Abstract
To provide information on the persistence and maintenance of colonization with Shiga toxin-producing Escherichia coli (STEC) in sheep, pulsed-field gel electrophoresis analysis of STEC isolates (n = 145) belonging to serogroups O5, O91, and O146 from 39 healthy animals was performed in a 12-month longitudinal study carried out with four sheep flocks. At the flock level as well as the individual-animal level, the same clones were obtained on sampling occasions separated by as much as 11 months.
Shiga toxin-producing Escherichia coli (STEC) strains have recently emerged as important food-borne pathogens. Human diseases ranging from mild diarrhea to hemorrhagic colitis and hemolytic-uremic syndrome, typically affecting children, the elderly, and immunocompromised patients, can be caused by STEC (6). Serogroup O157 especially represents a major public health concern worldwide. However, as non-O157 STEC strains are more prevalent than O157 strains in meat-producing animals and as contaminants in foods, humans are more likely to become exposed to such strains (3, 9, 23), and therefore, non-O157 STEC should not be overlooked in human disease investigations. Although healthy asymptomatic cattle are the best-recognized animal reservoir for STEC strains (5), sheep are an important source of these organisms for humans in some countries (2, 4, 8, 19). The ecology and epidemiology of STEC O157 in cattle appear to be very complex, often involving multiple clones on a single farm (14, 17). There is some previously published information about the on-farm persistence of specific clones in different cattle production systems (13, 15, 18, 21), including information about persistence in individual animals in a few cases. Nevertheless, apart from the findings of a study of STEC O157 (12), little is known about the natural colonization of sheep with STEC over long time periods and the ecology of STEC strains in sheep flocks. A longitudinal study was conducted to provide information on the persistence and maintenance of colonization with non-O157 STEC at the flock level as well as the individual-animal level in the context of a typical sheep flock in southwest Spain.
The longitudinal study was conducted with four epidemiologically unrelated sheep flocks in southwest Spain and started in November 2003. During the first sampling visit, 12 ewes around 1 year of age in each flock were randomly selected for sampling, and subsequent monthly sampling visits were carried out until October 2004. On each sampling occasion, one sample of rectal feces per animal was collected from selected individuals. Fecal samples were examined for STEC by both phenotypic (Vero cells) and genotypic (PCR) methods as described previously (19). The identification of O antigen in isolates (n = 521) was carried out in the Laboratorio de Referencia de E. coli (Lugo, Spain) as described by Guinée et al. (10) by using the full range of O antisera for serogroups O1 to O185. The first confirmed colony from a positive sample was selected as representative for that animal and stored at −80°C until further pulsed-field gel electrophoresis (PFGE) analysis. The preparation and XbaI digestion of DNA for PFGE were conducted as described previously (20). Bands below 33.3 kb were ignored in the analysis because of the difficult band-marking procedure for this region. A single-band difference was defined as significant to name the different PFGE types. Nevertheless, epidemiologically related isolates with PFGE patterns differing by three bands or fewer were considered to be of the same clone, since such differences are consistent with a single genetic event, i.e., a point mutation in a restriction site, a deletion, or an insertion, according to Tenover criteria (22).
