Abstract
As a consequence of recent developments in molecular phylogenomics, all extant orders of placental mammals have been grouped into 3 lineages: Afrotheria, Xenarthra, and Boreotheria, which originated in Africa, South America, and Laurasia, respectively. Despite this advancement, the order of divergence of these 3 lineages remains unresolved. Here, we performed extensive retroposon analysis with mammalian genomic data. Surprisingly, we identified a similar number of informative retroposon loci that support each of 3 possible phylogenetic hypotheses: the basal position for Afrotheria (22 loci), Xenarthra (25 loci), and Boreotheria (21 loci). This result indicates that the divergence of the placental common ancestor into the 3 lineages occurred nearly simultaneously. Thus, we examined whether these molecular data could be integrated into the geological context by incorporating recent geological data. We obtained firm evidence that complete separation of Gondwana into Africa and South America occurred 120 ± 10 Ma. Accordingly, the previous reported time frame (division of Pangea into Gondwana and Laurasia at 148–138 Ma and division of Gondwana at 105 Ma) cannot be used to validate mammalian divergence order. Instead, we use our retroposon results and the recent geological data to propose that near-simultaneous divisions of continents leading to isolated Africa, South America, and Laurasia caused nearly concomitant divergence of the ancient placental ancestor into 3 lineages, Afrotheria, Xenarthra, and Boreotheria, ≈120 Ma.
Keywords: continental divisions, incomplete lineage sorting, long interspersed element 1, mammalian phylogeny, paleobiogeography
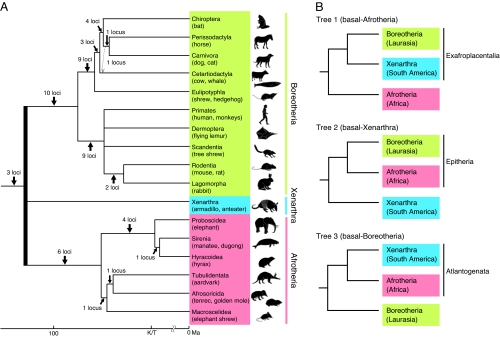
Recent phylogenetic analysis using dozens of gene sequences and other genomic markers such as insertions/deletions (indels) and retroposon insertions have largely revealed the interordinal relationships among placental mammals (eutherians) (1–7). These studies agree that 18 orders of placental mammals can be grouped into 3 major lineages, Afrotheria, Xenarthra, and Boreotheria (or Boreoeutheria; Fig. 1A). Afrotheria comprises 6 orders that include elephants, manatees, and aardvarks, and it is considered to have originated in Africa. Xenarthra includes armadillos, sloths, and anteaters and originated in South America. Boreotheria comprises 2 major mammalian groups, Laurasiatheria (e.g., carnivorans and bats) and Euarchontoglires (or Supraprimates; e.g., primates and rodents) and is considered to have originated in Laurasia of the Northern Hemisphere. Despite large-scale molecular phylogenetic analyses, the relationship among these 3 groups, which will finally establish the root of the placental mammalian tree, remains unresolved (4, 8). Three phylogenetic trees have been proposed as follows: tree 1, basal-Afrotheria (Boreotheria + Xenarthra; Exafroplacentalia); tree 2, basal-Xenarthra (Boreotheria + Afrotheria; Epitheria); and tree 3, basal-Boreotheria (Afrotheria + Xenarthra; Atlantogenata) (Fig. 1B).
Fig. 1.
Placental mammalian tree and 3 phylogenetic hypotheses of major lineages. (A) Mammalian interordinal phylogeny previously analyzed by retroposon insertion analysis (6) with the time scale (53). The bold vertical line denotes the root of placental mammals that was addressed in this study. (B) Three phylogenetic hypotheses to root the eutherian tree: tree 1, Exafroplacentalia (Afrotheria basal); tree 2, Epitheria (Xenarthra basal); and tree 3, Atlantogenata (Boreotheria basal).
Possible association between continental fission/fusion events and divergence of mammals was first proposed by Hedges et al. (9). They estimated molecular divergence times among many mammalian orders to be ≈100 Ma, which is inconsistent with fossil records suggesting that most mammals had diverged after 65 Ma. One of their interpretations was that the early divergence of mammals occurred because of several continental fissions. This hypothesis prompted many biologists to consider available geological data when estimating lineage divergence. One of the outcomes of this idea is Afrotheria, which is a good representative case for the association between continental drift and mammalian evolution (10, 11). Springer et al. (10) provided the first evidence that African-endemic mammals are monophyletic, and this hypothesis has been supported by subsequent molecular phylogenetic studies (1–3, 12) and analysis of retroposon insertions among orders (13, 14). Afrotherian mammals are considered to have evolved in Africa after it became isolated from other landmasses for several tens of million years (10, 11).
