Abstract
Mycobacterium tuberculosis (M. tb) is an intracellular pathogen possessing a complex mixture of cell wall lipids that are thought to modulate the activities of host macrophages. In this study, we employed two state-of-the-art quantitative proteomic approaches, metabolic labeling SILAC and chemical isobaric tagging iTRAQ, to study changes in macrophage protein expression in response to exposure to M. tb lipids. From a total of 1286 proteins identified, 463 were discovered by both isotope-labeling strategies at a high consistency, and the rest of proteins were detected by only one of the two approaches. Upon exposure to mycobacterial cell wall lipids, 166 macrophage proteins showed differential expression. These included proteins involved in the immune response, oxidation and reduction, and vesicle transport, as well as other cellular processes. The response of the macrophage proteome to M. tb lipids reflects the cell’s innate defense mechanisms as well as lipid-induced processes that may benefit the pathogen.
Keywords: Cell wall lipids, macrophage, iTRAQ, SILAC, differential expression
Introduction
Mycobacterium tuberculosis (M. tb), the causative agent of tuberculosis, is a major cause of mortality worldwide, killing two million people annually and establishing long term, persistent infections in approximately one-third of the world’s population.1 Most infected individuals are clinically asymptomatic for infection with M. tb and can harbor the bacteria for much of their lifetime, all the while at risk for conversion to an active disease state.2 In persistent, asymptomatic infections, M. tb exists intracellularly in host macrophages that are themselves encased within granulomas. M. tb is able to survive for an extended period within its host by modulating host bactericidal responses, including phagosome-lysosome fusion, apoptosis, and a robust pro-inflammatory immune response.3
M. tb possesses highly elaborated cell wall lipids that display potent biological activities and are considered central effectors in M. tb pathogenesis.4, 5 Importantly, it has been found that these cell wall lipids can be released into the host cell cytoplasm, where they accumulate in vesicular organelles such as lysosomes6 or exocytose and traffic into neighboring cells7. For example, lipoarabinomannan (ManLAM) is known to have a vast array of effector functions such as inhibition of macrophage activation, repression of cytokine production, regulation of apoptosis, and obstruction of phagosome maturation.8,9 Interestingly, M. tb also biosynthesizes another glycolipid, lipomannan (LAM), that counteracts some of the effects of ManLAM by stimulating a strong pro-inflammatory response in macrophages. To further our understanding of how M. tb modulates host responses, we investigated the global effect of constitutive mycobacterial lipids on macrophage protein expression patterns using state-of-the-art proteomics techniques.
Quantitative proteomics, employing stable-isotope labeling and high resolution mass spectrometry, has gained success in deciphering diverse biological processes due to its high level of coverage of the proteome, accuracy in quantification and high-throughput platforms.10–12 Metabolic labeling and chemical labeling are two major strategies used for relative quantification of protein abundance under different conditions. Stable-isotope labeling by amino acids in cell culture (SILAC) involves metabolic incorporation of isotope mass tags into proteins and has been widely applied to large-scale kinetic analysis of proteomes and post-translational modifications13–17. SILAC has been used to eliminate false-positives in protein interaction studies18–20 and to measure protein/peptide turnover via transient labeling21–23. Chemical labeling has also attracted widespread interest in proteomics community, the benchmark technique being ICAT.24–27 Recently, the technique termed “isobaric tag for relative and absolute quantitation” (iTRAQ) has emerged as a superior choice due to its high proteome coverage and labeling efficiency.28–35 However, SILAC and iTRAQ, the two most popular and promising techniques in quantitative proteomics have never been utilized in one system to compare their performance and verify quantification results. Therefore, in this study we employed both methods to study the macrophage proteome upon exposure to M. tb lipids.
Experimental Procedures
M. tb growth and lipid extraction
M. tb strain H37Rv was grown to stationary phase in liquid 7H9 medium (Difco) supplemented with 10% OADC, 0.5% glycerol, and 0.1% Tween80. Ten (10) mL of culture was pelleted by centrifugation (15,000 × g), washed with PBS and extracted with 2.5 mL of 2:1 chloroform:methanol by vigorous shaking for 2 h at rt. The organic phase was clarified by centrifugation and dried under nitrogen gas and reconstituted in DMEM cell culture medium (GIBCO #11965) by water-bath sonication to prepare low-dose (10 µg/mL) and high-dose (50 µg/mL) lipid suspensions.
Macrophage infection with M. tb Lipids
Murine macrophage cell line J774A.1 was cultured as a monolayer in DMEM medium (GIBCO #11965) supplemented with 10% fetal bovine serum, 100 units/mL penicillin/streptomycin, and 2 mM L-glutamine (GIBCO). Adherent cells were washed with PBS and treated with media alone, low-dose or high-dose M. tb lipid extracts for 24 h until 70–80% confluence was achieved. For SILAC labeling, unstimulated cells were grown in stable isotope-labeled medium lacking lysine and arginine yet supplemented with both 13C6-lysine and 13C6, 15N4-arginine (Cambridge Isotope Labs, Andover, MA). The cells were cultured for at least 6 doublings to allow full incorporation of labeled amino acids before exposure to M. tb lipids.
Sample treatment for iTRAQ and SILAC labeling
For iTRAQ labeling, 3 plates (4.5 × 107 cells) of J774A.1 cells treated with media alone, high dose or low dose lipid extracts were harvested by scraping and centrifuged at 1300 rpm to pellet the cells. The cells were rinsed three times with ice cold PBS, resuspended in 500 mM triethylammonium bicarbonate (TEAB, Sigma) containing 0.05% SDS and protease inhibitor cocktail (Sigma), and lysed using a micro-tip sonicator (VirSonic, Gardiner, NY). The cell lysates were clarified by centrifugation at 14,000 rpm for 30 min and protein concentrations were determined by BCA assay (Pierce, Rockford, IL). Protein (70 µg) from each sample was reduced with TCEP, cysteine-blocked by treatment with iodoacetamide, digested with trypsin, and labeled with an isobaric tag reagent as per manufacturer’s directions (Applied Biosystems, Foster City, CA) and as described by Ross et al28. Samples were labeled and pooled (Scheme 1). iTRAQ-labeled peptide samples were aliquoted, snap-frozen, and stored at −80 °C until further analysis. For SILAC labeling, isotope-labeled cells and cells exposed to high-dose M. tb lipid extract were harvested by scraping, resuspended in 8 M urea, 50 mM Tris-HCl (pH 8.0) containing protease inhibitor cocktail (Sigma), and lysed by sonication. Protein concentrations were measured using with the BCA assay. Protein lysate (100 µg) from isotope labeled cells or high-dose lipid treated normal cells were pooled, subjected to reduction, alkylation and trypsin digestion (Scheme 1). Both iTRAQ and SILAC-labeled peptides were extensively desalted using C18 MacroSpin columns (The Nest Group, Southbotough, MA) according to the manufacturer’s directions and dried via speed vacuum.
