Abstract
Plus-stranded RNA viruses coopt host proteins to promote their robust replication in infected hosts. Tomato bushy stunt tombusvirus (TBSV) is a model virus that can replicate a small replicon RNA in Saccharomyces cerevisiae and in plants. The tombusvirus replicase complex contains heat shock protein 70 (Hsp70), an abundant cytosolic chaperone, which is required for TBSV replication. To dissect the function of Hsp70 in TBSV replication, in this paper we use an Hsp70 mutant (ssa1 ssa2) yeast strain that supports a low level of TBSV replication. Using confocal laser microscopy and cellular fractionation experiments, we find that the localization of the viral replication proteins changes to the cytosol in the mutant cells from the peroxisomal membranes in wild-type cells. An in vitro membrane insertion assay shows that Hsp70 promotes the integration of the viral replication proteins into subcellular membranes. This step seems to be critical for the assembly of the viral replicase complex. Using a gene-silencing approach and quercetin as a chemical inhibitor to downregulate Hsp70 levels, we also confirm the significance of cytosolic Hsp70 in the replication of TBSV and other plant viruses in a plant host. Taken together, our results suggest that cytosolic Hsp70 plays multiple roles in TBSV replication, such as affecting the subcellular localization and membrane insertion of the viral replication proteins as well as the assembly of the viral replicase.
Plus-stranded RNA [(+)RNA] viruses replicate efficiently in host cells by assembling their own replicase complexes, consisting of virus- and host-encoded proteins, the viral RNA, and host membranes (1, 2, 10, 33, 34, 36, 41, 54, 56, 58). The replication of (+)RNA viruses takes place on the cytosolic surfaces of various cellular organelles, such as the endoplasmic reticulum (ER) and mitochondria, whereas some viruses actively induce the formation of novel cytoplasmic vesicular compartments (12, 16, 23, 24, 31, 53, 54). The emerging picture is that the mechanism of genome replication and the functions of viral and host factors might be analogous to some extent among various (+)RNA viruses in spite of their diverse genome organization and gene expression strategies. Also, most of the previously identified host factors are conserved genes, suggesting that (+)RNA viruses might selectively target conserved host functions as opposed to species-specific factors. This strategy can help viruses to have a broader host range and to expand infections to new host species. Host factors are also key determinants of virus pathology, host-virus interactions, and the evolution of the virus.
The tombusvirus replicase is among the best characterized for (+)RNA viruses due to the development of in vitro assays and Saccharomyces cerevisiae as a model host (33, 35, 44, 46, 48, 55). Replication of tombusvirus RNA depends on the virus-encoded RNA-dependent RNA polymerase (p92pol RdRp) and the p33 replication cofactor, the key protein in the recruitment of the viral RNA into replication (50). The two viral proteins, in combination with 4 to 10 host proteins, assemble on the peroxisomal membrane into the viral replicase complex in the yeast model host (36, 64). The list of host proteins identified within the functional replicase includes heat shock protein 70 (Hsp70; encoded by the SSA1 and SSA2 genes in yeast), whose downregulation leads to markedly low replication (55). An additional host factor is glyceraldehyde-3-phosphate dehydrogenase (encoded by the TDH2 and TDH3 genes in yeast), which binds to the minus-stranded Tomato bushy stunt tombusvirus (TBSV) RNA and affects plus-strand synthesis (61). Another defined component is the ubiquitin-conjugating enzyme Cdc34p, which is involved in ubiquitination of the p33 replication cofactor (27). The cytosolic transport protein Pex19p is only transiently associated with the replicase, likely during the transport of the replication proteins to the peroxisomal membranes, the site of replication (47). It is not known at present how these host factors might affect the assembly of the viral replicase complex.
The host-encoded heat shock proteins, such as the Hsp70 chaperone family, the J-domain chaperones, and Hsp90, are implicated in the replication of plus-stranded RNA viruses (such as hepatitis C virus and Flock House virus), minus-stranded RNA viruses (such as influenza virus and vesicular stomatitis virus), retroviruses (such as human immunodeficiency virus), hepatitis B virus, and other RNA viruses (9, 11, 13, 26, 32, 37, 38, 42, 51, 57, 60, 62). The proposed activities for these host chaperones during virus replication include the stimulation of polymerase (RdRp) activity (32), the enhancement of replication (22), the activation of reverse transcriptase for hepadnaviruses (18, 59), the formation of virus-induced inclusion bodies (9), and the assembly of closterovirus virions (3). The cytosolic Hsp70 proteins might also affect the stability/function of viral proteins during infections, since a subset of HSP70 genes are expressed at enhanced levels in plants infected by various plant viruses (4, 6, 65, 66).
To dissect the function of Hsp70 in TBSV replication, we tested the effect of Hsp70 on the subcellular distribution of the viral replication proteins, on replicase activity, and on in vitro membrane insertion of replication proteins. Using an HSP70 mutant yeast (ssa1 ssa2), we find that the viral replication proteins remain cytosolic at an early time point, suggesting that Hsp70 is involved in the subcellular localization of the viral replication proteins to intracellular membranes. An in vitro membrane insertion assay demonstrates that Hsp70 promotes the integration of the viral replication proteins into the subcellular membranes. We also show evidence that cytosolic Hsp70 is critical for the replication of TBSV and other plant viruses in a plant host.
MATERIALS AND METHODS
Yeast strains and expression plasmids.
Saccharomyces cerevisiae strain InvSc1 (Invitrogen) was used as the wild type (WT). The double-mutant (ssa1 ssa2) strain MW123 (his3 leu2 lys2 trp1 ura3 ssa1::HIS3 ssa2::LEU2) was kindly provided by Elizabeth A. Craig (University of Wisconsin) (7).
To express Ssa1-yellow fluorescent protein (YFP), we PCR amplified the full length of SSA1 from yeast genomic DNA using high-fidelity Taq polymerase (Invitrogen) and primers 1905 (CGCGCTCGAGATGTCAAAAGCTGTCGG) and 1906 (CGCGGGATCCATCAACTTCTTCAACGG). The PCR product was treated with XhoI and BamHI, followed by ligation into the corresponding sites of pGBK-MS2CP-enhanced YFP (EYFP) (43). The plasmid for expression of Ssa1-HF has been described elsewhere (55). To express the C-terminal fusion proteins Pex13-cyan fluorescent protein (CFP) and Pho86-CFP from the galactose-inducible GAL1 promoter, we used PCR with primer pair 1277 (CGGCAAGCTTACCATGTCATCCACAGCAGTACCACGA)-1278 (CGGGCTCGAGGTGTGTACGCGTTTCATCATCAACA) for PEX13 and primer pair1269 (CGGCAAGCTTACCATGGCGGTTCAACAAAGAAAGAAGA)-1270 (CGGGCTCGAGGTCCTTGTGTTCGGCTTTAAAATGGA) for PHO86, and with pGAD-Pex13-CFP and pGAD-Pho86-CFP as templates, respectively (43). The PCR products were treated with HindIII and BamHI and then ligated into the corresponding sites of pYES/NT-C. The N-terminal fusion proteins YFP-p33 and YFP-p92 have been described elsewhere (43).
Confocal laser microscopy.
To view yeast cells expressing different fluorescent fusion proteins, yeast strains were transformed with Ssa1-YFP, in combination with pGBKCFP-p33/pGAD-CFP-p92 or the subcellular markers Pex13-CFP/Pho86-CFP. Confocal laser microscopy was performed on an Olympus FV1000 microscope as described previously (21, 61).
Tombusvirus in vitro replicase assay.
The “membrane-enriched” tombusvirus replicase preparations, which are suitable for testing in vitro replicase activity on the endogenous templates bound to the replicase, were obtained from yeast as previously described (46).
Affinity purification of FLAG-Ssa1p and in vitro replicase assay.
For overexpression of Ssa1p in strain InvSc1 (Invitrogen), we used pYC-HFSSA, expressing FLAG-His6-tagged Ssa1p from the galactose-inducible GAL1 promoter (55). The same yeast also coexpressed His6-tagged p92 from the pGAD-His92 plasmid (46) and His6-tagged p33 and DI-72 replicon RNA (repRNA) from the dual expression plasmid pESC-HisY-p33-DI-72 (21). The yeast was grown at 23°C to an optical density of 0.6 and then harvested, and the membrane proteins were solubilized as described previously (46). The FLAG-His6-tagged Ssa1p was affinity purified using an anti-FLAG M2-agarose affinity gel (Sigma) (55). The in vitro replication assay was performed as described previously (46).