Based on the fact that placental mammal groups of Afrotheria, Xenarthra, and Boreotheria originated in Africa, South America, and Laurasia, respectively, the divergence of these mammals has been proposed to be linked to the separation of continents and subsequent land mass drift (3, 15–17). Paleogeological estimates reported by Smith et al. (18) indicate that Laurasia (where Boreotheria originated) separated first from Pangaea at 148–138 Ma and that the supercontinent Gondwana in the Southern Hemisphere divided into Africa and South America at 105 Ma. Indeed, molecular analyses have estimated the time of divergence among the 3 mammalian lineages to be ≈120–95 Ma (17, 19), approximately coincident with the geological estimation. If the phylogeny of these 3 mammalian ancestral lineages accurately reflects the order of continental divisions as proposed by Smith et al. (18), then tree 3 (basal-Boreotheria) should be validated by phylogenetic studies. To address this issue, many researchers have evaluated the 3 phylogenetic hypotheses (i.e., trees 1–3) (Table 1). Previous molecular studies using a few dozen gene sequences favored tree 1 (basal-Afrotheria), but the bootstrap probability was low (1–3, 8, 20–22). Molecular analyses using genomic data (phylogenomics) have recently been developed, and one such analysis using a several-hundred-kbp phylogenomic dataset favored tree 1 (23). However, more extensive phylogenomic analyses using mega-base sequence datasets strongly supported tree 3 (basal-Boreotheria) (16, 24), although it is possible that these results are misleading because of model misspecification (25).
Table 1.
Previous studies supporting trees 1–3
| Hypotheses | Dataset, sequence length (ref.) |
|---|---|
| Tree 1 | Nuclear + mitochondrial genes, 16.4 kb (3) |
| Nuclear + mitochondrial genes, 17.7 kb (21) | |
| Three nuclear genes, 5 kb (20) | |
| Mitochondrial genes, 3,516 residues (55) | |
| Nuclear + mitochondrial genes (22) | |
| Nuclear genes, 205 kb (23) | |
| Tree 2 | Nuclear + mitochondrial genes, 7,999 residues (55) |
| Two retroposon loci (5) | |
| Tree 3 | Three nuclear genes (55, 56) |
| Two retroposon loci (17) | |
| 1,700 nuclear genes, 1.4 Mb (16) | |
| 2,840 nuclear genes, 723,000 residues (24) | |
| Nuclear genes + conserved sequences, 1.9 Mb (57) |
Rare genomic changes such as retroposon insertions and indels are quite useful phylogenetic markers. The nearly homoplasy-free character of retroposons has been established (26, 27), and application of retroposons to reveal true phylogeny already has a credible scientific history (5, 6, 14, 17, 28–35). In addition, duplication of several nucleotides [target-site duplications (TSDs)], which often occurs upon retrotransposition (36), can also be used as a hallmark of homoplasy-free insertion of retroposons. Interestingly, retroposon analysis can also be applied to cases in which multiple species divergences occurred in a short evolutionary time. In these cases, ancestral polymorphisms based on the presence or absence of a retroposon should be retained after species divergence, and each retroposon would be randomly fixed or lost in a diverged lineage, resulting in distinct retroposon insertion patterns (6, 33, 37–39). Identification of such inconsistent insertions in a certain species group leads to the conclusion that the lineages diverged in a very short evolutionary time (38).
Kriegs et al. (5) reported 2 retroposon insertion loci that support tree 2 (basal-Xenarthra). Their data, however, were somewhat ambiguous because of ragged end points of the aligned sequences as described by Murphy et al. (17). Murphy et al. reported 2 unambiguous retroposon loci and 4 indels in support of tree 3 (basal-Boreotheria).
Overall, recent phylogenomic analyses of both mega-base gene sequence data and rare genomic changes have provided evidence that Boreotheria first diverged from the common ancestor of placental mammals and that the other placentals (Atlantogenata) diverged into Xenarthra in South America and Afrotheria in Africa along with the division of Gondwana (tree 3) (Table 1). This scenario is apparently consistent with the geological data by Smith et al. (18). In the present study, however, we present a different scenario from those proposed previously. We performed an extensive retroposon analysis using genomic data and provide strong evidence that the 3 lineages diverged nearly simultaneously. Our molecular data prompted us to re-examine recent geological data, incorporation of which suggests that the Africa–South America separation occurred at ≈120 Ma. This estimated time frame is much earlier than that (i.e., 105 Ma) proposed by Smith et al. (18). We also discuss caveats for applying Smith et al.'s geological data to biogeographic questions.
Results and Discussion
Isolation of Multiple Long Interspersed Element 1 (L1) Loci Supporting Each of the 3 Phylogenetic Hypotheses.
In this study, genomic data of human, armadillo, and elephant were used as representatives of Boreotheria, Xenarthra, and Afrotheria, respectively. There are 83,331, 178,357, and 135,739 L1MB elements in the genomes of human, armadillo, and elephant, respectively. Among them, 19,783 loci were compared among the 3 species by using the method shown in Fig. S1, revealing 1,382 possible informative loci (i.e., loci where L1MB was present in any 2 of the 3 species). After removing false-positive loci by referring to alignments available in the University of California Santa Cruz Genome Browser, 68 phylogenetically-informative loci were aligned, including the additional sequences of other placental mammals such as chimpanzee, rhesus macaque, mouse, rat, rabbit, dog, cow, and tenrec (Fig. 2 and Fig. S2). Among them, trees 1, 2, and 3 were supported by 22, 25, and 21 loci, respectively. Using our strategy, we successfully identified the 4 loci reported by Kriegs et al. (5) and Murphy et al. (17) (Table 2), an indication of the comprehensiveness of the strategy. Our data include some cases of ambiguous L1 insertions in which no TSD was observed. However, even considering only bona fide L1 insertions having a TSD in the flanking regions, we still found an obvious inconsistency involving 16, 21, and 16 unambiguous L1 insertions that support trees 1, 2, and 3, respectively (Table 2).
