Abstract
Phytate is the most abundant organic phosphorus compound in nature, and microbial mineralization of phytate by phytase is a key process for phosphorus recycling in the biosphere. In the present study, beta-propeller phytase (BPP) gene fragments were readily amplified from the intestinal contents of grass carp (Ctenopharyngodon idellus) directly or from phytate-degrading isolates from the same source, confirming the widespread occurrence of BPP in aquatic communities. The amounts of sequences collected using these two methods differed (88 distinct genes versus 10 isolates), but the sequences showed the same general topology based on phylogenetic analysis. All of the sequences fell in five clusters and were distinct from those of Anabaena, Gloeobacter, Streptomyces, Flavobacterium, Prosthecochloris, and Desulfuromonas, which have never been found in the grass carp intestine. Analysis of the microbial diversity by denaturing gradient gel electrophoresis demonstrated that unculturable bacteria were dominant bacteria in the grass carp intestine and thus the predominant phytate-degrading organisms. The predominant cultured species corresponding to the phytate-degrading isolates, Pseudomonas, Bacillus and Shewanella species, might be the main source of known BPPs. A phytase from Brevundimonas was first obtained from cultured species. Combining our results with Lim et al.'s inference that phytate-mineralizing bacteria are widely distributed and highly diverse in nature (B. L. Lim, P. Yeung, C. Cheng, and J. E. Hill, ISME J. 1:321-330, 2007), we concluded that BPP is the major phytate-degrading enzyme in nature, that most of this enzyme might originate from unculturable bacteria, and that the distribution of BPP may be related to the type of niche. To our knowledge, this is the first study to experimentally estimate BPP diversity in situ.
myo-Inositol hexakisphosphate, or phytate, is the most prevalent form of the inositol phosphates (IPx, where x is 1, 2, 3, 4, 5, or 6) that comprise a group of organic phosphorus compounds found widely in nature (37). In terrestrial ecosystems, phytate is synthesized by plants and represents a very significant amount (60 to 80%) of the organic phosphorus in the soil (18). In aquatic environments, however, phytate from terrestrial runoff, which is the major external source of organic phosphorus, is only a minor component, suggesting that it is rapidly hydrolyzed under aquatic conditions (34, 35). Thus, phytate may be one of the main source of available phosphorus in aquatic environments, and studies of it might be helpful for further understanding aquatic phosphorus cycling.
The phosphate ester linkages in phytic acid are quite stable. Natural degradation is almost impossible, and chemical hydrolysis in the laboratory is very slow (37). However, phytase (myo-inositol hexakisphosphate phosphohydrolase; EC 3.1.3.8 or EC 3.1.3.26), which is secreted by a variety of microbes, can rapidly break down phytate into phosphorus compounds, myo-inositol, and other nutrients that can be consumed and utilized by animals (23). To date, four classes of phytases have been characterized: histidine acid phosphatase (HAP), cysteine phytase, purple acid phosphatase, and beta-propeller phytase (BPP) (17, 23). BPPs are widely distributed in nature and play a major role in phytate-phosphorus cycling in both soil and aquatic microbial communities (17). However, no research has been conducted to assess BPP diversity in vivo with respect to distinct roles of different BPPs in phytate-phosphorus cycling systems.
To assess BPP diversity in nature, we selected the digestive tract of grass carp (Ctenopharyngodon idellus C. et V.) because the fish digestive tract, which contains a dense microbial population (25), is an open system that is constantly in contact with the surrounding water (30). To meet the need for dietary phosphorus, the grass carp consumes plants, in which the majority of phosphorus is stored as phytate. In fish intestines, phytate is hydrolyzed by numerous microbes. Thus, the fish digestive tract is an attractive system in which to study aquatic phosphorus cycling.
In the present work, grass carp, an herbivorous fish that can consume hundreds of aquatic plant species to meet its nutritional needs (20), was selected to study BPP diversity among intestinal microbes. We exploited the gene sequences of BPPs in the Protein Family Database (Pfam) to design degenerate BPP-specific primers and used these primers to construct a clone library from metagenomic DNA extracted directly from the intestinal contents of grass carp; this approach allowed us to assess BPP diversity in a culture-independent manner. Simultaneously, a culture-based assessment was performed by using a serial dilution culture technique to isolate phytate-degrading strains. Partial BPP genes were cloned from these isolates using the same BPP-specific primers and compared with fragments in the clone library. Furthermore, denaturing gradient gel electrophoresis (DGGE) of variable region V3 of the 16S rRNA gene was used to assess the microbial community in situ. These approaches are very helpful for determining BPP diversity in the fish intestine and provide information concerning the roles of phytases in phosphorus cycling.
MATERIALS AND METHODS
Sample collection, assay of total phytase activity, and DNA extraction.
Twelve specimens of grass carp (weight, 2.0 to 2.5 kg; length, 35 to 40 cm) were obtained from Jiaxing (Zhejiang, People's Republic of China) in April 2006; in the habitat examined, the carp have access to only Lolium perenne as food. The fish were randomly divided into four groups containing three specimens each. The intestine of the grass carp was collected and washed with sterile phosphate-buffered saline (0.1 mol liter−1; pH 7.2; Sigma, United States) and then split with sterile scissors. The intestinal contents of the fish in each group were pooled to obtain a representative sample. To measure the phytase activity in the intestinal contents, the method reported by Yanke et al. (41) was used, with a slight modification. Briefly, 15 g of intestinal contents was transferred to a fresh centrifuge tube (50 ml) and centrifuged at 20,000 × g and 4°C for 20 min. The phytase activity in the supernatant was determined using the standard method described below at pH 4.5 and pH 7.0. Genomic DNA was obtained using an extraction method described by Yu and Morrison (43), with some modifications. Specifically, 0.3 g of intestinal contents was transferred to a fresh 2-ml screw-cap tube containing 1 ml lysis buffer (500 mM NaCl, 50 mM Tris-HCl [pH 8.0], 50 mM EDTA, 4% sodium dodecyl sulfate) and 0.4 g of sterile glass beads (0.3 g of 0.1-mm-diameter beads and 0.1 g of 0.5-mm-diameter beads). Homogenization was performed with a mini-Beadbeater (BioSpec Products, United States) at the maximum speed for 90 s. Then the sample was incubated at 70°C for 20 min with gentle shaking every 5 min and centrifuged at 16,000 × g and 4°C for 5 min. The supernatant was transferred to a fresh 2-ml tube. Genomic DNA was recovered by precipitation with isopropanol and subjected to sequential digestion with RNase and proteinase K, followed by final purification using a Cycle-Pure DNA kit (Omega, United States). The presence and size of purified DNA were determined by agarose gel electrophoresis and ethidium bromide staining.
Primer design and testing.