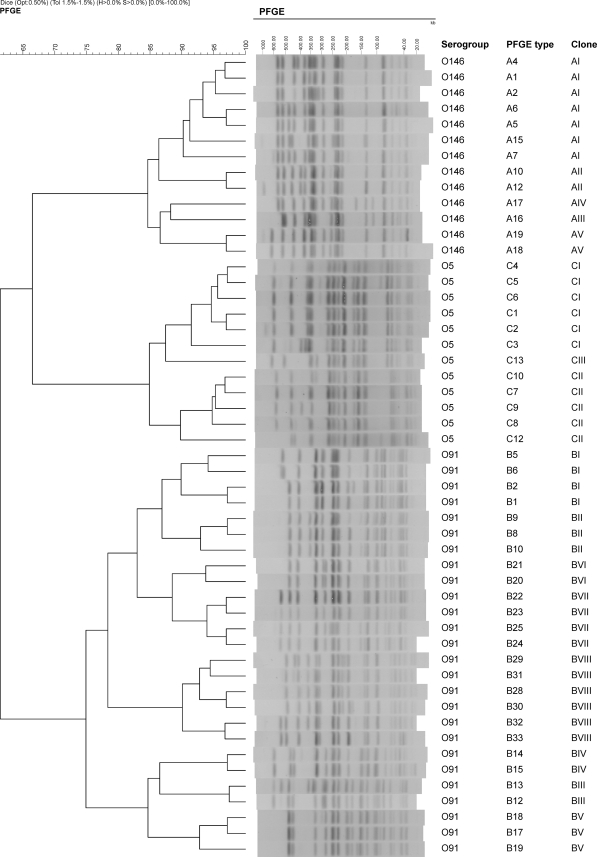
A total of 145 non-O157 STEC isolates from 39 healthy animals in four sheep flocks were characterized by PFGE (Table 1). This selection covered all isolates belonging to serogroups O5, O91, and O146, which are serogroups frequently represented among ovine STEC strains and associated with human strains that have caused hemolytic-uremic syndrome (4, 19). Overall, STEC O146 strains were detected in 19 (39.6%) of the animals, STEC O91 strains were detected in 26 (54.3%) of the animals, and STEC O5 strains were detected in 10 (20.8%) of the animals sampled. These 145 non-O157 STEC isolates produced 75 different PFGE types (Table 1). However, based on the previously described criteria, five persistent clones (AI to AV) were identified among O146 isolates, eight clones (BI to BVIII) were identified among O91 isolates, and three clones (CI to CIII) were identified among O5 isolates (Fig. 1; Table 1).
TABLE 1.
PFGE types and persistent clones among non-O157 STEC isolates
| Serogroup | No. of isolates | PFGE types (no. of isolates) | No. of types | Persistent clonesa (no. of types) | No. of clones |
|---|---|---|---|---|---|
| O146 | 50 | A1 (10), A2 (1), A3 (1), A4 (6), A5 (8), A6 (1), A7 (1), A8 (1), A9 (2), A10 (1), A11 (1), A12 (1), A13 (1), A14 (1), A15 (5), A16 (2), A17 (3), A18 (1), A19 (1), A20 (1), A21 (1) | 21 | AI (7), AII (2), AIII (1), AIV (1), AV (2) | 5 |
| O91 | 64 | B1 (9), B2 (2), B3 (1), B4 (1), B5 (2), B6 (1), B7 (1), B8 (2), B9 (1), B10 (1), B11 (1), B12 (2), B13 (1), B14 (1), B15 (1), B16 (1), B17 (4), B18 (2), B19 (4), B20 (4), B21 (1), B22 (1), B23 (1), B24 (3), B25 (1), B26 (1), B27 (1), B28 (3), B29 (1), B30 (2), B31 (1), B32 (3), B33 (1), B34 (1), B35 (1) | 35 | BI (4), BII (3), BIII (2), BIV (2), BV (3), BVI (2), BVII (4), BVIII (6) | 8 |
| O5 | 31 | C1 (7), C2 (2), C3 (1), C4 (1), C5 (1), C6 (1), C7 (4), C8 (1), C9 (2), C10 (1), C11 (1), C12 (1), C13 (2), C14 (1), C15 (1), C16 (1), C17 (1), C18 (1), C19 (1) | 19 | CI (6), CII (5), CIII (1) | 3 |
| Total | 145 | 75 | 16 |
Based on the criteria that isolates belong to the same serogroup and have PFGE patterns differing by three bands or fewer.
FIG. 1.
