Abstract
Positive-strand RNA viruses replicate their genomes on intracellular membranes, usually in conjunction with virus-induced membrane rearrangements. For the nodavirus flock house virus (FHV), we recently showed that multifunctional FHV replicase protein A induces viral RNA template recruitment to a membrane-associated state, but the site(s) and function of this recruitment were not determined. By tagging viral RNA with green fluorescent protein, we show here in Drosophila cells that protein A recruits FHV RNA specifically to the outer mitochondrial membrane sites of RNA replication complex formation. Using Drosophila cells and yeast cells, which also support FHV replication, we also defined the cis-acting regions that direct replication and template recruitment for FHV genomic RNA1. RNA1 nucleotides 68 to 205 were required for RNA replication and directed efficient protein A-mediated RNA recruitment in both cell types. RNA secondary structure prediction, structure probing, and phylogenetic comparisons in this region identified two stable, conserved stem-loops with nearly identical loop sequences. Further mutational analysis showed that both stem-loops and certain flanking sequences were required for RNA1 recruitment, negative-strand synthesis, and subsequent positive-strand amplification in yeast and Drosophila cells. Thus, we have shown that protein A recruits RNA1 templates to mitochondria, as expected for RNA replication, and identified a new RNA1 cis element that is necessary and sufficient for RNA1 template recognition and recruitment to these mitochondrial membranes for negative-strand RNA1 synthesis. These results establish RNA recruitment to the sites of replication complex formation as an essential, distinct, and selective early step in nodavirus replication.
All positive-strand RNA viruses replicate their genomes in virus-induced replication complexes associated with rearranged intracellular membranes (50). Different viruses replicate in association with different intracellular membranes, with endoplasmic reticulum-derived membranes used most frequently (50). Replication complexes may serve many purposes, including localizing all required viral and cellular factors in close proximity and high concentration to allow effective initiation and efficient progression of replication, protecting replication intermediates such as double-stranded RNA from antiviral responses, and providing a scaffold to organize sequential replication steps (50). Although these complexes are crucial for positive-strand RNA virus replication, their formation is not well understood.
For replication complexes to form and function, viral replication proteins, viral genomic RNA, and any required host factors must localize to the relevant intracellular membranes where replication complexes assemble. Some viral replication proteins such as hepatitis C virus (HCV) NS4B, brome mosaic virus (BMV) protein 1a, and tombusvirus p33 direct their own localization to the appropriate membrane (14, 49, 53). Other viral replication proteins are directed to the site of replication complex formation by interacting with such self-targeting viral proteins. For example, BMV RNA-dependent RNA polymerase (RdRp) 2aPol interacts via its N terminus with the C terminus of BMV 1a to localize to endoplasmic reticulum membranes (8). In the presence or absence of 2aPol, 1a also recruits BMV genomic RNAs to a membrane-associated, nuclease-resistant state by interacting with similar recruitment elements in each genomic RNA (9, 22, 56). Likewise, tombusvirus RNA replication protein p33 interacts with and recruits defective interfering RNAs to replication complex sites (39, 40, 42, 48).
In the present study, we characterized genomic RNA recruitment to membranes by a well-studied positive-strand RNA virus, flock house virus (FHV). FHV, the best-studied member of the nodavirus family, has a bipartite genome. RNA1 (3.1 kb) encodes the sole FHV RNA replication protein, the multifunctional protein A (110 kDa). Protein A has a central RdRp domain, multiple domains that direct protein A self-interaction in vivo, a putative guanylyl-transferase domain for viral RNA capping, and an N-terminal targeting signal and transmembrane domain that direct protein A insertion into outer mitochondrial membranes (12, 13, 16, 17, 23, 35). RNA1 also encodes subgenomic RNA3 (387 nucleotides [nt]) that translates protein B2 (12 kDa), an RNA silencing inhibitor (14, 20). FHV genomic RNA2 (1.4 kb) encodes protein α, the 47-kDa capsid precursor, and is thus required for virion formation but dispensable for RNA replication (11, 16, 18). Both genomic RNAs are copackaged into a single icosahedral virion with T=3 symmetry (52, 54).
Although originally isolated from the insect Costelytra zealandica, FHV also productively infects Drosophila cells (15, 46). If genomic RNA templates are provided, FHV can also replicate its RNA and produce infectious virions in the yeast Saccharomyces cerevisiae, plants, Caenorhabditis elegans, and mammalian cells (3, 15, 33, 45, 55). FHV replication in yeast reproduces nearly all characteristics of that in Drosophila cells including mitochondrial localization of RNA replication, subgenomic RNA3 synthesis, formation of infectious virions, and many other detailed interactions (31, 45, 46).
Recently, we showed that, even when bearing a polymerase-inactivating mutation, FHV nonstructural protein A recruits genomic RNA1 to a membrane-associated state in yeast and Drosophila cells (57). However, the location and possible function of this RNA1 membrane association remained unknown. For the positive-strand Flaviviridae, e.g., only a small fraction of the nonstructural protein copies that accumulate are involved in RNA synthesis (47), so that some or many interactions with these proteins may be nonproductive or directed to processes other than RNA replication, such as the crucial, RNA replication-independent roles of nonstructural proteins NS2A and replicative helicase NS3 in virion assembly (28, 41). Similarly, tomato mosaic virus nonstructural protein 130K, an essential RNA replication factor and BMV 1a homolog, also suppresses posttranscriptional gene silencing (27). Given the unusually small size of the FHV genome, the clearly multifunctional protein A may also have functions beyond its currently known roles in RNA replication.
Here we show by fluorescence microscopy of green fluorescent protein (GFP)-tagged RNAs that a protein A mutant lacking RdRp activity recruits FHV genomic RNA1 to mitochondria in Drosophila cells, as expected for an early step in FHV RNA replication complex assembly. We also use deletion and gain-of-function analysis to identify an ∼140-nt RNA1 region within the protein A open reading frame (ORF) that is necessary and sufficient to support protein A-mediated RNA1 recruitment in both yeast and Drosophila cells. Fine structure deletion analysis of this region showed that two conserved stem-loops and sequences between them were required not only for RNA1 recruitment but also for negative-strand RNA1 synthesis in yeast and Drosophila. Together, these results show that protein A recruitment of FHV RNA to mitochondrial membranes is a crucial early step in RNA replication that occurs prior to and can be studied independently of later replication steps such as negative-strand RNA synthesis. Moreover, our definition of conserved, cis-acting RNA sequences and structures that direct RNA1 recognition and recruitment complements our recent definition of the protein A domains required (57).
MATERIALS AND METHODS
Plasmids.
Standard molecular cloning techniques were used throughout (2, 51). All products generated by PCR were verified by sequencing. Detailed methods and primer sequences are available from the authors upon request. Numbering of FHV, BBV, BoV, and NoV RNA1 sequences are based on GenBank accession numbers NC_004146, NC_001411, NC_004142, and NC_002690, respectively. FHV RNA expression plasmids for RNA (pF1), RNA1fs (pF1fs), protein Awt (pFA), and protein AD692E (pFAD692E) were described previously (31, 35, 37, 45, 46, 57). pF1fs and pF1 are HIS3-selectable yeast centromeric plasmids that contain full-length RNA1 flanked by the GAL1 promoter and the hepatitis delta virus ribozyme.
A 4-nt insertion at nt 373 in pF1fs causes a frameshift in the protein A ORF. Protein A expression plasmids are LEU2-selectable yeast centromeric plasmids that contain protein A mRNA flanked by the GAL1 promoter and leader sequence at the 5′ end and a 3′ CYC1 poly(A) adenylation signal. A single point mutation in the polymerase active site (D692E) inhibits protein A polymerase activity (53). Addition of the yellow fluorescent protein ORF downstream of and separate from the protein A ORF in pFAwt and pFAD692E expressed in yeast increases the size of protein A mRNA to allow visualization by Northern blotting (53).
RNA1 and RNA1fs deletions.
All RNA1 and RNA1fs deletions were made by a two-step PCR process. First, the region surrounding the deletion site was amplified in two separate PCRs with overlapping primers. Next, the resulting PCR products were combined and amplified with bordering primers, digested, and ligated into common sites in pF1 or pF1fs.
Drosophila expression plasmids.