Fig. 2.
Three examples of L1 insertion loci supporting tree 1 (A), tree 2 (B), and tree 3 (C). Black and gray boxes denote the L1 and the TSD sequences, respectively. The latter were generated during L1 integration and indicate bona fide retrotransposition. The central region of the inserted L1 sequence has been omitted.
Table 2.
Informative L1MB loci supporting trees 1–3
| Tree 1 |
Tree 2 |
Tree 3 |
||||||
|---|---|---|---|---|---|---|---|---|
| Locus | L1 subfamily | TSD, nt | Locus | L1 subfamily | TSD, nt | Locus | L1 subfamily | TSD, nt |
| HDL1007 | L1MB5 | - | HDL2003 | L1MB4 | 4 | HDL3016† | L1MB5 | 19 |
| HDL1040 | L1MB5 | - | HDL2090 | L1MB5 | 10 | HDL3051 | L1MB7 | 7 |
| HDL1061 | L1MB5 | - | HDL2102 | L1MB4 | 13 | HDL3074 | L1MB7 | - |
| HDL1081 | L1MB2 | 8 | HDL2121 | L1MB5 | 6 | HDL3078 | L1MB5 | 15 |
| HDL1119 | L1MB8 | 7 | HDL2203 | L1MB5 | 4 | HDL3089 | L1MB8 | 13 |
| HDL1122 | L1MB5 | 7 | HDL2223 | L1MB8 | 9 | HDL3101 | L1MB5 | 13 |
| HDL1125 | L1MB7 | 12 | HDL2237 | L1MB5 | 14 | HDL3133 | L1MB7 | - |
| HDL1136 | L1MB2 | 15 | HDL2242 | L1MB5 | 11 | HDL3138 | L1MB5 | 5 |
| HDL1141 | L1MB7 | - | HDL2279 | L1MB8 | 15 | HDL3146 | L1MB5 | 10 |
| HDL1144 | L1MB8 | 18 | HDL2307 | L1MB5 | - | HDL3161 | L1MB5 | 6 |
| HDL1171 | L1MB5 | 5 | HDL2309 | L1MB5 | 10 | HDL3214 | L1MB5 | - |
| HDL1200 | L1MB5 | 9 | HDL2333 | L1MB5 | 16 | HDL3225 | L1MB8 | 14 |
| HDL1208 | L1MB5 | 14 | HDL2340 | L1MB5 | 10 | HDL3266† | L1MB5 | 15 |
| HDL1233 | L1MB7 | - | HDL2345 | L1MB7 | - | HDL3283 | L1MB5 | 8 |
| HDL1256 | L1MB4 | 14 | HDL2368 | L1MB5 | 15 | HDL3295 | L1MB5 | 6 |
| HDL1262 | L1MB4 | 14 | HDL2370 | L1MB4 | 8 | HDL3314 | L1MB5 | - |
| HDL1276 | L1MB5 | 16 | HDL2380 | L1MB5 | 9 | HDL3324 | L1MB4 | - |
| HDL1287 | L1MB8 | 11 | HDL2387 | L1MB5 | 13 | HDL3347 | L1MB4 | 7 |
| HDL1337 | L1MB5 | 7 | HDL2433* | L1MB5 | 6 | HDL3355 | L1MB8 | 6 |
| HDL1360 | L1MB5 | - | HDL2443 | L1MB5 | 15 | HDL3366 | L1MB5 | 7 |
| HDL1372 | L1MB5 | 8 | HDL2446 | L1MB7 | 10 | HDL3369 | L1MB5 | 10 |
| HDL1373 | L1MB5 | 14 | HDL2457 | L1MB4 | 8 | |||
| HDL2483 | L1MB8 | 6 | ||||||
| HDL2499* | L1MB5 | - | ||||||
| HDL2548 | L1MB8 | - | ||||||
Near-Simultaneous Divergence of a Common Ancestor of Placental Mammals into 3 Lineages.
As briefly described in the Introduction, inconsistency of retroposon insertions will be observed if species had diverged over a short evolutionary time span in which ancestral polymorphisms were followed by incomplete lineage sorting in a diverged lineage (38). Our group has already reported 3 cases, namely charr (40), cichlids (37, 41), and baleen whales (39), in which the order of species divergence could not be deduced because of extensive retroposon insertion inconsistency. In the present case, it is remarkable that such a large number of inconsistent loci could be characterized (Table 2). The fact that our L1 insertion data evenly support 3 different trees provides convincing evidence that 3 ancient lineages of placental mammals, namely Boreotheria, Xenarthra, and Afrotheria, diverged nearly simultaneously from a common ancestor.