Dendrogram generated with InfoQuestFP software showing the PFGE-XbaI digestion types for non-O157 STEC isolates of the same persistent clones. The bands generated were analyzed by using the Dice coefficient and the unweighted-pair group method with arithmetic averages. The scales at the top indicate the similarity indices (in percentages) and molecular sizes (in kilobases).
Table 2 illustrates the persistence of specific non-O157 STEC clones at the flock level. The maximum number of consecutive sampling occasions on which a single clone was isolated from a flock was 11 (for the AI clone from flock 1). The maximum time between the first and last recoveries of a single clone from a flock with the isolation of the clone during the intervening period was 11 months (for the CI clone from flock 2). The maximum time between the first and last recoveries of a single clone from a flock without the isolation of the clone during the intervening period was 5 months (for the BVII, BIV, and AII clones from flocks 2, 3, and 4, respectively).
TABLE 2.
Temporal isolation of non-O157 STEC clones at the flock level
| Flock | Clone isolated in (mo/yr)a:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 11/03 | 12/03 | 1/04 | 2/04 | 3/04 | 4/04 | 5/04 | 6/04 | 7/04 | 8/04 | 9/04 | 10/04 | |
| 1 | AI | AI | AI | AI | AI | AI | AI | AI | AI | NS | AI | AI |
| AIV | AIV | NS | AIV | |||||||||
| BI | BI | BI | BI | BI | BI | BI | BI | BI | NS | BI | ||
| BII | BII | BII | BII | NS | ||||||||
| CII | CII | CII | NS | CII | CII | |||||||
| 2 | AI | AI | ||||||||||
| BIII | BIII | BIII | ||||||||||
| BV | BV | BV | BV | BV | BV | |||||||
| BVI | BVI | BVI | BVI | |||||||||
| BVII | BVII | BVII | BVII | |||||||||
| CI | CI | CI | CI | CI | CI | CI | CI | CI | ||||
| CII | CII | CII | CII | |||||||||
| 3 | AI | AI | AI | NS | AI | AI | NS | AI | ||||
| AIII | AIII | NS | NS | |||||||||
| AV | AV | NS | NS | |||||||||
| NS | BIV | NS | BIV | |||||||||
| BVIII | BVIII | BVIII | BVIII | NS | BVIII | BVIII | BVIII | NS | BVIII | |||
| 4 | NS | AII | AII | |||||||||
| NS | CI | CI | CI | CI | ||||||||
| NS | CIII | CIII | ||||||||||
NS, no sample from the flock was obtained on that occasion. An empty cell indicates that STEC strains were not isolated from the premises or that the isolates obtained were genetically unrelated to the persistent clones.
Table 3 illustrates the persistence of specific non-O157 STEC clones at the individual-animal level. The maximum number of consecutive sampling occasions on which a single clone was isolated from an animal was seven (for the AI clone from animal 1-8). The maximum time between the first and last recoveries of a single clone from an animal with the isolation of the clone during the intervening period was 11 months (for the CI clone from animal 2-12). The maximum time between the first and last recoveries of a single clone from an animal without the isolation of the clone during the intervening period was 10 months (for the BVIII clone from animal 3-10).
TABLE 3.