All FHV expression plasmids were derivatives of pIE1hr/PA, which contains the baculovirus immediate-early 1 (IE1) promoter, the baculovirus transactivating hr5 enhancer and a poly(A) adenylation signal (7). Drosophila expression plasmids for RNA1fs (pIE1-F1fs), protein Awt (pIE1-FAwt), and AD692E (pIE1-FAD692E) were as described previously (57). pIE1-F1 encodes RNA1 flanked at the 3′ end by the hepatitis delta virus ribozyme like pIE1-F1fs. pIE1-F1 was made by amplifying RNA1 from pF1 and ligating the PstI- and HindIII-digested product into common sites in pIE1hr/PA. pIE1-FB2 expresses B2 and was made by amplifying the B2 ORF from pF1 and ligating the digested product into common sites in pIE1-F1. Deletions were made in pIE1-F1fs by the same two-step PCR process used to make the RNA1fs deletions described above except that the final digested products were ligated into common sites in pIE1-F1fs.
IE1-MS2CP-eGFP.
Sequences containing the MS2 coat protein ORF fused to enhanced GFP (eGFP) and a nuclear localization signal were digested from pG14-MS2-GFP (courtesy of the Long lab) with HindIII and ClaI and ligated into common sites in pBluescript II KS(+) (Stratagene, La Jolla, CA) to make pBlue(MS2CP-eGFP). The MS2 coat protein-eGFP sequence was amplified from pBlue(MS2CP-eGFP), digested with PstI and HindIII, and ligated into common sites in pIE1/PA to make pIE1-MS2CP-eGFP. The downstream activating region of the IE1 promoter was removed from pIE1-MS2CP-eGFP by amplifying the IE1 promoter, digestion with XbaI and PstI, and ligation into common sites in pIE1-MS2CP-eGFP.
RNA1-MS2 expression plasmids.
SnaBI and ClaI restriction enzyme sites were inserted between nt 376 and 2277 in pF1fsΔ(377-2276) by a two-step PCR process to make pF1Δ(SnaBI-ClaI). Briefly, two PCR products that contained and overlapped the restriction sites were amplified with bordering primers in a second PCR, digested and ligated into common sites in pF1fsΔ(377-2276). Sequence containing six MS2 coat protein binding sites was digested from pSL-MS2-6 (courtesy of the Singer lab [6]) with MscI and ClaI and ligated into pF1Δ(SnaBI-ClaI) digested with SnaBI and ClaI to make pF1Δ(MS2). pF1Δ(MS2) was digested and ligated into common sites in pIE1-F1fs to make pIE1-F1Δ(MS2) (1-M-4,5). pIE1-F1(1-377)-MS2 (1-MS2) and pIE1-F1-MS2-(2277-3107) (MS2-4,5) were made by, respectively, digesting pIE1-F1Δ(MS2) with ClaI or NcoI and ligating the resulting vector after treatment with T4 DNA polymerase (New England Biolabs, Beverly, MA). Deletions were made in pIE1-F1(1-377)-MS2 by a two-step PCR process that flanked the deletion site as explained above, except that the resulting digested product was ligated into common sites in pIE1-F1(1-377)-MS2.
RNA1 templates for cleavage studies.
RNA1 nt 1 to 3107 or nt 1 to 377 were amplified from pF1, digested, and ligated into common sites in pBluescript.
Cells.
The haploid yeast strain YPH500 (MATα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1) was transformed by using an E-Z transformation kit (Zymo Research, Orange, CA) (35). Yeast cells were grown as described previously (57). Drosophila melanogaster S2 cells were grown and transfected as described previously (57).
Antibodies.
Mouse monoclonal antibody against biotin and rabbit polyclonal antibody against protein A were described previously (36). Rabbit polyclonal antibody to GFP was obtained from Invitrogen (Carlsbad, CA). Rabbit polyclonal antibody to B2 was made by Harlan (Madison, WI) from an Escherichia coli-expressed B2-His6/T7 fusion protein (courtesy of B. Dye). Alexa Fluor 488 and Alexa Fluor 568 secondary antibodies were purchased from Invitrogen.
RNA extraction and Northern blot analysis.
Total RNA from yeast or Drosophila melanogaster cells was isolated and prepared by the hot phenol method (31). Northern blotting was performed as previously described (57). Multiple probes were used to detect positive- and negative-strand RNA1. Probes targeting RNA1 nt 1 to 377, nt 2287 to 2843, or nt 2847 to 3107 were transcribed from pBluescript II KS(+). Probes against 18S rRNA and positive- and negative-strand RNA1 nt 2718 to 3064 were described (45, 56, 57). Probes were synthesized with a Strip-EZ RNA kit from Ambion (Austin, TX) or an Epicentre Riboscribe probe synthesis kit (Madison, WI) with the appropriate enzyme: T7, T3, or SP6. Northern blots were imaged on a Typhoon 9200 (Amersham Biosciences, Piscataway, NJ). Band intensities were analyzed by using ImageQuant software (Molecular Dynamics, Piscataway, NJ).
The size of the protein A-stabilized pool of RNA1 is proportional to the level of protein A expressed (P. M. Van Wynsberghe, unpublished results; see also references 53 and 56) rather than being a constant fold increase of the level of RNA1 that accumulates in the absence of protein A. Accordingly, for RNA1 and its derivatives, we measured protein A stimulation of RNA1 accumulation as the difference rather than the ratio of RNA1 levels in the presence or absence of protein A, i.e., [the level of (+)RNA1 in the presence of protein AD692E, normalized to the level of 18s rRNA in the same sample] - [the level of (+)RNA1 in the absence of protein AD692E, normalized to the level of 18S rRNA], relative to a parallel measurement of the same stimulation parameter for wild-type RNA1fs in the presence or absence of protein A.
RNA secondary structure predictions.
The computer program M-fold version 3.2 was used to fold full-length RNA1 (34, 61). RNA1 sequences were aligned by using CLUSTAL W associated with Lasergene and MegAlign software (DNAStar, Madison, WI).
Confocal microscopy.
At 2 days after transfection, cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 1 h, permeabilized with 0.1% Triton X-100 for 10 min at room temperature, blocked in blocking buffer (PBS with 0.1% azide, 1% nonfat milk, 1% bovine serum albumin, and 0.5% Tween 20) for 1 h, and incubated with primary antibodies for at least 4 h and with secondary antibodies for 1 h. Cells were treated with 300 nM DAPI (4′,6′-diamidino-2-phenylindole; Invitrogen) for 5 min in PBS. Mitochondria were visualized with anti-biotin antibodies (36) and Alexa Fluor 568-conjugated secondary antibodies (Invitrogen). MS2-CP visualization was enhanced by anti-GFP primary antibodies and Alexa Fluor 488 secondary antibodies (Invitrogen). Images were acquired with a Bio-Rad (Hercules, CA) Radiance 2100 MP Rainbow confocal microscope at the W. M. Keck Laboratory for Biological Imaging at the University of Wisconsin, Madison.
Solution structure probing.
RNA was in vitro transcribed with an AmpliScribe transcription kit (Epicentre, Madison, WI) from pBluescript II KS(+) plasmids containing the appropriate RNA1 sequence. RNA (0.5 or 1 pmol) in structure probing buffer (10 mM MgCl2, 50 mM Tris-Cl; pH 8.0) (38) was heated to 75°C for 3 min and incubated at room temperature for 15 min. In separate reactions, 1 pmol of RNA was treated with 5 μg of tRNA (Sigma, St. Louis, MO) and 0.1 U of RNase I (Epicentre), 1 U of RNase T1 (Ambion, Austin, TX), 0.05 U of RNase V1 (Pierce, Milwaukee, WI) or no enzyme, or 0.5 pmol RNA was treated with 2.5 μg of tRNA and 0.02 U of RNase A before incubation at 37°C for 5 min. Cleavage was stopped with 5 μg of tRNA and 250 μl of 100% ethanol (EtOH). RNA was extracted with an RNeasy extraction kit (Qiagen, Valencia, CA) and EtOH precipitated.
Primer extension.
Primers (345 ng) were labeled by treatment with 1 μl of T4 polynucleotide kinase (New England Biolabs, Beverly, MA) at 37°C for 1 h in a 20-μl reaction containing 12.5 μl of [γ-32P]ATP (6,000 Ci/mmol, 10 μCi/μl) and 2 μl of 10× kinase buffer (New England Biolabs). Labeling was stopped by adding 1 μl of 0.5 M EDTA and purified with an Autoseq G-50 spin column (Amersham Biosciences). Primers (1.5 μl) and 2 μl of hybridization buffer (0.15 M KCl, 50 mM Tris-Cl [pH 8.0], and 1 mM EDTA) were added to treated RNA before incubation at 90°C for 5 min, 42°C for 60 to 90 min, and room temperature for 30 min. Reverse transcription mix (30 μl of 30 mM Tris-Cl [pH 8.0], 15 mM MgCl2, 10 mM dithiothreitol, 6.75 μg of actinomycin D [Calbiochem, San Diego, CA], 0.225 mM deoxynucleoside triphosphates, 5 U of RNase inhibitor [Ambion], and 100 U of SuperScript II reverse transcriptase [Invitrogen]) was added to each sample before incubation at 42°C for 1 to 2 h. Transcription was stopped with 1 μl of 0.5 M EDTA before EtOH precipitation. Sequencing ladders of pF1 with the same labeled primers were made with a SequiTherm cycle sequencing kit (Epicentre). Samples were run on an 8% acrylamide RapidGel (USB, Cleveland, OH). Sequencing gels were dried before imaging on a Typhoon 9200 (Amersham Biosciences).