Analysis of gene sequences alone generally is insufficient to provide convincing evidence that multiple divergences occurred nearly simultaneously. Even if the same analysis of 2 different genes in several lineages yields different trees, this is not necessarily an indication that they diverged nearly simultaneously because the result might be misleading owing to unusual gene characteristics or systematic errors such as long branch attraction, compositional bias, or heterotachy. Therefore, a comprehensive survey of retroposon loci from genomewide sequence data yields very useful markers for determining whether several ancient lineages diverged nearly simultaneously, as is demonstrated by our current results.
In general, acquisition of additional sequence data may allow resolution of a confounding phylogenetic relationship. As such, it could be argued that additional sequence data from the armadillo and elephant genomes could resolve the relationship among the 3 mammalian groups. However, a nearly equivalent number of L1 loci for the 3 trees was obtained from our extensive screen of all L1 loci from the complete human genome and the 2× genomic sequence coverage of armadillo and elephant data, corresponding to ≈70% of their whole genomes. Thus, given the data presented in Table 2 we consider that, even if additional sequence data were obtained once the armadillo and elephant genome sequences are completed, the number of L1 loci for the 3 trees still would not differ significantly. In any cases, the conclusion of nearly simultaneous divergence of 3 early mammalian lineages would not change.
Geological Evidence Supporting Multiple Continental Divisions, Occurring Over a Short Period ≈120 Ma, that Hindered Dispersion of Terrestrial Mammals.
As described in the Introduction, divergences and dispersal of mammals has been linked to continental separation and subsequent drift, as has often been suggested by evolutionary biologists (9–11). Also, in the case of the 3 ancient mammalian lineages, based on the origins of Boreotheria in Laurasia, Xenarthra in South America, and Afrotheria in Africa, the association between mammalian divergence and continental divisions has been discussed by many researchers (3, 15–17, 42), with many referring to the geological data collected by Smith et al. (18) who proposed division of Pangea into Laurasia and Gondwana at 148–138 Ma and division of Gondwana into Africa and South America at 105 Ma. Although the conclusions of Smith et al. are meaningful from a geological viewpoint, we propose that they may not be directly applicable to biogeographic questions, as discussed below.
First, we examined the physical connection between Africa and South America. The data by Smith et al. (18) were based on the spatiotemporal distribution of magnetic stripes on the Atlantic Ocean floor, which were identified by measuring magnetic patterns of crust at the deep sea bottom from a ship. Magnetic stripes are generated by the normal polarity reversals of the Earth's magnetism over time. Accordingly, these data indicate the appearance at 105 Ma of a new oceanic crust with a particular magnetic stripe, the depth of which is always ≈2,700 m above the active midoceanic ridge. Taking into account the time required to create an ocean of 2,700-m depth from a continental division, the geographical separation between Africa and South America that hindered terrestrial dispersion of mammals would have occurred considerably earlier than 105 Ma. Curiously, this time lag has never been discussed by any molecular evolutionist who cited Smith et al.'s work. In addition, the geomagnetic time scale that Smith et al. used was revised recently. Further, magnetic stripes must be at least tens or hundreds of kilometers wide to be measured accurately, and thus the resolution of time by this method is limited.
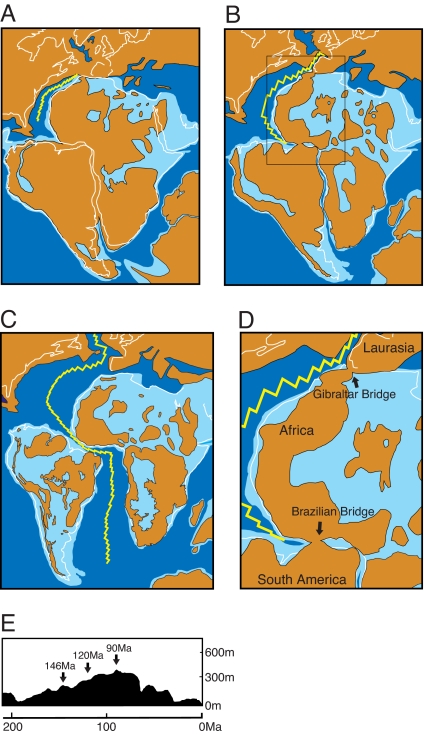
However, paleogeographic maps of Africa have recently been published by Guiraud and Maulin (43) and Guiraud et al. (44) based on extensive compilation of drilling results of Mesozoic shallow marine sediments in Africa. These studies show the spatiotemporal distribution of the shallow marine ocean, which progressively extended landward because of the global sea-level rise (transgression) of ≈300 m from the Jurassic to the Cretaceous. Using on-land drilling data by Milani and Thomaz Filho (45) and paleogeography by Guiraud et al. (44) and de Wit (46), we reconstructed the paleogeography of the southern half of Pangea (Fig. 3). Because the North and South American continents were not connected above sea level from the Jurassic until the Miocene (20 Ma) (47), we focused on the relationships between Africa and South America and between Africa and Laurasia. At 146 Ma (Fig. 3A), the Central Atlantic Ocean was born by the eastward drift of Africa. Ridge propagation to open the South Atlantic Ocean proceeded not only southward from the Central Atlantic Ocean but also from south to north. The final connection point between South America and Africa, designated here as the Brazilian Bridge, remained at 120 Ma (Fig. 3 B and D). At 90 Ma (Fig. 3C), the opening of the Central and South Atlantic Oceans was completed. We used the revised geomagnetic time scale (47, 48) to update the paleogeographic map. In generating these maps, we considered changes in sea level (Fig. 3E). As shown in Fig. 3, from 146 Ma to 90 Ma, the area of the landmass (brown regions in Fig. 3 A–C) gradually decreased because of an increase in the sea level (compare the light-blue regions, representing shallow oceans, in Fig. 3 A–C), resulting in fragmentation of the continents. Thus, according to the updated geological estimation, we propose that the Brazilian Bridge was the last connection point between Africa and South America and that 120 Ma (or slightly later) represents the most likely period when west and east Gondwana became separated. Indeed, unconformity in the basins of eastern South America ≈115–120 Ma (figures 10 and 15 in ref. 45) suggests that the connection was finally broken around that time.