A total of 66 amino acid sequences of BPPs were obtained from Pfam (release 18.0) (http://www.sanger.ac.uk/Software/Pfam/) and aligned using ClustalX software (ftp://ftp-igbmc.u-strasbg.fr/pub/ClustalX/) (12). Fifteen sequences representing different microbial species were selected and aligned using the boxshade service program (www.ch.embnet.org/software/BOX_form.html) for primer design. Based on the alignment, conserved regions were identified (Fig. 1) and primers were designed and tested further to check their specificity. To further identify the conserved regions, the phytase genes of Bacillus subtilis 168PhyA (accession no. CAB13871) and Shewanella oneidensis PhyS (accession no. AAN55555), which have been successfully expressed in a heterogeneous host (2), were used as putative probes to search online databases of microbial genomes and environmental metagenomes using BLAST.
FIG. 1.
Multiple-sequence alignment of 15 BPP sequences. The conserved lysine residue is indicated by an asterisk, and the two conserved sequence motifs are enclosed in boxes. Abbreviations (GenBank accession numbers are in parentheses): B. sub, Bacillus subtilis WHNB02 (CAB91845); B. lic, Bacillus licheniformis ATCC 14580 (YP_077686); S. wit, Sphingomonas wittichii RW1 (ABQ66899); C. cre, Caulobacter crescentus CB15 (AAK23276); V. ang, Vibrio angustum S14 (EAS63574); P. flu, Pseudomonas fluorescens Pf-5 (YP_260816); P. syr, Pseudomonas syringae pv. tomato strain DC3000 (NP_793025); A. vin, Azotobacter vinelandii AvOP (ZP_00418781); I. bal, Idiomarina baltica OS145 (ZP_01042413); S. sp., Shewanella sp. strain ANA-3 (YP_869843); O. sp., Oceanobacter sp. strain RED65 (EAT12633); D. ace, Desulfuromonas acetoxidans DSM 684 (ZP_01312505); F. joh, Flavobacterium johnsoniae UW101 (YP_001193718); C. lim, Chlorobium limicola DSM 245 (ZP_00512507); S. coe, Streptomyces coelicolor A3(2) (NP_631736).
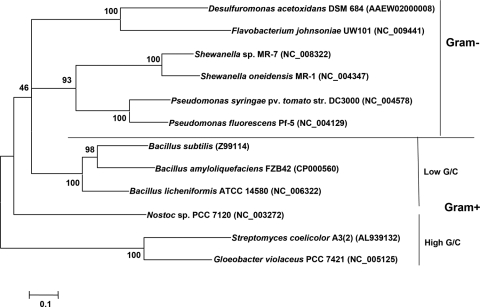
Twelve representative BPP-producing strains with distinct separation among Cyanobacteria, low-G+C- and high-G+C-content gram-positive bacteria, and gram-negative bacteria (Fig. 2) were selected to evaluate the feasibility of primers designed in our study. The genomic DNA of these strains were extracted for use as templates and subjected to a touchdown PCR. Briefly, the template DNA (∼50 to 100 ng) was added to a 50-μl reaction mixture containing 2.5 U Taq polymerase, 0.2 mM deoxynucleoside triphosphates, 1 mM of each primer, and 2 mM Mg2+ in the buffer supplied by the manufacturer (New England Biolabs, United States). The optimized PCR conditions for primers BPP-F and BPP-R were 4 min at 95°C, followed by eight cycles of 95°C for 30 s, 57°C (decreasing by 1°C after each cycle) for 30 s, and 72°C for 30 s, followed by 27 cycles of 95°C for 30 s, 48°C for 30 s, and 72°C for 30 s and then a final extension at 72°C for 5 min. The presence and size of amplification products were determined on agarose gels.
FIG. 2.
Phylogenetic tree showing the relationship between 12 complete BPP sequences from representative strains used for primer testing. The tree was constructed by the neighbor-joining method using MEGA 4.0. The sequences were selected from GenBank, and the strains showed distinct separation among Cyanobacteria, low-G+C- and high-G+C-content gram-positive bacteria, and gram-negative bacteria.
PCR and construction of a clone library.
BPP gene fragments were amplified by touchdown PCR as described above using the purified metagenomic DNA from the intestinal contents of grass carp as the template. PCR products were visualized on an agarose gel, the correct size was confirmed, and the bands were excised and purified using a QIAquick gel extraction kit (Qiagen, United States). To construct the clone library, the purified PCR products were ligated into the vector pGEM-T Easy (Promega, United States) and then electroporated into Escherichia coli DH5α (TaKaRa, Japan) using the procedure recommended by the manufacturer. Cells were grown on solid LB medium containing 100 mg ml−1 ampicillin, 80 mg ml−1 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), and 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 37°C for 15 h.
Partial sequencing and phylogenetic analysis.
Three hundred transformants (white colonies) were randomly picked and subjected to PCR using primers pGEM-F (5′-CCGACGTCGCATGCTCC-3′) and pGEM-R (5′-CTCCCATATGGTCGACCTG-3′) to verify the presence of full-length clone inserts. Sequencing was performed by Sunbiotech (China) using an automated sequencer (model 3730; Applied Biosystems, United Kingdom). Sequences were corrected by hand and compared to similar DNA sequences retrieved from the GenBank database using the BLASTx program. Nucleotide sequences of BPP gene fragments were translated into amino acid sequences by EMBOSS Transeq (http://www.ebi.ac.uk/emboss/transeq) and aligned at the protein level using ClustalX. Phylogenetic trees were constructed with MEGA version 4.0 (36) using the neighbor-joining method (29). Bootstrap analysis with 1,000 replications was performed to obtain confidence estimates for tree topologies.
Isolation of phytate-degrading strains.
Phytate-specific medium (PSM) (38), with some modifications, was used to isolate and culture bacteria that utilize phytic acid (P1036 from rice; Sigma) as the only source of phosphorus; this medium contained (per liter) 0.5 g glucose, 5 g phytic acid, 1 g (NH4)2SO4, 7 g KCl, 0.1 g MgSO4·7H2O, 0.1 g CaCl2·2H2O, 1 ml 0.1 M FeNa-EDTA, and 1 ml of a complete trace element solution (which contained [per liter] 15 g Na2EDTA·2H2O, 0.99 g MnCl2·4H2O, 0.43 g ZnSO4·7H2O, 0.24 g CoCl2·6H2O, 0.22 g Na2MoO4·2H2O, 0.19 g NiCl2·6H2O, 0.15 g H3BO4, and 0.08 g Na2SeO3·6H2O). The pH of PSM was adjusted to 7.2 using HCl, and solid PSM contained 1% agarose.