RESULTS
Identification of 5′- and 3′-proximal regions required for RNA1 replication.
Any RNA sequences required in cis for protein A-mediated recruitment of RNA1 replication templates to membrane-associated RNA replication complexes must be a subset of the sequences required for RNA1 replication. Accordingly, to focus our studies, we extended prior partial characterizations (4, 31) to determine what RNA1 regions were required for its replication in Drosophila cells and yeast. To study RNA1's cis functions separately from its protein coding, we used previously established trans replication systems for Drosophila and yeast cells, in which separate plasmids expressed protein A and a frameshifted RNA1 replication template (RNA1fs) (Fig. 1A) unable to translate protein A (57). The resulting RNA replication products were analyzed by Northern blotting. Since DNA-derived transcripts of RNA1fs and protein A mRNA contributed positive-strand RNA1 background even in the absence of RNA replication, the level of negative-strand RNA1 was used as a measure of RNA-dependent RNA synthesis. In addition to single-stranded negative-sense RNA3, a higher-molecular-weight band that comigrates with double-stranded RNA3 (dsRNA3) was seen, as in previous experiments (1, 31). This higher band is likely a consequence of the great excess, small size, and low sequence complexity of RNA3, which facilitates dsRNA3 renaturation (31). Moreover, this higher band likely also includes RNA3 dimers, which have been detected in association with FHV RNA replication in Drosophila, mammalian, and yeast cells (1).
FIG. 1.
5′ and 3′ proximal regions 1, 4, and 5 are required for RNA1 replication. (A) Schematic of known RNA1 and RNA3 cis elements and the deletions made within RNA1. Insertion of a 4-nt sequence causes a frameshift (fs) in RNA1 (F1fs) that prevents full-length protein A translation. Interaction between the distal subgenomic control element (DSCE) and the proximal subgenomic control element (PSCE) is required for RNA3 synthesis in yeast (31). RNA3 translates the RNA silencing inhibitor protein B2 (FB2). The internal replication element (intRE) and 3′ replication element (3′ RE) are required for RNA1 replication (31). Five RNA1 deletions were made and analyzed: 1, nt 1 to 377; 2, nt 377 to 1373; 3, nt 1377 to 2276; 4, nt 2287 to 2843; and 5, nt 2847 to 3107. (B and C) Total RNA was extracted from Drosophila cells (B) or yeast (C) expressing protein A and wild-type (wt) or deletions of RNA1fs. Total RNA (2 μg) was analyzed by Northern blotting with 32P-labeled cRNA probes for positive- or negative-strand RNA1 (nt 1 to 377 or nt 2718 to 3064) or 18S rRNA. The diamond in panel B represents a B2-specific, protein A-independent background band. The star in panel C represents a background band. The arrowheads in panels B and C mark the locations of negative-strand RNA1 bands. The band labeled dsRNA3 likely corresponds to both double-stranded RNA3 and RNA3 dimers (1, 31). Levels of negative-strand RNA1 were graphed as the percentage of wild-type RNA1fs levels. The measurements in panel B were made in the absence of B2.
Five sequential deletions, together encompassing the RNA1fs sequence (Fig. 1A), were tested in parallel. In both Drosophila cells (Fig. 1B) and yeast cells (Fig. 1C), deleting the 3′-proximal region 4 (nt 2287 to 2843) or region 5 (nt 2847 to 3107) severely inhibited negative-strand RNA1fs accumulation. This was as expected since regions 4 and 5 contain previously defined internal (intRE) and 3′ (3′ RE) replication elements required for RNA1 replication (31). Similarly, deleting 5′-terminal region 1 (nt 1 to 377) severely inhibited negative-strand RNA1fs accumulation in both cell types (Fig. 1B to C). This was also as expected since small 5′ and 3′ extensions or deletions inhibit RNA1 replication in BHK cells (4).
In contrast to regions 1, 4, and 5, deleting region 3 (nt 1377 to 2276) in either cell type had no effect on RNA1fs replication or even enhanced replication (Fig. 1B to C). Finally, deleting region 2 (nt 377 to 1373) showed cell type-specific effects. In yeast, deleting region 2 had no effect on RNA1fs replication (Fig. 1C). However, in Drosophila cells, deleting region 2 markedly inhibited negative- and positive-strand RNA1fs accumulation (Fig. 1B).
Such a Drosophila-specific effect of region 2 was anticipated due to effects on production of FHV subgenomic RNA3 and its encoded RNA silencing inhibitor, protein B2, which is required for normal FHV RNA accumulation in Drosophila cells (29; unpublished results) but not in yeast (46). Region 2 contains a small distal subgenomic control element (Fig. 1A) whose base pairing to the proximal subgenomic control element of region 4 is crucial for FHV subgenomic RNA3 synthesis (31). Accordingly, deleting region 2 blocked RNA3 production in both cell types (Fig. 1B and C). Expressing B2 in trans from a separate plasmid increased the accumulation of positive-strand RNA1fs Δ2 in Drosophila cells (Fig. 1B). However, this restoration was only partial since Western blotting showed that, despite use of a strong baculovirus IE1 promoter, plasmid-based B2 expression was weak compared to that from replicating wild-type RNA1. In yeast cells, however, which lack RNA interference pathways and show no effect of B2 on the accumulation of FHV RNA replication products (46), positive-strand RNA1fs Δ2 and RNA1fs Δ2,3 accumulated to wild-type or slightly higher levels (Fig. 1C). Thus, the Drosophila-specific effect of region 2 on RNA1fs accumulation appears not to be due to a direct cis-acting contribution to genomic RNA replication but to an indirect effect on viral suppression of Drosophila RNA silencing. Overall, then, the results show that the 5′ and 3′ proximal regions 1, 4, and 5, but not the internal regions 2 and 3, are required in cis for RNA1fs replication in yeast and Drosophila cells.
The 5′ proximal region 1 is sufficient to direct protein A-mediated RNA1 recruitment to mitochondria.
Since regions 1, 4, and 5 comprised the complete set of signals required in cis for RNA1 replication, we next tested the ability of these regions to direct protein A-mediated RNA recruitment in yeast and in Drosophila cells. To assay RNA recruitment independently of RNA replication, the relevant RNA1 fragments were analyzed in the presence or absence of protein A bearing a single D-to-E substitution in the conserved GDD motif of the polymerase domain. The resulting protein A derivative (AD692E) lacks RNA polymerase activity but retains all other known protein A functions (57).
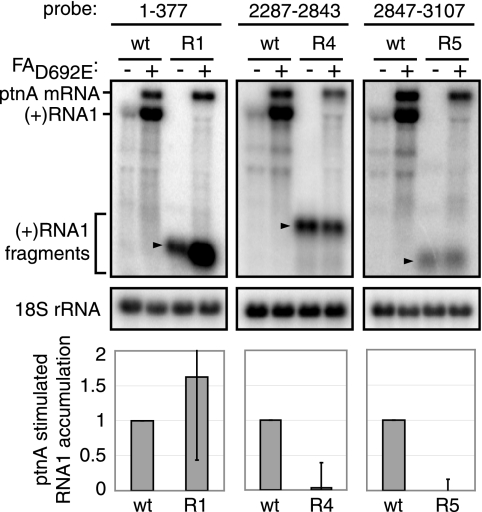
In yeast, RNA1 expressed alone fractionates in the cytoplasm and has a half-life of 4 to 5 min. When RNA1 is coexpressed with polymerase-inactive protein AD692E, this short-lived cytoplasmic pool is joined by a new membrane-associated pool of RNA1 with a half-life of >60 min, yielding a dramatic increase in RNA1 accumulation that directly correlates with RNA1 recruitment to membranes (57). When expressed in the absence of other RNA1 sequences, an RNA transcript comprising region 1 alone fully duplicated the wild-type level of this protein AD692E-induced increase in RNA1 accumulation (Fig. 2). Transcripts comprising region 4 or region 5 also were easily detected in yeast but, in contrast to region 1, showed no increase in accumulation when protein AD692E was expressed (Fig. 2). Equivalent results were obtained in six independent experiments, and representative Northern blots are shown. Thus, region 1 alone reproduced the full protein A responsiveness of wild-type RNA1, whereas regions 4 and 5 had no activity.
