Abstract
Esc2 is a member of the RENi family of SUMO-like domain proteins and is implicated in gene silencing in Saccharomyces cerevisiae. Here, we identify a dual role for Esc2 during S-phase in mediating both intra-S-phase DNA damage checkpoint signaling and preventing the accumulation of Rad51-dependent homologous recombination repair (HRR) intermediates. These roles are qualitatively similar to those of Sgs1, the yeast ortholog of the human Bloom's syndrome protein, BLM. However, whereas mutation of either ESC2 or SGS1 leads to the accumulation of unprocessed HRR intermediates in the presence of MMS, the accumulation of these structures in esc2 (but not sgs1) mutants is entirely dependent on Mph1, a protein that shows structural similarity to the Fanconi anemia group M protein (FANCM). In the absence of both Esc2 and Sgs1, the intra-S-phase DNA damage checkpoint response is compromised after exposure to MMS, and sgs1esc2 cells attempt to undergo mitosis with unprocessed HRR intermediates. We propose a model whereby Esc2 acts in an Mph1-dependent process, separately from Sgs1, to influence the repair/tolerance of MMS-induced lesions during S-phase.
INTRODUCTION
Homologous recombination repair (HRR) is a conserved cellular process that is required for the maintenance of genomic stability in all organisms. More specifically, HRR is required for the repair of double-strand DNA breaks and single-strand DNA (ssDNA) gaps that can arise in S-phase due to replication fork collapse or the bypass of lesions in the template. Many of the key proteins involved in HRR have been identified, but their roles are not yet fully understood (Paques and Haber, 1999; West, 2003; Krogh and Symington, 2004). Most HRR factors belong to the so-called “RAD52 epistasis group” of proteins, which are able to effect repair of DNA lesions via a variety of different mechanisms, including synthesis-dependent strand annealing, break-induced replication, single-stand annealing, and gene conversion (Paques and Haber, 1999). Precisely how cells regulate HRR and determine the optimal choice of repair pathway or course of action is poorly understood, but this probably depends on the type of initiating lesion, the cell cycle stage, and the availability of HRR proteins/mediators (Sung et al., 2003; West, 2003; Krogh and Symington, 2004). Increasing evidence also suggests that posttranslational modification of HRR proteins/mediators by phosphorylation, by ubiquitination, or by the covalent attachment of small ubiquitin-like modifiers (SUMO) may be an additional way in which HRR is regulated and coordinated with other cellular functions, such as cell cycle progression, DNA damage checkpoint signaling, and DNA replication (Branzei and Foiani, 2008).
In humans, deficiencies or defects in HRR may be associated with an increased predisposition to the development of cancer and/or premature ageing. This is best exemplified by the RecQ family of DNA helicases, which appear to perform evolutionarily conserved roles in HRR (Hanada and Hickson, 2007). In particular, the cancer-prone disorder Bloom's syndrome (BS) is caused by mutations in the human RecQ helicase gene, BLM (German, 1993; Ellis et al., 1995). BLM also associates with the Fanconi anemia (FA) complex, defects that lead to developmental abnormalities, progressive bone marrow failure, and a predisposition to the development of acute myeloid leukemia and some solid cancers (Wang, 2007). Therefore, BLM appears to be a central component of the DNA damage response network that is critical for the maintenance of genome stability and prevention of cancer in humans.
At the cellular level, BS cells classically demonstrate elevated levels of mitotic recombination, sister chromatid exchanges, and genome instability (Hanada and Hickson, 2007). Evidence that these phenotypes arise because of defects in HRR is suggested by the fact that the BLM protein (either alone or in conjunction with its associated proteins, hTOPOIIIα and RMI1; Johnson et al., 2000; Wu et al., 2000; Yin et al., 2005) can resolve different types of HRR intermediates, such as D-loops and single or double Holliday junctions in vitro (Karow et al., 2000; van Brabant et al., 2000; Wu and Hickson, 2003; Bachrati et al., 2006; Raynard et al., 2006; Wu et al., 2006; Mankouri and Hickson, 2007). Mutation of the Saccharomyces cerevisiae (budding yeast) or Schizosaccharomyces pombe (fission yeast) orthologues of BLM, called SGS1 and rqh1+, respectively, similarly causes genome instability, hyper-recombination and sensitivity to DNA-damaging agents (Gangloff et al., 1994; Watt et al., 1996; Murray et al., 1997; Stewart et al., 1997; Yamagata et al., 1998). More specifically, unprocessed HRR intermediates have been directly observed in methylmethanesulfonate (MMS)-treated sgs1, top3, and rmi1 mutants using two-dimensional (2D) gel electrophoresis (Liberi et al., 2005; Branzei et al., 2006; Mankouri and Hickson, 2006; Mankouri et al., 2007). Because many of the phenotypes of sgs1 or rqh1 mutants can be suppressed, or prevented, by the mutation of genes involved in the early steps of HRR (e.g., RAD51 in S. cerevisiae and rhp51+ in S. pombe; Gangloff et al., 2000; Fabre et al., 2002; Laursen et al., 2003; Hope et al., 2005; Miyabe et al., 2006), this suggests that the deleterious phenotypes of cells lacking RecQ helicases arises, at least in part, due to unregulated or incomplete HRR.
Because cells lacking RecQ helicases demonstrate genomic instability, and yet often retain near normal levels of viability, one challenge has been to identify proteins that cooperate with and/or compensate for deficiencies in RecQ helicases in HRR (Mullen et al., 2001; Tong et al., 2001). Several genes have been successfully identified in both S. cerevisiae and S. pombe that cause synthetic growth defects/lethality when combined with mutation of SGS1 or rqh1+. Examples of these include MUS81/mus81+, MMS4/eme1+, and SRS2/srs2+ (Gangloff et al., 2000; Boddy et al., 2000; Mullen et al., 2001; Wang et al., 2001). Because these synthetic growth defects can also be suppressed by mutation of genes involved in the early steps of HRR (Gangloff et al., 2000; Fabre et al., 2002; Maftahi et al., 2002; Bastin-Shanower et al., 2003; Hope et al., 2007), it seems likely that these proteins regulate HRR and/or function redundantly with RecQ helicases to process HRR intermediates.
When mutated, the S. pombe rad60+ gene demonstrates synthetic growth defects in conjunction with mutation of rqh1+ (Boddy et al., 2003). Similar findings have been reported for the homolog of rad60+ in S. cerevisiae, ESC2, in large-scale genetic analyses (Tong et al., 2001), suggesting that this genetic interaction is evolutionarily conserved. Rad60 has been implicated in the processing of DNA repair intermediates after DNA damage or replication fork arrest in S. pombe (Morishita et al., 2002; Miyabe et al., 2006). After release from replication arrest or exposure to UV in the G2 phase of the cell cycle, rad60-1 mutants undergo an aberrant mitosis in which septation occurs despite incomplete chromosomal segregation (Miyabe et al., 2006). Deletion of rhp51+ suppresses the aberrant mitosis of rad60-1 cells (Miyabe et al., 2006), suggesting that unprocessed, or aberrant, Rhp51-dependent HRR intermediates may be responsible for some of the abnormalities in rad60-1 mutants. Furthermore, consistent with the theory that Rad60 and Rqh1 cooperate in HRR, rad60-1 rqh1 synthetic lethality can be prevented by mutation of rhp51+ (Miyabe et al., 2006).
ESC2 was initially identified in a screen for genes that can restore defective sir1 silencing phenotypes when overexpressed in S. cerevisiae (Dhillon and Kamakaka, 2000). Consistent with a role for Esc2 in the control of gene silencing, Esc2 binds to Sir2, an NAD+-dependent histone deacetylase, and also restores certain sir2 silencing defects when overexpressed (Cuperus and Shore, 2002). A homolog of rad60+ and ESC2, called NIP45, has been identified in human cells (Novatchkova et al., 2005). Although no obvious biochemical function is apparent based on their primary sequences, these proteins contain two carboxy-terminal SUMO-like domains and comprise the so-called RENi (Rad60-Esc2-NIP45) family of SUMO-like domain proteins (Novatchkova et al., 2005). Because posttranslational modification of target proteins by SUMO has been implicated in the regulation of numerous cellular processes, including HRR (Branzei and Foiani, 2008), it is possible that RENi proteins might mimic SUMO and share some of its interaction partners. Intriguingly, both Sgs1 and BLM are posttranslationally modified by SUMO (Eladad et al., 2005; Branzei et al., 2006), and this modification appears to be important for the correct cellular localization of BLM after DNA damage (Eladad et al., 2005). However, the functional significance of SUMOylation of RecQ helicases, and their relationship to RENi proteins in general, remains poorly characterized. Furthermore, there is no current evidence to suggest that Esc2 or NIP45 function in HRR in their respective organisms.