Fig. 3.
A revised paleogeographic scenario based on recent geological data. (A–C) Paleogeographic reconstruction of Pangea at 146 Ma (A), 120 Ma (B), and 90 Ma (C) (modified after refs. 43–48 and 52). (D) A magnified map at 120 Ma (boxed area in B) is shown emphasizing 2 bridges that connect 3 continents. Blue and light blue represent deep and shallow ocean depths, respectively. Brown areas and the yellow jagged line represent landmasses and the mid-Atlantic ridge, respectively. Only deep ocean is represented for Laurasia because drilling data are currently unavailable. (E) Sea-level rise (54) to yield subsequent fragmentation of Africa and South America was taken into consideration for the reconstruction.
Interestingly, based on the presence of fossils of ammonites and other invertebrates from the north extending their range to the south through an open corridor, Jacobs et al. (49) proposed that the Atlantic Ocean was completed by the opening of the Equatorial Atlantic Gateway, which occurred in the Early Cretaceous (≈115 Ma). Therefore, terrestrial dispersion of an ancestral mammal between Africa and South America might have been blocked just before that time. Accordingly, our estimation of 120 Ma for that time is consistent with the estimate of Jacobs et al.
Regarding the relationship between Laurasia (Europe) and Africa, although conclusive geological evidence such as drilling data of the western Mediterranean Sea is not yet available, the geographic position of the west African promontory (Morocco) was close to the Iberian peninsula at 120 Ma, suggesting the possibility of a connection (i.e., Gibraltar Bridge) between Africa and Laurasia (see SI Text). As shown in Fig. 3D, landmasses (brown regions) of 3 continents (Laurasia, Africa, and South America) all can be connected through the Gibraltar and Brazilian Bridges. Traditionally, as shown in Table 3, the timing of the final separation of Africa from Europe has been variously defined by researchers as ranging from 148 Ma (18) to 110 Ma (50). Therefore, based on the updated drilling data supporting the final separation of Africa and South America ≈120 Ma (45), the possibility remains that the near-simultaneous separations of Laurasia, South America, and Africa might have occurred at ≈120 Ma. This geological estimation is not inconsistent with our present conclusion based on retroposon evidence.
Table 3.
Estimated ages (in Ma) of the divisions of Africa–Laurasia and Africa–South America as determined in several studies
Conclusion
Contrary to recent phylogenomic results supporting tree 3 (16, 17, 24), an extensive genome-scale retroposon analysis in the present study revealed that the common ancestor of placental mammals diverged into Boreotheria, Xenarthra, and Afrotheria nearly simultaneously. From a geological viewpoint, Smith et al. (18) proposed that Laurasia was first separated at 148–138 Ma, followed by division of Gondwana into South America and Africa at 105 Ma, the time difference of which has been used by evolutionary biologists as a rationale to support a biogeographic scenario that Boreotheria diverged first. Here, we show that the data of Smith et al. (18) cannot be directly applied to biogeographic questions, especially with regard to the terrestrial dispersion of mammals. By re-examining the geological data, including newly-accumulated drilling data, and considering the sea-level change during the Cretaceous, we provide evidence that the last connection between Africa and South America was through the Brazilian Bridge at 120 Ma, which is considerably earlier than the 105 Ma (18) that has been referred to by evolutionary biologists (3, 15, 17, 42). We note the possibility that Laurasia and Africa were connected by the Gibraltar Bridge until 120 Ma, although definitive evidence to confirm this hypothesis is currently lacking. Finally, by combining the updated geological data with our retroposon evidence, we provide a scenario in which the divergence of the eutherian ancestor into Boreotheria, Xenarthra, and Afrotheria occurred nearly simultaneously at 120 Ma, possibly with almost concomitant cut-off of 2 bridges that had connected the 3 ancient continents, Laurasia, South America, and Africa (Fig. S3).