Temporal recovery of non-O157 STEC clones isolated from individual animals on two or more occasions
| Flock | Animal | Clone isolated in (mo/yr)a:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 11/03 | 12/03 | 1/04 | 2/04 | 3/04 | 4/04 | 5/04 | 6/04 | 7/04 | 8/04 | 9/04 | 10/04 | ||
| 1 | 1-1 | AI | AI | AI | AI | AI | NS | ||||||
| 1-2 | BI | BI | NS | BI | |||||||||
| 1-4 | BI | BI | BI | BI | BI | NS | |||||||
| 1-6 | AIV | AIV | NS | AIV | |||||||||
| 1-7 | BI | BI | BI | NS | |||||||||
| 1-8 | AI | AI | AI | AI | AI | AI | AI/CII | NS | CII | ||||
| 1-9 | BII | BII | BII | BII | NS | NS | NS | NS | NS | ||||
| 1-10 | AI | AI | AI | CII | CII | NS | CII | ||||||
| 1-12 | BI | BI | AI | AI | AI | NS | AI | AI | |||||
| 2 | 2-2 | CII | CII | ||||||||||
| 2-5 | BVI/CII | BVI | BVI | BVI/CII | |||||||||
| 2-8 | BV | BV | BV | BV | BV | NS | NS | NS | |||||
| 2-9 | BIII | BIII | BIII | ||||||||||
| 2-10 | BVII | BVII | |||||||||||
| 2-11 | BV | BV | BV | BV | |||||||||
| 2-12 | CI | BVII/CI | CI | BVII/CI | CI | CI | CI | CI | CI | ||||
| 3 | 3-1 | BVIII | NS | BVIII | BVIII | NS | |||||||
| 3-3 | NS | BIV | NS | BIV | |||||||||
| 3-5 | AI | NS | AI | NS | |||||||||
| 3-6 | AIII | AIII | NS | BVIII | NS | BVIII | |||||||
| 3-7 | BVIII | NS | BVIII | BVIII | NS | ||||||||
| 3-8 | AV | AV | NS | NS | NS | NS | NS | NS | NS | NS | NS | ||
| 3-9 | AI | AI | AI | NS | AI | NS | AI | ||||||
| 3-10 | BVIII | BVIII | AI | AI | NS | AI | NS | BVIII | |||||
| 4 | 4-2 | NS | CI | CI | CI | CI | |||||||
| 4-9 | NS | CIII | CIII | ||||||||||
| 4-12 | NS | AII | AII | ||||||||||
NS, no sample from the animal was obtained on that occasion. An empty cell indicates that the sample from the animal was negative for STEC or that isolates were genetically unrelated to the persistent clones.
Previous works from the United States have indicated that individual STEC O157 strains can be isolated for as long as 2 years from some dairy herds (21), for as long as 10 months on cattle ranges (18), and over the entire feeding period on cattle feedlots (13). Other studies from Italy and the United Kingdom have referred to this persistence on cattle farms (7, 15). In the only study performed with sheep, Kudva et al. (12) isolated individual STEC O157 strains from a single flock for as long as 2 months. To our knowledge, our study is the first report of the persistence and maintenance of colonization with non-O157 STEC in sheep flocks. LeJeune et al. (13) suggested that strain type stability observed on dairy farms for periods from months to years may be the result of the persistent infection of individual animals because the rate of animal turnover in these cattle production systems is vastly lower than is typical of feedlots, where the presence of other, nonanimal stable reservoirs is the most probable explanation for this situation. Actually, some studies describing the on-farm persistence of PFGE types over long time periods have also noted this persistence in individual animals (7, 15, 21). In addition, the persistence of some non-O157 STEC clones at the individual-animal level has been reported in a recent study on dairy goats (16).
Small variations in the PFGE types of isolates obtained from the same individuals in experimental and natural infections, similar to the variations observed in all serogroups in this study, have been reported before (1, 11). We are aware that the storage of only one colony per positive animal may have influenced our results, since individual animals may shed multiple strains simultaneously and the spectrum of types changes over time (12). However, in spite of these limitations, we have successfully demonstrated that some non-O157 STEC clones can be isolated from the same sheep flock, as well as the same animal, over a period of at least 11 months and from the same animal on consecutive sampling occasions for as long as 7 months. In our opinion, given the low rate of animal turnover which is typical of these sheep production systems, persistent individual-animal colonization by specific clones is the most probable explanation for the persistence of PFGE types at the flock level over long time periods. Nevertheless, we could not evaluate a possible role of residual contamination of the sheep flock environment and recycling through the host in this study.