Here, we have characterized the phenotypic effects of deletion of the S. cerevisiae ESC2 gene and reveal a role for Esc2 in the repair/tolerance of MMS-induced lesions during S-phase. Furthermore, we demonstrate that Esc2 acts in a process distinct from that of Sgs1 to influence HRR, because the accumulation of unprocessed HRR intermediates in esc2 (but not sgs1) mutants requires Mph1, the yeast ortholog of the Fanconi anemia group M protein (FANCM; Meetei et al., 2005). In the absence of both Esc2 and Sgs1, MMS-treated cells fail to fully activate the intra-S-phase DNA damage checkpoint, and sgs1esc2 double mutants attempt to traverse mitosis with unprocessed HRR intermediates. We propose that RENi proteins perform evolutionarily conserved roles during S-phase that are critical for viability in the absence of RecQ helicases.
MATERIALS AND METHODS
S. cerevisiae Strains
Unless stated otherwise, all strains used in this study were W303 strains derived from YMK165 (EUROSCARF, University of Frankfurt, Germany). Key findings were verified in the BY4741 or T344 strain backgrounds. All strains carrying gene deletions were constructed using a PCR-based gene disruption method (Wach et al., 1994). A table of strains used in this study and their relevant genotypes can be found in Supplemental Table S1.
Gene Conversion and Gross Chromosomal Rearrangement Assays
To determine rates of gene conversion, stationary phase (RDKY3615) cells were washed and spread onto plates lacking arginine for selecting Arg+ recombinants. To determine rates of gross chromosomal rearrangements (GCRs), stationary phase (D325–7D) cells were washed and spread onto plates lacking arginine and uracil and supplemented with canavanine and 5-fluoro-orotic acid (5FOA) (Sigma-Aldrich, Poole, UK), for the selection of can1 ura3 recombinants. Viability of cultures was determined in both cases by plating cells onto YPD plates without any selection pressure. Plates were incubated at 30°C. Spontaneous rates of gene conversion and GCRs were measured by fluctuation analysis (Spell and Jinks-Robertson, 2004). Experiments were repeated three times using at least five independent cultures in each case, and the average value of the median from the experiments was calculated.
Growth Conditions, Cell Synchronization, and Flow Cytometry Analysis
Strains were grown and synchronized with α-factor mating pheromone, as described previously (Mankouri and Hickson, 2006; Mankouri et al., 2007). After release from α-factor, all subsequent experiments were performed at 25°C. For the analysis of recovery of S-phase cells from exposure to 0.0167% MMS, cells were incubated at 30°C, and medium was supplemented with α-factor to arrest cells that had traversed mitosis in the G1 stage of the cell cycle, and hence prevent initiation of a second round of DNA synthesis. Release of cells from MMS treatment was achieved by centrifugation, washing, and resuspension in drug-free medium. For analysis of the G2/M checkpoint, cells were arrested with 10 μg/ml nocodazole (Sigma-Aldrich, Poole, UK) for 2 h at 30°C, and 0.15% MMS was added for the final 30 min of the nocodazole arrest to elicit a G2/M checkpoint response. After release from nocodazole arrest, cells were incubated at 25°C in fresh medium supplemented with α-factor. Cell cycle progression was monitored using flow cytometry, as described previously (Mankouri and Hickson, 2006; Mankouri et al., 2007).
2D Gel Electrophoresis
The hexadecyltrimethylammonium bromide (CTAB) method of DNA extraction and 2D gel procedures were described previously (Brewer and Fangman, 1987; Allers and Lichten, 2000; Lopes et al., 2003; Liberi et al., 2005; Mankouri and Hickson, 2006; Mankouri et al., 2007). DNA was digested with NciI and NcoI before running the 1D gels. Quantification of X-molecules on 2D gels was performed as described previously (Mankouri et al., 2007).
Western Blot Analysis
Protein extraction, SDS-PAGE, and Western blot analysis were performed as described previously (Mankouri and Hickson, 2006). The Rad53 mouse mAb (EL7) was a gift from Dr. Marco Foiani (Institute of Molecular Oncology Foundation, Milan, Italy).
Microscopy
Cells were harvested and fixed with 70% ethanol. The fixed cells were rehydrated, washed in distilled water, and stained with Vectashield mounting medium (Vector Laboratories, Burlingame, CA) with DAPI before microscopic analysis. Images were captured with a Nikon Eclipse 80i fluorescence microscope (Melville, NY), using Lucia G/F software.
RESULTS
Mutation of ESC2 Causes Genomic Instability
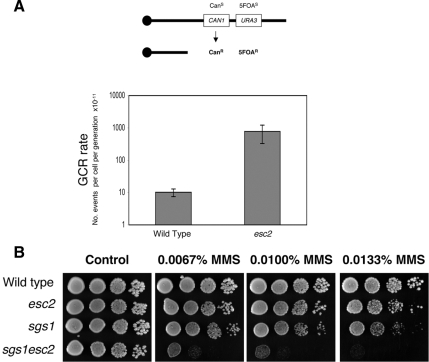
Rad60 (S. pombe), Esc2 (S. cerevisiae), and NIP45 (humans) belong to the RENi family of SUMO-like domain proteins (Novatchkova et al., 2005). Previous studies in S. pombe suggested that Rad60 is required for the maintenance of genome stability and viability, particularly in strains lacking the RecQ helicase, Rqh1 (Morishita et al., 2002; Boddy et al., 2003). ESC2 was initially identified in S. cerevisiae in a screen for genes that restore certain sir1-silencing defects when overexpressed (Dhillon and Kamakaka, 2000). To further examine the physiological role(s) of RENi proteins in eukaryotes, we examined the effects of mutating ESC2 in S. cerevisiae. Although rad60+ is an essential gene in S. pombe (Morishita et al., 2002), mutation of ESC2 does not compromise viability in S. cerevisiae (Figure 1 and Supplemental Figure S1). To investigate if Esc2 is required for the maintenance of genome integrity in S. cerevisiae, we analyzed if esc2 mutants demonstrate chromosomal instability. For this, we utilized an established assay for the measurement of rates of GCRs (Myung and Kolodner, 2002). This assay measures the stability of a chromosome arm containing both the URA3 and CAN1 genes, loss of which confer resistance to 5FOA and canavanine, respectively. Consistent with previous analyses (Myung and Kolodner, 2002), we observed that the wild-type (RDKY3615) strain demonstrated a GCR rate of ∼10−10 per cell per generation. Interestingly, deletion of ESC2 caused a 77-fold increase in GCRs (Figure 1A). Similar findings were reported in an independent study that examined GCRs at a different genomic region (Kanellis et al., 2007). We conclude that Esc2 is required for the suppression of GCRs, which implies that Esc2, like Rad60 in S. pombe, normally functions to preserve genomic integrity in vivo.
Figure 1.
Mutation of ESC2 causes genomic instability in S. cerevisiae. (A) Deletion of ESC2 causes an increase in gross chromosomal rearrangements (GCRs). GCR rate was measured by the simultaneous loss of two marker genes, URA3 and CAN1. The average and SEs of three fluctuation tests are shown for wild-type (RDKY3615) and esc2 strains. (B) Deletion of ESC2 causes a synergistic increase in sensitivity to MMS when combined with mutation of SGS1. Growth of wild-type (W303), esc2, sgs1, and sgs1esc2 strains was compared on plates containing the indicated doses of MMS. Tenfold serial dilutions of each strain were spotted onto plates, and plates were incubated at 30°C for 3 d. Genotypes of strains are indicated on the left.
ESC2 Genetically Interacts with the S. cerevisiae RecQ Helicase, SGS1
Next, we examined if esc2 mutants demonstrate any sensitivity to DNA-damaging agents. We observed that esc2 mutants were slightly sensitive to the DNA alkylating agent MMS (Figure 1B). This phenotype was more pronounced in the T344 or BY4741 genetic backgrounds (Supplemental Figure S1 and data not shown, respectively) than in the W303 background in which most of our analyses were performed. However, esc2 mutants were not sensitive to hydroxyurea (HU), UV light, camptothecin, or ionizing radiation (data not shown). Therefore, we propose that Esc2 likely acts to prevent, or process, only limited types of DNA damage in S. cerevisiae that, in this case, are elicited by treatment with MMS.