Subcellular fractionation.
Yeast cells were grown to an optical density at 600 nm of 0.8 to 1.0. One hundred milligrams of cells was broken in 600 μl of yeast lysis buffer (200 mM sorbitol, 50 mM Tris-HCl [pH 7.5], 15 mM MgCl2, 10 mM KCl, 10 mM β-mercaptoethanol, yeast protease inhibitor mix; Sigma), followed by centrifugation for 5 min at 100 × g to pellet unbroken cells (48, 49). For sucrose flotation gradient analysis, samples were adjusted to 52% (wt/wt) sucrose in lysis buffer, and 1,500 μl was loaded into the bottoms of ultraclear polycarbonate ultracentrifuge tubes (Beckman), overlaid with 2,700 μl of 45% sucrose in lysis buffer, topped with 600 μl of 10% sucrose in lysis buffer, and subsequently centrifuged at 40,000 rpm at 4°C for >16 h by using an SW55 Ti rotor in a Beckman L8-55M ultracentrifuge. Gradients were manually fractionated into eight fractions of 480 μl each. Then 15-μl samples from each fraction were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting procedures as described previously (20).
RNA and protein analysis.
Total-RNA isolation and Northern blot analysis were performed as described previously (44). Protein analysis was performed as described previously using an anti-His6 antibody (46) or anti-Hsc70 (Stressgen Bioreagents, MI) as the primary antibody for the detection of Nicotiana benthamiana Hsp70, whereas the secondary antibody was alkaline phosphatase-conjugated anti-mouse immunoglobulin G (Sigma).
Whole plant and protoplast experiments.
Preparation of N. benthamiana protoplasts, electroporation with TBSV RNA, and viral RNA analysis were performed as described previously (45).
Plasmids pTRV1, pTRV2, pYL619, pYL622, and pPDS for virus-induced gene silencing (VIGS) were kindly provided by S. Dinesh-Kumar (Yale University) (15). Agrobacterium tumefaciens strain C58C1 carrying one of the VIGS plasmids was infiltrated into leaves of N. benthamiana as described previously (14). The mRNA levels of HSP70-1, which expresses the major cytosolic Hsc70 protein, in the upper leaves were examined by reverse transcription-PCR (RT-PCR) using primers 2731 (CGTGCCAGATTTGAGGAGTTGAAC) and 2732 (CTGTGATCTGTGGCACACCCCTAGGTG) 6 days postinfiltration. The leaves above the agroinfiltrated leaves were inoculated with virion preparations of TBSV, Cucumber necrosis virus (CNV), Turnip crinkle virus (TCV), or Tobacco mosaic virus (TMV) 6 days after infiltration. Total-RNA extracts from the upper (systemically infected) leaves were prepared at 3 days after inoculation.
Quercetin (Sigma) was dissolved in dimethyl sulfoxide (DMSO) and used at concentrations of 100 μM to 1 mM for infiltration into N. benthamiana leaves by using a syringe. For the protoplast treatment, quercetin was added to the isolated N. benthamiana protoplasts 30 min prior to electroporation. After electroporation, the protoplasts were cultured in protoplast culture medium (45) containing different amounts of quercetin. For the whole-plant treatment, we first inoculated N. benthamiana leaves with a TBSV, TMV, or TCV virion preparation, followed by infiltration with quercetin, used between concentrations of 100 μM to 1 mM, by using a syringe. The same concentration of DMSO was infiltrated into leaves as a control. Note that the inoculated leaves were infiltrated with either quercetin or DMSO ∼10 min after inoculation.
In vitro assay for the insertion of p33 into the membrane.
For the in vitro membrane insertion assay, p33, p92, p33C, and TDH2 mRNAs were synthesized, using the T7 transcription kit (TaKaRa Bio Inc.), from PCR products obtained with primers 2144 (GTAATACGACTCACTATAGGGAAGCTATA) and 2145 (TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTAAAACCT AAGAGTCAC) and with pGBK-HisCNV33, pGAD-His92, and pGBK-His-Tdh as templates. Using these mRNAs, we obtained 35S-labeled p33, p92, and Tdh2p in wheat germ extract according to the manufacturer's instructions (Promega). The membrane insertion reaction was carried out in a 10-μl reaction volume containing 2 μl of yeast membrane (48), 2.5 μl of buffer A (30 mM HEPES, 150 mM potassium acetate, 5 mM magnesium acetate, 130 mM sorbitol), 1 mM (each) ribonucleotides (ATP, CTP, GTP, UTP), 20 mM creatine phosphate, 0.1 μl of creatine kinase, and 0.05 μl of dithiothreitol. As a negative control, we used the soluble fraction of the yeast extract (the supernatant fraction was obtained by centrifugation at 40,000 × g for 1 h at 4°C) lacking membranes.
For the protease protection assay, we used 4 μl of the reaction assay product described above plus 1 μl of buffer A (48) and 1 μl of water or 1 μl of 10% Triton X-100, followed by the addition of 1 μl of protease K (70 μg/ml). The reaction mixtures were incubated at room temperature for 30 or 45 min, and the reaction was stopped with 50 μl of 13% trichloroacetic acid for protein precipitation. The samples were analyzed using 13% sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
RESULTS
Partial relocalization of Hsp70 to the sites of TBSV replication.
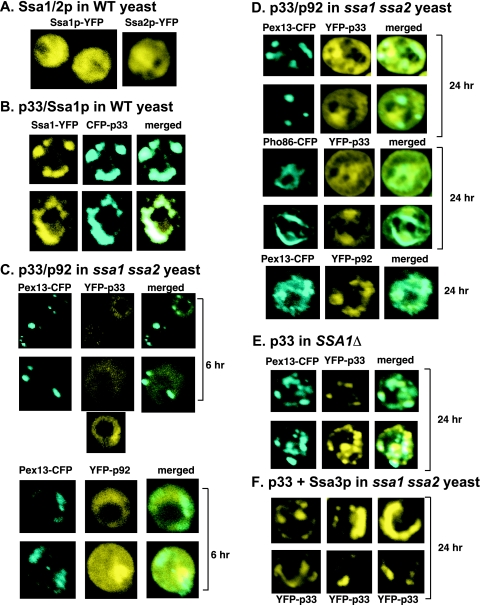
A previous proteomics approach revealed that the constitutively expressed Hsp70 proteins, termed Ssa1p and Ssa2p (Ssa1/2p) in yeast, are present in the highly purified tombusvirus replicase complex (55). To determine if the interaction between Ssa1/2p and the viral replicase is cytoplasmic (since a large portion of Ssa1/2p is normally localized in the cytoplasm of yeast cells in the absence of viral replication) (Fig. 1A) (19) or if it occurs at the sites of TBSV replication, we coexpressed YFP-tagged Ssa1p and CFP-tagged p33 replication protein in yeast cells. Confocal laser microscopy of yeast cells 16 h after the induction of Ssa1p-YFP and CFP-p33 expression revealed partial relocalization of Ssa1p-YFP from the cytosol to punctate structures (Fig. 1B), which is a characteristic feature of TBSV replication proteins (21, 30, 43). The redistributed Ssa1p-YFP colocalized with CFP-p33, suggesting that Ssa1p-YFP is located in the same subcellular compartment as CFP-p33, which is present in the viral replicase (46, 55). As expected on the basis of earlier studies (30, 43, 52), this subcellular compartment is the peroxisomal membrane. The redistribution of Ssa1p-YFP to punctate structures was also observed during the replication of TBSV DI-72 repRNA, when we coexpressed the p33 and p92pol replication proteins in yeast (data not shown). These data support the idea that Ssa1/2p are recruited to the site of TBSV replication through interaction with the p33 replication protein.
FIG. 1.
Cytosolic localization of the p33 and p92pol replication proteins in ssa1 ssa2 yeast. (A) Cytosolic localization of Ssa1-YFP and Ssa2-YFP in the absence of TBSV replication. Each experiment was repeated, and 20 or more cells were analyzed. (B) Relocalization of Ssa1-YFP to punctate structures containing viral CFP-p33 in yeast. (C) Cytosolic localization of YFP-p33 or YFP-p92 in ssa1 ssa2 yeast 6 h after induction from pGBK-YFP-p33 or pGBK-YFP-p92, respectively, by the addition of galactose to the growth medium. The peroxisomal marker protein Pex13-CFP was expressed from the GAL1 promoter. (D) Localization of YFP-p33 or YFP-p92 in ssa1 ssa2 yeast 24 h after induction. Pex13-CFP and Pho86-CFP were used as marker proteins for the peroxisomal and ER membranes, respectively. (E) Peroxisomal localization of YFP-p33 in SSA1Δ yeast. See further details in panel B. (F) Overexpression of Ssa3p in ssa1 ssa2 yeast leads to the formation of large punctate structures containing YFP-p33. These structures are reminiscent of those found in WT yeast (panel B).
