 10.1016/j.tcb.2006.08.003. [DOI] [PubMed] [Google Scholar]
- White I. J., Bailey L. M., Aghakhani M. R., Moss S. E., Futter C. E. EGF stimulates annexin 1-dependent inward vesiculation in a multivesicular endosome subpopulation. EMBO J. 2006;25:1–12. doi: 10.1038/sj.emboj.7600759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R. L., Urbe S. The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell Biol. 2007;8:355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- Woodman P. G., Futter C. E. Multivesicular bodies: co-ordinated progression to maturity. Curr. Opin. Cell Biol. 2008;20:408–414. doi: 10.1016/j.ceb.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. T., Castle J. D. Tyrosine phosphorylation of selected secretory carrier membrane proteins, SCAMP1 and SCAMP3, and association with the EGF receptor. Mol. Biol. Cell. 1998;9:1661–1674. doi: 10.1091/mbc.9.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagida-Ishizaki Y., Takei T., Ishizaki R., Imakagura H., Takahashi S., Shin H. W., Katoh Y., Nakayama K. Recruitment of Tom1L1/Srcasm to endosomes and the midbody by Tsg101. Cell Struct. Funct. 2008;33:91–100. doi: 10.1247/csf.07037. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Doray B., Poussu A., Lehto V. P., Kornfeld S. Binding of GGA2 to the lysosomal enzyme sorting motif of the mannose 6-phosphate receptor. Science. 2001;292:1716–1718. doi: 10.1126/science.1060896. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.