Acknowledgments
We thank Raquel Rubio and Nohelia Tejero for their skillful technical assistance. Sergio Sánchez acknowledges the Ministerio de Educación y Ciencia for his research fellowship (AP2002-3286).
This study was supported by grants from the Fondo de Investigación Sanitaria (grant FIS G03-025-COLIRED-O157), from the Xunta de Galicia (grant PGIDIT065TAL26101PR), and from the Junta de Extremadura and FEDER (grant 3PR05A009-III Plan Regional de Investigación).
Footnotes
Published ahead of print on 23 January 2009.
REFERENCES
- 1.Akiba, M., T. Sameshima, and M. Nakazawa. 2000. Clonal turnover of enterohemorrhagic Escherichia coli O157:H7 in experimentally infected cattle. FEMS Microbiol. Lett. 184:79-83. [DOI] [PubMed] [Google Scholar]
- 2.Bettelheim, K. A. 2003. Non-O157 verotoxin-producing Escherichia coli: a problem, paradox, and paradigm. Exp. Biol. Med. 228:333-344. [DOI] [PubMed] [Google Scholar]
- 3.Blanco, J. E., M. Blanco, M. P. Alonso, A. Mora, G. Dahbi, M. A. Coira, and J. Blanco. 2004. Serotypes, virulence genes, and intimin types of Shiga toxin (verotoxin)-producing Escherichia coli isolates from human patients: prevalence in Lugo, Spain, from 1992 through 1999. J. Clin. Microbiol. 42:311-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanco, M., J. E. Blanco, A. Mora, J. Rey, J. M. Alonso, M. Hermoso, J. Hermoso, M. P. Alonso, G. Dahbi, E. A. González, M. I. Bernárdez, and J. Blanco. 2003. Serotypes, virulence genes, and intimin types of Shiga toxin (verotoxin)-producing Escherichia coli isolates from healthy sheep in Spain. J. Clin. Microbiol. 41:1351-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caprioli, A., S. Morabito, H. Brugere, and E. Oswald. 2005. Enterohaemorrhagic Escherichia coli: emerging issues on virulence and modes of transmission. Vet. Res. 36:289-311. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2001. Outbreaks of Escherichia coli O157:H7 infections among children associated with farm visits—Pennsylvania and Washington, 2000. MMWR Morb. Mortal. Wkly. Rep. 50:293-296. [PubMed] [Google Scholar]
- 7.Conedera, G., P. A. Chapman, S. Marangon, E. Tisato, P. Dalvit, and A. Zuin. 2001. A field survey of Escherichia coli O157 ecology on a cattle farm in Italy. Int. J. Food Microbiol. 66:85-93. [DOI] [PubMed] [Google Scholar]
- 8.Cookson, A. L., S. C. Taylor, J. Bennett, F. Thomson-Carter, and G. T. Attwood. 2006. Serotypes and analysis of distribution of Shiga toxin producing Escherichia coli from cattle and sheep in the lower North Island, New Zealand. N. Z. Vet. J. 54:78-84. [DOI] [PubMed] [Google Scholar]
- 9.Djordjevic, S. P., V. Ramachandran, K. A. Bettelheim, B. A. Vanselow, P. Holst, G. Bailey, and M. A. Hornitzky. 2004. Serotypes and virulence gene profiles of Shiga toxin-producing Escherichia coli strains isolated from feces of pasture-fed and lot-fed sheep. Appl. Environ. Microbiol. 70:3910-3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guinée, P. A. M., W. H. Jansen, T. Wadström, and R. Sellwood. 1981. Escherichia coli associated with neonatal diarrhoea in piglets and calves, p. 126-162. In P. W. Leeww and P. A. M. Guinée (ed.), Laboratory diagnosis in neonatal calf and pig diarrhoea: current topics in veterinary and animal science, vol. 13. Martinus-Nijhoff, The Hague, The Netherlands. [Google Scholar]