Cytosolic distribution of the p33 and p92pol replication proteins in ssa1 ssa2 yeast.
To test if Ssa1/2p are involved in the formation of the characteristic punctate structures driven by p33/p92pol, we used ssa1 ssa2 double-mutant yeast, which is viable due to the expression of additional Hsp70 proteins, termed Ssa3p and Ssa4p (7). Six hours after the expression of YFP-p33, the localization of YFP-p33 in ssa1 ssa2 yeast looked dramatically different from that in WT yeast. For example, the characteristic punctate structures did not form, and the YFP-p33 remained cytosolic, frequently showing a “doughnut”-like distribution (Fig. 1C). In contrast, the CFP-tagged Pex13p, a cellular marker of peroxisomal membranes (19), localized to punctate structures as in WT yeast cells. We found a cytosolic distribution similar to that of YFP-p33 for YFP-p92pol in ssa1 ssa2 yeast (Fig. 1C), suggesting a defect in the subcellular localization of both TBSV replication proteins shortly after their expression in the absence of functional Ssa1/2p.
Testing of the subcellular distribution of YFP-p33 and YFP-p92pol 24 h after their induction in ssa1 ssa2 yeast revealed a distribution different from that at the 6-h time point. Both p33 and p92pol formed a few punctate structures in ssa1 ssa2 yeast cells (Fig. 1D). However, these punctate structures in ssa1 ssa2 yeast showed less-defined borders and were present in lower numbers than the punctate structures formed in WT yeast. Also, a major portion of p33 and p92pol was still localized in the cytosol in ssa1 ssa2 yeast (Fig. 1D). Colocalization studies with the peroxisomal protein Pex13-CFP and the ER-specific protein Pho86-CFP (19) showed that p33 localized partly to the peroxisomal and ER membranes in ssa1 ssa2 yeast (Fig. 1D). This is in contrast with the strong colocalization of p33 with the peroxisomal marker in ssa1Δ yeast (Fig. 1E), which still expresses WT Ssa2p. Taken together, these data suggest that the subcellular localization of p33 and p92pol is greatly affected by Ssa1/2p. Moreover, there are host factors present in ssa1 ssa2 yeast that can partially complement the missing functions of Ssa1/2p, leading to peroxisomal and ER localization of a portion of p33 at the 24-h time point.
Since the host factors capable of partial complementation during p33/p92pol localization are likely the stress-inducible Hsp70 proteins Ssa3p and Ssa4p, we overexpressed Ssa3p from an expression vector in ssa1 ssa2 yeast. Coexpression of YFP-p33 in this yeast revealed mostly punctate structures (Fig. 1F), reminiscent of those seen in WT or ssa1Δ yeast (Fig. 1B and E). Based on the Ssa3p overexpression experiment, we conclude that Ssa3p overexpression can complement the function of Ssa1/2p in the subcellular localization of p33 and likely in that of p92pol.
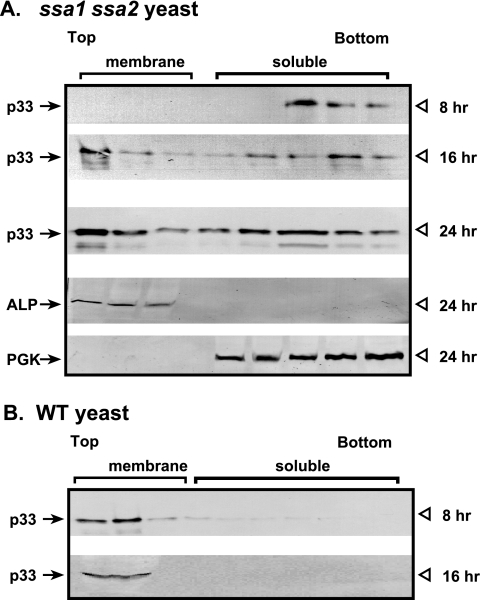
We also tested the subcellular distribution of p33 in ssa1 ssa2 yeast by fractionation experiments. Cellular extracts from WT and ssa1 ssa2 yeast strains expressing His6-tagged p33 from the galactose-inducible GAL1 promoter were subjected to membrane flotation studies, followed by Western blotting to identify the fractions containing p33. When the extract was obtained from ssa1 ssa2 yeast 8 h after the induction of p33 expression, we detected p33 in the bottom fractions, representing the soluble, cytosolic proteins (Fig. 2A), whereas p33 was present in the top, membrane-containing fractions in the WT extract (Fig. 2B). However, the distribution of p33 changed in extracts obtained from ssa1 ssa2 yeast at the 16- and 24-h time points. We found that p33 was present in both the top and bottom fractions, suggesting that it was partitioned between soluble and membrane-bound forms at the later time points when expressed in ssa1 ssa2 yeast (Fig. 2A). On the other hand, p33 was present exclusively in the top fractions at the 16-h time point when expressed in WT yeast (Fig. 2B). Taken together, these data from the fractionation experiments are in agreement with the observations from confocal microscopy studies that the subcellular localization of p33 is dramatically affected by Ssa1/2p in yeast cells.
FIG. 2.
Altered subcellular localization of p33 in ssa1 ssa2 yeast based on fractionation experiments. (A) Membrane flotation experiments followed by Western blotting reveal that p33 is distributed in the fractions containing soluble proteins at the early time point (8 h) and is partitioned between fractions containing soluble and membrane proteins at the late time points (16 and 24 h) in ssa1 ssa2 yeast. We used alkaline phosphatase (ALP), a vacuolar membrane protein, and 3-phosphoglycerate kinase (PGK), a soluble protein, as internal controls in the Western blots. (B) In WT yeast, in contrast, p33 is localized in fractions containing membrane proteins at both early and late time points.
Hsp70 is an integral component of the functional tombusvirus replicase.
To test if the Hsp70 proteins Ssa1p and Ssa2p affect the activity of the tombusvirus replicase, we obtained membrane-enriched fractions from WT and ssa1 ssa2 yeast strains expressing p33, p92pol, and TBSV repRNA. An in vitro replication assay with the endogenous TBSV repRNA revealed that the tombusvirus replicase prepared from the WT yeast was ∼5-fold more active (Fig. 3A, lanes 1 to 3) than that prepared from the ssa1 ssa2 yeast (lanes 4 to 6). This effect is not due to the expression of replication proteins, since they were present in comparable amounts (Fig. 3B). These data support the idea that Hsp70 directly affects the activity of the tombusvirus replicase.
FIG. 3.
Hsp70 affects the in vitro activity of the tombusvirus replicase and binds to the functional replicase. (A) In vitro replicase assay using the membrane-enriched fraction carrying the copurified repRNA. Gels show in vitro synthesis of the repRNA products by the tombusvirus replicase preparations obtained from WT (lanes 1 to 3) and ssa1 ssa2 (lanes 4 to 6) yeast. Yeast cells were harvested 16 h after the induction of TBSV replication. Note that p33 and p92 were expressed from the galactose-inducible GAL1 promoter, resulting in similar amounts of p33 and p92 proteins. (B) Western blot analysis of p33 and p92pol levels in the replicase preparations. Both proteins were expressed from the GAL1 promoter. (C) Copurification of the functional tombusvirus replicase with FLAG-tagged Ssa1p. The membrane-enriched fraction of yeast coexpressing His6-tagged p33, p92pol, and DI-72 (+)repRNA with FLAG-tagged Ssa1p was solubilized with a detergent, followed by FLAG affinity purification. The FLAG-Ssa1p preparations obtained were tested by a standard replicase assay with an added minus-stranded repRNA template for the presence of copurified tombusvirus replicase. The control preparations were also prepared using FLAG affinity purification, but from yeast expressing His6-tagged Ssa1p.