Materials and Methods
Genomic sequences of human (hg18), 9-banded armadillo (Dasypus novemcinctus) (dasNov1; 2× genome coverage), and African elephant (Loxodonta africana) (loxAfr1; 2× genome coverage) were obtained from the University of California Santa Cruz Genome Bioinformatics database (http://genome.ucsc.edu). We compared patterns based on the presence or absence of L1, a mammalian retroposon. Among >50 L1 subfamilies in mammalian genomes (51), L1 elements belonging to the L1MB subgroup are considered to be the most appropriate subfamily for analyses of mammalian interordinal relationships immediately before and after the divergence of the 3 major lineages of mammalian ancestors, the availability of which was shown in our previous study (6). In addition, the 4 L1 elements reported by Kriegs et al. (5) and Murphy et al. (17) also belong to the L1MB subgroup. Therefore, we used only elements of the L1MB subgroup as phylogenetic markers. The genomic data for human, armadillo, and African elephant were screened for L1MB elements by using the RepeatMasker program (www.repeatmasker.org). From the 17-way genomic sequence alignment in the University of California Santa Cruz database, human–armadillo and human–elephant alignments were constructed and used to compare the orthologs of each L1MB locus. As shown in Fig. S1, 4 sites located 50, 100, 200, and 500 nt upstream and downstream of the L1MB sequence were set as queries for ortholog searches with the D. novemcinctus and L. Africana genomes. Orthologs of the 8 sites were sought in the 2 species by using the genomic alignment data. If at least 1 orthologous site was found on both sides of the L1 in the other 2 species (armadillo and elephant; Fig. S1), the presence or absence of a L1MB element between the 2 nearest orthologous sites was verified by referring to the RepeatMasker results. Among the loci compared, we used only those that were potentially phylogenetically informative (i.e., L1MB was present in 2 of the species and absent in the other species). Next, we checked alignments of these loci in the University of California Santa Cruz Genome Browser to remove clear false positive and paralogous loci. Most of the false positive results were caused by independent insertions of L1 within the genomic region compared. For the remaining 68 loci, orthologous sequences of various mammals were collected and aligned by using the GENETYX version 5.2 program.
Supplementary Material
Acknowledgments.
This work was supported by research grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to N.O.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809297106/DCSupplemental.
References
- 1.Madsen O, et al. Parallel adaptive radiations in two major clades of placental mammals. Nature. 2001;409:610–614. doi: 10.1038/35054544. [DOI] [PubMed] [Google Scholar]
- 2.Murphy WJ, et al. Molecular phylogenetics and the origins of placental mammals. Nature. 2001;409:614–618. doi: 10.1038/35054550. [DOI] [PubMed] [Google Scholar]
- 3.Murphy WJ, et al. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science. 2001;294:2348–2351. doi: 10.1126/science.1067179. [DOI] [PubMed] [Google Scholar]
- 4.Springer MS, Stanhope MJ, Madsen O, de Jong WW. Molecules consolidate the placental mammal tree. Trends Ecol Evol. 2004;19:430–438. doi: 10.1016/j.tree.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 5.Kriegs JO, et al. Retroposed elements as archives for the evolutionary history of placental mammals. PLoS Biol. 2006;4:e91. doi: 10.1371/journal.pbio.0040091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishihara H, Hasegawa M, Okada N. Pegasoferae, an unexpected mammalian clade revealed by tracking ancient retroposon insertions. Proc Natl Acad Sci USA. 2006;103:9929–9934. doi: 10.1073/pnas.0603797103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janecka JE, et al. Molecular and genomic data identify the closest living relative of primates. Science. 2007;318:792–794. doi: 10.1126/science.1147555. [DOI] [PubMed] [Google Scholar]
- 8.Murphy WJ, Pevzner PA, O'Brien SJ. Mammalian phylogenomics comes of age. Trends Genet. 2004;20:631–639. doi: 10.1016/j.tig.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 9.Hedges SB, Parker PH, Sibley CG, Kumar S. Continental breakup and the ordinal diversification of birds and mammals. Nature. 1996;381:226–229. doi: 10.1038/381226a0. [DOI] [PubMed] [Google Scholar]
- 10.Springer MS, et al. Endemic African mammals shake the phylogenetic tree. Nature. 1997;388:61–64. doi: 10.1038/40386. [DOI] [PubMed] [Google Scholar]