A dilution method was used to isolate phytate-degrading strains from the intestinal contents of grass carp using PSM agarose plates. After 3 days of cultivation, the colonies were repeatedly picked and streaked on the same medium to obtain pure cultures. Each subculture was transferred at least three times to ensure that it was pure, and all cultivation steps were performed at 28°C for 5 days. The pure cultures were then incubated in liquid PSM, and each culture supernatant was used for measurement of the inorganic phosphate content and phytase activity. To identify the phytate-degrading strains, partial 16S rRNA gene sequences of pure cultures were amplified with primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-CGGYTACCTTGTTACGACTT-3′) and were sequenced using the same primers to authenticate the isolates.
Orthophosphate content and phytase activity assay.
Orthophosphate content was determined by the ferrous sulfate-molybdenum blue method (10). Phytase activity was measured using the same method, with minor modifications. Briefly, 50 μl of an enzyme solution was incubated with 950 μl of a substrate solution (1.5 mM sodium phytate in 0.1 M Tris-HCl containing 1 mM Ca2+, pH 7.2) at 37°C for 30 min. The reaction was stopped by adding 1 ml of 10% (wt/vol) trichloroacetic acid. The amount of released inorganic phosphate was determined by adding 2 ml of coloring reagent C (1% [wt/vol] ammonium molybdate, 3.2% [vol/vol] sulfuric acid, and 7.2% [wt/vol] ferrous sulfate), and the optical density at 700 nm was measured. One unit of phytase activity was defined as the amount of enzyme that released 1 μmol of phosphate per min at 37°C. All phytase activity determinations were performed in triplicate using phosphate-free glassware.
Cloning and expression of phytase genes from phytate-degrading strains.
To obtain BPP gene fragments from phytate-degrading strains, touchdown PCR was performed with degenerate primers BPP-F and BPP-R and genomic DNA of phytate-degrading strains as templates using the same procedure that was used for library construction. The amplified fragment of the appropriate size was ligated into the pGEM-T Easy vector for sequencing. Based on sequence novelty, the phytase gene from Brevundimonas sp. strain Gc-2-c was selected for gene cloning and expression. The 5′ and 3′ flanking regions of the gene fragment were obtained using thermal asymmetric interlaced PCR (19). Amplification was performed using a genome walking kit (TaKaRa). The Brevundimonas sp. BPP gene without the signal peptide-encoding sequence was cloned into the NcoI-NotI site of pET-22b(+) and then transformed into E. coli BL21(DE3) competent cells. Positive transformants were cultured in 25 ml LB medium containing 100 μM ampicillin at 37°C to an optic density at 600 nm of 0.5 to 0.7. Protein expression was induced by addition of IPTG at a final concentration of 1 mM and incubation for an additional 4 h at 37°C. The phytase activities in the pellet and culture supernatant were determined.
DGGE analysis.
To explore the diversity of microflora in the intestinal contents of grass carp, DGGE was performed using a DCode universal mutation detection system (Bio-Rad). Variable region 3 of the 16S rRNA gene was amplified by PCR using primers 338f (5′-ACTCCTACGGGAGGCAGCAG-3′) and 519r (5′-ATTACCGCGGCTGCTGG-3′) (16). The 338f primer had a 40-bp GC clamp (5′-CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGG-3′) at its 5′ end. The thermocycling program was as follows: 95°C for 4 min, followed by 20 cycles of 95°C for 30 s, 65°C (decreasing by 0.5°C after each cycle) for 30 s, and 72°C for 30 s, followed by 15 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s and then a final extension of 72°C for 5 min. PCR products were analyzed on agarose gels to confirm the presence of a single amplicon of the expected size. The DGGE gels were 8% polyacrylamide gels with a 40 to 60% denaturant gradient; 100% denaturant was defined as 7 M urea and 40% formamide (21). Each 1 to 2 μg of purified PCR product was loaded, and gels were run in 1× Tris-acetate-EDTA buffer for 3.5 h at 200 V at 60°C. Gels were stained with 5 mg ml−1 ethidium bromide for 20 min, washed with deionized water, and visualized by UV transillumination.
Prominent bands appearing in the DGGE profile were purified and cloned into E. coli TOP10 using the pGEM-T vector system (Promega) according to the manufacturer's instructions. Plasmids containing DNA inserts were sequenced by Sunbiotech. The sequences for representative bands were checked for chimeric constructs using the CHECK_CHIMERA (Classifier) program at the Ribosomal Database Project website (http://rdp.cme.msu.edu/index.jsp) (4). After unreliable sequences at the 3′ and 5′ ends were removed, all sequences were subjected to similarity searches with the BLAST program.
Nucleotide sequence accession numbers.
All of the gene sequences determined in this study have been deposited in the GenBank database. Accession numbers EU822342 to EU822429, FJ539245 to FJ539340, and FJ548651 to FJ548746 have been assigned to the BPP gene fragments cloned from intestinal contents of grass carp, pond sediments and the aquatic environment, and soil collected from forests, vegetables, and glacier carp, respectively. Accession numbers of FJ548747, FJ548748, and FJ529821 have been assigned to the phytase gene from Citrobacter sp., Klebsiella sp., and Brevundimonas sp., respectively. Accession numbers EF669485 to EF669504 have been assigned to representative sequences obtained by DGGE. The accession numbers of the 16S rRNA gene sequences for the 10 strains isolated are FJ159432 to FJ159441.
RESULTS
BPP-specific primers.
Based on amino acid sequence alignment for 15 BPPs representing different microbial species and BLAST analysis, two conserved regions of BPPs, D-A-(A/T/E)-D-D-P-A-(I/L/V)-W and N-N-(V/I)-D-(I/L/V)-R-(Y/D/Q), were identified (Fig. 1). CODEHOPs primers (26, 27) were designed to amplify part of the BPP gene fragments. The sequence of forward primer BPP-F was 5′-GACGCAGCCGAYGAYCCNGCNITNTGG-3′, and the sequence of reverse primer BPP-R was 5′-CAGGSCGCANRTCIACRTTRTT-3′. Using these primers, PCR products of the expected size (160 to 200 bp) were obtained when DNA from 12 representative BPP-producing strains were used as templates (Fig. 2). All of the fragments were confirmed to be part of BPP genes by sequencing and BLAST analysis.
BPP gene fragment clone library.
BPP-specific primers were used to amplify phytase gene fragments directly from the metagenomic DNA extracted from the intestinal contents of grass carp with touchdown PCR. The amplified fragments ranged from 160 to 200 bp long and were ligated into the pGEM-T Easy vector. Over 3,000 clones were obtained after incubation at 37°C overnight. Three hundred positive clones were randomly picked for further confirmation by PCR with primers pGEM-F (5′-CCGACGTCGCATGCTCC-3′) and pGEM-R (5′-CTCCCATATGGTCGACCTG-3′). Of the 300 positive clones, 264 with an insert of the correct size were sequenced for analysis. Based on BLASTx analysis, 231 clones showed the highest level of identity (45 to 98%) to the validly published BPP protein sequences in the GenBank database, indicating that these clones were BPPs and that some of them might be novel.