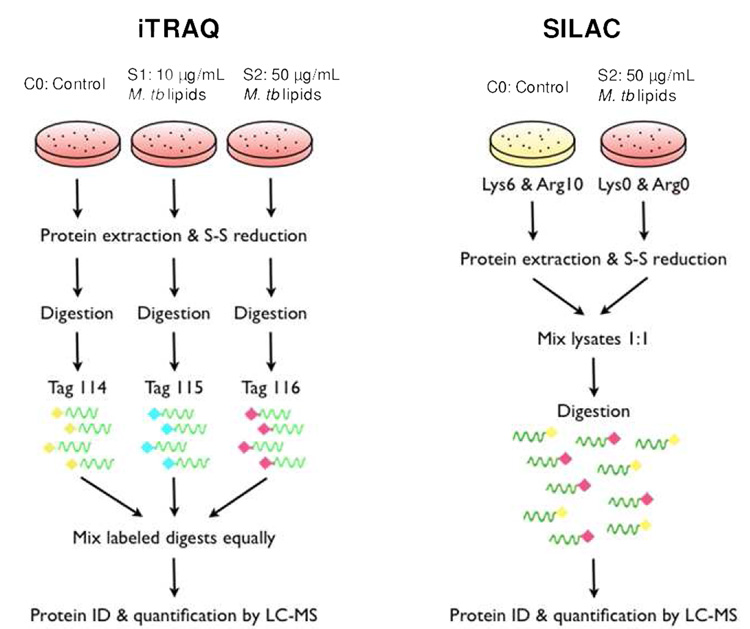
Scheme 1.
Schematic of the iTRAQ and SILAC strategy for labeling and quantifying the macrophage proteome under stimulation of a low (S1) or high (S2) dose of M. tb lipid extract.
2D LC-MS/MS analysis
iTRAQ- and SILAC-labeled peptide mixtures were separated by two-dimensional liquid chromatography and analyzed by electrospray ionization tandem MS. Briefly, the peptide mixture was separated by off-line strong cation exchange (SCX) chromatography using an Ultimate HPLC with a UV detector (Dionex-LC Packings, Sunnyvale, CA). Labeled samples were resuspended in SCX running buffer (5 mM KH2PO4, 25% acetonitrile (ACN), 0.1% formic acid (FA)) and loaded onto a PolyLC Polysulfoethyl A column (2.1 mm × 200 mm, The Nest Group, Southboro, MA). Peptides were eluted with increasing concentrations of 800 mM KCl, 5 mM KH2PO4, 25% ACN, and 0.1% FA using a three step gradient: 0–10% over 10 min, 10–25% over 30 min, and 25–100% over 10 min. Fifteen fractions were collected at a flow rate of 300 µL/min according to UV trace at 214 nm.
Fractions were partially evaporated to remove ACN on a speed vacuum and desalted using C18 MacroSpin columns. Desalted fractions were dried and reconstituted in 0.1% FA. Half of each fraction was injected onto a PepMap100 trapping column (0.3 mm × 5 mm). Reversed-phase separation was completed on a LC Packings PepMap C18 column (3 µm, 0.075 × 150 mm) at a flow rate of 200 nL/min using buffers 2% ACN, 0.1% FA (A) and 80% ACN, 0.1% FA (B). The gradient was 0–30% B in 100 min, 30–100% B in 10 min, and 100% B for 10 min. The samples were directly injected into an ESI Q-TOF Mass Analyzer (QSTAR® Hybrid Quadrupole TOF, Applied Biosystems, Framingham, MA) via nanoelectrospray ionization. The QSTAR system carried out a survey scan in the mass range of m/z 350–1600. Excising dynamic exclusion, up to three precursor ions exceeding a total ion current of 30 counts were selected for fragmentation in MS/MS analysis. Product ions were detected in the range of m/z 70–2000. The instrument was calibrated and tuned following each batch of five injections. Both iTRAQ- and SILAC-labeled samples were analyzed twice on the same platform.
Database search and relative quantification
LC-MS/MS data from both labeling routes were analyzed with Protein Pilot 2.0 software (Applied Biosystems MDS Sciex) as described by Wolff et al.36 The ‘Search Effort’ parameter ‘Thorough ID’, which provides a broad search of various protein modifications and multiple mass cleavages, was chosen. The Paragon™ algorithm used in Protein Pilot requires no definition of peptide/fragment mass tolerance, as it iteratively searches for the optimal mass error for a dataset. It also examines a number of common modification forms included in a generic workup set. Searches were performed against the International Protein Index (IPI) database (mouse, version 8–25–2006) downloaded to the local engine. The majority of average protein ratios reported by Protein Pilot have a p-value (evaluating the statistical difference between the observed ratio and unity) and EF (error factor) for each protein ID. The EF term indicates the actual average value lies between (reported ratio)/(EF) and (reported ratio) × (EF) at a 95% confidence. Only those protein matches having a p-value < 0.05 and a meaningful EF (<2), and at least two unique peptides identified, were saved for protein identification and relative quantification. The false positive rates of the aforementioned filter criteria were all below 5%, estimated by using an individual reversed (decoy) sequence database of the entire mouse genome as described previously.37 In brief, false positive rates are calculated by dividing the number of decoy hits by that of hits acquired in search against forward sequence database. All statistical tests and analyses were performed using Excel and R programs.
ELISA for TNF-α
Normal or SILAC-labeled J774.A1 cells were seeded in 24-well tissue culture plates at 5 × 106 cells/well with DMEM media or DMEM isotope enriched media, respectively. After 1 h of cultivation, 10 µg/mL or 50 µg/mL of M.tb lipid extract, or 0.1 µg/mL LPS were added to the wells in duplicate. After 24 h, supernatants were collected and levels of TNF-α were measured using a cytokine kit (R&D Systems, Minneapolis, MN) according to the manufacturer’s directions. The experiment was performed three times to calculate the mean TNF-α concentration.
Results
Isotope-labeled macrophages mount a similar pro-inflammatory response as unlabeled cells
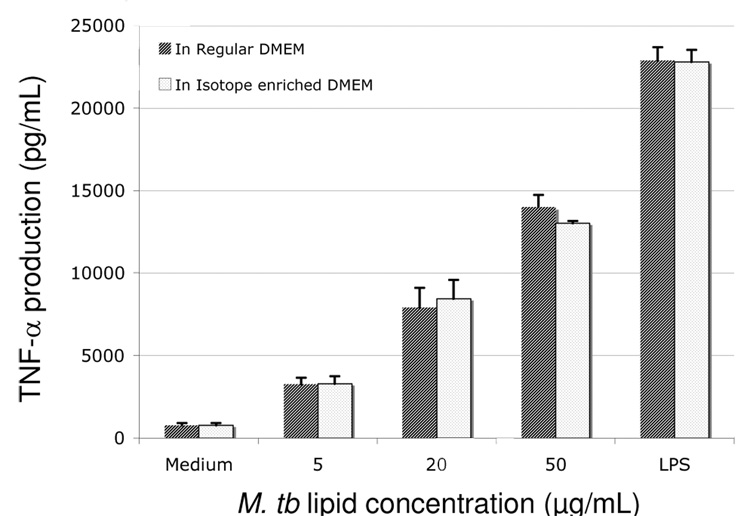
Once stimulated by agonists such as the gram negative bacterial cell wall component lipopolysaccharide (LPS) or M. tb lipid extracts, macrophages undergo a potent pro-inflammatory response generating cytokines such as TNF-α.38 To confirm that isotope-labeled macrophages are able to mount an immune response similar to unlabled cells, unlabeled and labeled macrophages were treated with increasing amounts of M. tb lipid extract or LPS, and TNF-α released was quantified via ELISA (Fig. 1). Comparable levels of TNF-α were observed from cells grown in stable isotope-enriched media and regular DMEM medium, suggesting that isotopic labeling of the cellular proteome does not alter the characteristic inflammatory response pathways.