FIG. 2.
The 5′-proximal region 1 is sufficient to mediate RNA1 recruitment in yeast. Total RNA was extracted from yeast expressing RNA1 regions 1 (R1), 4 (R4), or 5 (R5) in the absence or presence of FAD692E. Total RNA (2 μg) was analyzed by Northern blotting with 32P-labeled cRNA probes corresponding to positive-strand RNA1 nt 1 to 377, nt 2287 to 2843, or nt 2847 to 3107 or 18S rRNA. The levels of positive-strand RNA1 were determined from six experiments after normalization to 18S rRNA and are expressed as the level of protein A-stimulated RNA1 accumulation relative to wild-type RNA1 (see Materials and Methods). The arrowheads denote the locations of positive-strand RNA1 fragments.
To more directly analyze the importance of regions 1, 4, and 5 for RNA recruitment in Drosophila cells and to identify the site(s) to which protein A induces RNA1 recruitment, we used confocal fluorescence microscopy to image the intracellular localization of RNA1 derivatives (Fig. 3). For such visualization, selected portions of RNA1 were fused to six tandem copies of the MS2 coat protein binding site (Fig. 3A) and examined in cells coexpressing an MS2 coat protein-GFP fusion (MS2CP-GFP) (Fig. 3B). Nuclei were visualized by DAPI staining, while mitochondria were viewed with anti-biotin antibodies (36). Each RNA-protein combination tested was analyzed in numerous cells during multiple independent experiments, and representative results are shown in Fig. 3.
FIG. 3.
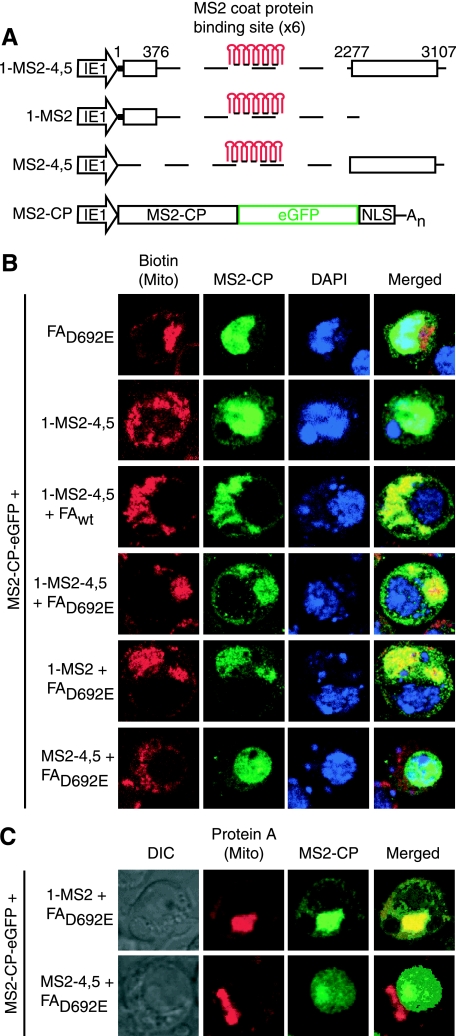
Region 1 is sufficient to mediate RNA1 recruitment in Drosophila. (A) Six MS2 stem-loops were inserted into RNA1fsΔ(2,3) (1-MS2-4,5), 3′ to RNA1 region 1 (1-MS2) or 5′ to RNA1 regions 4 and 5 (MS2-4,5). All of these RNA1-MS2 stem-loop fusions and the MS2 coat protein GFP fusion protein (MS2-CP) were under the control of the baculovirus IE1 promoter. (B and C) Confocal microscopy was performed on Drosophila cells expressing MS2-CP and protein AD692E or 1-MS2-4,5 in the absence or presence of protein Awt or AD692E or of protein AD692E and 1-MS2 or MS2-4,5. Mitochondria were visualized by using anti-biotin antibodies and Alexa Fluor 568 secondary antibodies (B). Protein A was visualized by using anti-protein A antibodies and Alexa Fluor 568-conjugated secondary antibodies (C). MS2-CP signal was amplified with anti-GFP antibodies and Alexa Fluor 488 secondary antibodies. DAPI staining (B) or differential interference contrast (DIC) images (C) were used to visualize nuclei.
Since MS2CP-GFP contained a nuclear localization signal, the default distribution of this fusion protein when expressed in the absence of an MS2CP binding site-tagged RNA was wholly nuclear, either in the presence or absence of protein Awt or AD692E (Fig. 3B, top row, and other results). We first examined the effect of coexpressing MS2CP-GFP with RNA 1-MS2-4,5, which retains all RNA1 replication signals while replacing replication-dispensable regions 2 and 3 (Fig. 1) with MS2CP binding sites (Fig. 3A). When RNA 1-MS2-4,5 (Fig. 3A) or other RNAs similarly tagged with MS2CP binding sites were expressed with MS2CP-GFP in the absence of protein A, the GFP signal predominantly localized to the nucleus (Fig. 3B, row 2), with a small number of cells showing various degrees of diffuse cytoplasmic localization. Since RNA1 and most of its derivatives, such as 1-MS2-4,5, are very stable in Drosophila cells with or without protein A (57), this suggests that, in the absence of protein A, the nuclear localization signal of MS2CP-GFP shuttled most 1-MS2-4,5 transcripts and similar MS2CP binding site-tagged transcripts back into the nucleus.
Coexpressing wild-type protein A and RNA 1-MS2-4,5 dramatically retargeted the MS2CP-GFP signal to mitochondria (Fig. 3B, row 3), the sites of FHV RNA replication (36). Importantly, in the absence of FHV RNA replication, the MS2CP-GFP signal also localized to mitochondria when RNA 1-MS2-4,5 and protein AD692E were expressed (Fig. 3B, row 4).
To further subdivide 1-MS2-4,5, we fused the six MS2CP binding sites 3′ to region 1 or 5′ to regions 4 and 5 to make the RNAs 1-MS2 and MS2-4,5 (Fig. 3A). Coexpressing RNA 1-MS2, protein AD692E and MS2CP-GFP in Drosophila cells showed that region 1 was sufficient for protein A-mediated recruitment of MS2CP-GFP-tagged RNA to mitochondria (Fig. 3B, row 5). Immunofluorescence with antibodies to protein A confirmed that MS2CP-GFP-tagged RNA 1-MS2 colocalized with protein AD692E (Fig. 3C, row 1). In contrast, RNA MS2-4,5 was never observed to colocalize with mitochondria (Fig. 3B, row 6) or protein A (Fig. 3C, row 2). Rather, the GFP signal in Drosophila cells expressing MS2-4,5 was unresponsive to coexpressed protein AD692E, and, like 1-MS2-4,5 in the absence of protein A, remained localized to the nucleus (Fig. 3B, rows 2 and 6, and Fig. 3C, row 2). Thus, protein A-directed localization of the MS2CP-GFP signal to mitochondria was completely dependent on the presence of RNA1 region 1 sequences linked to MS2CP binding sites.
RNA1 recruitment signals are between nt 40 and 290.
To further define the sequences within region 1 contributing to protein A-mediated RNA1 recruitment, we next tested progressive 5′ and 3′ truncation mutants that collectively encompassed all of region 1 (Fig. 4). In yeast, the protein A responsiveness of each deletion mutant was analyzed, as in Fig. 2, as the increase in RNA accumulation induced by coexpressing protein AD692E in trans. Deleting the 5′ 39 nt, comprising the full RNA1 5′ untranslated region, had no effect on RNA1 recruitment (Fig. 4B, lane 2). Deleting the 5′ 72 nt or longer 5′ regions extending into the protein A ORF, however, significantly reduced protein A responsiveness (Fig. 4B, lanes 3 to 5).
FIG. 4.
nt 40 to 290 mediate RNA1 recruitment. (A) Four 5′ deletions and five 3′ deletions of RNA1 region 1 (R1) were made and labeled in pairs with respect to the nucleotide deletion boundary of RNA1. (B) Total RNA was extracted from yeast expressing protein AD692E and wild-type or deletions of RNA1fs, and 2 μg was analyzed by Northern blotting with 32P-labeled cRNA probes for positive-strand RNA1 (nt 1 to 377) or 18S rRNA. The levels of positive-strand RNA1 were determined from three experiments after normalization to 18S rRNA and are expressed as the level of protein A-stimulated RNA1 accumulation relative to RNA1 region 1 (see Materials and Methods).