Alignment of BPP amino acid sequences.
According to the correct reading frame, all 231 BPP gene fragments were translated into amino acid sequences by EMBOSS Transeq (http://www.ebi.ac.uk/Tools/emboss/transeq) and aligned using ClustalX. A total of 88 of these sequences showed divergence with levels of identity less than 95%. A highly conserved lysine residue, corresponding to Lys76 of the B. subtilis sequence, was identified in all of the sequences (Fig. 1). B. subtilis Lys76 has been reported to be an active site residue for phosphate binding (2, 7). These results further confirmed that the PCR products amplified directly from extracted metagenomic DNA were closely related to members of the BPP family.
Phylogenetic analysis.
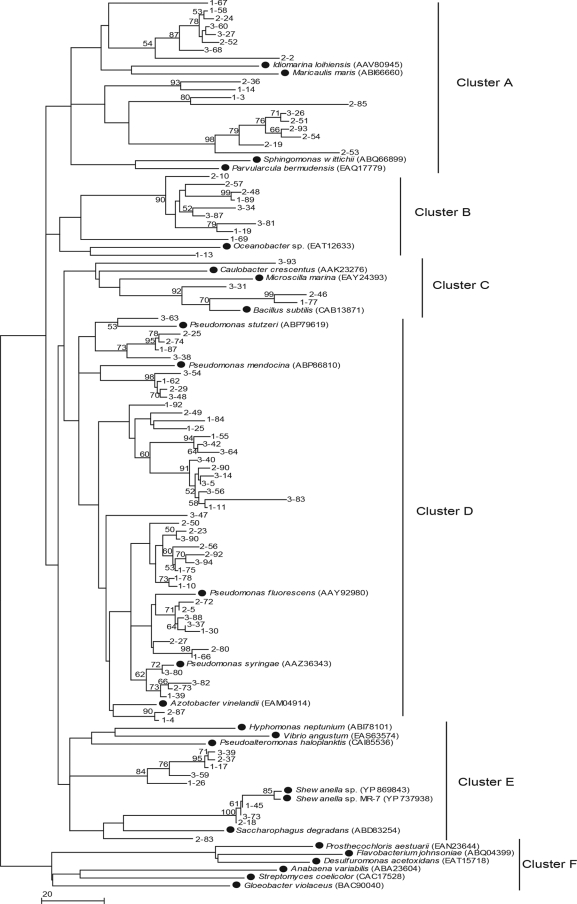
There are 66 microbial BPPs that were derived from 19 genera and 48 species and are validated in the Pfam database, and 25 representative sequences from 19 genera were randomly selected and used as references. A phylogenetic tree was constructed using the 88 clones and 25 reference sequences. High bootstrap values separated the partial sequences of BPPs into six distinct clusters, designated clusters A, B, C, D, E, and F (Fig. 3). The BPP gene fragments from the grass carp intestine were distributed in five of the six clusters (clusters A to E), indicating the substantial diversity of the clone sequences and their distinct separation from some of the known BPPs.
FIG. 3.
Phylogenetic analysis based on partial amino acid sequences of BPPs encoded by the metagenomic DNA from the intestinal contents of grass carp and the relationship of these sequences to the sequences of strains containing a BPP gene (indicated by dots) from the Protein Family Database (Pfam). The tree was constructed using the neighbor-joining method (MEGA 4.0), and the phytases were assigned to clusters A to F based on partial amino acid sequences. The lengths of the branches indicate the relative divergence among the amino acid sequences. Scale bar = 0.20 substitution per base position. The numbers at the nodes indicate bootstrap values based on 1,000 bootstrap replications.
There were 41 (46.6%) clones and 14 (56%) reference sequences in clusters A to C and E (Fig. 3). The reference sequences represented 13 genera, namely, Alteromonas, Bacillus, Caulobacter, Hyphomonas, Idiomarina, Maricaulis, Microscilla, Pseudoalteromonas, Parvularcula, Oceanobacter, Shewanella, Sphingomonas, and Vibrio. More than one-half of the clones (53.4%) fell in cluster D (Fig. 3), which included five reference sequences derived from Pseudomonadaceae. Most clone sequences in cluster D were highly similar (62 to 98%) to the phytases from Pseudomonas, Shewanella, and Bacillus, bacteria that are common in fish intestines.
Cluster F (Fig. 3) contained only six reference sequences, which were derived from Anabaena, Desulfuromonas, Flavobacterium, Gloeobacter, Prosthecochloris, and Streptomyces. Except for Flavobacterium, these genera are widely distributed in soil and/or aquatic systems, but they have never been reported to be in fish intestines (25, 33). No clone fell in cluster F, probably due to a low level or lack of these bacteria in the grass carp intestine. This result also indicated that BPP sequences might be correlated with their microbial sources.
Isolation of phytate-degrading strains.
Ten phytate-degrading bacteria (Table 1) were isolated using PSM (38), in which phytic acid was as the only phosphate source added. Based on 16S rRNA gene sequences, these strains were classified into six genera: Pseudomonas (four strains), Brevundimonas (two strains), Shewanella (two strains), Bacillus (one strain), Klebsiella (one strain), and Citrobacter (one strain). BPPs have been found in Bacillus (13), Shewanella (2), and Pseudomonas based on GenBank sequences. HAPs have also been isolated from Klebsiella and Citrobacter and characterized (6, 15). This is the first report, however, of a phytase in Brevundimonas.
TABLE 1.
Phytate-degrading strains isolated from the intestinal contents of grass carp, their closest relatives based on 16S rRNA gene sequence identity, amount of phosphate released by each culture, phytase activity in the supernatant, length of phytase gene fragment, and clone number
| Strain | Closest relative (% 16S rRNA gene identity) | Amt of phosphate released (μmol ml−1)a | Phytase activity (U ml−1)b | Fragment length (bp) | Clone |
|---|---|---|---|---|---|
| Gc-1-a | Bacillus subtilis (99) | 45 | 0.63 | 163 | 1-77 |
| Gc-2-c | Brevundimonas aurantiaca (99) | 83 | 0.37 | 181 | 2-19 |
| Gc-3-a | Brevundimonas aurantiaca (99) | 79 | 0.31 | 181 | 2-19 |
| Gc-4-d | Shewanella sp. strain Hac353 (99) | 111 | NMc | 163 | 3-73 |
| Gc-4-c | Pseudomonas rhodesiae (98) | 130 | NM | 163 | 2-74 |
| Gc-5-a | Pseudomonas pseudoalcaligenes (99) | 103 | NM | 163 | 1-10 |
| Gc-6-b | Pseudomonas sp. strain MACL14 (98) | 114 | NM | 163 | 2-27 |
| Gc-7-a | Pseudomonas marginalis (99) | 98 | NM | 163 | 3-48 |
| Gc-7-c | Klebsiella variicola (99) | 38 | NM | NDd | ND |
| Gc-8-a | Citrobacterfreundii (99) | 59 | NM | ND | ND |
Amount of phosphate released into the culture supernatant after cultivation for 120 h.