- 11.Hedges SB. Afrotheria: Plate tectonics meets genomics. Proc Natl Acad Sci USA. 2001;98:1–2. doi: 10.1073/pnas.98.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stanhope MJ, et al. Molecular evidence for multiple origins of Insectivora and for a new order of endemic African insectivore mammals. Proc Natl Acad Sci USA. 1998;95:9967–9972. doi: 10.1073/pnas.95.17.9967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nikaido M, Nishihara H, Fukumoto Y, Okada N. Ancient SINEs from African endemic mammals. Mol Biol Evol. 2003;20:522–527. doi: 10.1093/molbev/msg052. [DOI] [PubMed] [Google Scholar]
- 14.Nishihara H, et al. A retroposon analysis of Afrotherian phylogeny. Mol Biol Evol. 2005;22:1823–1833. doi: 10.1093/molbev/msi179. [DOI] [PubMed] [Google Scholar]
- 15.Eizirik E, Murphy WJ, O'Brien SJ. Molecular dating and biogeography of the early placental mammal radiation. J Hered. 2001;92:212–219. doi: 10.1093/jhered/92.2.212. [DOI] [PubMed] [Google Scholar]
- 16.Wildman DE, et al. Genomics, biogeography, and the diversification of placental mammals. Proc Natl Acad Sci USA. 2007;104:14395–14400. doi: 10.1073/pnas.0704342104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy WJ, Pringle TH, Crider TA, Springer MS, Miller W. Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res. 2007;17:413–421. doi: 10.1101/gr.5918807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith AG, Smith DG, Funnell BM. Atlas of Cenozoic and Mesozoic Coastlines. Cambridge, UK: Cambridge Univ Press; 1994. [Google Scholar]
- 19.Springer MS, Murphy WJ, Eizirik E, O'Brien SJ. Placental mammal diversification and the Cretaceous–Tertiary boundary. Proc Natl Acad Sci USA. 2003;100:1056–1061. doi: 10.1073/pnas.0334222100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delsuc F, et al. Molecular phylogeny of living xenarthrans and the impact of character and taxon sampling on the placental tree rooting. Mol Biol Evol. 2002;19:1656–1671. doi: 10.1093/oxfordjournals.molbev.a003989. [DOI] [PubMed] [Google Scholar]
- 21.Amrine-Madsen H, Koepfli KP, Wayne RK, Springer MS. A new phylogenetic marker, apolipoprotein B, provides compelling evidence for eutherian relationships. Mol Phylogenet Evol. 2003;28:225–240. doi: 10.1016/s1055-7903(03)00118-0. [DOI] [PubMed] [Google Scholar]
- 22.Waddell PJ, Shelley S. Evaluating placental interordinal phylogenies with novel sequences including RAG1, gamma-fibrinogen, ND6, and mt-tRNA, plus MCMC-driven nucleotide, amino acid, and codon models. Mol Phylogenet Evol. 2003;28:197–224. doi: 10.1016/s1055-7903(03)00115-5. [DOI] [PubMed] [Google Scholar]
- 23.Nikolaev S, et al. Early history of mammals is elucidated with the ENCODE multiple species sequencing data. PLoS Genet. 2007;3:e2. doi: 10.1371/journal.pgen.0030002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hallstrom BM, Kullberg M, Nilsson MA, Janke A. Phylogenomic data analyses provide evidence that Xenarthra and Afrotheria are sister groups. Mol Biol Evol. 2007;24:2059–2068. doi: 10.1093/molbev/msm136. [DOI] [PubMed] [Google Scholar]
- 25.Nishihara H, Okada N, Hasegawa M. Rooting the eutherian tree: The power and pitfalls of phylogenomics. Genome Biol. 2007;8:R199. doi: 10.1186/gb-2007-8-9-r199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rokas A, Holland PW. Rare genomic changes as a tool for phylogenetics. Trends Ecol Evol. 2000;15:454–459. doi: 10.1016/s0169-5347(00)01967-4. [DOI] [PubMed] [Google Scholar]
- 27.Shedlock AM, Okada N. SINE insertions: Powerful tools for molecular systematics. BioEssays. 2000;22:148–160. doi: 10.1002/(SICI)1521-1878(200002)22:2<148::AID-BIES6>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 28.Murata S, Takasaki N, Saitoh M, Okada N. Determination of the phylogenetic relationships among Pacific salmonids by using short interspersed elements (SINEs) as temporal landmarks of evolution. Proc Natl Acad Sci USA. 1993;90:6995–6999. doi: 10.1073/pnas.90.15.6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimamura M, et al. Molecular evidence from retroposons that whales form a clade within even-toed ungulates. Nature. 1997;388:666–670. doi: 10.1038/41759. [DOI] [PubMed] [Google Scholar]
- 30.Nikaido M, Rooney AP, Okada N. Phylogenetic relationships among cetartiodactyls based on insertions of short and long interspersed elements: Hippopotamuses are the closest extant relatives of whales. Proc Natl Acad Sci USA. 1999;96:10261–10266. doi: 10.1073/pnas.96.18.10261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmitz J, Ohme M, Zischler H. SINE insertions in cladistic analyses and the phylogenetic affiliations of Tarsius bancanus to other primates. Genetics. 2001;157:777–784. doi: 10.1093/genetics/157.