At the 3′ end, truncating region 1 from 377 nt to 340 or 290 nt had no effect or even stimulated protein A responsiveness relative to full-length region 1 (Fig. 4B, lanes 6 to 7). Truncating region 1 to 240 nt or shorter, however, inhibited protein A responsiveness to a level similar to 5′ deletions extending into the protein A ORF (Fig. 4B, lanes 8 to 10). Thus, the cis-acting signals for protein A-mediated recruitment are encoded within the 5′ proximal region of the protein A ORF, between nt 40 and 290. Below, we confirm and extend these results by further analyzing the detailed contributions of region 1 sequences to RNA1 recruitment and replication in Drosophila cells.
Secondary structure of 5′ proximal nt 40 to 290.
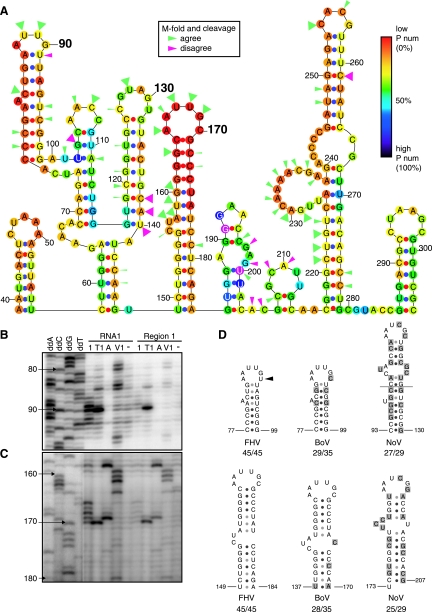
The secondary structure of full-length RNA1 was predicted with M-fold version 3.2 (34, 61), and nt 40 to 290 of the lowest energy structure are shown in Fig. 5A. Each nucleotide is color-coded according to its P-num, a measure of the number of alternate base pairs formed by that nucleotide in all RNA foldings within a specific energy range (62). Nucleotides with low P-nums, colored red to orange, have few alternate pairings and are said to be structurally well determined, i.e., likely to be found in the indicated base pairing. Less well-determined nucleotides with average or high numbers of alternate pairings, relative to the P-num distribution across a given energy range of RNA foldings, are colored blue to indigo. Notably, RNA1 nt 40 to 290 contains two predicted stem-loops, centered near nt 90 and 170, whose nt all have low P-num (Fig. 5A).
FIG. 5.
Secondary structure analysis of RNA1 nt 40 to 290. (A) M-fold version 3.2 by Zuker and coworkers (34, 61) predicted the secondary structure of full-length RNA1. The region from nt 40 to 290 is shown. The P-num of each nucleotide is represented by a colored dot. Nucleotides with low (0%), medium (50%), and high (100%) P-num values are colored red, light blue, and black, respectively (62). Arrowheads correspond to cleavages from in vitro RNA secondary structure determination. Solution structure probing results that agreed or disagreed with the predicted secondary structure are colored green or pink, respectively. The arrowhead size represents the primer extension band intensity after RNA cleavage. (B and C) In vitro-transcribed full-length RNA1 or region 1 were treated with RNase 1, RNase T1, RNase A, RNase V1, or no enzyme; extracted; and transcribed into cDNA by primer extension with various 32P-labeled primers. Primer extension products and dideoxy-terminated sequencing ladders made with the same 32P-labeled primers were analyzed on 8% acrylamide sequencing gels. Structures centered around nt 90 (B) and nt 170 (C) are shown. The strength of the four sequence ladder lanes in panel C was increased relative to the other lanes to allow visualizing the reference sequence ladder without obscuring the signals in the remainder of the blot. (D) The secondary structure of FHV, BoV, and NoV RNA1 was predicted with M-fold version 3.2 by Zuker and coworkers (34, 61), and the predicted stem-loops corresponding to the FHV RNA1 nt 90 and 170 stem-loops, after alignment by CLUSTAL W, are shown. The number of lowest energy structures that agree with the shown structure out of the total number of lowest-energy structures analyzed is listed below each stem-loop. Shaded nucleotides highlight sequence differences with FHV. The arrowhead represents the location where RNA solution and predicted secondary structure data differed.
To test the predicted secondary structure of Fig. 5A, we performed RNA solution structure probing with RNases 1, T1, and A, which cut ssRNA 3′ to all bases, guanosines, or pyrimidines, respectively, and RNase V1, which cuts 3′ to nucleotides in helical regions (32). These cleavage specificities, however, can be locally modified by various factors, including nearest neighbors, base stacking, steric hindrance, or RNA structure alteration after initial cut(s), resulting in occasional deviations from single base specificity predictions (25). Structure probing of full-length RNA1 and transcripts of region 1 alone gave similar patterns for nt 40 to 290 (Fig. 5B to C and additional results), implying that the folding of these sequences is relatively independent of other RNA1 sequences. Distinct cleavages were observed for many nucleotides in region 1, and cleavages that matched or differed from the predicted secondary structure were, respectively, marked with green and pink arrowheads (Fig. 5A). As predicted, short segments of single-stranded nucleotides, centered at nt 90 and 170, were surrounded by regions of complementary double-stranded nucleotides in both RNA1 substrates probed (Fig. 5B to C). A predicted single-stranded bulge on the nt 170 stem-loop was also confirmed (Fig. 5A and C). In contrast and as discussed further below, nt 91 was predicted to be base paired at the top of the low P-num stem-loop but was shown to be unpaired by cleavage with RNase 1 (Fig. 5A to B).
Sequences and secondary structures important for FHV RNA1 replication are likely to show some conservation in other alphanodaviruses. Consequently, RNA1 sequences of FHV, black beetle virus (BBV), boolarra virus (BoV), and nodamura virus (NoV) were aligned by using CLUSTAL W (DNASTAR programs; Lasergene). The BBV, BoV, or NoV RNA1s are 99, 78, and 51% identical, respectively, to FHV RNA1. Despite these primary sequence differences, M-fold predicted that the BBV, BoV, and NoV RNA1 secondary structures each contained two low P-num stem-loops in 5′-proximal regions that aligned with the sequences of the FHV RNA1 nt 90 and 170 stem-loops (Fig. 5D). The only nucleotide sequence and structural differences between the FHV stem-loops and predicted BBV stem-loops occurred at two nucleotides in the nt 90 stem-loop. In BBV RNA1, a U-to-C change at the position corresponding to FHV RNA1 nt 92 had no effect on stem base pairing, while a U-to-C change at nt 91 caused a 6-nt loop to form, matching the FHV solution structure results that diverged from the M-fold prediction (Fig. 5A and B). Among the nt 90 and 170 stem-loop equivalents of FHV, BBV, BoV, and NoV RNA1, differences were found in stem sequences, but the loops always were 6 nt long and matched the consensus sequence AAUYGB, where Y = U or C and B = C, G, or U (Fig. 5D).
Fine-scale deletion analysis of the 5′-proximal region in RNA1 recruitment and replication.