Phytase activity in the culture supernatant at pH 7.2.
NM, not measurable.
ND, no data collected.
Orthophosphate content and phytase activity.
The total phytase activity of grass carp intestinal contents was 0.29 ± 0.06 U ml−1 at pH 7.0, and no phytase activity was detected at pH 4.5, suggesting that the phytase in the grass carp intestine functions only at neutral pH. The orthophosphate content and phytase activity in the culture supernatant of each isolate were determined after growth for 120 h in PSM. In a culture that was not inoculated there was a negligible level of phosphate in the medium after 120 h, and thus any phosphate measured in the supernatant was the result of bacterial activity. The concentration of orthophosphate ranged from 45 to 114 μmol ml−1, and the highest phytase activity detected was the activity in a Bacillus sp. strain (Gc-1-a) (0.63 ± 0.11 U ml−1) (Table 1).
Cloning and expression of phytase genes from phytate-degrading strains.
Two HAP gene fragments have been cloned from Citrobacter sp. and Klebsiella sp. using the degenerate primers specific for HAP (10), and they showed the highest levels of identity to the phytases from Citrobacter freundii (86%) and Klebsiella pneumoniae (98%), respectively. The other eight ∼160-bp amplicons were obtained using degenerate primers BBP-F and BBP-R with the same PCR procedure that was used for the genomes of the corresponding phytate-degrading strains. These PCR products were sequenced and showed 45 to 84% sequence identity to BPP genes in the GenBank database. Importantly, all eight sequences could be accounted for by the clones in the library (Table 1).
The full-length BPP gene of Brevundimonas sp. strain Gc-2-c was obtained by thermal asymmetric interlaced PCR. The complete gene contained a 1,125-bp open reading frame encoding 374 amino acid residues with a typical signal peptide (residues 1 to 20). The mature protein has a theoretical molecular mass of 37.5 kDa. Sequence homology searches showed that the phytase exhibited the highest level of identity (42%) with a 3-phytase from Oceanicaulis alexandrii (accession no. ZP_00953252). The gene encoding the mature protein was successfully expressed in E. coli. After induction with 1 mM IPTG at 37°C for 4 h, the cell lysate and culture supernatant showed 0.11 ± 0.04 and 0.05 ± 0.05 U ml−1 phytase activity at pH 7.0, respectively. No phytase activity was detected in the supernatant of an uninduced culture or an induced transformant harboring an empty plasmid.
PCR-DGGE analysis.
The diversity of microflora in the intestinal contents of grass carp was determined by using a touchdown PCR approach based on variable region 3 of the16S rRNA gene and DGGE analysis. Figure 4 shows more than 24 bands for each group, indicating that there is microbial diversity in the intestinal contents of grass carp.
FIG. 4.
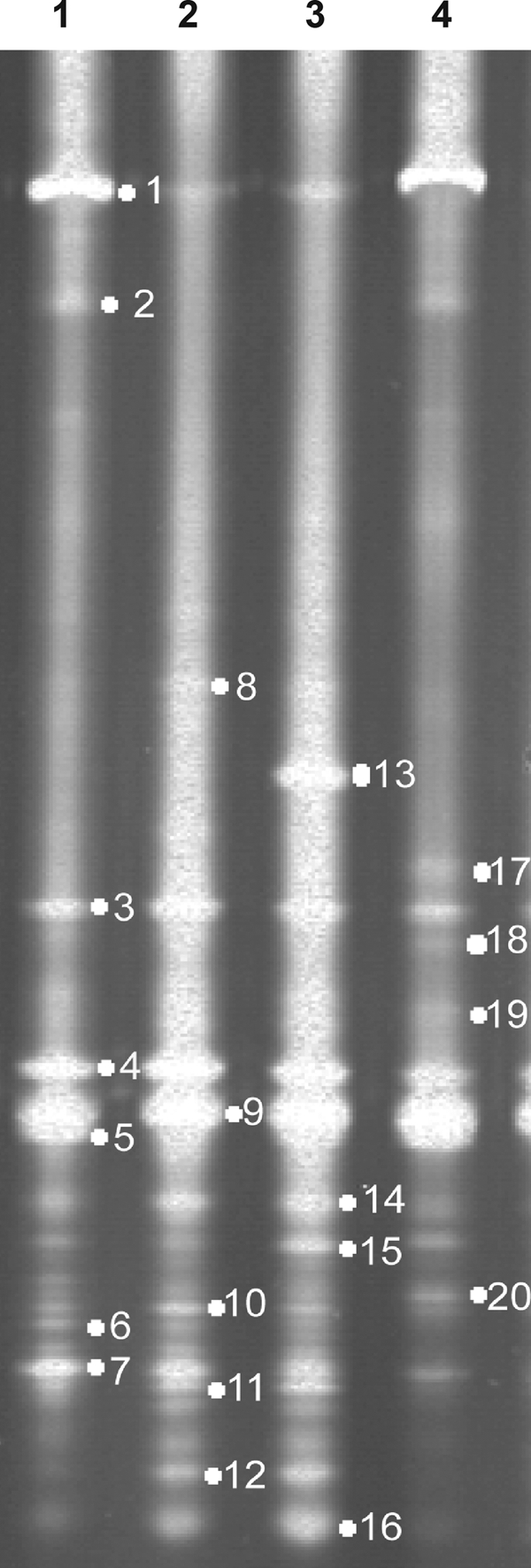
DGGE analysis based on sequence differences in the V3 region of the 16S rRNA genes extracted from the intestinal contents of grass carp. Lanes 1 to 4 contained representatives of the intestinal microflora community from four independent groups. The gel used was a 8% polyacrylamide gel with a 40 to 60% denaturant gradient (10% acrylamide).
To identify some of the bands and compare the DGGE profile to the diversity assessments based on culture-independent and traditional techniques, 20 distinct bands were collected for sequencing (Fig. 4 and Table 2). Most bands (14/20) showed high levels of sequence identity (93 to 100%) to sequences of uncultured bacteria, suggesting that unculturable microbes are the predominant microbes in the intestinal contents of grass carp. Bands 8, 10, 11, and 15 were most closely related to BPPs from Bacillus thuringiensis (99% identity), Pseudomonas aeruginosa (98% identity), Citrobacter sp. (100% identity), and a betaproteobacterium (98% identity), respectively, all of which were predominantly cultured species in fish intestine contents.
TABLE 2.