Previous large-scale genetic analyses suggested that mutation of ESC2 and SGS1 causes a synthetic growth defect (Tong et al., 2001). In the W303 and T344 strain backgrounds, we found that the sgs1esc2 double mutant was viable, but slower growing compared with either single mutant control strain (data not shown). The W303 and T344 sgs1esc2 double mutants also demonstrated a synergistic increase in sensitivity to MMS compared with either an sgs1 or an esc2 single mutant (Figure 1B and Supplemental Figure S1, respectively). We propose that there is a strong dependence on Esc2 for robust growth and the repair/tolerance of MMS-induced DNA damage when Sgs1 is mutated and vice versa. Furthermore, a similar genetic relationship is observed between rad60+ and rqh1+ in S. pombe (Boddy et al., 2003), which suggests that the genetic interactions between RENi proteins and RecQ helicases are likely to be conserved in all organisms.
RAD51, ESC2, and SGS1 Are Epistatic for MMS Sensitivity
RecQ helicases have been strongly implicated in HRR (Karow et al., 2000; van Brabant et al., 2000; Wu and Hickson, 2003; Bachrati et al., 2006; Raynard et al., 2006; Wu et al., 2006; Mankouri and Hickson, 2007). Rad60 has also been implicated in some aspect of HRR in S. pombe, because mutations in rhp51+, which is required for HRR strand invasion events, and rad60+ are epistatic (Morishita et al., 2002). We therefore examined the relationship between ESC2 and HRR in S. cerevisiae by mutating RAD51 (the S. cerevisiae homolog of rhp51+). Mutation of RAD51 causes a pronounced sensitivity to MMS, indicating that Rad51-dependent HRR is important for cell survival after exposure to MMS (Figure 2A). In contrast to the data shown in Figure 1B using sgs1 strains, mutation of ESC2 did not exacerbate the MMS sensitivity of a rad51 mutant. Rather, the rad51esc2 mutant was phenotypically indistinguishable from a rad51 mutant on plates containing MMS (Figure 2A). Similar findings were observed in the T344 and BY4741 genetic backgrounds (Supplemental Figure S1 and data not shown, respectively). Therefore, in agreement with the findings in S. pombe (Morishita et al., 2002), rad51 is epistatic to mutation of ESC2 for MMS sensitivity, suggesting that the genetic relationship between RENi proteins and HRR is evolutionarily conserved. A similar genetic (epistatic) relationship was also observed between rad51 and sgs1 (Figure 2A and Supplemental Figure S1). Furthermore, the rad51sgs1esc2 triple mutant closely resembles the rad51 single mutant on plates containing MMS (Figure 2A and Supplemental Figure S1). Taken together, we propose that Esc2 and Sgs1 both act in, or influence, Rad51-dependent HRR of MMS-induced DNA damage.
Figure 2.
Esc2 influences Rad51-dependent homologous recombination repair. (A) Mutations in RAD51, ESC2 and SGS1 are epistatic for MMS sensitivity. Growth of wild-type (W303), esc2, sgs1, sgs1esc2, rad51, rad51esc2, rad51sgs1, and rad51sgs1esc2 strains was compared on plates containing the indicated doses of MMS. Tenfold serial dilutions of each strain were spotted onto plates, and plates were incubated at 30°C for 3 d. Genotypes of strains are indicated on the left. (B) Deletion of ESC2 or SGS1 causes an additive increase in gene conversion rate. (i) Gene conversion rate was measured between two arg4 alleles located on chromosomes V and VIII. (ii) The average and SD of three fluctuation tests are shown for wild-type (D325-7D), sgs1, esc2, and sgs1esc2 strains. The fold increase (relative to the wild-type strain) is stated below each strain in parentheses. (C) esc2 mutants accumulate Rad51-dependent X-molecules after DNA damage. Wild-type (W303) and esc2 and rad51esc2 strains were released from G1 arrest into fresh medium containing 0.033% MMS. DNA replication intermediates were analyzed around ARS305 by 2D gel electrophoresis at the times indicated. The key on the right denotes DNA structures that can be identified by the 2D gel technique. The arrow denotes X-shaped structures present in esc2 mutants at 150 min. (D) Mutation of ESC2 and/or SGS1 causes the accumulation of MMS-induced X-molecules at ARS305 after DNA damage. DNA replication intermediates at ARS305 were analyzed in wild-type (W303) and esc2, sgs1, and sgs1esc2 strains by 2D gel electrophoresis after a 150-min exposure to 0.033% MMS.
Mutation of ESC2 and SGS1 Reveals Two Distinct Mechanisms That Affect Recombination
Previous analyses have demonstrated that esc2 and sgs1 mutants demonstrate an increased rate of inter-homolog recombination in diploid cells (Alvaro et al., 2007). To analyze whether this was a general effect of Esc2 on recombination rates, we analyzed gene conversion rates in haploid esc2, sgs1, and sgs1esc2 mutants using a different recombination assay. For this, we measured the rate of ectopic recombination between arg4-Bg and arg4-RV heteroalleles in the D325-7D genetic background (Robert et al., 2006). In agreement with previous findings (Robert et al., 2006), we observed an elevated rate of gene conversion (Arg+ recombinants) in sgs1 mutants (Figure 2B). Consistent with a role for Esc2 in influencing HRR, we also observed an elevated rate of gene conversion in the esc2 mutant (Figure 2B). We note, however, that the effect was less pronounced in the esc2 mutant than in the sgs1 mutant (Figure 2B). Interestingly, in our assay, the sgs1esc2 double mutant showed an additive increase in gene conversion rate, compared with either the esc2 or sgs1 single mutant (Figure 2B). We propose, therefore, that Esc2 and Sgs1 may function via two separate mechanisms to influence HRR reactions. Furthermore, Esc2, like Sgs1, probably plays a general role in HRR, because both proteins affect the rate of interhomolog recombination in diploid cells (Alvaro et al., 2007), as well as gene conversion rate in haploid cells.
esc2 Mutants Accumulate Unprocessed Rad51-dependent Recombination Intermediates after DNA Damage
Previous studies demonstrated an accumulation of unresolved HRR intermediates in MMS-treated sgs1 mutants using neutral-neutral 2D gel electrophoresis (Liberi et al., 2005; Branzei et al., 2006; Mankouri et al., 2007). In S. pombe rad60-1 mutants, Rhp51-dependent structures that are not recognized by the checkpoint cause an aberrant mitosis after exposure to HU or UV (Miyabe et al., 2006). However, the putative abnormal structures arising in rad60-1 mutants have not yet been detected. Therefore, we examined if esc2 mutants accumulate any abnormal HRR intermediates after exposure to MMS using 2D gel electrophoresis. Wild-type and esc2 and rad51esc2 strains were synchronized in G1 with α-factor and then released into medium containing 0.033% MMS. Samples were taken at fixed intervals to observe DNA replication intermediates on 2D gels originating from an early firing replication origin, ARS305. Genomic DNA was prepared using the CTAB method of DNA extraction to restrain branch migration of joint (X-shaped) molecules (Lopes et al., 2003). Origin firing at ARS305 was detectable after 20 min in all strains by the appearance of bubbles, Y-molecules, and origin-associated X-spikes (Figure 2C). Previous analyses have demonstrated that the origin-associated X-spikes are normal DNA replication intermediates that are not dependent on HRR proteins for their formation and are not, therefore, HRR intermediates (Lopes et al., 2003). By 150 min, the majority of replication intermediates detectable at ARS305 had disappeared in wild-type strains, consistent with completion of DNA replication at this genomic locus (in the presence of MMS) by this time. In contrast, although esc2 mutants also demonstrated comparable kinetics of origin firing, bulk DNA replication (see Figure 4A below), and bubble and Y-molecule disappearance (at 150 min), a structure corresponding to a joint (X-shaped) molecule was evident after 150 min (Figure 2C). Similar kinetics of DNA replication and the persistence of X-shaped molecules in MMS-treated esc2 mutants were also observed in a second (BY4741) yeast background (Supplemental Figure S2), and independently verified in the companion article by Sollier et al. (2009). Therefore, mutation of ESC2 causes abnormal (X-shaped) replication intermediates to accumulate at ARS305 after exposure to MMS. Furthermore, because these structures do not arise/persist in rad51esc2 mutants (Figure 2C), we propose that Rad51 is required for their formation and/or stabilization and that these structures are likely to be Rad51-dependent HRR intermediates. Similar types of RAD51-dependent structures are detectable in MMS-treated sgs1 and sgs1esc2 mutants (Figure 2D and Supplemental Figure S3; Liberi et al., 2005; Mankouri et al., 2007). We note, however, that the intensity of the X-molecule signal was less pronounced in esc2 mutants compared with sgs1 or sgs1esc2 mutants. Taken together with the results from Figure 2A, our data are consistent with Esc2 and Sgs1 acting to prevent the accumulation and/or persistence of Rad51-dependent HRR intermediates.