To obtain evidence that Hsp70 is bound to the functional replicase complex and not to misfolded replication proteins, we affinity purified FLAG-tagged Ssa1p from the solubilized membrane fraction of WT yeast cells expressing p33, p92pol, and the TBSV repRNA. An in vitro replicase assay performed with exogenously added viral RNA using the FLAG affinity-purified Ssa1p preparation revealed the presence of the active tombusvirus replicase (Fig. 3C, lanes 1 to 3). This suggests that the active replicase can be copurified with FLAG-Ssa1p. The control experiment, based on similar FLAG affinity purification of His6-tagged Ssa1p, revealed a lack of replicase activity, excluding the possibility that the replicase bound nonspecifically to the affinity column (Fig. 3C, lanes 4 to 6). Overall, these data convincingly demonstrated that Hsp70 (Ssa1p) is an integral component of the functional tombusvirus replicase.
Hsp70 is involved in the insertion of the replication proteins to the intracellular membranes.
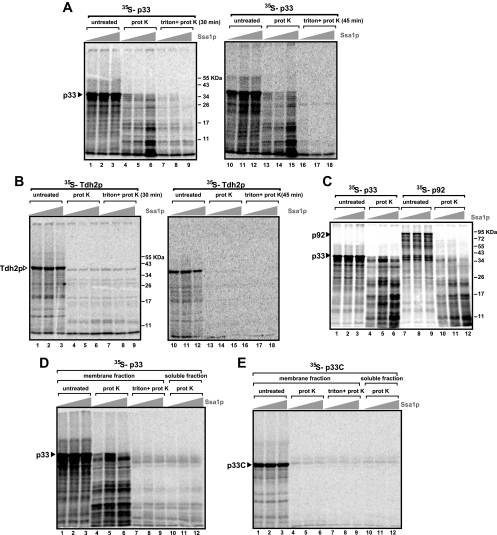
The cytosolic location of p33 and p92pol in ssa1 ssa2 yeast indicates that Ssa1/2p could be involved in the intracellular targeting of the replication proteins and/or in the insertion of the replication proteins, which are integral membrane proteins with two transmembrane-domains (30, 39, 46), into the membrane. To test the latter model, we have developed an in vitro assay including purified recombinant Ssa1p, 35S-labeled p33, and 35S-labeled p92pol, as well as a yeast cell extract containing cellular membranes (48, 49). First, we synthesized 35S-labeled p33 and p92pol using the wheat germ in vitro translation assay. Then we performed the in vitro membrane insertion assay with either 35S-p33 or 35S-p92pol and added purified recombinant Ssa1p. Finally, to test the insertion of 35S-p33 into the membranes in the extract, we washed the isolated membranes with a high-salt buffer to remove peripheral membrane proteins. Then we treated the samples with proteinase K, since membrane insertion of p33 should provide increased protection against protease digestion. These experiments showed that the added purified recombinant Ssa1p increased the amount of proteinase K-resistant p33 fragments in the membrane fraction ∼5-fold (Fig. 4A, lanes 5 and 6 and 14 and 15) over that in the sample without added Ssa1p (lanes 4 and 13). Treatment of the membrane fraction with Triton X-100 made p33 sensitive to proteinase K regardless of the presence or absence of added Ssa1p (Fig. 4A, lanes 7 to 9 and 16 to 18). The simplest interpretation of these data is that treatment of the membranes with Triton X-100 rendered the integral p33 exposed and as susceptible to proteinase K as the nonintegral p33. Similar treatment of the 35S-labeled cytosolic protein Tdh2p with proteinase K did not show significant differences between the presence and the absence of Ssa1p (Fig. 4B, lanes 4 to 6 and 13 to 15). Also, treatment of the membranes with Triton X-100 and proteinase K did not alter the sensitivity of Tdh2p to proteinase K (Fig. 4B, lanes 7 to 9 and 16 to 18), indicating that Tdh2p was not associated with or protected by the membranes present in the samples.
FIG. 4.
Increased protection of p33 and p92pol fragments from proteinase K (prot K) digestion in the presence of yeast membranes and added purified recombinant Hsp70. (A) 35S-labeled p33 was added to a yeast membrane preparation in the absence of Ssa1p (lanes 1, 4, 7, 10, 13, and 16) or in the presence of 1.5 (lanes 2, 5, 8, 11, 14, and 17) or 2.5 (lanes 3, 6, 9, 12, 15, and 18) μg of purified Ssa1p, followed by proteinase K digestion (for 30 min [lanes 4 to 6] or 45 min [lanes 13 to 15]) or by solubilization with 1% Triton X-100 and subsequent proteinase K digestion (for 30 min [lanes 7 to 9] or 45 min [lanes 16 to 18]). The amount of proteinase-resistant fragments increased ∼5-fold (compare lanes 4 and 6). Note that a basic level of Hsp70, from the in vitro translation assay performed in wheat germ extract, is present in each sample. (B) Similar experiments were also performed with soluble 35S-labeled Tdh2p as a control in the presence of yeast membranes and added purified recombinant Hsp70, as described for panel A. Note that the added Ssa1p does not increase the protection of Tdh2p fragments. (C) A ∼5-fold increase in the amount of proteinase-resistant fragments of p92pol was also observed in the presence of yeast membranes when purified recombinant Hsp70 was added. Note that p33 and p92pol give rise to similar-sized fragments, suggesting that the overlapping segments with the two membrane-spanning domains constitute the protease-resistant fragments. (D) Ssa1p does not protect p33 from proteinase K in the absence of the membrane fraction. Experiments were carried out as described for panel A, except that the soluble fraction of the yeast extract was added to the samples in lanes 10 to 12. (E) The C-terminal half of p33, lacking the N-terminal hydrophobic membrane-spanning domains, is not protected from proteinase K by the addition of Ssa1p or the membrane fraction. For further details, see the legend to panel A.
To test if the membrane integration of p92pol was also affected by Hsp70, we used a similar in vitro assay with 35S-p92pol and proteinase K (Fig. 4C). The patterns of the proteinase K-resistant fragments for 35S-p33 and 35S-p92pol were strikingly similar, indicating that the overlapping portion of p92pol was also protected by the membrane. The most prevalent fragment was an ∼11-kDa fragment, likely including the two transmembrane domains present in both replication proteins. Addition of recombinant Ssa1p to the assay mixture enhanced the protection of p92pol fragments ∼4- to-5-fold (Fig. 4C, lanes 10 to 12). Overall, these data support the model that Ssa1p is involved in the insertion of p33 and p92pol into membranes.
To provide further support for the role of membranes in the protection of p33 from proteinase K digestion, we tested if the soluble fraction of the yeast extract in combination with Ssa1p could confer resistance against proteinase K on p33. This experiment demonstrated that p33 was as sensitive to proteinase K digestion in the presence of Ssa1p and the soluble fraction (Fig. 4D, lanes 10 to 12) as after the Triton treatment of the membrane fraction-containing samples (lanes 7 to 9). We also tested the sensitivity of p33 to proteinase K in the absence of the membrane-spanning N-terminal domain in p33. The purified recombinant p33C, containing the C-terminal hydrophilic half of p33, was as sensitive to proteinase K digestion in the presence of the membrane fraction and Ssa1p (Fig. 4E, lanes 4 to 6) as in the presence of the soluble fraction and Ssa1p (lanes 10 to 12). Altogether, the sensitivity of p33 in the presence of the soluble fraction of the yeast extract and the sensitivity of p33C in the presence of the membrane fraction suggest that Ssa1p does not make p33 insensitive to proteinase K in the absence of the membrane fraction and does not protect p33C, which lacks the membrane-spanning domain. Thus, these results support the model that Ssa1p is involved in the insertion of p33 into the membrane. However, it is likely that additional host proteins also contribute to p33 insertion into the membrane, as has been shown for some cellular proteins (67, 68).
Inhibition of TBSV gRNA accumulation by downregulation of HSP70 via gene silencing in the Nicotiana host.