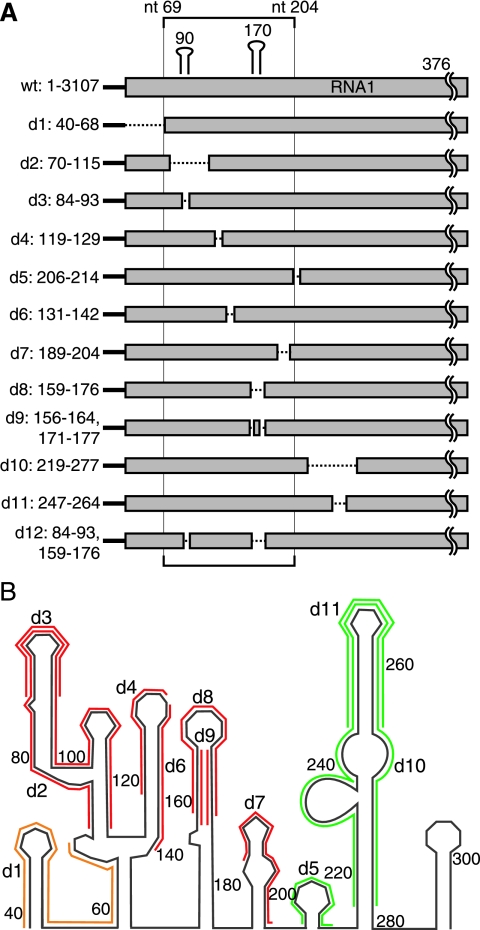
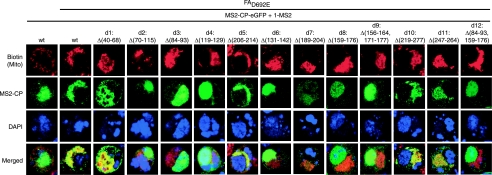
To further define the region 1 sequences required for RNA1 recruitment, 12 small deletions between nt 40 and nt 290 of RNA1 region 1 were made based on the secondary structure shown in Fig. 5 (Fig. 6). Deletions d2 (Δ70-115) and d3 (Δ84-93) eliminated all or the top of stem-loop 90. Other deletions targeted stem-loop 170 by eliminating both the stem and loop sequence (d8, Δ159-176) or decreasing the stem size while maintaining the loop sequence [d9, Δ(156-164, 171-177)]. Deletion d12 [Δ(84-93, 159-176)] combined d3 and d8 to remove both stem-loops 90 and 170. Sequences 5′ to stem-loop 90 were removed in d1 (Δ40-68), while sequences between stem-loops 90 and 170 were targeted by d4 (Δ119-129) and d6 (Δ131-142). All or the top portion of a predicted stem-loop centered at nt 250 was eliminated by d10 (Δ219-277) and d11 (Δ247-264). Other sequences 3′ to stem-loop 170 were targeted by d5 (Δ206-214) and d7 (Δ189-204). The importance of these sequences and secondary structure elements in RNA1 recruitment then were analyzed by testing the effect of these deletions in Drosophila cells using the RNA localization assay of Fig. 3.
FIG. 6.
Twelve small deletions made in RNA1 region 1. (A) Diagram of 12 small deletions made in RNA1 region 1. (B) Twelve small deletions overlaid on predicted secondary structure of RNA1 region 1 from Fig. 5. Red deletions were required for both RNA1 recruitment and negative-strand RNA1 synthesis, while green deletions were not. Yellow deletions inhibited negative-strand RNA1 synthesis but had no effect on RNA1 recruitment.
As before, the MS2CP-GFP-tagged region 1 RNA (1-MS2) colocalized with mitochondria only when expressed in the presence of protein AD692E (Fig. 7, columns 1 and 2). When RNA 1-MS2 and MS2CP-GFP were coexpressed in the absence of protein AD692E, 1-MS2 predominantly colocalized with DAPI-stained nuclei, with some cells displaying diffuse cytoplasmic localization as seen previously in Fig. 3 (Fig. 7, column 1). In the presence of protein A, deleting sequences 5′ to stem-loop 90 (d1) or most sequences 3′ to stem-loop 170 (d5, d10, and d11) had no effect on RNA 1-MS2 colocalization with the mitochondria (Fig. 7). Deleting sequences immediately 3′ to stem-loop 170 (d7) largely dispersed the RNA 1-MS2 signal, while allowing a fraction to colocalize with mitochondria (Fig. 7). All other 1-MS2 deletion mutants that eliminated stem-loops 90 and/or 170 (d2, d3, d8, d9, and d12) or sequences in between stem-loops 90 and 170 (d4 and d6) never colocalized with mitochondria (Fig. 7). Instead, the GFP signal in these deletion mutant samples was localized to the nucleus or, occasionally, diffusely throughout the cytoplasm as seen for 1-MS2 when expressed alone (Fig. 7). Thus, deletions within nt 68 to 205 inhibited protein AD692E-induced RNA1 recruitment to mitochondria, while deletions outside of nt 68 to 205 did not.
FIG. 7.
RNA1 nt 40 to 68 and nt 206 to 277 are not important for RNA1 recruitment in Drosophila. Confocal microscopy was performed on Drosophila cells expressing MS2-CP and wild-type (wt) or mutant 1-MS2 (as depicted in Fig. 6) in the presence or absence of protein AD692E, and representative results are shown. Mitochondria, GFP, and nucleus staining was performed as described in the legend to Fig. 3.
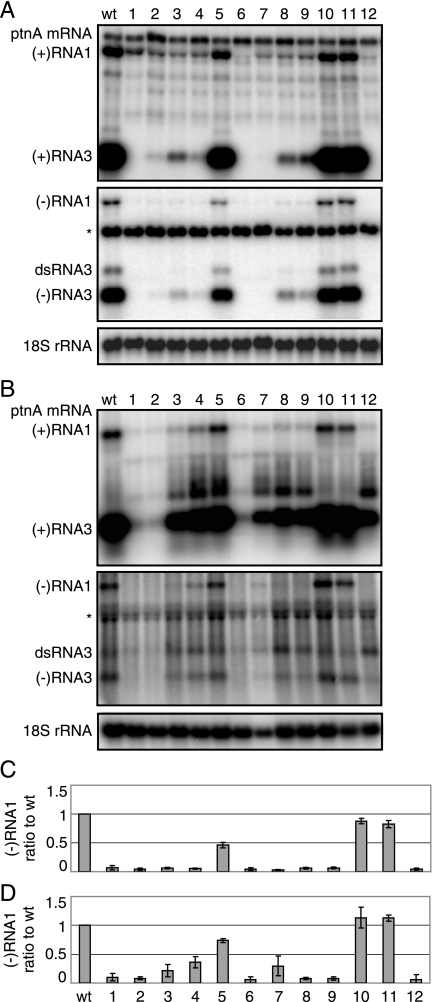
Since RNA1 recruitment to the site of replication complex formation at the outer mitochondrial membrane is an early step in FHV RNA replication, inhibiting RNA1 recruitment should inhibit later RNA replication steps, including negative-strand RNA1 synthesis and positive-strand RNA1 amplification. Consequently, we also introduced these same small region 1 deletions into full-length RNA1fs and studied their impact on RNA replication in yeast and Drosophila cells (Fig. 8). Of particular interest as measures of FHV RNA replication were the levels of negative-strand RNA1fs, since this RNA is produced only by RNA-dependent RNA synthesis and, unlike positive-strand RNA1fs, lacks a background of DNA-dependent transcripts used to initiate RNA replication.
FIG. 8.
RNA1 nt 206 to 277 are not important for RNA1 replication in yeast and Drosophila. Total RNA from yeast (A) or Drosophila cells (B) expressing protein Awt and wild-type (wt) or deletion mutants of RNA1, depicted in Fig. 6, was collected and analyzed by Northern blotting with 32P-labeled probes against RNA1 nt 2718 to 3064. The asterisk represents a background band. The ratios of negative-strand RNA1 to full-length RNA1fs levels in yeast (C) and Drosophila cells (D) are shown.
As expected, deletions 3′ to stem-loop 170 that retained strong activity for RNA recruitment (d5, d10, and d11; Fig. 7) also retained substantial RNA replication (Fig. 8). Specifically, more distal 3′ deletions d10 (Δ219-277) and d11 (Δ247-264) caused only minor reductions in RNA1 replication in yeast (Fig. 8A and C) and no detectable change in Drosophila cells (Fig. 8B and D). Similarly, the more proximal 3′ deletion d5 (Δ206-214) retained half of the wild-type negative-strand RNA synthesis in yeast and ∼75% of the wild-type synthesis in Drosophila cells (Fig. 8). In contrast, immediately flanking the 3′ deletion d7, which retained only a fraction of mitochondrial localization (Fig. 7), showed only weak levels of negative-strand RNA1 accumulation in Drosophila cells and no negative-strand activity in yeast (Fig. 8). The other deletions in RNA1fs, including those that eliminated stem-loop 90 and/or 170 (d2, d3, d8, d9, and d12) or sequences in between stem-loop 90 and 170 (d6), all severely inhibited negative-strand RNA1 synthesis in yeast cells (Fig. 8A and C) and in Drosophila cells (Fig. 8B and D). One deletion between stem-loop 90 and 170, d4, showed ∼35% of wild-type negative-strand RNA accumulation in Drosophila cells, but none in yeast. Deleting sequences 5′ to stem-loop 90 (d1) also eliminated detectable negative-strand RNA1 synthesis in both cell types (Fig. 8). Since d1 had little or no effect on RNA1 recruitment in Drosophila cells (Fig. 7), this region likely contains a cis element required for a later step of RNA1 replication, such as initiation of negative-strand RNA synthesis. In most cases, RNA3 replication was inhibited in parallel with RNA1 replication (Fig. 8A to B). Interestingly, for deletions d3, d4, d5, and d7, it was notable that inhibition of RNA1 and RNA3 replication was not quite as severe in Drosophila cells as in yeast. The reasons for this differential are considered further in the Discussion.
DISCUSSION
FHV protein A recruits RNA1 to mitochondria for RNA replication.