Taxa, closest relatives as determined by sequence identity, and accession numbers for the bands on the 16S rRNA gene region V3 DGGE gels
| Phylogenetic group | Banda | Closest relative (obtained by BLAST search) | % Identity | Accession no. |
|---|---|---|---|---|
| Firmicutes | 1 | Uncultured bacterium clone PeM37 | 96 | EF669485 |
| 13 | Uncultured bacterium isolate BF0001C129 | 100 | EF669486 | |
| 3 | Uncultured bacterium clone PI48L1A11 | 96 | EF669490 | |
| 16 | Uncultured bacterium clone J5-01 | 98 | EF669499 | |
| 9 | Uncultured bacterium clone J5-01 | 99 | EF669501 | |
| 8 | Bacillus thuringiensis strain N21 | 99 | EF669502 | |
| Cyanobacteria | 19 | Uncultured cyanobacterium clone Lk1Mc-1 | 100 | EF669487 |
| 6 | Uncultured bacterium clone FCPT677 | 98 | EF669489 | |
| 14 | Uncultured cyanobacterium clone 1PN75 | 97 | EF669491 | |
| 2 | Uncultured bacterium clone FCPT677 | 99 | EF669495 | |
| 4 | Uncultured Dunaliella sp. clone At18AugA12 | 93 | EF669500 | |
| 5 | Uncultured bacterium clone FCPT677 | 99 | EF669503 | |
| Proteobacteria | 18 | Escherichia coli strain BETD | 100 | EF669492 |
| 15 | Betaproteobacterium strain IMCC1721 | 98 | EF669493 | |
| 7 | Uncultured bacterium isolate BF0001C047 | 100 | EF669494 | |
| 11 | Citrobacter sp. strain F3-3 | 100 | EF669496 | |
| 20 | Afipia sp. SNRB6-4 | 99 | EF669497 | |
| 12 | Uncultured bacterium isolate BF0002C056 | 97 | EF669498 | |
| 10 | Pseudomonas aeruginosa strain 6702 | 98 | EF669504 | |
| Archaea | 17 | Uncultured bacterium clone M3a | 98 | EF669488 |
Bands in the 16S rRNA gene region V3 DGGE gel shown in Fig. 4.
DISCUSSION
To date, four classes of phytases have been identified. A comparison of the properties of these phytases indicated that only BPP has a neutral optimum pH (pH 6.0 to 7.5) for activity, whereas the other enzymes have an acidic optimum pH (pH 2.5 to 5.5) (2, 23). In terms of structure and function, HAP, purple acid phosphatase, and cysteine phytase require binding of free phytic acid; however, under neutral conditions negatively charged phytic acid could bind positively charged ions, such as Ca2+, Mg2+, etc., to form stable but insoluble complexes, thereby precluding direct hydrolysis by these three classes of phytases. In contrast, BPP is the only phytase that hydrolyzes phytate and liberates inorganic phosphate, when phytic acid forms an insoluble complex under neutral conditions in the presence of Ca2+ (2, 13). Therefore, because most terrestrial and aquatic environments have a neutral pH, we suggest that BPP might be the major (and most widespread) phytate-degrading enzyme in nature and that it may play a major role in phytate-phosphorus cycling. The same hypothesis has been proposed by Lim et al. (18) based on an analysis of all the microbial and environmental sequence databases available online.
For many aquatic animals, such as the common carp (1), Atlantic salmon (39), red sea bream (42), juvenile grass shrimp (32), and others, the main hydrolysis products of phytate (phosphorus and myo-inositol) are essential for normal growth. Monogastric or agastric aquatic animals do not produce intestinal phytase, and thus they require phytases derived from intestinal microbes for efficient phytate hydrolysis in the neutral pH environment of intestine. Microbial populations are rather dense in the fish digestive tract, and the numbers of microorganisms are much higher than the numbers in the surrounding water (25). These environmental factors suggest that both dense microbial populations and BPP production in the fish intestine may be necessary to hydrolyze sufficient amounts of phytate to meet the nutritional needs of the fish. Grass carp, an herbivorous fish, can consume hundreds of aquatic plant species to meet its nutritional needs (20). To meet the nutrient requirement for phosphorus and myo-inositol, phytate in plants could be degraded in the digestive tract of grass carp by intestinal microbial phytases. Therefore, the digestive tract of grass carp is a good model with which to study the diversity of BPP, phosphorus nutrition, and aquatic phosphorus cycling.
In this study, a set of degenerate primers specific for BPP was used to amplify BPP gene fragments from metagenomic DNA of intestinal contents of grass carp, and 88 distinct sequences (levels of identity, <95%) of BPP gene fragments were obtained (Fig. 3). Currently, HAP phytases (40) with high specific activity are the most widely studied phytases and have been used in animal feed and for environmental protection; however, no correct PCR products were amplified from the same source using degenerate primers specific for HAP (primers FI and RI) (10). This result indicated that BPPs might be more widely distributed in intestinal contents of grass carp than HAPs. Furthermore, the primer set specific for BPP also amplified a number of BPP gene fragments from pond sediments (54 distinct sequences) and water (42 distinct sequences) directly, confirming the great diversity and high density of BPPs in pond sediments and the aquatic environment. Also, some of the sequences in the three different environments have very high levels of identity (>95%). Generally, the diverse BPPs in the intestine of grass carp showed a good level of similarity with the BPPs obtained from the pond sediments and water, indicating that BPPs are widely distributed in nature at a high density. Similar results were also obtained by amplifying BPP gene fragments using the specific degenerate primers from the metagenomic DNA of soil collected from forests (40 distinct sequences), vegetables (23 distinct sequences), and glaciers (33 distinct sequences).
The BLAST and conserved lysine residue (7) analysis of 88 independent fragment sequences confirmed that all of the fragments belonged to the BPP family. Then the phylogenetic analysis of independent fragments and reference sequences illustrated the substantial diversity of BPPs in the grass carp intestine (Fig. 3). However, BPPs have previously been classified into seven clusters based on the domain structure and the position of the cysteine residue (18), but the makeup of these clusters is not consistent with that of our six clusters. In our phylogenetic tree, BPP gene fragments from Pseudomonadaceae and Shewanellaceae, which have similar domain structures, were classified into two clusters. This difference between the clusters of Lim et al. (18) and our clusters may be due to the difference in the sequence lengths used to construct the phylogenetic trees. A partial sequence of the phytase domain structure was used in our study, whereas complete protein sequences, which generally contain other domains, were used by Lim et al. (18). Both trees, however, indicated the substantial diversity of BPPs.