Figure 4.
sgs1esc2 double mutants fail to fully activate Rad53 in response to DNA damage and attempt to traverse mitosis with unresolved recombination intermediates. (A) sgs1esc2 mutants undergo an aberrant mitosis after recovery from 0.0167% MMS. Wild-type (W303) and esc2, sgs1, and sgs1esc2 strains were released from G1 arrest into fresh medium containing 0.0167% MMS. After 90 min, cells were harvested, washed, and resuspended in fresh medium. α-factor was added to all cultures to prevent initiation of a second round of DNA replication. Cell cycle progression after the removal of MMS was monitored at fixed intervals by flow cytometry. The shaded peaks represent experimental data, whereas the unshaded peak is a reference to indicate a normal G2/M peak (at 90 min release from G1 arrest). The positions of the 1N (G1) and 2N (G2/M) peaks are indicated below. (B) sgs1esc2 mutants fail to fully activate Rad53 after DNA damage. Protein extracts were prepared from wild-type, esc2, sgs1, and sgs1esc2 strains 90 min after release from α-factor arrest in the presence or absence of 0.0167% MMS. Rad53 phosphorylation status was monitored by Western blotting. The positions of the unphosphorylated Rad53 and slower migrating phosphorylated forms of Rad53 are shown on the right. (C) sgs1esc2 mutants exhibit morphological defects. (i) Representative image of DAPI-stained wild-type and sgs1esc2 cells after recovery from exposure to 0.0167% MMS. Examples of common abnormalities observed in sgs1esc2 populations are denoted by white arrows. These include (ii) cells with little or no DNA, (iii) cells exhibiting morphological defects and evidence of DNA fragmentation, and (iv) multibudded cells exhibiting gross morphological abnormalities. (D) Onset of the aberrant mitosis in sgs1esc2 mutants correlates with the persistence of MMS-induced X-molecules. DNA replication intermediates at ARS305 were analyzed by 2D gel electrophoresis after a 90-min exposure to 0.0167% MMS and after 6 h of recovery after the removal of MMS. X-shaped structures are denoted by arrows.
X-Molecules Arising in MMS-treated esc2 or sgs1 Mutants Are Attenuated by the Mutation of SHU1
Our previous findings demonstrated that the MMS-induced X-molecules arising in sgs1 mutants are attenuated by mutation of any one of the SHU genes (comprising CSM2, PSY3, SHU1, and SHU2; Shor et al., 2005; Mankouri et al., 2007). We therefore investigated if the MMS-induced X-molecules arising in esc2 mutants were also affected by mutation of SHU1. Consistent with our previous findings, we observed that shu1 was epistatic to mutation of SGS1 for MMS sensitivity (Supplemental Figure S4A; Mankouri et al., 2007). Similarly, mutation of ESC2 did not exacerbate the MMS sensitivity of a shu1 mutant strain, and shu1esc2 mutants resembled shu1 mutants on plates containing MMS (Supplemental Figure S4). To test if the MMS-induced X-molecules arising in esc2 or sgs1 mutants (Figure 2D) were affected by mutation of SHU1, we compared the intensity of X-molecules at ARS305 after a 150-min exposure to 0.033% MMS in esc2, shu1esc2, sgs1, and shu1sgs1 strains. Interestingly, we observed that MMS-induced X-molecules were attenuated in both the shu1esc2 and shu1sgs1 double mutants, compared with either the esc2 or the sgs1 single mutants, respectively (Supplemental Figure S4B). Taken together, our data are consistent with both Esc2 and Sgs1 acting downstream of Shu1 in Rad51-dependent HRR of MMS-induced lesions.
MMS-induced X-Molecules That Accumulate in esc2 or sgs1 Mutants Can Be Genetically Distinguished by Mutation of MPH1
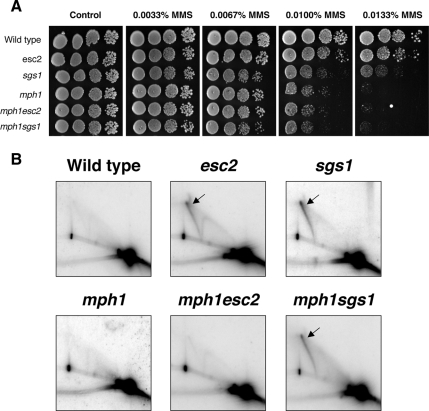
To further examine the roles of Esc2 and Sgs1 in influencing HRR, we analyzed the genetic relationship between ESC2, SGS1, and other genes implicated in HRR. One such candidate gene analyzed was MPH1, which encodes a 3′–5′ DNA helicase homologous to human FANCM protein (Meetei et al., 2005), that is proposed to act downstream of the Shu complex in HRR (Schurer et al., 2004; St. Onge et al., 2007). Consistent with this proposal, we observed that mutation of MPH1 caused MMS sensitivity that was hypostatic to mutation of RAD51 (data not shown). We also observed that mph1 was epistatic to mutation of ESC2 or SGS1 for MMS sensitivity, suggesting that Mph1, Esc2, and Sgs1 may act in a common pathway to process MMS-induced DNA lesions (Figure 3A).
Figure 3.
Mutation of MPH1 prevents the accumulation of MMS-induced X-molecules at ARS305 in esc2, but not sgs1, mutants. (A) Growth of wild-type (W303), esc2, sgs1, mph1, mph1esc2, and mph1sgs1 strains was compared on plates containing the indicated doses of MMS. Tenfold serial dilutions of each strain were spotted onto plates, and plates were incubated at 30°C for 3 d. Genotypes of strains are indicated on the left. (B) DNA replication intermediates at ARS305 were analyzed in the indicated strains by 2D gel electrophoresis after a 3-h exposure to 0.033% MMS. X-shaped structures are denoted by arrows. Note the lack of MMS-induced X-molecules in mph1esc2 mutants.
We then examined if the MMS-induced X-molecules arising in esc2 or sgs1 mutants (Figure 2D) were affected by mutation of MPH1. Wild-type, mph1, esc2, mph1esc2, sgs1, and mph1sgs1 strains were released from G1 arrest into medium containing 0.033% MMS, and DNA was harvested for 2D gel analysis of ARS305 after 3 h. At this time point, all replication intermediates normally associated with origin firing had largely disappeared from ARS305 in wild-type and mph1 cells, consistent with the completion of DNA replication at or around ARS305 by this time (Figures 2, C and D, and 3B). In esc2 or sgs1 mutants, we again observed persistent MMS-induced X-molecules at ARS305 after MMS exposure (Figure 3B). Interestingly, however, we observed that the MMS-induced X-molecules normally observed in esc2 mutants at this time were not detectable in mph1esc2 mutants (Figure 3B). Therefore, Mph1 is required for the acute formation and/or stabilization of MMS-induced X-molecules in esc2 mutants. Interestingly, we observed that mph1sgs1 mutants largely retained MMS-induced X-molecules (Figure 3B). Nevertheless, quantification of the average intensity of X-molecules arising in sgs1 and mph1sgs1 mutants revealed that X-molecule intensity was reduced in mph1sgs1 mutants by ∼30%, suggesting that a fraction of the X-molecules arising in sgs1 mutants were Mph1-dependent. We propose that the X-molecules that arise in esc2 and sgs1 mutants after 2–3 h of MMS treatment can be distinguished genetically by mutation of MPH1, suggesting that, although they appear qualitatively similar on 2D gels, they likely represent different types of HRR intermediates. Taken together with the data in Figures 1 and 2, we propose that mutation of ESC2 or SGS1 reveals two distinct processes that influence Rad51-dependent HRR of MMS-induced DNA lesions.