In the present study, we characterized early steps in positive-strand RNA virus genome replication by using the well-studied FHV model system. Previously, we showed that, even in the presence of RdRp-inactivating mutations, FHV protein A induces genomic RNA1 into a membrane-associated state in both yeast and Drosophila cells (57). However, the membrane site involved and the functional consequences of such RNA recruitment were not determined (57). Such recruitment might be important not only for RNA replication but also for virion assembly, RNA interference suppression, or other processes, since such nonreplication functions have been previously associated with replication proteins (27, 28, 41).
Here we addressed this in multiple ways, including developing a new assay to image RNA1 localization in Drosophila cells by confocal microscopy. The results showed that, in a step prior to negative-strand RNA synthesis, protein A induces RNA1 to localize to mitochondria (Fig. 3), whose outer membranes are the sites of FHV RNA replication complex formation (26, 36). These findings, together with our identification of the cis-acting RNA1 signals involved and the consequences of their mutation (see below), confirm that protein A-mediated recruitment of viral RNA to membranes is an early step in RNA replication, as opposed to an aspect of virion assembly or a separate strategy against host antiviral responses such as RNA interference. Since protein A itself localizes to outer mitochondrial membranes via an N-proximal transmembrane domain and mitochondrial targeting signal (26, 36), these findings also suggest that RNA1 relocalization likely involves the formation of a complex between RNA1 and protein A, rather than some more indirect effect. However, these results do not show whether the interaction between protein A and RNA1 is direct or indirect, i.e., mediated by one or more other factors. Future studies, such as RNA-binding assays with purified protein A, could be pursued to address this and related issues.
During early phases of infection, prior to genome recruitment to replication complexes, the presence of only one or a few infecting genomes per cell makes the virus particularly vulnerable. This fragility is highlighted in Fig. 8, which shows that for the same panel of deletions in the cis-acting RNA recruitment signals, the gradient of function losses is steeper in yeast than in Drosophila. These enhanced deletion effects correlate with the observation that unrecruited RNA1, like most cellular mRNAs (58, 60), has a shorter half-life in yeast than in Drosophila cells (57), so that delays in recruitment are even more likely to result in RNA1 degradation and loss of replication. Similarly, pharmacological or genetic strategies to inhibit or delay initial template recruitment into the protective RNA replication complex could provide significant antiviral effects against FHV and other positive-strand RNA viruses.
cis-Acting signals for RNA1 recruitment.
Previously, we mapped the regions of protein A required to induce membrane association of FHV genomic RNAs, finding that multiple domains within and flanking the RdRp domain, although not RdRp activity itself, were required (57). Here, to complement these results, we mapped the cis-acting regions of RNA1 required for protein A-dependent RNA localization to mitochondrial membranes.
Initial results from the RNA1 localization system established here and other assays showed that, out of all sequences required for RNA1 replication, 5′-proximal nt 1 to 376 of RNA1 (region 1) contained the full cis activity for directing protein A-mediated RNA1 recruitment to mitochondria in Drosophila cells (Fig. 3) and for protein A-stimulated accumulation of RNA1 in yeast (Fig. 2), which is closely linked to RNA1-membrane association (57). Within this region 1, secondary structure predictions and solution structure probing identified two stem-loops (stem-loops 90 and 170; Fig. 5A to C). Sequence alignment and secondary structure analysis showed that the RNA1's of the additional alphanodaviruses BBV, BoV, and NoV also contained predicted stem-loops corresponding to FHV RNA1 stem-loops 90 and 170, all with nearly identical AAUYGB loop sequences (Fig. 5D). These conserved hexaloops may be stabilized by internal interactions similar to the G-A pairing and R stacking of GNRA tetraloops and some related pentaloops involved in other RNA-protein interactions (10, 21, 43, 59). Such non-Watson-Crick interactions might explain why, as revealed by structure probing, a predicted A-U pair at the top of the stem-loop 90 stem does not form, preserving the hexaloop conformation of the related loops in other nodaviruses (Fig. 5). Alternatively, the loops of stem-loops 90 and 170 may interact by pairing their respective AAUU sequences in a kissing-loop or pseudoknot interaction.
Within the RNA1 region corresponding to stem-loops 90 and 170 and their intervening sequences (nt 68 to 184), each of a series of small deletions targeted to specific structural features (d2, d3, d4, d6, d8, d9, and d12; Fig. 6) markedly inhibited RNA 1-MS2 colocalization with mitochondria in Drosophila (Fig. 7) and negative-strand RNA1 synthesis in Drosophila and yeast cells (Fig. 8). Although stem-loops 90 and 170 have nearly identical loop sequences, they are not redundant structures since deletion of either stem-loop (d2, d3, d8, or d9) inhibits RNA 1-MS2 colocalization with mitochondria and negative-strand RNA1 synthesis (Fig. 7 and 8). A kissing-loop interaction between these two stem-loops may explain this lack of redundancy.
Among the strongly inhibiting deletions in the set that spans nt 68 to 184, d4 and d6 cover the two sides of a stem-loop centered near nt 130 (Fig. 5, 6). Interestingly, the loop sequence of this stem-loop is complementary to the conserved loops of stem-loops 90 and 170 and thus may be involved in tertiary interactions with one or both of these flanking stem-loops. Computer analysis predicts that all related nodaviruses contain a stem-loop at this position. However, while the 3′ stem sequence of stem-loop 130 is conserved among other nodaviruses, the loop sequence is only conserved in BBV and BoV.
Sequences immediately 3′ to stem-loop 170 (d7 [Δ189-204]) also contributed significantly to RNA1 recruitment and negative-strand synthesis, although more distal 3′ sequences (d5 [Δ206-214], d10 [Δ219-277], and d11 [Δ246-264]) did not (Fig. 7 and 8). Since 5′-proximal sequences extending to nt 205 were important for RNA recruitment, it is interesting that characterized FHV RNA1 defective interfering RNAs all preserve these sequences, often with deletion boundaries closely approaching nt 205 (30; R. Dasgupta, unpublished data). 5′ to the region of stem-loops 90 and 170, deleting the 5′ untranslated region (nt 1 to 39) or first nine codons of the protein A ORF (d1, Δ40-68) had no effect on RNA1 recruitment (Fig. 4 and 7). However, sequences in this region were required for detectable negative-strand RNA1 synthesis in Drosophila and yeast cells (Fig. 8) and thus are essential for some replication step(s), such as negative-strand RNA1 initiation, occurring after RNA1 recruitment to mitochondria.
Emerging general features of replication template recruitment by positive-strand RNA viruses.
Although template recruitment steps and signals have been defined for only a few positive-strand RNA viruses, the results revealed here for FHV show parallels with several of these, including BMV, tombusviruses, and poliovirus. The emergence of such similarities in the presence of other substantial differences among these viruses suggests that these common features may have parallels in many positive-strand RNA viruses. For example, for FHV, as well as for BMV and tombusviruses, a single viral protein directs recruitment of the genomic RNA to a membrane corresponding to the site of replication complex formation, i.e., mitochondrial, endoplasmic reticulum, and peroxisomal membranes, respectively (9, 22, 39, 42, 53, 56). In addition, recruitment of the cognate viral genomic RNA requires specific, highly structured recruitment cis elements usually found near the 5′ ends of the genomic RNAs (5, 9, 19, 39, 42, 56). For both FHV and tomato bushy stunt virus, these template recruitment cis elements are found within protein coding sequences, while for BMV RNA1 and RNA2 they are in 5′ untranslated regions (5, 9, 39, 42, 56). For poliovirus genomic RNA, a tetraloop stem-loop in the 5′ untranslated region regulates the switch between translation and negative-strand synthesis and likely plays a role in RNA recruitment (19). As in bacteriophage QB (24, 44), this overlap of template recruitment cis elements with translation initiation sites and ORFs likely provides for necessary coregulation of competing early use of viral RNA templates in translation and RNA replication.
Overall, our results show that FHV replicase protein A induces genomic RNA1 recruitment specifically to mitochondrial membranes, the site of FHV RNA replication complex assembly, in a distinct, early step that is essential for negative-strand RNA synthesis and requires cis-acting RNA recruitment elements between nt 68 and 205. Within this region, two phylogenetically conserved stem-loops with nearly identical loop sequences, and the sequences between these stem-loops, are necessary and sufficient for RNA1 recruitment. These results highlight the complex, sequential nature of RNA replication complex assembly, the highly multifunctional nature of protein A, and the specificity involved in selectively recruiting viral over cellular RNA templates for replication and provide foundations for further study of these important features. Moreover, identifying this RNA step as a crucial early event in infection suggests that it may be a valuable target for antiviral interference.