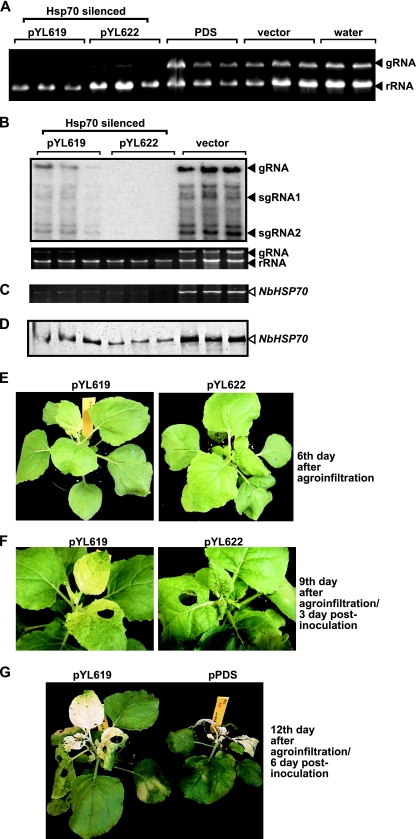
To demonstrate that Hsp70 plays a role in TBSV replication in a natural plant host, we used a gene-silencing approach with Nicotiana benthamiana as the host. Although the number of Hsp70 genes in N. benthamiana is currently unknown, there are 14 Hsp70 genes in Arabidopsis thaliana, and five of those Hsp70 genes code for cytosolic chaperones, which share high sequence homology (28). We tested two regions of the HSP70 gene cloned from N. benthamiana, which were inserted separately into a Tobacco rattle virus (TRV)-based gene-silencing vector in the Dinesh-Kumar laboratory (14, 15). The TRV-YL619 and TRV-YL622 vectors carrying the HSP70 sequences were expressed in N. benthamiana via agroinfiltration (14). The control constructs were the empty vector (TRV-RNA2) and TRV-PDS, carrying a sequence to silence N. benthamiana phytoene desaturase (PDS) gene expression (15). Another control was water infiltration into the leaves. Gene silencing with TRV-YL619 and TRV-YL622 affected the growth of the treated plants 14 days after infiltration (data not shown). Therefore, we aimed at testing TBSV RNA accumulation in the plants in which HSP70 had been silenced before the day 14 time point. Briefly, we inoculated the new leaves of agroinfiltrated (with TRV-YL619 and TRV-YL622) plants with a TBSV virion preparation 6 days after infiltration, followed by total-RNA isolation from both the TBSV-inoculated and uninoculated (systemically infected) leaves 3 days postinoculation (dpi). RNA gel analysis revealed that TBSV RNA accumulation decreased markedly in TRV-YL619- and TRV-YL622-silenced plants (Fig. 5A, 1st to 6th lanes), whereas TBSV RNA accumulation was as high in plants in which PDS had been silenced (7th to 9th lanes) or in empty-vector-treated plants (10th to 12th lanes) as in the nonsilenced (water-treated) plants (13th to 14th lanes). We confirmed by semiquantitative RT-PCR that N. benthamiana HSP70 (NbHSP70) mRNA levels were reduced in TRV-YL619- and TRV-YL622-silenced plants (Fig. 5C). Western blotting of total-protein extracts with an anti-Hsp70 antibody revealed that the levels of Hsp70 protein accumulation in TRV-YL619- and TRV-YL622-silenced plants were reduced to 32 and 21% of the control level, respectively (Fig. 5D). Northern blot analysis revealed 83 and 96% inhibition of TBSV genomic RNA (gRNA) accumulation in the uninoculated leaves of TRV-YL619- and TRV-YL622-silenced plants, respectively (Fig. 5B). Similar levels of inhibition of viral RNA accumulation were observed when the plants in which NbHSP70 was silenced were inoculated with CNV, a closely related tombusvirus. These results demonstrate that downregulation of NbHSP70 significantly reduces tombusvirus RNA accumulation. The TRV2-infected plants (Fig. 5A, 10th to 12th lanes) did not inhibit TBSV accumulation, excluding the possibility that either agroinfiltration or TRV infection could inhibit TBSV accumulation in N. benthamiana, as shown previously (61).
FIG. 5.
Knockdown of HSP70 mRNA levels by VIGS reduces the accumulation of TBSV RNA in N. benthamiana. (A) Total-RNA samples obtained from N. benthamiana leaves silenced as shown were analyzed by gel electrophoresis. Two different segments of HSP70 (pYL619 and pYL622) were used in the TRV silencing vectors. The control experiments included PDS in the TRV vector and the empty TRV vector, as well as infiltration with water. We chose the 6th day after VIGS to inoculate the upper, systemically silenced leaves with TBSV virions. Samples for RNA extraction were taken from the inoculated leaves 3 days postinoculation. (B) (Top) Northern blotting shows the accumulation of TBSV gRNA and subgenomic RNAs (sgRNAs). (Bottom) The ethidium bromide-stained gel shows rRNA and TBSV gRNA levels. These results shown here and in panel A are from different experiments. (C) VIGS reduced NbHSP70 mRNA levels in treated leaves. Semiquantitative RT-PCR was performed with primers that allow the amplification of a region of HSP70 not present in the pYL619 and pYL622 VIGS constructs. (D) Western blotting of silenced N. benthamiana leaves with an anti-Hsp70 antibody 6 days postinoculation. (E) Phenotypes of N. benthamiana plants in which Hsp70 was silenced, shown 6 days after agroinfiltration with pYL619 or pYL622. The pictures were taken just before the inoculation of the upper extended leaves with a TBSV virion preparation. (F) Phenotypes of N. benthamiana plants in which Hsp70 was silenced, shown 9 days after agroinfiltration with pYL619 or pYL622 and 3 days after inoculation with TBSV. Note that no symptoms of TBSV infection are visible at this early time point. The pictures were taken after the samples were collected for TBSV RNA analysis. (G) Symptoms of TBSV-infected N. benthamiana plants in which Hsp70 or PDS was silenced, shown 12 days after agroinfiltration with pYL619 or pPDS. The white/yellow coloring of the leaves is due to the silencing of Hsp70 or PDS after agroinfiltration with pYL619 or pPDS, respectively. The PDS-silenced plants died as quickly as the control plants treated with the empty TRV vector (data not shown).
Silencing of NbHSP70 with pYL622 caused severe phenotypes, leading ultimately to the death of the plants (data not shown), whereas silencing with pYL619 resulted in a white/yellow leaf phenotype (Fig. 5F and G), an effect somewhat similar to that caused by the silencing of PDS (data not shown). However, the inhibition of TBSV RNA accumulation due to knockdown of NbHSP70 was more dramatic than that due to changes in the rRNA level (Fig. 5A), suggesting that TBSV replication is especially sensitive to the level of NbHSP70. The apparent changes in the phenotype of the plants caused by the silencing of NbHSP70 were visible during the 3-day period of infection (Fig. 5E and F), though no viral symptoms were visible. The young leaves went through rapid necrosis in nonsilenced or PDS-silenced plants after CNV or TBSV infections (Fig. 5G), followed by death of the whole plants.
Inhibition of TBSV genomic RNA accumulation in the Nicotiana host by quercetin, an inhibitor of HSP70.
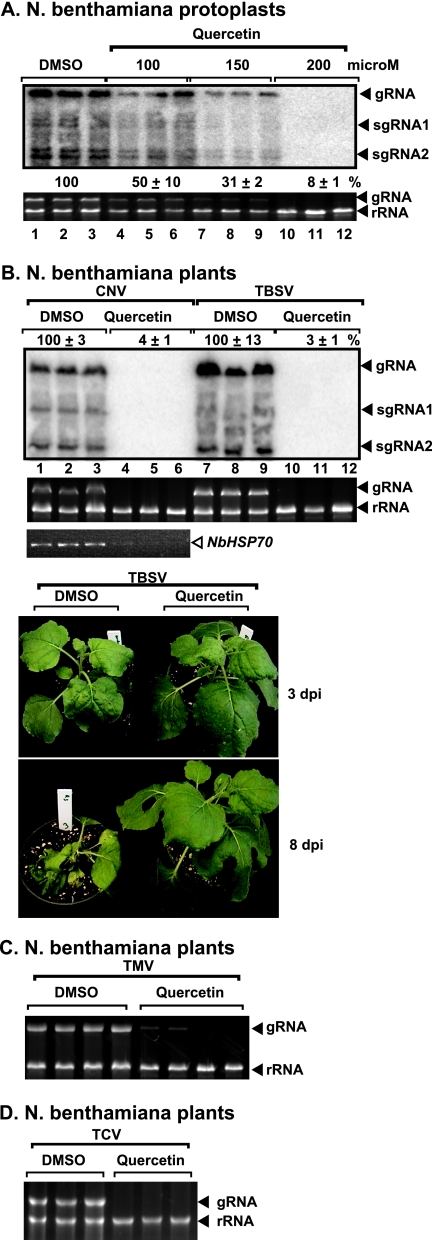
To further test the effect of NbHsp70 on TBSV replication, we used quercetin (17), a chemical inhibitor of HSP70 expression, in single cells (protoplasts lacking cell walls) and whole plants. Quercetin is a bioflavonoid that is known to specifically inhibit HSP70 mRNA expression in animal cells (5, 28, 29). The application of 200 μM quercetin inhibited TBSV gRNA accumulation by 92% in protoplasts (Fig. 6A, lanes 10 to 12) without affecting host rRNA levels or causing visible damage to the cells (data not shown).