sgs1esc2 Double Mutants Exhibit Defects in the intra-S-Phase Checkpoint Response after MMS-induced DNA Damage
Because esc2 and sgs1 mutants apparently accumulate distinct types of unprocessed HRR intermediates after exposure to MMS, we analyzed other aspects of the sgs1esc2 double mutant phenotype. Wild-type, esc2, sgs1, and sgs1esc2 strains were synchronized in G1 with α-factor and then released into medium containing 0.0167% MMS. This lower dose of MMS was used to allow visualization of any additive/synergistic consequences of mutating ESC2 and SGS1. Samples were taken to analyze bulk DNA replication by fluorescence-activated cell sorting (FACS; Figure 4A), as well as DNA damage checkpoint activation, as monitored by the phosphorylation status of the Rad53 checkpoint effector kinase (Figure 4B). Similar kinetics of S-phase progression in the absence of MMS were observed in all strains (data not shown). In the presence of MMS, wild-type cells apparently completed bulk DNA replication by ∼2–3 h (Supplemental Figure S5A). Consistent with this prolongation of S-phase occurring as a consequence of MMS-induced activation of the intra-S DNA damage checkpoint (Branzei and Foiani, 2007), DNA damage-induced Rad53 phosphorylation was detectable in wild-type cells that did not occur during an unperturbed S-phase (Figure 4B). Both esc2 and sgs1 single mutants demonstrated comparable (and wild-type-like) kinetics of S-phase progression in the presence of MMS (Figure 4A). However, both mutants demonstrated a mild reduction in the extent of Rad53 activation in the presence of MMS (Figure 4B; note the reduced proportion of the uppermost, hyperphosphorylated Rad53 band in esc2 and sgs1 extracts). Interestingly, the sgs1esc2 double mutant traversed S-phase slightly faster than wild-type or esc2 or sgs1 cells in the presence of 0.0167% MMS (Figure 4A and Supplemental Figure S5A). Consistent with this observation, the sgs1esc2 double mutant also demonstrated a failure to fully activate Rad53 in the presence of MMS (Figure 4B and Supplemental Figure S5B). We note, however, that sgs1esc2 mutants do retain the ability to partially activate Rad53 in response to a higher dose (0.033%) of MMS, suggesting that the intra-S-phase checkpoint is not completely defective in the sgs1esc2 double mutant (Supplemental Figure S6). Taken together, our data are consistent with Esc2 and Sgs1 defining two distinct mechanisms for the activation of Rad53 during S-phase after MMS exposure. Previous studies have identified mild checkpoint signaling defects in sgs1 mutants after treatment with MMS (Frei and Gasser, 2000; Bjergbaek et al., 2005; Liberi et al., 2005), but a role for Esc2 in checkpoint signaling has not been demonstrated previously. Intriguingly, Sgs1 interacts with Rad53 (Bjergbaek et al., 2005), and Rad60 interacts with Cds1 (the Rad53 ortholog) in S. pombe (Boddy et al., 2003). Therefore, it is conceivable that both proteins directly modulate checkpoint status while engaged in HRR via direct protein–protein interactions with checkpoint kinases or their associated factors.
The sgs1esc2 Double Mutant Undergoes an Aberrant Mitosis with Unresolved Recombination Intermediates
Next, we investigated the fate of wild-type, esc2, sgs1, and sgs1esc2 strains during a 6-h recovery period after a 90-min exposure to 0.0167% MMS. MMS-treated cells were harvested, washed, and resuspended in fresh medium. α-factor was added to the fresh medium to arrest any cells in G1 that had successfully traversed mitosis during the recovery period in order to prevent initiation of a second round of DNA replication. Under these experimental conditions, wild-type cells began to traverse mitosis (as revealed by FACS as a decrease in the 2N population of cells coupled with a concomitant build-up of 1N cells) by 2 h after removal of MMS (Figure 4A). Verification of this phenomena being due to mitotic progression was provided by the observation that the mitotic spindle poison, nocodazole, delayed the build-up of cells with a 1N DNA content (data not shown). By 4 h of recovery from MMS, the majority of wild-type cells had successfully traversed mitosis (Figure 4A). In esc2 and sgs1 mutants, we observed that a population of cells with 1N DNA content began to accumulate ∼2–4 h after removal of MMS (Figure 4A). Although their recovery from MMS was slightly delayed, it was clear that a significant proportion of esc2 and sgs1 cells had successfully traversed mitosis after 6 h of recovery from MMS (Figure 4A). Interestingly, however, a much more drastic phenotype was observed in the sgs1esc2 double mutant. Although the population of sgs1esc2 cells with a 2N DNA content began to decrease between 2 and 4 h after release from MMS (i.e., with similar kinetics to wild-type cells), there was no corresponding build-up of cells with a 1N DNA content (Figure 4A). Instead, the FACS traces revealed a broadened peak of DNA content, indicative of cell segregation defects occurring during an aberrant mitosis (Miyabe et al., 2006). Consistent with this hypothesis, microscopic analysis of DAPI-stained sgs1esc2 cells after 6 h of recovery from MMS revealed a significant proportion of cells with little or no DNA, as well as a number of cells exhibiting unusual cellular morphologies, and evidence of DNA fragmentation (Figure 4C).
To determine if mitotic progression is influenced by the MMS-induced X-molecules that accumulate in esc2 or sgs1 mutants (Figures 2, C and D, and 3B), we analyzed DNA replication intermediates at ARS305 in cells after 6 h of recovery from MMS (Figure 4D). After a 90-min exposure to 0.0167% MMS, we could detect MMS-induced X-molecules at ARS305 in esc2, sgs1, and sgs1esc2 mutants (Figure 4D). We note, however, that the MMS-induced X-molecules were reduced in intensity in esc2 mutants relative to those observed in Figures 2, C and D, and 3B, presumably because of the lower dose of MMS used in this protocol. After 6 h of recovery from MMS, we could no longer detect MMS-induced X-molecules in esc2 or sgs1 mutants (Figure 4D), indicating that mitotic progression of esc2 and sgs1 mutants (Figure 4A) correlates with the level of unprocessed HRR intermediates falling below a detection threshold level. Therefore, the MMS-induced X-molecules that arise in esc2 and sgs1 cells during S-phase can be resolved after removal of MMS, presumably via redundant repair pathways operating during late S-phase and/or G2/M. Interestingly, in contrast to esc2 or sgs1 mutants, unprocessed HRR intermediates did not significantly decline in intensity at ARS305 in the sgs1esc2 double mutant after 6 h of recovery from MMS (Figure 4D). Given that the absolute levels of unprocessed HRR intermediates arising during exposure to MMS appear very similar in sgs1 and sgs1esc2 cells (Figure 4D), we propose that the processing of HRR intermediates is impaired in sgs1esc2 mutants, relative to that in esc2 or sgs1 single mutants. Taken together with the data in Figures 4, A–C, we propose that MMS-treated sgs1esc2 double mutant cells fail to adequately activate Rad53 during S-phase, and then attempt to traverse mitosis with unresolved HRR intermediates. Furthermore, because the G2/M checkpoint response appears intact in the sgs1esc2 double mutant (Supplemental Figure S7), we propose that the X-molecules that arise, and persist, in the sgs1esc2 double mutant, are not recognized by the G2/M checkpoint machinery.