- 11.Karch, H., H. Rüssmann, H. Schmidt, A. Schwarzkopf, and J. Heesemann. 1995. Long-term shedding and clonal turnover of enterohemorrhagic Escherichia coli O157 in diarrheal diseases. J. Clin. Microbiol. 33:1602-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kudva, I. T., P. G. Hatfield, and C. J. Hovde. 1997. Characterization of Escherichia coli O157:H7 and other Shiga toxin-producing E. coli serotypes isolated from sheep. J. Clin. Microbiol. 35:892-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LeJeune, J. T., T. E. Besser, D. H. Rice, J. L. Berg, R. P. Stilborn, and D. D. Hancock. 2004. Longitudinal study of fecal shedding of Escherichia coli O157:H7 in feedlot cattle: predominance and persistence of specific clonal types despite massive cattle population turnover. Appl. Environ. Microbiol. 70:377-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liebana, E., R. P. Smith, E. Lindsay, I. McLaren, C. Cassar, F. A. Clifton-Hadley, and G. A. Paiba. 2003. Genetic diversity among Escherichia coli O157:H7 isolates from bovines living on farms in England and Wales. J. Clin. Microbiol. 41:3857-3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liebana, E., R. P. Smith, M. Batchelor, I. McLaren, C. Cassar, F. A. Clifton-Hadley, and G. A. Paiba. 2005. Persistence of Escherichia coli O157 isolates on bovine farms in England and Wales. J. Clin. Microbiol. 43:898-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orden, J. A., C. Cortés, P. Horcajo, R. De la Fuente, J. E. Blanco, A. Mora, C. López, J. Blanco, A. Contreras, A. Sánchez, J. C. Corrales, and G. Domínguez-Bernal. 2008. A longitudinal study of verotoxin-producing Escherichia coli in two dairy goat herds. Vet. Microbiol. 132:428-434. [DOI] [PubMed] [Google Scholar]
- 17.Renter, D. G., and J. M. Sargeant. 2002. Enterohemorrhagic Escherichia coli O157: epidemiology and ecology in bovine production environments. Anim. Health Res. Rev. 3:83-94. [PubMed] [Google Scholar]
- 18.Renter, D. G., J. M. Sargeant, R. D. Oberst, and M. Samadpour. 2003. Diversity, frequency, and persistence of Escherichia coli O157 strains from range cattle environments. Appl. Environ. Microbiol. 69:542-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rey, J., J. E. Blanco, M. Blanco, A. Mora, G. Dahbi, J. M. Alonso, M. Hermoso, J. Hermoso, M. P. Alonso, M. A. Usera, E. A. González, M. I. Bernárdez, and J. Blanco. 2003. Serotypes, phage types and virulence genes of Shiga-producing Escherichia coli isolated from sheep in Spain. Vet. Microbiol. 94:47-56. [DOI] [PubMed] [Google Scholar]
- 20.Sánchez, S., A. García-Sánchez, R. Martínez, J. Blanco, J. E. Blanco, M. Blanco, G. Dahbi, A. Mora, J. Hermoso de Mendoza, J. M. Alonso, and J. Rey. Detection and characterisation of Shiga toxin-producing Escherichia coli other than Escherichia coli O157:H7 in wild ruminants. Vet. J., in press. doi: 10.1016/j.tvjl.2008.01.011. [DOI] [PubMed]
- 21.Shere, J. A., K. J. Bartlett, and C. W. Kaspar. 1998. Longitudinal study of Escherichia coli O157:H7 dissemination on four dairy farms in Wisconsin. Appl. Environ. Microbiol. 64:1390-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zweifel, C., S. Schumacher, M. Blanco, J. E. Blanco, T. Tasara, J. Blanco, and R. Stephan. 2005. Phenotypic and genotypic characteristics of non-O157 Shiga toxin-producing Escherichia coli (STEC) from Swiss cattle. Vet. Microbiol. 105:37-45. [DOI] [PubMed] [Google Scholar]