Figure 1.
Effects of M. tb lipid extract on TNF-α production of macrophages grown in normal (black bar) and isotope-enriched (white bar) media (containing 13C6-lysine and 13C6, 15N4-arginine). Cells were incubated for 24h with different amount of M. tb lipid extract or LPS (100 ng/mL) and the cell-free supernatants were collected for measurements of TNF-α secretion via ELISA. The experiment was repeated in triplicate and error bars indicate the standard deviation of the mean.
Identification and relative quantification of macrophage proteins via SILAC and iTRAQ labeling strategies
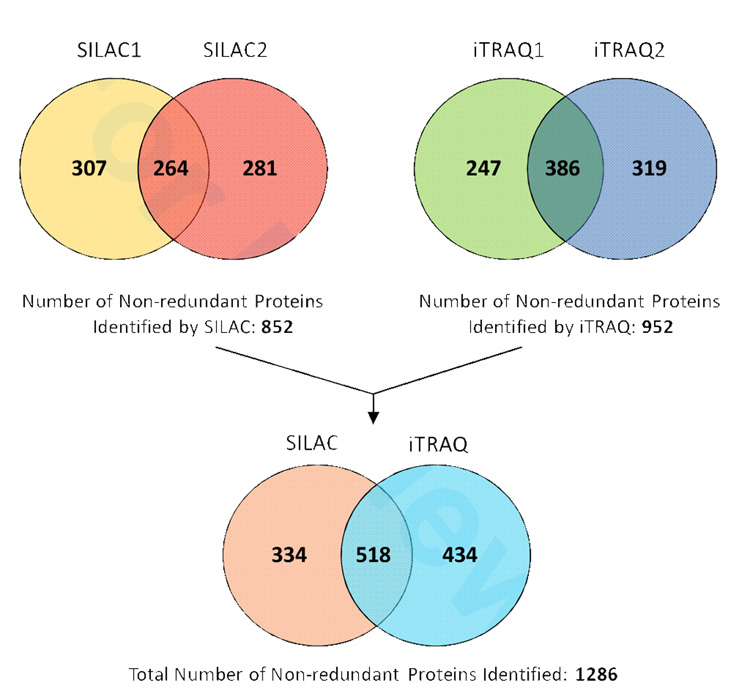
Scheme 1 illustrates our protocols for using the iTRAQ and SILAC techniques to study the response of macrophages to M. tb lipid exposure. Two injection replicates were performed for analyzing the peptide mixture obtained from each method. Only those protein IDs meeting the aforementioned three filtering criteria (as described in experimental procedures) were saved for further analysis. The number of non-redundant macrophage proteins identified by SILAC was 852 and by iTRAQ was 952 (Fig. 2), representing a total of 1286 unique proteins that were found by combining all four datasets. The use of injection replicates and biological replicates (i.e. cells treated identically with the exception of metabolic isotope-labeled biomass for SILAC experiment and protein digests chemically labeled for iTRAQ experiment) significantly increased the number of proteins identified and quantified (Fig. 2). Approximately 50% more unique proteins were found by analyzing a second injection for both labeling approaches. This is higher than the 5~33% increment in protein IDs from duplicate injections reported by Chong et al39. We assume the larger genome of murine cells dramatically increases the sample complexity compared with the smaller genome organisms used in previous proteomic studies, thus allowing for more variation in precursor selection and peptide sequencing during MS analysis. Therefore, multiple injection of the same labeling sample achieves higher proteome coverage and provides reproducible data for quantification.
Figure 2.
Venn diagram of protein identification via either iTRAQ or SILAC approach. Two injection replicates were made for each method, which are indicated by 1 and 2.
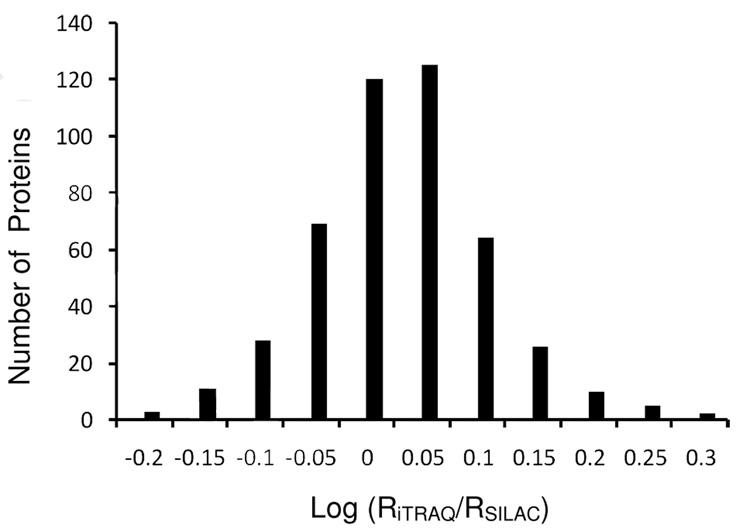
SILAC and iTRAQ identified 518 proteins in common. For each protein ratio reported by the software, an EF (error factor) value is calculated by the software based on the variance of peptide ratios to indicate a 95% confidence limit of the measurement error term of a given ratio (see Experimental Procedures for details). We selected those hits with EF<2 as ‘quantifiable entries’ with a variation below 20%35,36. Specifically, if the protein was identified in the injection duplicates with EF<2, an average of the two ratios was calculated to represent its differential expression; otherwise, only the ratio with EF<2 was saved for that protein. This data filtration resulted in 463 pairs of quantifiable proteins whose expression ratios (RiTRAQ and RSILAC) were measured by both labeling approaches. The protein IDs and expression ratios are listed in Supplementary Table 1. To determine the quantification variance, we plotted the log ratio distribution of RiTRAQ/RSILAC for each quantified protein (Figure 3) and found a normal distribution with narrow variability in standard deviation (SD) of only 0.08. Notably, this SD was smaller than the typically reported 0.1 in iTRAQ experiments28,32,40, which indicates a good consistency between the two measurements. We also did the paired-t test on log-transformed RiTRAQ and RSILAC to evaluate the statistical difference between the two large datasets. The p-value (0.90) was considerably larger than 0.05, suggesting that the distribution of these two data series does not significantly differ from each other. As a direct demonstration of the reproducible results obtained by both SILAC and iTRAQ, several mass spectra from representative peptides and their relative ratios are shown in Supplementary Fig. 1. For the 463 co-quantified proteins, we then calculated the average of each pair of RiTRAQ and RSILAC. Based on the standard deviation model (2 SD, P=0.05), any protein with an average ratio either above 1.23 or below 0.81, can be considered showing a significant degree of up- or down-regulation compared with the mean (1.00).
Figure 3.
Distribution of protein ratios in a log scale. RiTRAQ and RSILAC refer to the expression ratio measured by iTRAQ and SILAC for a specific protein. The SD (0.08) reflects a narrow variability.