Mutation of RAD51 Causes a Checkpoint Arrest and Prevents the Aberrant Mitosis Normally Observed in sgs1esc2 Cells
Because the unresolved HRR intermediates that persist in MMS-treated esc2, sgs1 or sgs1esc2 cells are Rad51-dependent (Figure 2C and Supplemental Figure S3; Liberi et al., 2005; Branzei et al., 2006; Mankouri et al., 2007), we examined if mutation of RAD51 prevented any of the abnormal phenotypes of sgs1esc2 mutants. Wild type, sgs1esc2, rad51, and rad51sgs1esc2 strains were treated with 0.0167% MMS for 90 min and were then allowed to recover in fresh medium as described in Figure 4A. Samples were taken at regular intervals to analyze cell cycle progression (Figure 5A) and Rad53 phosphorylation status (Figure 5B). Mutation of RAD51 has previously been demonstrated to cause impaired S-phase progression in the presence of MMS due to persistent activation of the DNA damage checkpoint (Supplemental Figure S5A; Mankouri and Hickson, 2006; Mankouri et al., 2007). Interestingly, we observed that the FACS profile of the rad51sgs1esc2 triple mutant was indistinguishable from that of the rad51 single mutant (Figure 5A and Supplemental Figure S5A). This is consistent with our observation that mutation of RAD51 predominates over mutation of ESC2 and/or SGS1 (Figure 2A and Supplemental Figure S1). During the period of recovery from MMS exposure, we again observed that the majority of wild-type cells successfully traversed mitosis by 4 h, whereas the sgs1esc2 double mutant underwent an aberrant mitosis (Figure 5A). Although rad51 and rad51sgs1esc2 mutants appeared to traverse S-phase more slowly than did wild-type or sgs1esc2 mutants (Supplemental Figure S5A), they do not attempt to traverse mitosis even after 6 h of recovery from MMS (Figure 5A). Rather, rad51 and rad51sgs1esc2 mutants appeared to undergo a prolonged arrest in late S or G2 phase. To test this further, the phosphorylation status of Rad53 was analyzed at various time points in the strains shown in Figure 5A. We observed that Rad53 was phosphorylated in wild-type cells after exposure to 0.0167% MMS, whereas the DNA damage checkpoint response was defective in sgs1esc2 mutants (Figure 5B and Supplemental Figure S5B). Interestingly, the progression of wild-type cells through mitosis (Figure 5A) correlated with the resetting (dephosphorylation) of Rad53 (Figure 5B), suggesting that the DNA damage checkpoint becomes deactivated either before the onset of, or during, mitosis. In sgs1esc2 mutants, however, robust Rad53 activation was not evident either during S-phase (in the presence of 0.0167% MMS; Supplemental Figure S5B), or during the subsequent recovery period (Figure 5B). In the presence of MMS, mutation of RAD51 reversed the checkpoint deficiency of sgs1esc2 mutants (Figure 5A and Supplemental Figure S5). Consistent with the mitotic delay observed in rad51 and rad51sgs1esc2 mutants being due to a checkpoint mediated arrest, we observed persistent phosphorylation of Rad53 in both rad51 and rad51sgs1esc2 mutants after 6 h of recovery from MMS exposure (Figure 5B). We propose that loss of RAD51 abolishes HRR at a early stage, thus preventing the accumulation of the unprocessed HRR intermediates in sgs1esc2 mutants, causing prolonged checkpoint activation, and thereby preventing the aberrant mitosis normally observed in sgs1esc2 mutants.
Figure 5.
Mutation of RAD51 causes a checkpoint-mediated arrest and prevents sgs1esc2 double mutants undergoing an aberrant mitosis after recovery from MMS. (A) Wild-type (W303), sgs1esc2, rad51, and rad51sgs1esc2 strains were released from G1 arrest into fresh medium containing 0.0167% MMS for 90 min and then resuspended in fresh medium to allow recovery as described in Figure 4A. (B) Mutation of RAD51 results in the persistent activation of Rad53 after treatment with MMS. Protein extracts were prepared from wild-type, sgs1esc2, rad51, and rad51sgs1esc2 strains at the indicated times. Rad53 phosphorylation status was monitored by Western blotting.
DISCUSSION
Mutation of ESC2 and SGS1 has revealed two genetically distinct mechanisms that influence Rad51-dependent HRR and Rad53 activation after DNA damage, leading us to propose a model in which Esc2 and Sgs1 act in two functionally separate processes to repair/tolerate MMS-induced lesions during S-phase (Figure 6). Inactivation of either Esc2 or Sgs1 results in accumulation of unprocessed HRR intermediates, and a mild defect in DNA damage checkpoint activation. Although we do not currently know the precise nature of the HRR intermediates arising in either single mutant, we propose that they likely represent different forms of X-shaped structures, because the X-molecules arising in esc2 mutants after 2–3 h of MMS exposure, were prevented by mutation of MPH1, whereas only a proportion (∼30%) of sgs1 X-molecules appeared to be Mph1-dependent. A corollary of this observation is that it raises the possibility that the X-shaped molecules detectable in sgs1 mutants may not be homogeneous in structure. Indeed, Sgs1 has been implicated in the processing of various different HRR intermediates (reviewed in Mankouri and Hickson, 2007), and the structures we detect on 2D gels could theoretically be a mixture of any two (or more) of these. Although both Mph1 and Sgs1 are able to branch migrate Holliday junctions in vitro (Karow et al., 2000; Prakash et al., 2005), it remains undetermined if this also holds true in vivo, where the local chromatin environment is likely to be more complex. Future studies should therefore be aimed at further investigating, and comparing, the relative substrate specificities of Mph1 and Sgs1.
Figure 6.
Proposed model for the role of Mph1, Esc2, and Sgs1 in processing recombination intermediates and mediating checkpoint signaling. MMS-induced DNA lesions cause discontinuous DNA synthesis, leading to the accumulation of postreplicative ssDNA gaps. These ssDNA gaps become coated with RPA, which directly activates the DNA damage checkpoint. ssDNA gaps can be repaired by Rad51-dependent homologous recombination repair (HRR) or by other redundant repair pathways (e.g., translesion synthesis). Esc2 and Sgs1 act downstream of Rad51 to process distinct HRR intermediates (denoted as 1 and 2, respectively) and communicate with the checkpoint machinery. Mph1 may act at an intermediate step, downstream of Rad51, to control the overall balance between the putative Esc2- and Sgs1-dependent processes. The Esc2 arm of the pathway (left) is entirely Mph1-dependent, whereas the Sgs1 pathway (right) is largely independent of Mph1, although a fraction of Sgs1 substrates are Mph1-dependent. In the absence of both Esc2 and Sgs1, checkpoint activation is impaired after DNA damage, and cells attempt to traverse mitosis with unprocessed HRR intermediates, leading to an aberrant mitosis. Mutation of RAD51 abolishes HRR of MMS-induced DNA lesions and leads to the accumulation of unrepaired RPA-coated ssDNA gaps. These cause prolonged activation of the DNA damage checkpoint, leading to a checkpoint-mediated arrest.
Further evidence that Esc2 and Sgs1 may act separately to influence HRR and checkpoint signaling is also suggested by the observation in the accompanying manuscript by Sollier et al. (2009) that Esc2 and Sgs1 exhibit distinct elution profiles when yeast protein extracts are fractioned by gel filtration after MMS treatment. Taken together with our data, this may explain why the combined loss of both of the Esc2- and Sgs1-dependent functions results in the poor growth phenotype of the sgs1esc2 double mutant. More specifically, we propose that the failure of the intra-S-phase checkpoint machinery to recognize unprocessed HRR structures that arise in the absence of Esc2 and Sgs1 results in a breakdown in the coordination between cell cycle progression and DNA damage repair/tolerance in sgs1esc2 double mutants. As a consequence of this, sgs1esc2 mutants attempt to traverse mitosis with unprocessed HRR intermediates, and the aberrant mitosis that ensues leads to chromosome segregation defects, extensive genome rearrangements and poor growth. However, we cannot rule out the alternative possibility that an entirely new species of X-molecule(s) arises in sgs1esc2 double mutants, which is distinct (and more persistent/toxic) from those arising in esc2 or sgs1 mutants, although we consider this unlikely. When RAD51 is mutated, it is likely that the DNA damage checkpoint becomes persistently activated because of an accumulation of RPA-coated ssDNA gaps that activate Mec1 and Rad53 (Branzei and Foiani, 2007). The failure to initiate HRR-mediated postreplication gap filling also prevents any accumulation of later-stage unprocessed HRR intermediates in esc2, sgs1, or sgs1esc2 mutants. As a consequence of this, rad51sgs1esc2 triple mutants do not undergo an aberrant mitosis and demonstrate a prolonged checkpoint-mediated arrest, similar to that observed in rad51 mutants. This prolonged arrest period possibly provides the time necessary for alternative repair processes, such as direct gap filling DNA synthesis, to take place.
Previous studies suggested a role for Esc2 in the establishment of silent chromatin (Dhillon and Kamakaka, 2000; Cuperus and Shore, 2002), indicating that Esc2 may affect chromatin structure. We do not currently know if this function of Esc2 is related, or distinct, from its putative roles in HRR and the maintenance of genome stability reported here. However, one speculative possibility is that Esc2 could modify the chromatin structure of Mph1 products to make these HRR intermediates accessible for late-stage processing activities. Although no obvious biochemical activity is evident based on primary sequence analysis, one intriguing feature of Esc2, and of RENi proteins in general, is the presence of two carboxy-terminal SUMO-like domains (Novatchkova et al., 2005). Additionally, Esc2 interacts with both SUMO and Ubc9 (a SUMO-conjugating enzyme), and Esc2 also contains several consensus sites that might be targets for SUMO-conjugation; see companion article by Sollier et al. (2009). Given that the posttranslational modification of Sgs1 and BLM by SUMO has also been implicated in the maintenance of genome integrity (Eladad et al., 2005; Branzei et al., 2006), it is likely that SUMOylation of Esc2 and Sgs1 is an important mechanism required for regulating and/or coordinating the abilities of these proteins to recognize key substrates and/or protein-binding partners. Consistent with this hypothesis, it is interesting to note that mutation of ESC2 causes synthetic growth defects when combined with mutations in UBC9 or MMS21 (which encodes a SUMO-conjugating ligase; Supplemental Figure S8). Furthermore, mutation of UBC9 or MMS21 also results in the accumulation of X-shaped structures in the presence of MMS (Branzei et al., 2006). Taken together, these findings further highlight the importance of SUMOylation of target proteins as a central theme in the regulation of HRR (Branzei and Foiani, 2008).