Notably, no clones fell in cluster F (Fig. 3). In this cluster, six reference sequences were derived from Anabaena, Gloeobacter, Streptomyces, Flavobacterium, Prosthecochloris, and Desulfuromonas. Anabaena and Gloeobacter, belonging to the Cyanobacteria, are found in various aquatic environments worldwide. Streptomyces is widely distributed in nature, especially in soil (28). Prosthecochloris is found only on sick coral that is infected with this pathogen (5). Desulfuromonas, a sulfur-reducing organism, can be found in marine sediments (3). Flavobacterium has been reported to occur in the intestines of salmonid fish (25), and it is common in soil and freshwater (22). However, strains belonging to these genera have never been reported to be related to the strains in the grass carp intestine (33). Lim et al. (18) also reported that no marine BPP-like sequences were found in the freshwater database by BLAST analysis. These results suggest that BPP diversity is related to the distribution of microflora in the environment. The diversity and phylogenetic distribution of BPP fragment sequences may therefore differ according to the niche type.
Ten phytate-degrading strains isolated from the intestinal contents of grass carp (Table 1) were classified into six genera: Bacillus, Brevundimonas, Shewanella, Pseudomonas, Klebsiella, and Citrobacter. Previously, several HAP genes have been cloned from Klebsiella and Citrobacter (16). In this study, the relevant HAP gene fragments were obtained using degenerate primers specific for HAP (10) from the two phytate-degrading strains of Klebsiella and Citrobacter. In addition, eight BPP fragments were cloned from the other strains, and six of them were cloned from Bacillus, Shewanella, and Pseudomonas strains. A large number of strains belonging to these three genera are common in fish intestines (25, 33), and several BPP genes have been reported (2, 9, 13). Our phylogenetic tree indicated that a large number of BPP fragments have high levels of identity (62 to 98%) with the phytases of these three genera, suggesting that common intestinal bacteria might be the prevalent phytate-degrading organisms. The other two phytase fragments from Brevundimonas were identical, so one of them was selected for gene cloning and expression. The phytase gene from Brevundimonas contained a 1,125-bp open reading frame and encoded a functional BPP. This is the first report of cloning and expression of a Brevundimonas BPP gene.
Based on the results of the isolation and phylogenetic analysis of phytate-degrading strains in cluster D, the genus Pseudomonas appears to be an important microbial source of BPP in the grass carp intestine. However, more experiments and analysis are required to determine whether Pseudomonas spp. are important sources of BPPs elsewhere in nature. Indeed, several Pseudomonas isolates have been shown to produce phytase (11, 14, 24). Hill et al. (9) once screened 10 typical soil bacteria that released orthophosphate from phytate into phosphate-free phytate medium after cultivation for 21 h, and 6 of them belonged to the genus Pseudomonas.
DGGE profiles showed that 14 of 20 bands (70%) were from unculturable microorganisms in the intestinal contents of grass carp, suggesting that unculturable bacteria are the predominant bacteria in situ. Six culturable strains were strains of Bacillus, Citrobacter, Afipia, Pseudomonas, Escherichia, and a betaproteobacterium. Three of these strains were isolated by culturing in PSM, and the corresponding BPP gene fragments were cloned. To some extent, the results of DGGE correlated well with those obtained by culture-based methods. Only 10 phytate-degrading strains were isolated with the selective medium; however, 88 distinct BPP gene fragments were cloned. BPP genes from isolates accounted for only 10% of the clonal sequences. Considering the DGGE results, this difference might be ascribed to the fact that most BPP-producing strains were unculturable.
Compared with the traditional culture-based methods, metagenomics has become a powerful tool to discover new genes because of its ability to assess environmental samples comprehensively (8, 31). In this study, a set of degenerate primers specific for BPP genes successfully amplified diverse BPP gene fragments from the metagenomic DNA extracted from the intestinal contents of grass carp. This degenerate primer set could also be used to screen clones containing BPP genes from cosmid or bacterial artificial chromosome libraries constructed by using metagenomic DNA from various environments. This culture-independent method could be more useful for cloning novel genes from extreme environments. In future studies, a highly effective method to isolate full-length functional genes might be established based on other degenerate primers and metagenomic libraries.
Acknowledgments
This research was supported by the National High Technology Research and Development Program of China (863 Program, grant 2007AA100601) and the National Basic Research Program of China (973 Program, grant 2004CB719606).
Footnotes
Published ahead of print on 16 January 2009.
REFERENCES
- 1.Aoe, H., and I. Masuda. 1967. Water soluble vitamin requirements of carp—II. Requirements for para-aminobenzoic acid and inositol. Bull. Jpn. Soc. Sci. Fish. 33:674-685. [Google Scholar]
- 2.Cheng, C., and B. L. Lim. 2006. Beta-propeller phytases in the aquatic environment. Arch. Microbiol. 185:1-13. [DOI] [PubMed] [Google Scholar]
- 3.Coates, J. D., D. J. Lonergan, E. J. Philips, H. Jenter, and D. R. Lovley. 1995. Desulfuromonas palmitatis sp. nov., a marine dissimilatory Fe(III) reducer that can oxidize long-chain fatty acids. Arch. Microbiol. 164:406-413. [PubMed] [Google Scholar]
- 4.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, and G. M. Garrity. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorlenko, V. M. 1970. A new phototrophic green sulphur bacterium, Prosthecochloris aestuarii nov. gen. nov. spec. Z. Allg. Mikrobiol. 10:147-149. [PubMed] [Google Scholar]
- 6.Greiner, R., E. Haller, U. Konietzny, and K. D. Jany. 1997. Purification and characterization of a phytase from Klebsiella terrigena. Arch. Biochem. Biophys. 341:201-206. [DOI] [PubMed] [Google Scholar]
- 7.Ha, N. C., B. C. Oh, S. Shin, H. J. Kim, T. K. Oh, Y. O. Kim, K. Y. Choi, and B. H. Oh. 2000. Crystal structures of a novel, thermostable phytase in partially and fully calcium-loaded states. Nat. Struct. Biol. 7:147-153. [DOI] [PubMed] [Google Scholar]
- 8.Hårdeman, F., and S. Sjöling. 2007. Metagenomic approach for the isolation of a novel low-temperature-active lipase from uncultured bacteria of marine sediment. FEMS Microbiol. Ecol. 59:524-534. [DOI] [PubMed] [Google Scholar]
- 9.Hill, J. E., D. Kysela, and M. Elimelech. 2007. Isolation and assessment of phytate-hydrolysing bacteria from the DelMarVa Peninsula. Environ. Microbiol. 9:3100-3107. [DOI] [PubMed] [Google Scholar]
- 10.Huang, H., H. Luo, P. Yang, K. Meng, Y. Wang, T. Yuan, Y. Bai, and B. Yao. 2006. A novel phytase with preferable characteristics from Yersinia intermedia. Biochem. Biophys. Res. Commun. 350:884-889. [DOI] [PubMed] [Google Scholar]
- 11.In, M. J., E. S. Jang, Y. J. Kim, and N. S. Oh. 2004. Purification and properties of an extracellular acid phytase from Pseudomonas fragi Y9451. J. Microbiol. Biotechnol. 14:1004-1008. [Google Scholar]
- 12.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23:403-405. [DOI] [PubMed] [Google Scholar]
- 13.Kerovuo, J., M. Lauraeus, P. Nurminen, N. Kalkkinen, and J. Apajalahti. 1998. Isolation, characterization, molecular gene cloning, and sequencing of a novel phytase from Bacillus subtilis. Appl. Environ. Microbiol. 64:2079-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, Y. H., M. N. Gwon, S. Y. Yang, T. K. Park, C. G. Kim, C. W. Kim, and M. D. Song. 2002. Isolation of phytase-producing Pseudomonas sp. and optimization of its phytase production. J. Microbiol. Biotechnol. 12:279-285. [Google Scholar]
- 15.Kim, Y. O., H. W. Kim, J. H. Lee, K. K. Kim, and S. J. Lee. 2006. Molecular cloning of the phytase gene from Citrobacter braakii and its expression in Saccharomyces cerevisiae. Biotechnol. Lett. 28:33-38. [DOI] [PubMed] [Google Scholar]
- 16.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, NY.