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nikaido M, et al. Retroposon analysis of major cetacean lineages: The monophyly of toothed whales and the paraphyly of river dolphins. Proc Natl Acad Sci USA. 2001;98:7384–7389. doi: 10.1073/pnas.121139198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salem AH, et al. Alu elements and hominid phylogenetics. Proc Natl Acad Sci USA. 2003;100:12787–12791. doi: 10.1073/pnas.2133766100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roos C, Schmitz J, Zischler H. Primate jumping genes elucidate strepsirrhine phylogeny. Proc Natl Acad Sci USA. 2004;101:10650–10654. doi: 10.1073/pnas.0403852101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kriegs JO, Churakov G, Jurka J, Brosius J, Schmitz J. Evolutionary history of 7SL RNA-derived SINEs in Supraprimates. Trends Genet. 2007;23:158–161. doi: 10.1016/j.tig.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 36.Kazazian HH., Jr Mobile elements: Drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 37.Takahashi K, Terai Y, Nishida M, Okada N. Phylogenetic relationships and ancient incomplete lineage sorting among cichlid fishes in Lake Tanganyika as revealed by analysis of the insertion of retroposons. Mol Biol Evol. 2001;18:2057–2066. doi: 10.1093/oxfordjournals.molbev.a003747. [DOI] [PubMed] [Google Scholar]
- 38.Shedlock AM, Takahashi K, Okada N. SINEs of speciation: Tracking lineages with retroposons. Trends Ecol Evol. 2004;19:545–553. doi: 10.1016/j.tree.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 39.Nikaido M, et al. Baleen whale phylogeny and a past extensive radiation event revealed by SINE insertion analysis. Mol Biol Evol. 2006;23:866–873. doi: 10.1093/molbev/msj071. [DOI] [PubMed] [Google Scholar]
- 40.Hamada M, et al. Detection of the ongoing sorting of ancestrally polymorphic SINEs toward fixation or loss in populations of two species of charr during speciation. Genetics. 1998;150:301–311. doi: 10.1093/genetics/150.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Terai Y, Takahashi K, Nishida M, Sato T, Okada N. Using SINEs to probe ancient explosive speciation: “Hidden” radiation of African cichlids? Mol Biol Evol. 2003;20:924–930. doi: 10.1093/molbev/msg104. [DOI] [PubMed] [Google Scholar]
- 42.Archibald JD. Timing and biogeography of the eutherian radiation: Fossils and molecules compared. Mol Phylogenet Evol. 2003;28:350–359. doi: 10.1016/s1055-7903(03)00034-4. [DOI] [PubMed] [Google Scholar]
- 43.Guiraud R, Maurin JC. Early Cretaceous rifts of Western and Central Africa: An overview. Tectonophysics. 1992;213:153–168. [Google Scholar]
- 44.Guiraud R, Bosworth W, Thierry J, Delplanque A. Phanerozoic geological evolution of northern and central Africa: An overview. J Afr Earth Sci. 2005;43:83–143. [Google Scholar]
- 45.Milani EJ, Thomaz Filho A. In: Tectonic Evolution of South America. Cordani UG, Milani EJ, Thomaz Filho A, Campos DA, editors. Rio de Janiero: International Geological Congress; 2000. pp. 389–449. [Google Scholar]
- 46.de Wit MJ. Madagascar: Heads it's a continent, tails it's an island. Annu Rev Earth Planet Sci. 2003;31:213–248. [Google Scholar]
- 47.Scotese CR. Atlas of Earth History 2001: Paleomap Progress Report 90-0497. University of Texas, Arlington: Department of Geology; 2001. [Google Scholar]
- 48.Collins WJ. Slab pull, mantle convection, and Pangaean assembly and dispersal. Earth Planet Sci Lett. 2003;205:225–237. [Google Scholar]
- 49.Jacobs LL, et al. Cretaceous paleogeography, paleoclimatology, and amniote biogeography of the low and mid-latitude South Atlantic ocean. Bull Geol Soc France. 2009 in press. [Google Scholar]
- 50.Frisch W. Tectonic progradation and plate tectonic evolution of the Alps. Tectonophysics. 1979;60:121–139. [Google Scholar]
- 51.Smit AF, Toth G, Riggs AD, Jurka J. Ancestral, mammalian-wide subfamilies of LINE-1 repetitive sequences. J Mol Biol. 1995;246:401–417. doi: 10.1006/jmbi.1994.0095. [DOI] [PubMed] [Google Scholar]
- 52.Owen HG. Atlas of Continental Displacement, 200 Million Years to the Present. New York: Cambridge Univ Press; 1983. [Google Scholar]
- 53.Hasegawa M, Thorne JL, Kishino H. Time scale of eutherian evolution estimated without assuming a constant rate of molecular evolution. Genes Genet Syst. 2003;78:267–283. doi: 10.1266/ggs.78.267. [DOI] [PubMed] [Google Scholar]
- 54.Morrow JR, Schindler E, Walliser OH. In: Global Events and Event Stratigraphy in the Phanerozoic. Walliser OH, editor. Berlin: Springer; 1995. pp. 53–61. [Google Scholar]
- 55.Waddell PJ, Kishino H, Ota R. A phylogenetic foundation for comparative mammalian genomics. Genome Inform Ser Workshop Genome Inform. 2001;12:141–154. [PubMed] [Google Scholar]
- 56.Huchon D, et al. Rodent phylogeny and a time scale for the evolution of Glires: Evidence from an extensive taxon sampling using three nuclear genes. Mol Biol Evol. 2002;19:1053–1065. doi: 10.1093/oxfordjournals.molbev.a004164. [DOI] [PubMed] [Google Scholar]
- 57.Prasad AB, Allard MW, Green ED. Confirming the phylogeny of mammals by use of large comparative sequence data sets. Mol Biol Evol. 2008;25:1795–1808. doi: 10.1093/molbev/msn104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.