Given that the genetic interaction between RENi proteins and RecQ helicases appears to be evolutionarily conserved, we propose that NIP45 (the human Rad60/Esc2 ortholog) may similarly be essential for the viability of Bloom's syndrome cells. Therefore, it will be of interest to investigate the effects of mutating BLM and NIP45 in mammalian cells in future studies. Another important ramification of this study comes from the demonstration that MPH1, the putative yeast homolog of the human FANCM protein (Meetei et al., 2005), is required for the accumulation of HRR intermediates in MMS-treated esc2 mutants. Although Mph1 in yeast and the FA proteins in humans have been implicated previously in some aspect of HRR (Wang, 2007), our results are the first demonstration of a direct role of Mph1 in the in vivo processing of HRR intermediates. Based on our findings, further analysis of the relationship between MPH1, SGS1, and ESC2 homologues in human cells may therefore reveal novel avenues for alleviating symptoms in BS and/or FA individuals, including cancer predisposition.
Supplementary Material
ACKNOWLEDGMENTS
We thank G. Blobel, S. Gangloff, S. Jentsch, and R. D. Kolodner for strains. We also thank D. Branzei, M. Foiani, P. McHugh, S. P. Jackson, L. Wu and various members of the Hickson laboratory for helpful discussions. α-factor was provided by the Cancer Research UK peptide synthesis laboratory. This work was funded by Cancer Research UK and the Bloom's Syndrome Foundation.
Glossary
Abbreviations used:
- BS
Bloom's syndrome
- FACS
fluorescent-activated cell sorting
- GCR
gross chromosomal rearrangement
- HHR
homologous recombination repair
- MMS
methylmethanesulfonate.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-08-0877) on January 21, 2009.
REFERENCES
- Allers T., Lichten M. A method for preparing genomic DNA that restrains branch migration of Holliday junctions. Nucleic Acids Res. 2000;28:e6. doi: 10.1093/nar/28.2.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvaro D., Lisby M., Rothstein R. Genome-wide analysis of Rad52 foci reveals diverse mechanisms impacting recombination. PLoS Genet. 2007;3:2439–2449. doi: 10.1371/journal.pgen.0030228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachrati C. Z., Borts R. H., Hickson I. D. Mobile D-loops are a preferred substrate for the Bloom's syndrome helicase. Nucleic Acids Res. 2006;34:2269–2279. doi: 10.1093/nar/gkl258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastin-Shanower S. A., Fricke W. M., Mullen J. R., Brill S. J. The mechanism of Mus81-Mms4 cleavage site selection distinguishes it from the homologous endonuclease Rad1-Rad10. Mol. Cell. Biol. 2003;23:3487–3496. doi: 10.1128/MCB.23.10.3487-3496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjergbaek L., Cobb J. A., Tsai-Pflugfelder M., Gasser S. M. Mechanistically distinct roles for Sgs1p in checkpoint activation and replication fork maintenance. EMBO J. 2005;24:405–417. doi: 10.1038/sj.emboj.7600511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy M. N., Lopez-Girona A., Shanahan P., Interthal H., Heyer W. D., Russell P. Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol. Cell. Biol. 2000;20:8758–8766. doi: 10.1128/mcb.20.23.8758-8766.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy M. N., Shanahan P., McDonald W. H., Lopez-Girona A., Noguchi E., Yates I. J., Russell P. Replication checkpoint kinase Cds1 regulates recombinational repair protein Rad60. Mol. Cell. Biol. 2003;23:5939–5946. doi: 10.1128/MCB.23.16.5939-5946.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D., Foiani M. Interplay of replication checkpoints and repair proteins at stalled replication forks. DNA Rep. 2007;6:994–1003. doi: 10.1016/j.dnarep.2007.02.018. [DOI] [PubMed] [Google Scholar]
- Branzei D., Foiani M. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- Branzei D., Sollier J., Liberi G., Zhao X., Maeda D., Seki M., Enomoto T., Ohta K., Foiani M. Ubc9- and mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell. 2006;127:509–522. doi: 10.1016/j.cell.2006.08.050. [DOI] [PubMed] [Google Scholar]
- Brewer B. J., Fangman W. L. The localization of replication origins on ARS plasmids in. S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- Cuperus G., Shore D. Restoration of silencing in Saccharomyces cerevisiae by tethering of a novel Sir2-interacting protein, Esc8. Genetics. 2002;162:633–645. doi: 10.1093/genetics/162.2.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon N., Kamakaka R. T. A histone variant, Htz1p, and a Sir1p-like protein, Esc2p, mediate silencing at HMR. Mol. Cell. 2000;6:769–780. doi: 10.1016/s1097-2765(00)00076-9. [DOI] [PubMed] [Google Scholar]
- Eladad S., Ye T. Z., Hu P., Leversha M., Beresten S., Matunis M. J., Ellis N. A. Intra-nuclear trafficking of the BLM helicase to DNA damage-induced foci is regulated by SUMO modification. Hum. Mol. Genet. 2005;14:1351–1365. doi: 10.1093/hmg/ddi145. [DOI] [PubMed] [Google Scholar]
- Ellis N. A., Groden J., Ye T. Z., Straughen J., Lennon D. J., Ciocci S., Proytcheva M., German J. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- Fabre F., Chan A., Heyer W. D., Gangloff S. Alternate pathways involving Sgs1/Top3, Mus81/ Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl. Acad. Sci. USA. 2002;99:16887–16892. doi: 10.1073/pnas.252652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei C., Gasser S. M. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev. 2000;14:81–96. [PMC free article] [PubMed] [Google Scholar]
- Gangloff S., McDonald J. P., Bendixen C., Arthur L., Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell. Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S., Soustelle C., Fabre F. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat. Genet. 2000;25:192–194. doi: 10.1038/76055. [DOI] [PubMed] [Google Scholar]
- German J. Bloom syndrome: a mendelian prototype of somatic mutational disease. Medicine. 1993;72:393–406. [PubMed] [Google Scholar]
- Hanada K., Hickson I.D. Molecular genetics of RecQ helicase disorders. Cell. Mol. Life Sci. 2007;64:2306–2322. doi: 10.1007/s00018-007-7121-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope J. C., Cruzata L. D., Duvshani A., Mitsumoto J., Maftahi M., Freyer G. A. Mus81-Eme1-dependent and -independent crossovers form in mitotic cells during double-strand break repair in Schizosaccharomyces pombe. Mol. Cell. Biol. 2007;27:3828–3838. doi: 10.1128/MCB.01596-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope J. C., Maftahi M., Freyer G. A. A postsynaptic role for Rhp55/57 that is responsible for cell death in Deltarqh1 mutants following replication arrest in Schizosaccharomyces pombe. Genetics. 2005;170:519–531. doi: 10.1534/genetics.104.037598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson F. B., Lombard D. B., Neff N. F., Mastrangelo M. A., Dewolf W., Ellis N. A., Marciniak R. A., Yin Y., Jaenisch R., Guarente L. Association of the Bloom syndrome protein with topoisomerase IIIalpha in somatic and meiotic cells. Cancer Res. 2000;60:1162–1167. [PubMed] [Google Scholar]
- Kanellis P., Gagliardi M., Banath J. P., Szilard R. K., Nakada S., Galicia S., Sweeney F. D., Cabelof D. C., Olive P. L., Durocher D. A screen for suppressors of gross chromosomal rearrangements identifies a conserved role for PLP in preventing DNA lesions. PLoS Genet. 2007;3:e134. doi: 10.1371/journal.pgen.0030134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karow J. K., Constantinou A., Li J. L., West S. C., Hickson I. D. The Bloom's syndrome gene product promotes branch migration of holliday junctions. Proc. Natl. Acad. Sci. USA. 2000;97:6504–6508. doi: 10.1073/pnas.100448097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh B. O., Symington L. S. Recombination proteins in yeast. Annu. Rev. Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- Laursen L. V., Ampatzidou E., Andersen A. H., Murray J. M. Role for the fission yeast RecQ helicase in DNA repair in G2. Mol. Cell. Biol. 2003;23:3692–3705. doi: 10.1128/MCB.23.10.3692-3705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberi G., Maffioletti G., Lucca C., Chiolo I., Baryshnikova A., Cotta-Ramusino C., Lopes M., Pellicioli A., Haber J. E., Foiani M. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev. 2005;19:339–350. doi: 10.1101/gad.322605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M., Cotta-Ramusino C., Liberi G., Foiani M. Branch migrating sister chromatid junctions form at replication origins through Rad51/Rad52-independent mechanisms. Mol. Cell. 2003;12:1499–1510. doi: 10.1016/s1097-2765(03)00473-8. [DOI] [PubMed] [Google Scholar]