Differential expression of macrophage proteins upon stimulation with M. tb lipids
Supplementary Table 2 includes all of the protein IDs from four individual datasets (SILAC-1/-2 and iTRAQ-1/-2) and statistical parameters for quantification. Among the strictly identified proteins, we defined three additional thresholds for selecting proteins that had significantly altered expression when macrophages were exposed to M. tb lipid extract: 1) P-value for that protein ratio (S1:C0 or S2:C0) given by the software is < 0.05, which indicates the ratio statistically differs from unity (>95% confidence); 2) The ratio is either above 1.23 or below 0.81 (using the same cutoff for analyzing iTRAQ and SILAC co-quantified pairs); and 3) Must be a ‘quantifiable entry’ with EF (error factor) <2. Applying these filtering criteria, a subset of regulated proteins were identified (see Supplementary Table 3 for quantification details), and then the total number of protein IDs (with P-value and EF), the number of proteins with P<0.05, and the number of regulated hits (meeting all three requirements) found by two labeling approaches were determined. SILAC and iTRAQ found a similar number of protein IDs and hits with P<0.05 (Table 1). But SILAC revealed a larger population of significant changes in protein expression than did iTRAQ. Many of the iTRAQ-determined ratios that surpass the p-value threshold show a small magnitude of change (no more than 20% beyond unity). Thus, it appears that the iTRAQ method may be more sensitive for identifying minor changes in protein expression level.
Table 1.
Comparison of the number of proteins identified in each dataset selected by different criteria
| Dataset No. | Total number of protein IDs1 | Number of hits with P<0.052 | Number of differentially expressed hits3 |
|---|---|---|---|
| SILAC-1 | 571 | 149 | 72 (48.3%) |
| SILAC-2 | 545 | 181 | 75 (41.4%) |
| iTRAQ-1 | 633 | 123 | 10 (8.1%) |
| iTRAQ-2 | 705 | 167 | 20 (12.0%) |
Refers to hits with both P-value (indicating at least two unique peptides contributing to ID) and EF
Selected from the pool of protein IDs and having P-value <0.05 (indicating significant difference of the ratio beyond unity)
Selected from the pool of protein IDs and meeting the additional standards stated in the text for differential expression. The percentage in the parentheses refers to the proportion of differentially expressed hits over all those with P<0.05
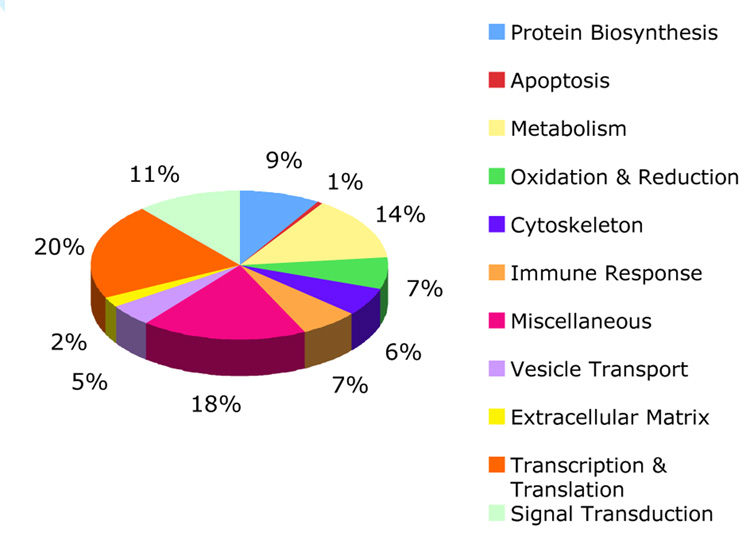
In response to high-dose M. tb lipid stimulation, a total of 51 and 103 non-redundant proteins were found to be down-regulated (ratio <0.81) or up-regulated (ratio >1.23), respectively (Supplementary Table 4). The low-dose lipid stimulation significantly downregulated the expression of one protein and upregulated the expression of 15 proteins. According to the functional description (see details in Supplementary Table 4), all significantly changed proteins fell into 11 major groups: immune response, oxidation and reduction, signal transduction, vesicle transport, apoptosis, cytoskeleton, extracellular matrix (ECM), protein biosynthesis, metabolism, transcription and translation, and miscellaneous (unclassified). Their relative distribution is shown in Fig.4.
Figure 4.
Relative distribution of macrophage protein subsets regulated by M. tb lipid exposure. Proteins are grouped by their cellular functions. The up- and down-regulated hits are combined in each group. The ‘Miscellaneous’ group consists mostly of proteins of unknown function.
Discussion
Comparison of SILAC and iTRAQ
In recent years, the proteomics field has increasingly focused on relative protein quantification via labeling multiple samples with distinct stable isotope-enriched mass tags. Stable isotope labeling approaches fall into three main categories: metabolic, chemical, or enzymatic. Currently, SILAC is the most commonly used metabolic isotope labeling approach, while iTRAQ is a newly emerging technique that labels peptide mixtures via a chemical reaction. iTRAQ confers higher sensitivity, better proteome coverage, and allows for multiple sample comparison, making it a more promising technique than the widely used chemical labeling approach ICAT.41
However, SILAC and iTRAQ both possess distinct strengths and weaknesses (summarized in Table 2). iTRAQ-based quantification uses isotope tags that are only detectable in MS/MS, so it requires high quality MS/MS spectra and sufficient LC resolution to avoid errors in precursor ion selection.11, 42 In contrast, SILAC quantifies peptide isoforms in MS scans and requires fewer chemical processes, which reduces variation introduced by sample preparation.42 Further, SILAC is compatible with gel-based protein separation, while iTRAQ is usually used to label in-solution digests. SILAC has been applied mostly to eukaryotic cell line studies, whereas iTRAQ has no limitation on sample sources and offers a relatively large multiplexing scale.
Table 2.
Distinct features of metabolic and chemical isotope mass tagging strategy for quantitative proteomics
| SILAC (metabolic) | iTRAQ (chemical) | |
|---|---|---|
| Tagging molecule | Protein | Peptide |
| Tag detection mode | MS | MS/MS |
| Tag residue(s) | H, C, N atoms of any amino acid | Primary amines |
| Specificity | High (via targeting one amino acid) | Low |
| Proteome coverage | High (via labeling appropriate amino acids) | High |
| Requirement on LC separation and MS/MS quality | Medium | High1 |
| Compatible mass analyzers | All types | Except ion trap |
| Compatible digestion | Both in-gel and in-solution | In-solution |
| Multiplexing scale | 2~3 | 4 or 8 |
| Sample source | Mainly mammalian cell lines2 | Cells & tissues |
| Cost | Medium | High |
iTRAQ is more susceptible to errors in precursor selection due to insufficient LC separation41; and low-quality MS/MS leads to less accuracy in quantification.
Until now, no direct comparison between SILAC and iTRAQ has been reported, though a comparison of quantification between 2D-gel and different stable isotope labeling methods has been made on small sets of proteins36, 43. Notably, comparisons between 2D-gel and ICAT (the well-established isotope-labeling approach) demonstrate quantitative differences in certain protein ratios due to imperfect protein separation and assignment of post-translational modifications in both platforms44, 45.