FIG. 6.
Inhibition of NbHSP70 mRNA levels by quercetin treatment reduces the accumulation of TBSV and other plant viruses in N. benthamiana. (A) Protoplasts from N. benthamiana were electroporated with TBSV gRNA and treated with various concentrations of quercetin, as shown. Total-RNA samples were obtained 40 h postelectroporation. Northern blotting (top) shows the accumulation of TBSV gRNA and subgenomic RNAs (sgRNAs), and the ethidium-bromide stained gel (bottom) shows rRNA and TBSV gRNA levels. Note that treatment with DMSO, which is used to dissolve quercetin, was chosen as the control. (B) Treatment of N. benthamiana leaves with quercetin interferes with the accumulation of CNV and TBSV RNAs. (Top and second panels) Total-RNA samples from the inoculated leaves were obtained 3 days postinoculation and used for Northern blotting and for gel analysis to show rRNA and TBSV gRNA levels, respectively. (Third panel) Semiquantitative RT-PCR of NbHSP70 mRNA levels in treated leaves. (Bottom panel) Symptoms developed on quercetin- and DMSO-treated N. benthamiana plants 3 and 8 days postinoculation. (C) Effect of quercetin treatment on TMV accumulation. See further details in the legend to panel B. (D) Inhibition of TCV accumulation by quercetin treatment in N. benthamiana. See further details in the legend to panel B.
Similar treatment of plant leaves infected with TBSV using 200 μM quercetin had no effect on TBSV RNA accumulation in N. benthamiana (data not shown). However, increasing the concentration to 1,000 μM quercetin led to 97% inhibition of TBSV RNA accumulation in infected N. benthamiana plants at 3 dpi (Fig. 6B, lanes 10 to 12). The quercetin treatment inhibited the accumulation of cytosolic NbHSP70 mRNA by ∼80% (Fig. 6B) without visibly affecting the plants (Fig. 6B, bottom panels). Also, quercetin treatment inhibited the formation of TBSV symptoms and the death of infected plants at 8 dpi (Fig. 6B). In contrast, the control plants treated with DMSO died following TBSV infection, which causes systemic necrosis at 8 dpi (Fig. 6B). The DMSO treatment does not have any phenotypic effect on the host plants (data not shown). These experiments further demonstrate the role of NbHsp70 in tombusvirus replication in planta.
To determine whether quercetin offers broad protection against plant viruses, we tested the accumulation of CNV; TCV, a more distant member of the family Tombusviridae; and TMV, which is a very different virus belonging to the Alphavirus supergroup (25). Interestingly, the accumulation of CNV, TCV, and TMV was inhibited as much as 90% by the quercetin treatment (Fig. 6B, C, and D). Thus, the replication of several plant viruses might be dependent on Hsp70.
DISCUSSION
Replication of (+)RNA viruses of plants and animals greatly depends on coopted host proteins, which provide many different functions not supplied by the few virus-encoded replication proteins. Among the host proteins that have been identified within the tombusviral replicase complex, the Hsp70 chaperone seems to play multiple and essential roles. Downregulation of Hsp70 inhibited TBSV accumulation, whereas overexpression of Hsp70 increased TBSV accumulation, in the yeast model host (27, 55, 61), suggesting that Hsp70 is present in limiting amounts in the cell for TBSV replication. The function of Hsp70 is likely through its ability to bind the p33 and p92pol replication proteins, resulting in partial redistribution of Hsp70 (Fig. 1B, Ssa1p) from the cytosol to the peroxisomal membrane, the site of TBSV replication (30, 43, 47). It is possible that the binding of p33 and p92pol replication proteins to Hsp70 results in shielding the hydrophobic transmembrane domains in the replication protein, which could prevent their aggregation and promote binding to the Pex19p transport protein. The latter interaction is needed for peroxisomal targeting of the replication proteins (47).
In addition to the protein chaperone function of Hsp70 discussed above, we have now demonstrated that Hsp70 is involved in controlling the subcellular localization of the viral replication proteins. This is based on the finding that mutations in both SSA1 and SSA2, two genes coding for Hsp70, affected the intracellular distribution of the viral replication proteins and inhibited replicase activity in vitro. Confocal laser microscopy and subcellular fractionation experiments have shown that the viral replication proteins are mostly cytosolic in ssa1 ssa2 yeast at the early time point, while the distribution of the viral proteins becomes partly membrane bound at a latter time point. This partial complementation over time is likely due to the cytosolic, stress-inducible proteins Ssa3p and Ssa4p, which operate at higher levels in ssa1 ssa2 cells (7, 63). Yeast in which the four SSA genes are deleted is not viable; thus, we could not test viral replication in the absence of these cytosolic Hsp70 proteins. It is worth noting that Hsp70 has been shown to affect the intracellular localization of cellular proteins as well (67, 68).
We also find that Hsp70 affects the insertion of the replication proteins into intracellular membranes in vitro. The integration events were tested with proteinase K treatment, which leaves portions of p33 and p92 protected from digestion. Addition of recombinant Ssa1p to the in vitro membrane insertion assay containing yeast extract increased the amount of the protected p33 and p92pol fragments four- to-fivefold (Fig. 4C). Solubilization of yeast membranes with 1% Triton X-100 prevented the protection of a fragment of the TBSV p33 replication protein against proteinase K digestion in the presence of added Ssa1p, confirming that Ssa1p enhances the insertion of the replication protein into the membranes. Integration of the p33 replication protein into subcellular membranes, such as peroxisomal and ER membranes (21), is thought to be critical for tombusvirus replication, since p33 mutants localized in the cytosol do not support TBSV replication in yeast or in plant cells (30, 43). Also, ssa1 ssa2 yeast cells, which show partial cytosolic localization of p33 and p92, support reduced levels of TBSV replication (Fig. 1 and 3). Based on these data, we propose that the insertion of p33 and p92pol proteins into the intracellular membrane by the cytosolic Hsp70 is critical for TBSV replication in yeast cells. However, the insertion of the replication proteins into the membrane might require additional cellular factors, as shown for cellular membrane-associated proteins (8, 67).
The functions of Hsp70 discovered in this work might be connected, since membrane insertion of p33 and p92pol could be critical for their intracellular localization to membranes. It is possible that those p33/p92pol proteins that are not integrated remain cytosolic in ssa1 ssa2 cells. The cellular peroxisomal transporter protein Pex19p, which binds to p33 and targets it to the peroxisomal membrane (47), might require Hsp70 for delivering/unloading/inserting the cargo p33/p92pol proteins to the peroxisomal membrane.
Interestingly, the subcellular localization data (Fig. 1 to 2) and copurification experiments (Fig. 3) (55) indicate that Hsp70 remains associated with the viral replicase even after the insertion of the replication proteins into the membranes, suggesting an additional function for Hsp70 during tombusvirus replication. Indeed, we were able to copurify functional replicase complex with Hsp70 after solubilization of the membrane fraction (Fig. 3C), suggesting that the activated replicase still contains Hsp70. Moreover, a recent study of in vitro assembly of the TBSV replicase based on a yeast cell extract, purified replication proteins, and recombinant Hsp70 revealed that Hsp70 is required in order to reconstitute the fully functional TBSV replicase (49).
The cytosolic Hsp70 is also required for TBSV genomic RNA replication in plant cells and whole-plant hosts, based on knockdown and inhibition experiments (Fig. 5 and 6). VIGS using two different regions from HSP70 (15) resulted in strong inhibition of TBSV RNA replication in N. benthamiana plants (Fig. 5). However, knockdown of the NbHSP70 level also had detrimental effects on the plants, including yellowing (pYL619) and necrosis (pYL622). Therefore, we performed the experiments within a short period (a total of 9 to 12 days from agroinfiltration to sample collection), before the appearance of phenotypes. Furthermore, we used a gentle approach based on quercetin, which inhibits HSP70 mRNA accumulation (17). The treatment with quercetin led to greatly reduced TBSV gRNA accumulation, confirming that the cytosolic Hsp70 is critical for TBSV replication in a host plant. The quercetin treatment also inhibited CNV, TCV, and TMV accumulation, suggesting that other plant viruses also depend on Hsp70 during their infection cycles. Indeed, Hsp70 was shown to be part of the CNV and TMV replicase complexes (40, 55). The interaction between Hsp70 and the tombusvirus replication proteins likely takes place in the functional replicase, since FLAG affinity purification of Ssa1p from the solubilized membrane fraction of yeast resulted in copurification of tombusvirus replicase activity (Fig. 3C). Taken together, the data support a functional role for the cytosolic Hsp70 in several steps of tombusvirus replication, including subcellular localization, membrane insertion, and assembly of the viral replicase complex (49).