- Maftahi M., Hope J. C., Delgado-Cruzata L., Han C. S., Freyer G. A. The severe slow growth of Deltasrs2 Deltarqh1 in Schizosaccharomyces pombe is suppressed by loss of recombination and checkpoint genes. Nucleic Acids Res. 2002;30:4781–4792. doi: 10.1093/nar/gkf581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankouri H. W., Hickson I. D. Top3 processes recombination intermediates and modulates checkpoint activity after DNA damage. Mol. Biol. Cell. 2006;17:4473–4483. doi: 10.1091/mbc.E06-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankouri H. W., Hickson I. D. The RecQ helicase-topoisomerase III-Rmi1 complex: a DNA structure-specific ‘dissolvasome’? Trends Biochem. Sci. 2007;32:538–546. doi: 10.1016/j.tibs.2007.09.009. [DOI] [PubMed] [Google Scholar]
- Mankouri H. W., Ngo H. P., Hickson I. D. Shu proteins promote the formation of homologous recombination intermediates that are processed by Sgs1-Rmi1-Top3. Mol. Biol. Cell. 2007;18:4062–4073. doi: 10.1091/mbc.E07-05-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meetei A. R., et al. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat. Genet. 2005;37:958–963. doi: 10.1038/ng1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyabe I., Morishita T., Hishida T., Yonei S., Shinagawa H. Rhp51-dependent recombination intermediates that do not generate checkpoint signal are accumulated in Schizosaccharomyces pombe rad60 and smc5/6 mutants after release from replication arrest. Mol. Cell. Biol. 2006;26:343–353. doi: 10.1128/MCB.26.1.343-353.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita T., Tsutsui Y., Iwasaki H., Shinagawa H. The Schizosaccharomyces pombe rad60 gene is essential for repairing double-strand DNA breaks spontaneously occurring during replication and induced by DNA-damaging agents. Mol. Cell. Biol. 2002;22:3537–3548. doi: 10.1128/MCB.22.10.3537-3548.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen J. R., Kaliraman V., Ibrahim S. S., Brill S. J. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2001;157:103–118. doi: 10.1093/genetics/157.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J. M., Lindsay H. D., Munday C. A., Carr A. M. Role of Schizosaccharomyces pombe RecQ homolog, recombination, and checkpoint genes in UV damage tolerance. Mol. Cell. Biol. 1997;17:6868–6875. doi: 10.1128/mcb.17.12.6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K., Kolodner R. D. Suppression of genome instability by redundant S-phase checkpoint pathways in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2002;99:4500–4507. doi: 10.1073/pnas.062702199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novatchkova M., Bachmair A., Eisenhaber B., Eisenhaber F. Proteins with two SUMO-like domains in chromatin-associated complexes: the RENi (Rad60-Esc2-NIP45) family. BMC Bioinformatics. 2005;6:22. doi: 10.1186/1471-2105-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F., Haber J. E. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash R., Krejci L., Van Komen S., Schurer K.A., Kramer W., Sung P. Saccharomyces cerevisiae MPH1 gene, required for homologous recombination-mediated mutation avoidance, encodes a 3′ to 5′ DNA helicase. J. Biol. Chem. 2005;280:7854–7860. doi: 10.1074/jbc.M413898200. [DOI] [PubMed] [Google Scholar]
- Raynard S., Bussen W., Sung P. A double Holliday junction dissolvasome comprising BLM, topoisomerase IIIalpha, and BLAP75. J. Biol. Chem. 2006;281:13861–13864. doi: 10.1074/jbc.C600051200. [DOI] [PubMed] [Google Scholar]
- Robert T., Dervins D., Fabre F., Gangloff S. Mrc1 and Srs2 are major actors in the regulation of spontaneous crossover. EMBO J. 2006;25:2837–2846. doi: 10.1038/sj.emboj.7601158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurer K. A., Rudolph C., Ulrich H. D., Kramer W. Yeast MPH1 gene functions in an error-free DNA damage bypass pathway that requires genes from Homologous recombination, but not from postreplicative repair. Genetics. 2004;166:1673–1686. doi: 10.1534/genetics.166.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shor E., Weinstein J., Rothstein R. A genetic screen for top3 suppressors in Saccharomyces cerevisiae identifies SHU1, SHU2, PSY3 and CSM 2, four genes involved in error-free DNA repair. Genetics. 2005;169:1275–1289. doi: 10.1534/genetics.104.036764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollier J., Driscoll R., Castellucci F., Foiani M., Jackson S. P., Branzei D. The Saccharomyces cerevisiae Esc2 and Smc5-6 proteins promote sister chromatid junction–mediated intra-S repair. Mol. Biol. Cell. 2009;20:1671–1682. doi: 10.1091/mbc.E08-08-0875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spell R. M., Jinks-Robertson S. Determination of mitotic recombination rates by fluctuation analysis in Saccharomyces cerevisiae. Methods Mol. Biol. 2004;262:3–12. doi: 10.1385/1-59259-761-0:003. [DOI] [PubMed] [Google Scholar]
- St. Onge R. P., Mani R., Oh J., Proctor M., Fung E., Davis R. W., Nislow C., Roth F. P., Giaever G. Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nat. Genet. 2007;39:199–206. doi: 10.1038/ng1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart E., Chapman C. R., Al-Khodairy F., Carr A. M., Enoch T. rqh1+, a fission yeast gene related to the Bloom's and Werner's syndrome genes, is required for reversible S phase arrest. EMBO J. 1997;16:2682–2692. doi: 10.1093/emboj/16.10.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P., Krejci L., Van Komen S., Sehorn M. G. Rad51 recombinase and recombination mediators. J. Biol. Chem. 2003;278:42729–42732. doi: 10.1074/jbc.R300027200. [DOI] [PubMed] [Google Scholar]
- Tong A. H., et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- van Brabant A. J., Ye T., Sanz M., German I. J., Ellis N. A., Holloman W. K. Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry. 2000;39:14617–14625. doi: 10.1021/bi0018640. [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat A., Pohlmann R., Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Wang S. W., Goodwin A., Hickson I. D., Norbury C. J. Involvement of Schizosaccharomyces pombe Srs2 in cellular responses to DNA damage. Nucleic Acids Res. 2001;29:2963–2972. doi: 10.1093/nar/29.14.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat. Rev. Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- Watt P. M., Hickson I. D., Borts R. H., Louis E. J. SGS1, a homologue of the Bloom's and Werner's syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics. 1996;144:935–945. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S. C. Molecular views of recombination proteins and their control. Nat. Rev. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- Wu L., Bachrati C. Z., Ou J., Xu C., Yin J., Chang M., Wang W., Li L., Brown G. W., Hickson I. D. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc. Natl. Acad. Sci. USA. 2006;103:4068–4073. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Davies S. L., North P. S., Goulaouic H., Riou J. F., Turley H., Gatter K. C., Hickson I. D. The Bloom's syndrome gene product interacts with topoisomerase III. J. Biol. Chem. 2000;275:9636–9644. doi: 10.1074/jbc.275.13.9636. [DOI] [PubMed] [Google Scholar]
- Wu L., Hickson I. D. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- Yamagata K., Kato J., Shimamoto A., Goto M., Furuichi Y., Ikeda H. Bloom's and Werner's syndrome genes suppress hyperrecombination in yeast sgs1 mutant: implication for genomic instability in human diseases. Proc. Natl. Acad. Sci. USA. 1998;95:8733–8738. doi: 10.1073/pnas.95.15.8733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J., Sobeck A., Xu C., Meetei A. R., Hoatlin M., Li L., Wang W. BLAP75, an essential component of Bloom's syndrome protein complexes that maintain genome integrity. EMBO J. 2005;24:1465–1476. doi: 10.1038/sj.emboj.7600622. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.