- 17.Lei, X., J. M. Porres, E. J. Mullaney, and H. Pedersen. 2007. Phytase: source, structure and application, p. 505-529. In J. Polaina and A. P. MacCabe (ed.), Industrial enzymes: structure, function and applications. Springer, Dordrecht, The Netherlands.
- 18.Lim, B. L., P. Yeung, C. Cheng, and J. E. Hill. 2007. Distribution and diversity of phytate-mineralizing bacteria. ISME J. 1:321-330. [DOI] [PubMed] [Google Scholar]
- 19.Liu, Y. G., and R. F. Whittier. 1995. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25:674-681. [DOI] [PubMed] [Google Scholar]
- 20.Masser, M. P. 2002. Using grass carp in aquaculture and private impoundments. Southern Regional Aquaculture Center publication no. 3600. Southern Regional Aquaculture Center, Stoneville, MS.
- 21.McCaig, A. E., L. A. Glover, and J. I. Prosser. 2001. Numerical analysis of grassland bacterial community structure under different land management regimens by using 16S ribosomal DNA sequence data and denaturing gradient gel electrophoresis banding patterns. Appl. Environ. Microbiol. 67:4554-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKay, A. M. 1991. Isolation of spontaneous mutant strains of Flavobacterium spp. Lett. Appl. Microbiol. 13:265-267. [Google Scholar]
- 23.Mullaney, E. J., and A. H. Ullah. 2003. The term phytase comprises several different classes of enzymes. Biochem. Biophys. Res. Commun. 312:179-184. [DOI] [PubMed] [Google Scholar]
- 24.Richardson, A. E., and P. A. Hadobas. 1997. Soil isolates of Pseudomonas spp. that utilize inositol phosphates. Can. J. Microbiol. 43:509-516. [DOI] [PubMed] [Google Scholar]
- 25.Ringe, E., E., E. Strom, and J. A. Tabachek. 1995. Intestinal microflora of salmonids: a review. Aquacult. Res. 26:773-789. [Google Scholar]
- 26.Rose, T. M., J. G. Henikoff, and S. Henikoff. 2003. CODEHOP (COnsensus-DEgenerate Hybrid Oligonucleotide Primer) PCR primer design. Nucleic Acids Res. 31:3763-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rose, T. M., E. R. Schultz, J. G. Heinkoff, S. Pietrokovski, C. M. McCallum, and S. Heinkoff. 1998. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 26:1628-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saadoun, I., and E. Al-Momani. 1997. Studies on soil streptomycetes from Jordan. Actinomycetes 8:42-48. [Google Scholar]
- 29.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 30.Sangbrita, S., R. R. Narayan, S. S. Kumar, and R. A. Kumar. 2006. Characterization of cellulase-producing bacteria from the digestive tract of tilapia, Oreochromis mossambica (Peters) and grass carp, Ctenopharyngodon idella (Valenciennes). Aquacult. Res. 37:380-388. [Google Scholar]
- 31.Schmeisser, C., H. Steele, and W. R. Streit. 2007. Metagenomics, biotechnology with non-culturable microbes. Appl. Microbiol. Biotechnol. 75:955-962. [DOI] [PubMed] [Google Scholar]
- 32.Shiau, S. Y., and S. L. Su. 2004. Dietary inositol requirement for juvenile grass shrimp, Penaeus monodon. Aquaculture 241:1-8. [Google Scholar]
- 33.Sugita, H., K. Tokuyama, and Y. Deguchi. 1985. The intestinal microflora of carp Cyprinus carpio, grass carp Ctenopharyngodon idella and tilapia Sarotherodon niloticus. Bull. Jpn. Soc. Sci. Fish. 51:1325-1329. [Google Scholar]
- 34.Suzumura, M., and A. Kamatani. 1995. Origin and distribution of inositol hexaphosphate in estuarine and coastal sediments. Limnol. Oceanogr. 40:1254-1261. [Google Scholar]
- 35.Suzumura, M., and A. Kamatani. 1995. Mineralization of inositol hexaphosphate in aerobic and anaerobic marine sediments: implications for the phosphorus cycle. Geochim. Cosmochim. Acta 59:1021-1026. [Google Scholar]
- 36.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 37.Turner, B. L., M. J. Papházy, P. M. Haygarth, and I. D. McKelvie. 2002. Inositol phosphates in the environment. Philos. Trans. R. Soc. Lond. B 357:449-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Unno, Y., K. Okubo, J. Wasaki, T. Shinano, and M. Osaki. 2005. Plant growth promotion abilities and microscale bacterial dynamics in the rhizosphere of lupin analyzed by phytate utilization ability. Environ. Microbiol. 7:396-404. [DOI] [PubMed] [Google Scholar]
- 39.Waagbø, R., K. Sandnes, and Ø. Lie. 1998. Effects of inositol supplementation on growth, chemical composition and blood chemistry in Atlantic salmon, Salmo salar L., fry. Aquacult. Nutr. 4:53-59. [Google Scholar]
- 40.Wodzinski, R., and A. H. Ullah. 1996. Phytase. Adv. Appl. Microbiol. 42:263-302. [DOI] [PubMed] [Google Scholar]
- 41.Yanke, L. J., H. D. Bae, L. B. Selinger, and K. J. Cheng. 1998. Phytase activity of anaerobic ruminal bacteria. Microbiology 144:1565-1573. [DOI] [PubMed] [Google Scholar]
- 42.Yone, Y., M. Furuichi, and K. Shitanda. 1971. Vitamin requirements of red sea bream. I. Relationship between inositol requirements and glucose levels in the diet. Bull. Jpn. Soc. Sci. Fish. 37:149-155. [Google Scholar]
- 43.Yu, Z., and M. Morrison. 2004. Improved extraction of PCR quality community DNA from digesta and fecal samples. BioTechniques 36:808-814. [DOI] [PubMed] [Google Scholar]