Acknowledgments
We thank S. Dinesh-Kumar for the pYL619, pYL622, pYL-PDS, pTRV1, and pTRV2 VIGS vectors, D. Lewandowski for the infectious TMV clone, and Judit Pogany, Daniel Barajas, Zhenghe Li, and Zsuzsanna Sasvari for critical reading of the manuscript and for very helpful suggestions.
This work was supported by NIH-NIAID and by the Kentucky Tobacco Research and Development Center at the University of Kentucky (to P.D.N.).
Footnotes
Published ahead of print on 19 January 2009.
REFERENCES
- 1.Ahlquist, P. 2002. RNA-dependent RNA polymerases, viruses, and RNA silencing. Science 2961270-1273. [DOI] [PubMed] [Google Scholar]
- 2.Ahlquist, P., A. O. Noueiry, W. M. Lee, D. B. Kushner, and B. T. Dye. 2003. Host factors in positive-strand RNA virus genome replication. J. Virol. 778181-8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alzhanova, D. V., A. J. Napuli, R. Creamer, and V. V. Dolja. 2001. Cell-to-cell movement and assembly of a plant closterovirus: roles for the capsid proteins and Hsp70 homolog. EMBO J. 206997-7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aparicio, F., C. L. Thomas, C. Lederer, Y. Niu, D. Wang, and A. J. Maule. 2005. Virus induction of heat shock protein 70 reflects a general response to protein accumulation in the plant cytosol. Plant Physiol. 138529-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aquino, D. A., D. Peng, C. Lopez, and M. Farooq. 1998. The constitutive heat shock protein-70 is required for optimal expression of myelin basic protein during differentiation of oligodendrocytes. Neurochem. Res. 23413-420. [DOI] [PubMed] [Google Scholar]
- 6.Aranda, M. A., M. Escaler, D. Wang, and A. J. Maule. 1996. Induction of HSP70 and polyubiquitin expression associated with plant virus replication. Proc. Natl. Acad. Sci. USA 9315289-15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becker, J., W. Walter, W. Yan, and E. A. Craig. 1996. Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol. Cell. Biol. 164378-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brodsky, J. L., and G. Chiosis. 2006. Hsp70 molecular chaperones: emerging roles in human disease and identification of small molecule modulators. Curr. Top. Med. Chem. 61215-1225. [DOI] [PubMed] [Google Scholar]
- 9.Brown, G., H. W. Rixon, J. Steel, T. P. McDonald, A. R. Pitt, S. Graham, and R. J. Sugrue. 2005. Evidence for an association between heat shock protein 70 and the respiratory syncytial virus polymerase complex within lipid-raft membranes during virus infection. Virology 33869-80. [DOI] [PubMed] [Google Scholar]
- 10.Buck, K. W. 1996. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res. 47159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castorena, K. M., S. A. Weeks, K. A. Stapleford, A. M. Cadwallader, and D. J. Miller. 2007. A functional heat shock protein 90 chaperone is essential for efficient Flock House virus RNA polymerase synthesis in Drosophila cells. J. Virol. 818412-8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cherry, S., A. Kunte, H. Wang, C. Coyne, R. B. Rawson, and N. Perrimon. 2006. COPI activity coupled with fatty acid biosynthesis is required for viral replication. PLoS Pathog. 2e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Connor, J. H., M. O. McKenzie, G. D. Parks, and D. S. Lyles. 2007. Antiviral activity and RNA polymerase degradation following Hsp90 inhibition in a range of negative strand viruses. Virology 362109-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dinesh-Kumar, S. P., R. Anandalakshmi, R. Marathe, M. Schiff, and Y. Liu. 2003. Virus-induced gene silencing. Methods Mol. Biol. 236287-294. [DOI] [PubMed] [Google Scholar]
- 15.Dong, Y., T. M. Burch-Smith, Y. Liu, P. Mamillapalli, and S. P. Dinesh-Kumar. 2007. A ligation-independent cloning tobacco rattle virus vector for high-throughput virus-induced gene silencing identifies roles for NbMADS4-1 and -2 in floral development. Plant Physiol. 1451161-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egger, D., and K. Bienz. 2005. Intracellular location and translocation of silent and active poliovirus replication complexes. J. Gen. Virol. 86707-718. [DOI] [PubMed] [Google Scholar]
- 17.Hosokawa, N., K. Hirayoshi, A. Nakai, Y. Hosokawa, N. Marui, M. Yoshida, T. Sakai, H. Nishino, A. Aoike, K. Kawai, et al. 1990. Flavonoids inhibit the expression of heat shock proteins. Cell Struct. Funct. 15393-401. [DOI] [PubMed] [Google Scholar]
- 18.Hu, J., D. Flores, D. Toft, X. Wang, and D. Nguyen. 2004. Requirement of heat shock protein 90 for human hepatitis B virus reverse transcriptase function. J. Virol. 7813122-13131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425686-691. [DOI] [PubMed] [Google Scholar]
- 20.Jaag, H. M., J. Stork, and P. D. Nagy. 2007. Host transcription factor Rpb11p affects tombusvirus replication and recombination via regulating the accumulation of viral replication proteins. Virology 368388-404. [DOI] [PubMed] [Google Scholar]
- 21.Jonczyk, M., K. B. Pathak, M. Sharma, and P. D. Nagy. 2007. Exploiting alternative subcellular location for replication: tombusvirus replication switches to the endoplasmic reticulum in the absence of peroxisomes. Virology 362320-330. [DOI] [PubMed] [Google Scholar]
- 22.Kampmueller, K. M., and D. J. Miller. 2005. The cellular chaperone heat shock protein 90 facilitates Flock House virus RNA replication in Drosophila cells. J. Virol. 796827-6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirkegaard, K., and W. T. Jackson. 2005. Topology of double-membraned vesicles and the opportunity for non-lytic release of cytoplasm. Autophagy 1182-184. [DOI] [PubMed] [Google Scholar]
- 24.Knoops, K., M. Kikkert, S. H. Worm, J. C. Zevenhoven-Dobbe, Y. van der Meer, A. J. Koster, A. M. Mommaas, and E. J. Snijder. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 6e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koonin, E. V., and V. V. Dolja. 1993. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 28375-430. [DOI] [PubMed] [Google Scholar]
- 26.Kumar, M., and D. Mitra. 2005. Heat shock protein 40 is necessary for human immunodeficiency virus-1 Nef-mediated enhancement of viral gene expression and replication. J. Biol. Chem. 28040041-40050. [DOI] [PubMed] [Google Scholar]
- 27.Li, Z., D. Barajas, T. Panavas, D. A. Herbst, and P. D. Nagy. 2008. Cdc34p ubiquitin-conjugating enzyme is a component of the tombusvirus replicase complex and ubiquitinates p33 replication protein. J. Virol. 826911-6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, B. L., J. S. Wang, H. C. Liu, R. W. Chen, Y. Meyer, A. Barakat, and M. Delseny. 2001. Genomic analysis of the Hsp70 superfamily in Arabidopsis thaliana. Cell Stress Chaperones 6201-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manwell, L. A., and J. J. Heikkila. 2007. Examination of KNK437- and quercetin-mediated inhibition of heat shock-induced heat shock protein gene expression in Xenopus laevis cultured cells. Comp. Biochem. Physiol. A 148521-530. [DOI] [PubMed] [Google Scholar]
- 30.McCartney, A. W., J. S. Greenwood, M. R. Fabian, K. A. White, and R. T. Mullen. 2005. Localization of the tomato bushy stunt virus replication protein p33 reveals a peroxisome-to-endoplasmic reticulum sorting pathway. Plant Cell 173513-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, S., and J. Krijnse-Locker. 2008. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 6363-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Momose, F., T. Naito, K. Yano, S. Sugimoto, Y. Morikawa, and K. Nagata. 2002. Identification of Hsp90 as a stimulatory host factor involved in influenza virus RNA synthesis. J. Biol. Chem. 27745306-45314. [DOI] [PubMed] [Google Scholar]
- 33.Nagy, P. D. 2008. Yeast as a model host to explore plant virus-host interactions. Annu. Rev. Phytopathol. 46217-242. [DOI] [PubMed] [Google Scholar]