In this study, we sought to directly compare the SILAC and iTRAQ labeling approaches. Statistical analysis suggests high agreement between SILAC and iTRAQ quantification. Therefore, for the 463 pairs of co-identified proteins, the ratios determined by one approach were verified using the other. Notably, we observed a small number of proteins with discrepant ratios measured by two methods. Manual inspection of the raw data for these hits revealed that a few have poorly identified peptides contributing to the average protein ratio. However, in other cases where the raw MS and MS/MS spectra were of high quality, the variation between SILAC and iTRAQ ratios reflected intrinsic differences between the two technologies with respect to labeled moieties, chemical treatment, and MS signals used for quantification, all of which should be investigated more closely.
Interestingly, we also noticed that among the 1286 unique proteins identified with two labeling approaches, 768 proteins were distinctly identified by only one labeling route. This could be due to the complexity of the total cell lysate, which also leads to a low duplication of protein IDs between technical replicates (Fig. 2), or to differences in sample preparation such as lysis buffer composition.
Very consistent peak ratios were observed for different peptides from the same protein identified in either SILAC replicates or by iTRAQ analysis. In Supplementary Fig. 1, the first three proteins are members of Rab family, which are small GTPases mediating vesicular trafficking and fusion. These proteins tend to co-migrate in 2D gels, rendering them impossible to quantify individually by densitometry analysis alone. In contrast, we were able to determine the relative ratios of each Rab protein using isotope labeling approaches and found they were regulated to different extent.
We also observed interesting features of SILAC vs iTRAQ regarding the discovery of significant changes in protein expression. Among 166 proteins of differential expressions, 40 were quantified by iTRAQ and 134 by SILAC. In fact, iTRAQ labeling identified a significant number of proteins of modest expression changes at a high certainty, whereas SILAC was more capable of tracking dramatic changes of protein expression (Table 1). In the extreme cases, a few proteins reported by SILAC were only present in either control or lipid-treated cells, with the ratio S2:C0 to be 0 or infinite. Therefore, the double-standard quantification method described here combines the strengths of both techniques to identify far more significant changes in protein expression than either one would alone.
Significant protein regulation occurred upon macrophage exposure to M. tb lipid extract
The exotic lipids of the mycobacterial cell wall comprise 60% of the dry weight of the bacterium and serve as mediators of host-pathogen interactions. Purified M. tb cell wall components have been shown to modulate the host immune response as well as pathogenic processes5,46–48. Interestingly, recent studies have shown that lipids and lipid conjugates are actively trafficking out of the mycobacterium-containing phagosomes into the host cell cytosol, or even transferred to nearby cells.6, 7, 49 This finding further expands the possible mechanisms by which mycobacterial lipids might exert their immunomodulatory functions.
The pathways by which M. tb lipids might directly affect host cells have not yet been characterized via proteomic approaches. Notably, a previous proteomic study examined changes in macrophage protein levels upon infection with live M. tb.50, yet only found three differentially expressed proteins. In contrast, our MudPIT platform coupled with isotopic labeling uncovered a total of 166 unique macrophage proteins that are differentially expressed upon treatment with M. tb lipid extract. The majority of proteins detected in our study remained virtually unchanged, indicating that the lipids released by pathogenic mycobacteria may target and regulate a specific small set of host proteins.
The significantly changed proteins engage in diverse cellular processes (Fig. 4 and Table 3), which suggests a complex interaction between the bacterium and the host. That is, while M. tb lipids appear to act as immunostimulants, they may simultaneously undermine host cell processes that would otherwise limit pathogenesis.
Table 3.
Global modulation of macrophage cellular functions upon M. tb lipid stimulation
| Cellular Function of Macrophages | Global Effect of M. tb Lipid Stimulation |
|---|---|
| Immune response | Receptor binding, cytokine production, protease activation and MHC class I and II processing may be affected |
| Oxidation & reduction | A few proteins important for respiratory burst are induced, while others involved in protection against oxidative stress are upregualted as well |
| Signal transduction | Protein components of multiple pathways (PKC, MAPK, Ca2+, etc.) change expression |
| Vesicle transport | Proteins involved in endocytosis and endosome/lysosome fusion & maturation are regulated |
| Metabolism | Many proteins involved in glycolysis are induced |
Several immunologically relevant proteins changed upon M. tb lipid stimulation include CD14, Fc gamma receptor type IIb (FcRIIb), TAP binding protein isoform1 (tapasin 1), cathepsin L, and dipeptidyl-peptidase 1 precursor (cathepsin C). CD14 is a GPI-anchored, leucine-rich repeat (LRR) protein expressed mainly on the cell surface of myeloid lineage cells.51 CD14 has been shown to bind bacterial lipopolysaccharides (LPS) in conjunction with TLR452, 53, which leads to proinflammatory response54. Although the M. tb cell wall does not contain LPS, our study found CD14 upregulated by 25% upon exposure to a high-dose of M. tb lipids (50 µg/mL). Previous microarray studies reported a 2-fold increase of CD14 transcripts in macrophages exposed to heat-killed M. tb.55 The modest increase in CD14 protein that we observed might reflect less potency of the isolated lipids compared to intact bacilli. Alternatively, post-transcriptional regulation could account for the discrepancy in RNA versus protein abundances.56–58
In addition to CD14, we found FcRIIb to be upregulated 1.7-fold upon high-dose lipid treatment. This low affinity inhibitory IgG receptor, when engaged, blocks calcium influx and inhibits phagocytosis, cytokine release and proinflammatory activation against pathogens.59
Cathepsin L and tapasin 1, both important for antigen processing and loading onto MHC molecules, were upregulated by 2.1 fold and 1.4 fold, respectively. Thus, lipids released by the pathogen may activate the host’s machinery for antigen presentation. Our observed induction of cathepsin L agrees with microarray data on M. tb-infected host response55, while upregulation of tapasin is a new finding. Cathepsin C, whose expression is suppressed upon lipid treatment, is a protease involved in activating granule-associated serine proteases that degrade pathogen proteins. Its down-regulation could reflect a host anti-bactericidal response to M. tb lipid treatment.
The oxidative burst via reactive oxygen and nitrogen intermediates (ROI and RNI) is a critical mechanism by which macrophages kill intracellular mycobacteria.60, 61 Not surprisingly, we observed significant changes in redox-related protein levels upon mycobacterial lipid treatment. P67phox, a subunit of the NADPH oxidase Phox, is necessary for the generation of ROI and their delivery to the pathogen-contained vacuole.60 This protein was up-regulated 44% by high-dose lipid stimulation. Another protein NCF1, required for activation of latent Phox, was upregulated 1.5-fold, presumably to coordinate the process. Macrophage migration inhibitory factor (MIF), a cytokine important for macrophage activation and production of nitric oxide,62 underwent a lipid dose-dependent increase in its cytosolic expression (1.3-fold change at high-dose stimulation), indicating RNI burst might have been still triggered as a result of M. tb lipid stimulation.
Interestingly, we observed upregulation of several macrophage proteins that are able to counteract the effect of oxidative stress, including Mn-dependent superoxide dismutase (MnSOD), peroxiredoxin-1 (prdx-1), peroxiredoxin 5 (prdx5), and heme oxygenase 1. MnSOD and prdx-1 are both antioxidants important for quenching ROI and hydrogen peroxides to prevent cellular damage.63 The induction of MnSOD was observed in a proteomic study of a human monocyte cell line infected with live M. tb.50 The authors speculated that its elevated expression might protect the host cell and pathogen from O2−-mediated toxicity, a potential function of specific M. tb lipids.
With respect to endocytosis and vesicle transport, the observed upregulation of vacuolar ATP synthase (H+ transporter) upon low-dose lipid stimulation attracted our attention. It is known that live M. tb or microbeads heavily coated with ManLAM are able to block acidification of the endocytic compartment containing the pathogen or its constituents.1 Therefore, it is insightful to note that H+-ATPase induction is weakened at high-dose lipid exposure, perhaps to favor pathogen survival inside the host. However, further experiments should be carried out to investigate whether the dose of lipid extract inversely lowers the pH of endosomes/phagosomes, and which specific lipid components are inducers or suppressors.
Conclusion
SILAC and iTRAQ, the two most predominantly used isotope-labeling approaches for quantitative proteomics, provided accurate and reproducible values for relative expression of proteins under cellular stresses. The two methods allowed mutual validation of identified proteins, while expanding our knowledge of the macrophage response to M. tb cell wall lipids. A number of macrophage proteins were found to be regulated by M. tb lipids. The observed response suggests that M. tb lipids can both induce host anti-bacterial responses while also modulating host processes to favor bacterial survival.
Supplementary Material
All Pairs of Expression Ratios (S2:C0) Measured via iTRAQ or SILAC
Expression Ratios of Proteins of Significant Changes upon M. tb Lipid Stimulation
Typical spectra for relative protein quantification by metabolic and chemical isotope labeling. Peptides on each row are derived from one protein: (A) Ras-related Rab-14 (B) Ras-related Rab-1B (C) Ras-related Rab-7 (D) Brian acid soluble protein 1 On the left, typical ion pairs of peptides derived from one protein are identified in two injection replicates of SILAC samples. Peptide sequences and relative peak ratios R (S2:C0) are shown below. On the right, the iTRAQ reporter ion region in the MS/MS spectrum of another peptide from the same protein was expanded and the reporter peak ratios R (116:114) are presented below. Both the two ratios reflect the change of protein expression under high-dose M. tb lipid exposure.
This information is available free of charge via the Internet at http://pubs.acs.org.
Protein IDs, Expression Ratios and Quantitative Parameters of four datasets
Differential Expression and Functional Annotation of Proteins Significantly Changed upon M. tb Lipid Stimulation
Acknowledgement
We thank Kangyu Zhang and Ding Chen for important help with statistical tests and proteomic data analysis. This research was supported by a grant to C. R. B. from the National Institutes of Health (AI51622).
Abbreviations
- SILAC
Stable Isotopes Labeling by Amino Acids in Cell Culture
- iTRAQ
Isobaric Tags for Relative and Absolute Quantitation
- ICAT
Isotope-code Affinity Tag
References
- 1.Koul A, Herget T, Klebl B, Ullrich A. Interplay between mycobacteria and host signalling pathways. Nat Rev Microbiol. 2004;2(3):189–202. doi: 10.1038/nrmicro840. [DOI] [PubMed] [Google Scholar]
- 2.Monack DM, Mueller A, Falkow S. Persistent bacterial infections: the interface of the pathogen and the host immune system. Nat Rev Microbiol. 2004;2(9):747–765. doi: 10.1038/nrmicro955. [DOI] [PubMed] [Google Scholar]
- 3.Rosenberger CM, Finlay BB. Phagocyte sabotage: disruption of macrophage signalling by bacterial pathogens. Nat Rev Mol Cell Biol. 2003;4(5):385–396. doi: 10.1038/nrm1104. [DOI] [PubMed] [Google Scholar]
- 4.Daffe M, Draper P. The envelope layers of mycobacteria with reference to their pathogenicity. Adv Microb Physiol. 1998;39:131–203. doi: 10.1016/s0065-2911(08)60016-8. [DOI] [PubMed] [Google Scholar]
- 5.Russell DG, Mwandumba HC, Rhoades EE. Mycobacterium and the coat of many lipids. J Cell Biol. 2002;158(3):421–426. doi: 10.1083/jcb.200205034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beatty WL, Rhoades ER, Ullrich HJ, Chatterjee D, Heuser JE, Russell DG. Trafficking and release of mycobacterial lipids from infected macrophages. Traffic. 2000;1(3):235–247. doi: 10.1034/j.1600-0854.2000.010306.x. [DOI] [PubMed] [Google Scholar]
- 7.Beatty WL, Ullrich HJ, Russell DG. Mycobacterial surface moieties are released from infected macrophages by a constitutive exocytic event. Eur J Cell Biol. 2001;80(1):31–40. doi: 10.1078/0171-9335-00131. [DOI] [PubMed] [Google Scholar]
- 8.Chatterjee D, Khoo KH. Mycobacterial lipoarabinomannan: an extraordinary lipoheteroglycan with profound physiological effects. Glycobiology. 1998;8(2):113–120. doi: 10.1093/glycob/8.2.113. [DOI] [PubMed] [Google Scholar]
- 9.Fratti RA, Chua J, Vergne I, Deretic V. Mycobacterium tuberculosis glycosylated phosphatidylinositol causes phagosome maturation arrest. Proc Natl Acad Sci U S A. 2003;100(9):5437–5442. doi: 10.1073/pnas.0737613100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Julka S, Regnier F. Quantification in proteomics through stable isotope coding: a review. J Proteome Res. 2004;3(3):350–363. doi: 10.1021/pr0340734. [DOI] [PubMed] [Google Scholar]
- 11.Ong SE, Mann M. Mass spectrometry-based proteomics turns quantitative. Nat Chem Biol. 2005;1(5):252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 12.Mann M. Functional and quantitative proteomics using SILAC. Nat Rev Mol Cell Biol. 2006;7(12):952–958. doi: 10.1038/nrm2067. [DOI] [PubMed] [Google Scholar]
- 13.Kratchmarova I, Blagoev B, Haack-Sorensen M, Kassem M, Mann M. Mechanism of divergent growth factor effects in mesenchymal stem cell differentiation. Science. 2005;308(5727):1472–1477. doi: 10.1126/science.1107627. [DOI] [PubMed] [Google Scholar]
- 14.Zhang G, Spellman DS, Skolnik EY, Neubert TA. Quantitative phosphotyrosine proteomics of EphB2 signaling by stable isotope labeling with amino acids in cell culture (SILAC) J Proteome Res. 2006;5(3):581–588. doi: 10.1021/pr050362b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M. Nucleolar proteome dynamics. Nature. 2005;433(7021):77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- 16.Blagoev B, Ong SE, Kratchmarova I, Mann M. Temporal analysis of phosphotyrosine-dependent signaling networks by quantitative proteomics. Nat Biotechnol. 2004;22(9):1139–1145. doi: 10.1038/nbt1005. [DOI] [PubMed] [Google Scholar]
- 17.Ong SE, Mittler G, Mann M. Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nat Methods. 2004;1(2):119–126. doi: 10.1038/nmeth715. [DOI] [PubMed] [Google Scholar]