- 34.Nagy, P. D., and J. Pogany. 2008. Multiple roles of viral replication proteins in plant RNA virus replication. Methods Mol. Biol. 45155-68. [DOI] [PubMed] [Google Scholar]
- 35.Nagy, P. D., and J. Pogany. 2000. Partial purification and characterization of Cucumber necrosis virus and Tomato bushy stunt virus RNA-dependent RNA polymerases: similarities and differences in template usage between tombusvirus and carmovirus RNA-dependent RNA polymerases. Virology 276279-288. [DOI] [PubMed] [Google Scholar]
- 36.Nagy, P. D., and J. Pogany. 2006. Yeast as a model host to dissect functions of viral and host factors in tombusvirus replication. Virology 344211-220. [DOI] [PubMed] [Google Scholar]
- 37.Naito, T., F. Momose, A. Kawaguchi, and K. Nagata. 2007. Involvement of Hsp90 in assembly and nuclear import of influenza virus RNA polymerase subunits. J. Virol. 811339-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakagawa, S., T. Umehara, C. Matsuda, S. Kuge, M. Sudoh, and M. Kohara. 2007. Hsp90 inhibitors suppress HCV replication in replicon cells and humanized liver mice. Biochem. Biophys. Res. Commun. 353882-888. [DOI] [PubMed] [Google Scholar]
- 39.Navarro, B., L. Rubino, and M. Russo. 2004. Expression of the Cymbidium ringspot virus 33-kilodalton protein in Saccharomyces cerevisiae and molecular dissection of the peroxisomal targeting signal. J. Virol. 784744-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishikiori, M., K. Dohi, M. Mori, T. Meshi, S. Naito, and M. Ishikawa. 2006. Membrane-bound tomato mosaic virus replication proteins participate in RNA synthesis and are associated with host proteins in a pattern distinct from those that are not membrane bound. J. Virol. 808459-8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Noueiry, A. O., and P. Ahlquist. 2003. Brome mosaic virus RNA replication: revealing the role of the host in RNA virus replication. Annu. Rev. Phytopathol. 4177-98. [DOI] [PubMed] [Google Scholar]
- 42.Okamoto, T., Y. Nishimura, T. Ichimura, K. Suzuki, T. Miyamura, T. Suzuki, K. Moriishi, and Y. Matsuura. 2006. Hepatitis C virus RNA replication is regulated by FKBP8 and Hsp90. EMBO J. 255015-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Panavas, T., C. M. Hawkins, Z. Panaviene, and P. D. Nagy. 2005. The role of the p33:p33/p92 interaction domain in RNA replication and intracellular localization of p33 and p92 proteins of Cucumber necrosis tombusvirus. Virology 33881-95. [DOI] [PubMed] [Google Scholar]
- 44.Panavas, T., and P. D. Nagy. 2003. Yeast as a model host to study replication and recombination of defective interfering RNA of Tomato bushy stunt virus. Virology 314315-325. [DOI] [PubMed] [Google Scholar]
- 45.Panaviene, Z., J. M. Baker, and P. D. Nagy. 2003. The overlapping RNA-binding domains of p33 and p92 replicase proteins are essential for tombusvirus replication. Virology 308191-205. [DOI] [PubMed] [Google Scholar]
- 46.Panaviene, Z., T. Panavas, S. Serva, and P. D. Nagy. 2004. Purification of the cucumber necrosis virus replicase from yeast cells: role of coexpressed viral RNA in stimulation of replicase activity. J. Virol. 788254-8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pathak, K. B., Z. Sasvari, and P. D. Nagy. 2008. The host Pex19p plays a role in peroxisomal localization of tombusvirus replication proteins. Virology 379294-305. [DOI] [PubMed] [Google Scholar]
- 48.Pogany, J., and P. D. Nagy. 2008. Authentic replication and recombination of Tomato bushy stunt virus RNA in a cell-free extract from yeast. J. Virol. 825967-5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pogany, J., J. Stork, Z. Li, and P. D. Nagy. 2008. In vitro assembly of the Tomato bushy stunt virus replicase requires the host heat shock protein 70. Proc. Natl. Acad. Sci. USA 10519956-19961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pogany, J., K. A. White, and P. D. Nagy. 2005. Specific binding of tombusvirus replication protein p33 to an internal replication element in the viral RNA is essential for replication. J. Virol. 794859-4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qanungo, K. R., D. Shaji, M. Mathur, and A. K. Banerjee. 2004. Two RNA polymerase complexes from vesicular stomatitis virus-infected cells that carry out transcription and replication of genome RNA. Proc. Natl. Acad. Sci. USA 1015952-5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rubino, L., and M. Russo. 1998. Membrane targeting sequences in tombusvirus infections. Virology 252431-437. [DOI] [PubMed] [Google Scholar]
- 53.Rust, R. C., L. Landmann, R. Gosert, B. L. Tang, W. Hong, H. P. Hauri, D. Egger, and K. Bienz. 2001. Cellular COPII proteins are involved in production of the vesicles that form the poliovirus replication complex. J. Virol. 759808-9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salonen, A., T. Ahola, and L. Kaariainen. 2005. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 285139-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Serva, S., and P. D. Nagy. 2006. Proteomics analysis of the tombusvirus replicase: Hsp70 molecular chaperone is associated with the replicase and enhances viral RNA replication. J. Virol. 802162-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi, S. T., and M. M. Lai. 2005. Viral and cellular proteins involved in coronavirus replication. Curr. Top. Microbiol. Immunol. 28795-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sohn, S. Y., S. B. Kim, J. Kim, and B. Y. Ahn. 2006. Negative regulation of hepatitis B virus replication by cellular Hsp40/DnaJ proteins through destabilization of viral core and X proteins. J. Gen. Virol. 871883-1891. [DOI] [PubMed] [Google Scholar]
- 58.Strauss, J. H., and E. G. Strauss. 1999. Viral RNA replication. With a little help from the host. Science 283802-804. [DOI] [PubMed] [Google Scholar]
- 59.Tavis, J. E., B. Massey, and Y. Gong. 1998. The duck hepatitis B virus polymerase is activated by its RNA packaging signal, epsilon. J. Virol. 725789-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tomita, Y., T. Mizuno, J. Diez, S. Naito, P. Ahlquist, and M. Ishikawa. 2003. Mutation of host DnaJ homolog inhibits brome mosaic virus negative-strand RNA synthesis. J. Virol. 772990-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang, R. Y., and P. D. Nagy. 2008. Tomato bushy stunt virus co-opts the RNA-binding function of a host metabolic enzyme for viral genomic RNA synthesis. Cell Host Microbe 3178-187. [DOI] [PubMed] [Google Scholar]
- 62.Weeks, S. A., and D. J. Miller. 2008. The heat shock protein 70 cochaperone YDJ1 is required for efficient membrane-specific Flock House virus RNA replication complex assembly and function in Saccharomyces cerevisiae. J. Virol. 822004-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Werner-Washburne, M., D. E. Stone, and E. A. Craig. 1987. Complex interactions among members of an essential subfamily of hsp70 genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 72568-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.White, K. A., and P. D. Nagy. 2004. Advances in the molecular biology of tombusviruses: gene expression, genome replication, and recombination. Prog. Nucleic Acid Res. Mol. Biol. 78187-226. [DOI] [PubMed] [Google Scholar]
- 65.Whitham, S. A., S. Quan, H. S. Chang, B. Cooper, B. Estes, T. Zhu, X. Wang, and Y. M. Hou. 2003. Diverse RNA viruses elicit the expression of common sets of genes in susceptible Arabidopsis thaliana plants. Plant J. 33271-283. [DOI] [PubMed] [Google Scholar]
- 66.Whitham, S. A., C. Yang, and M. M. Goodin. 2006. Global impact: elucidating plant responses to viral infection. Mol. Plant-Microbe Interact. 191207-1215. [DOI] [PubMed] [Google Scholar]
- 67.Young, J. C., V. R. Agashe, K. Siegers, and F. U. Hartl. 2004. Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 5781-791. [DOI] [PubMed] [Google Scholar]
- 68.Young, J. C., N. J. Hoogenraad, and F. U. Hartl. 2003. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 11241-50. [DOI] [PubMed] [Google Scholar]