- 18.Wang T, Gu S, Ronni T, Du YC, Chen X. In vivo dual-tagging proteomic approach in studying signaling pathways in immune response. J Proteome Res. 2005;4(3):941–949. doi: 10.1021/pr050031z. [DOI] [PubMed] [Google Scholar]
- 19.Blagoev B, Kratchmarova I, Ong SE, Nielsen M, Foster LJ, Mann M. A proteomics strategy to elucidate functional protein-protein interactions applied to EGF signaling. Nat Biotechnol. 2003;21(3):315–318. doi: 10.1038/nbt790. [DOI] [PubMed] [Google Scholar]
- 20.Selbach M, Mann M. Protein interaction screening by quantitative immunoprecipitation combined with knockdown (QUICK) Nat Methods. 2006;3(12):981–983. doi: 10.1038/nmeth972. [DOI] [PubMed] [Google Scholar]
- 21.Milner E, Barnea E, Beer I, Admon A. The turnover kinetics of major histocompatibility complex peptides of human cancer cells. Mol Cell Proteomics. 2006;5(2):357–365. doi: 10.1074/mcp.M500241-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Meiring HD, Soethout EC, Poelen MC, Mooibroek D, Hoogerbrugge R, Timmermans H, Boog CJ, Heck AJ, de Jong AP, van Els CA. Stable isotope tagging of epitopes: a highly selective strategy for the identification of major histocompatibility complex class I-associated peptides induced upon viral infection. Mol Cell Proteomics. 2006;5(5):902–913. doi: 10.1074/mcp.T500014-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Doherty MK, Whitehead C, McCormack H, Gaskell SJ, Beynon RJ. Proteome dynamics in complex organisms: using stable isotopes to monitor individual protein turnover rates. Proteomics. 2005;5(2):522–533. doi: 10.1002/pmic.200400959. [DOI] [PubMed] [Google Scholar]
- 24.Brand M, Ranish JA, Kummer NT, Hamilton J, Igarashi K, Francastel C, Chi TH, Crabtree GR, Aebersold R, Groudine M. Dynamic changes in transcription factor complexes during erythroid differentiation revealed by quantitative proteomics. Nat Struct Mol Biol. 2004;11(1):73–80. doi: 10.1038/nsmb713. [DOI] [PubMed] [Google Scholar]
- 25.Ranish JA, Yi EC, Leslie DM, Purvine SO, Goodlett DR, Eng J, Aebersold R. The study of macromolecular complexes by quantitative proteomics. Nat Genet. 2003;33(3):349–355. doi: 10.1038/ng1101. [DOI] [PubMed] [Google Scholar]
- 26.Han DK, Eng J, Zhou H, Aebersold R. Quantitative profiling of differentiation-induced microsomal proteins using isotope-coded affinity tags and mass spectrometry. Nat Biotechnol. 2001;19(10):946–951. doi: 10.1038/nbt1001-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17(10):994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 28.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3(12):1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Wolf-Yadlin A, Ross PL, Pappin DJ, Rush J, Lauffenburger DA, White FM. Time-resolved mass spectrometry of tyrosine phosphorylation sites in the epidermal growth factor receptor signaling network reveals dynamic modules. Mol Cell Proteomics. 2005;4(9):1240–1250. doi: 10.1074/mcp.M500089-MCP200. [DOI] [PubMed] [Google Scholar]
- 30.Chen X, Walker AK, Strahler JR, Simon ES, Tomanicek-Volk SL, Nelson BB, Hurley MC, Ernst SA, Williams JA, Andrews PC. Organellar proteomics: analysis of pancreatic zymogen granule membranes. Mol Cell Proteomics. 2006;5(2):306–312. doi: 10.1074/mcp.M500172-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Cong YS, Fan E, Wang E. Simultaneous proteomic profiling of four different growth states of human fibroblasts, using amine-reactive isobaric tagging reagents and tandem mass spectrometry. Mech Ageing Dev. 2006;127(4):332–343. doi: 10.1016/j.mad.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 32.Melanson JE, Avery SL, Pinto DM. High-coverage quantitative proteomics using amine-specific isotopic labeling. Proteomics. 2006;6(16):4466–4474. doi: 10.1002/pmic.200600112. [DOI] [PubMed] [Google Scholar]
- 33.Unwin RD, Smith DL, Blinco D, Wilson CL, Miller CJ, Evans CA, Jaworska E, Baldwin SA, Barnes K, Pierce A, Spooncer E, Whetton AD. Quantitative proteomics reveals posttranslational control as a regulatory factor in primary hematopoietic stem cells. Blood. 2006;107(12):4687–4694. doi: 10.1182/blood-2005-12-4995. [DOI] [PubMed] [Google Scholar]
- 34.Gan CS, Chong PK, Pham TK, Wright PC. Technical, Experimental, and Biological Variations in Isobaric Tags for Relative and Absolute Quantitation (iTRAQ) J Proteome Res. 2007;6(2):821–827. doi: 10.1021/pr060474i. [DOI] [PubMed] [Google Scholar]
- 35.Lund TC, Anderson LB, McCullar V, Higgins L, Yun GH, Grzywacz B, Verneris MR, Miller JS. iTRAQ Is a Useful Method To Screen for Membrane-Bound Proteins Differentially Expressed in Human Natural Killer Cell Types. J Proteome Res. 2007;6(2):644–653. doi: 10.1021/pr0603912. [DOI] [PubMed] [Google Scholar]
- 36.Wolff S, Otto A, Albrecht D, Zeng JS, Buttner K, Gluckmann M, Hecker M, Becher D. Gel-free and gel-based proteomics in Bacillus subtilis: a comparative study. Mol Cell Proteomics. 2006;5(7):1183–1192. doi: 10.1074/mcp.M600069-MCP200. [DOI] [PubMed] [Google Scholar]
- 37.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res. 2003;2(1):43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 38.Zuckerman SH, Evans GF, Guthrie L. Transcriptional and post-transcriptional mechanisms involved in the differential expression of LPS-induced IL-1 and TNF mRNA. Immunology. 1991;73(4):460–465. [PMC free article] [PubMed] [Google Scholar]
- 39.Chong PK, Gan CS, Pham TK, Wright PC. Isobaric tags for relative and absolute quantitation (iTRAQ) reproducibility: Implication of multiple injections. J Proteome Res. 2006;5(5):1232–1240. doi: 10.1021/pr060018u. [DOI] [PubMed] [Google Scholar]
- 40.Wiese S, Reidegeld KA, Meyer HE, Warscheid B. Protein labeling by iTRAQ: A new tool for quantitative mass spectrometry in proteome research. Proteomics. 2007;7(3):340–350. doi: 10.1002/pmic.200600422. [DOI] [PubMed] [Google Scholar]
- 41.Zieske LR. A perspective on the use of iTRAQ reagent technology for protein complex and profiling studies. J Exp Bot. 2006;57(7):1501–1508. doi: 10.1093/jxb/erj168. [DOI] [PubMed] [Google Scholar]
- 42.Wu WW, Wang G, Baek SJ, Shen RF. Comparative study of three proteomic quantitative methods, DIGE, cICAT, and iTRAQ, using 2D gel- or LC-MALDI TOF/TOF. J Proteome Res. 2006;5(3):651–658. doi: 10.1021/pr050405o. [DOI] [PubMed] [Google Scholar]
- 43.Kolkman A, Dirksen EH, Slijper M, Heck AJ. Double standards in quantitative proteomics: direct comparative assessment of difference in gel electrophoresis and metabolic stable isotope labeling. Mol Cell Proteomics. 2005;4(3):255–266. doi: 10.1074/mcp.M400121-MCP200. [DOI] [PubMed] [Google Scholar]
- 44.Schmidt F, Dahlmann B, Janek K, Kloss A, Wacker M, Ackermann R, Thiede B, Jungblut PR. Comprehensive quantitative proteome analysis of 20S proteasome subtypes from rat liver by isotope coded affinity tag and 2-D gel-based approaches. Proteomics. 2006;6(16):4622–4632. doi: 10.1002/pmic.200500920. [DOI] [PubMed] [Google Scholar]
- 45.Schmidt F, Donahoe S, Hagens K, Mattow J, Schaible UE, Kaufmann SH, Aebersold R, Jungblut PR. Complementary analysis of the Mycobacterium tuberculosis proteome by two-dimensional electrophoresis and isotope-coded affinity tag technology. Mol Cell Proteomics. 2004;3(1):24–42. doi: 10.1074/mcp.M300074-MCP200. [DOI] [PubMed] [Google Scholar]
- 46.Geisel RE, Sakamoto K, Russell DG, Rhoades ER. In vivo activity of released cell wall lipids of Mycobacterium bovis bacillus Calmette-Guerin is due principally to trehalose mycolates. J Immunol. 2005;174(8):5007–5015. doi: 10.4049/jimmunol.174.8.5007. [DOI] [PubMed] [Google Scholar]
- 47.Russell DG. Who puts the tubercle in tuberculosis? Nat Rev Microbiol. 2007;5(1):39–47. doi: 10.1038/nrmicro1538. [DOI] [PubMed] [Google Scholar]
- 48.Rhoades ER, Geisel RE, Butcher BA, McDonough S, Russell DG. Cell wall lipids from Mycobacterium bovis BCG are inflammatory when inoculated within a gel matrix: characterization of a new model of the granulomatous response to mycobacterial components. Tuberculosis (Edinb) 2005;85(3):159–176. doi: 10.1016/j.tube.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 49.Rhoades E, Hsu F, Torrelles JB, Turk J, Chatterjee D, Russell DG. Identification and macrophage-activating activity of glycolipids released from intracellular Mycobacterium bovis BCG. Mol Microbiol. 2003;48(4):875–888. doi: 10.1046/j.1365-2958.2003.03473.x. [DOI] [PubMed] [Google Scholar]
- 50.Ragno S, Romano M, Howell S, Pappin DJ, Jenner PJ, Colston MJ. Changes in gene expression in macrophages infected with Mycobacterium tuberculosis: a combined transcriptomic and proteomic approach. Immunology. 2001;104(1):99–108. doi: 10.1046/j.0019-2805.2001.01274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ferrero E, Hsieh CL, Francke U, Goyert SM. CD14 is a member of the family of leucine-rich proteins and is encoded by a gene syntenic with multiple receptor genes. J Immunol. 1990;145(1):331–336. [PubMed] [Google Scholar]
- 52.Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282(5396):2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 53.Wright SD, Ramos RA, Tobias PS, Ulevitch RJ, Mathison JC. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990;249(4975):1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 54.Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 55.Ehrt S, Schnappinger D, Bekiranov S, Drenkow J, Shi S, Gingeras TR, Gaasterland T, Schoolnik G, Nathan C. Reprogramming of the macrophage transcriptome in response to interferon-gamma and Mycobacterium tuberculosis: signaling roles of nitric oxide synthase-2 and phagocyte oxidase. J Exp Med. 2001;194(8):1123–1140. doi: 10.1084/jem.194.8.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nie L, Wu G, Zhang W. Correlation of mRNA expression and protein abundance affected by multiple sequence features related to translational efficiency in Desulfovibrio vulgaris: a quantitative analysis. Genetics. 2006;174(4):2229–2243. doi: 10.1534/genetics.106.065862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nissom PM, Sanny A, Kok YJ, Hiang YT, Chuah SH, Shing TK, Lee YY, Wong KT, Hu WS, Sim MY, Philp R. Transcriptome and proteome profiling to understanding the biology of high productivity CHO cells. Mol Biotechnol. 2006;34(2):125–140. doi: 10.1385/mb:34:2:125. [DOI] [PubMed] [Google Scholar]
- 58.Xia Q, Hendrickson EL, Zhang Y, Wang T, Taub F, Moore BC, Porat I, Whitman WB, Hackett M, Leigh JA. Quantitative proteomics of the archaeon Methanococcus maripaludis validated by microarray analysis and real time PCR. Mol Cell Proteomics. 2006;5(5):868–881. doi: 10.1074/mcp.M500369-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ravetch JV, Bolland S. IgG Fc receptors. Annu Rev Immunol. 2001;19:275–290. doi: 10.1146/annurev.immunol.19.1.275. [DOI] [PubMed] [Google Scholar]
- 60.Nathan C, Shiloh MU. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc Natl Acad Sci U S A. 2000;97(16):8841–8848. doi: 10.1073/pnas.97.16.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Flynn JL, Chan J. Immune evasion by Mycobacterium tuberculosis: living with the enemy. Curr Opin Immunol. 2003;15(4):450–455. doi: 10.1016/s0952-7915(03)00075-x. [DOI] [PubMed] [Google Scholar]
- 62.Calandra T, Roger T. Macrophage migration inhibitory factor: a regulator of innate immunity. Nat Rev Immunol. 2003;3(10):791–800. doi: 10.1038/nri1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Imlay JA. Pathways of oxidative damage. Annu Rev Microbiol. 2003;57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- 64.Ishihama Y, Sato T, Tabata T, Miyamoto N, Sagane K, Nagasu T, Oda Y. Quantitative mouse brain proteomics using culture-derived isotope tags as internal standards. Nat Biotechnol. 2005;23(5):617–621. doi: 10.1038/nbt1086. [DOI] [PubMed] [Google Scholar]
- 65.Nirmalan N, Sims PF, Hyde JE. Quantitative proteomics of the human malaria parasite Plasmodium falciparum and its application to studies of development and inhibition. Mol Microbiol. 2004;52(4):1187–1199. doi: 10.1111/j.1365-2958.2004.04049.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All Pairs of Expression Ratios (S2:C0) Measured via iTRAQ or SILAC
Expression Ratios of Proteins of Significant Changes upon M. tb Lipid Stimulation
Typical spectra for relative protein quantification by metabolic and chemical isotope labeling. Peptides on each row are derived from one protein: (A) Ras-related Rab-14 (B) Ras-related Rab-1B (C) Ras-related Rab-7 (D) Brian acid soluble protein 1 On the left, typical ion pairs of peptides derived from one protein are identified in two injection replicates of SILAC samples. Peptide sequences and relative peak ratios R (S2:C0) are shown below. On the right, the iTRAQ reporter ion region in the MS/MS spectrum of another peptide from the same protein was expanded and the reporter peak ratios R (116:114) are presented below. Both the two ratios reflect the change of protein expression under high-dose M. tb lipid exposure.
This information is available free of charge via the Internet at http://pubs.acs.org.
Protein IDs, Expression Ratios and Quantitative Parameters of four datasets
Differential Expression and Functional Annotation of Proteins Significantly Changed upon M. tb Lipid Stimulation