Acknowledgments
We thank Ben Kopek for helpful comments, David Miller for various plasmids, Nick Zaban for technical assistance, Ann Palmenberg and Jean-Yves Sgro for assistance with RNA1 secondary structure folding predictions, and Lance Rodenkirch at the W. M. Keck Laboratory for Biological Imaging for assistance with confocal microscopy.
This study was supported by NIH grant GM35072. P.M.V.W. gratefully acknowledges support from a Howard Hughes Medical Institute Predoctoral Fellowship and NIH Predoctoral Training Grant T32 GM07215 in Molecular Biosciences. P.A. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Published ahead of print on 14 January 2009.
REFERENCES
- 1.Albarino, C. G., B. D. Price, L. D. Eckerle, and L. A. Ball. 2001. Characterization and template properties of RNA dimers generated during flock house virus RNA replication. Virology 289269-282. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1987. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, NY.
- 3.Ball, L. A. 1992. Cellular expression of a functional nodavirus RNA replicon from vaccinia virus vectors. J. Virol. 662335-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ball, L. A. 1995. Requirements for the self-directed replication of flock house virus RNA 1. J. Virol. 69720-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumstark, T., and P. Ahlquist. 2001. The brome mosaic virus RNA3 intergenic replication enhancer folds to mimic a tRNA TpsiC-stem-loop and is modified in vivo. RNA. 71652-1670. [PMC free article] [PubMed] [Google Scholar]
- 6.Bertrand, E., P. Chartrand, M. Schaefer, S. M. Shenoy, R. H. Singer, and R. M. Long. 1998. Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2437-445. [DOI] [PubMed] [Google Scholar]
- 7.Cartier, J. L., P. A. Hershberger, and P. D. Friesen. 1994. Suppression of apoptosis in insect cells stably transfected with baculovirus p35: dominant interference by N-terminal sequences p35(1-76). J. Virol. 687728-7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, J., and P. Ahlquist. 2000. Brome mosaic virus polymerase-like protein 2a is directed to the endoplasmic reticulum by helicase-like viral protein 1a. J. Virol. 744310-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, J., A. Noueiry, and P. Ahlquist. 2001. Brome mosaic virus protein 1a recruits viral RNA2 to RNA replication through a 5′ proximal RNA2 signal. J. Virol. 753207-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Correll, C. C., and K. Swinger. 2003. Common and distinctive features of GNRA tetraloops based on a GUAA tetraloop structure at 1.4 Å resolution. RNA 9355-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dasgupta, R., A. Ghosh, B. Dasmahapatra, L. A. Guarino, and P. Kaesberg. 1984. Primary and secondary structure of black beetle virus RNA2, the genomic messenger for BBV coat protein precursor. Nucleic Acids Res. 127215-7223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dasmahapatra, B., R. Dasgupta, A. Ghosh, and P. Kaesberg. 1985. Structure of the black beetle virus genome and its functional implications. J. Mol. Biol. 182183-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dye, B. T., D. J. Miller, and P. Ahlquist. 2005. In vivo self-interaction of nodavirus RNA replicase protein a revealed by fluorescence resonance energy transfer. J. Virol. 798909-8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elazar, M., P. Liu, C. M. Rice, and J. S. Glenn. 2004. An N-terminal amphipathic helix in hepatitis C virus (HCV) NS4B mediates membrane association, correct localization of replication complex proteins, and HCV RNA replication. J. Virol. 7811393-11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friesen, P., P. Scotti, J. Longworth, and R. Rueckert. 1980. Black beetle virus: propagation in Drosophila line 1 cells and an infection-resistant subline carrying endogenous black beetle virus-related particles. J. Virol. 35741-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friesen, P. D., and R. R. Rueckert. 1982. Black beetle virus: messenger for protein B is a subgenomic viral RNA. J. Virol. 42986-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallagher, T. M., P. D. Friesen, and R. R. Rueckert. 1983. Autonomous replication and expression of RNA 1 from black beetle virus. J. Virol. 46481-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallagher, T. M., and R. R. Rueckert. 1988. Assembly-dependent maturation cleavage in provirions of a small icosahedral insect ribovirus. J. Virol. 623399-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gamarnik, A. V., and R. Andino. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 122293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guarino, L. A., A. Ghosh, B. Dasmahapatra, R. Dasgupta, and P. Kaesberg. 1984. Sequence of the black beetle virus subgenomic RNA and its location in the viral genome. Virology 139199-203. [DOI] [PubMed] [Google Scholar]
- 21.Heus, H. A., and A. Pardi. 1991. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science 253191-194. [DOI] [PubMed] [Google Scholar]
- 22.Janda, M., and P. Ahlquist. 1998. Brome mosaic virus RNA replication protein 1a dramatically increases in vivo stability but not translation of viral genomic RNA3. Proc. Natl. Acad. Sci. USA 952227-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson, K. N., K. L. Johnson, R. Dasgupta, T. Gratsch, and L. A. Ball. 2001. Comparisons among the larger genome segments of six nodaviruses and their encoded RNA replicases. J. Gen. Virol. 821855-1866. [DOI] [PubMed] [Google Scholar]
- 24.Klovins, J., V. Berzins, and J. Van Duin. 1998. A long-range interaction in Qbeta RNA that bridges the thousand nucleotides between the M-site and the 3′ end is required for replication. RNA 4948-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knapp, G. 1989. Enzymatic approaches to probing of RNA secondary and tertiary structure. Methods Enzymol. 180192-212. [DOI] [PubMed] [Google Scholar]
- 26.Kopek, B. G., G. Perkins, D. J. Miller, M. H. Ellisman, and P. Ahlquist. 2007. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 51-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kubota, K., S. Tsuda, A. Tamai, and T. Meshi. 2003. Tomato mosaic virus replication protein suppresses virus-targeted posttranscriptional gene silencing. J. Virol. 7711016-11026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leung, J., G. Pijlman, N. Kondratieva, J. Hyde, J. Mackenzie, and A. Khromykh. 2008. Role of nonstructural protein NS2A in flavivirus assembly. J. Virol. 824731-4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, H., W. X. Li, and S. W. Ding. 2002. Induction and suppression of RNA silencing by an animal virus. Science 2961319-1321. [DOI] [PubMed] [Google Scholar]
- 30.Li, Y., and A. Ball. 1993. Nonhomologous RNA recombination during negative-strand synthesis of flock house virus RNA. J. Virol. 673854-3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindenbach, B. D., J. Y. Sgro, and P. Ahlquist. 2002. Long-distance base pairing in flock house virus RNA1 regulates subgenomic RNA3 synthesis and RNA2 replication. J. Virol. 763905-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lowman, H. B., and D. E. Draper. 1986. On the recognition of helical RNA by cobra venom V1 nuclease. J. Biol. Chem. 2615396-5403. [PubMed] [Google Scholar]
- 33.Lu, R., M. Maduro, F. Li, H. W. Li, G. Broitman-Maduro, W. X. Li, and S. W. Ding. 2005. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature 4361040-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288911-940. [DOI] [PubMed] [Google Scholar]
- 35.Miller, D. J., and P. Ahlquist. 2002. Flock house virus RNA polymerase is a transmembrane protein with amino-terminal sequences sufficient for mitochondrial localization and membrane insertion. J. Virol. 769856-9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, D. J., M. D. Schwartz, and P. Ahlquist. 2001. Flock house virus RNA replicates on outer mitochondrial membranes in Drosophila cells. J. Virol. 7511664-11676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, D. J., M. D. Schwartz, B. T. Dye, and P. Ahlquist. 2003. Engineered retargeting of viral RNA replication complexes to an alternative intracellular membrane. J. Virol. 7712193-12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller, W. A., and S. L. Silver. 1991. Alternative tertiary structure attenuates self-cleavage of the ribozyme in the satellite RNA of barley yellow dwarf virus. Nucleic Acids Res. 195313-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Panavas, T., C. M. Hawkins, Z. Panaviene, and P. D. Nagy. 2005. The role of the p33:p33/p92 interaction domain in RNA replication and intracellular localization of p33 and p92 proteins of cucumber necrosis tombusvirus. Virology 33881-95. [DOI] [PubMed] [Google Scholar]
- 40.Panaviene, Z., T. Panavas, and P. D. Nagy. 2005. Role of an internal and two 3′-terminal RNA elements in assembly of tombusvirus replicase. J. Virol. 7910608-10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patkar, C., and R. Kuhn. 2008. Yellow fever virus NS3 plays an essential role in virus assembly independent of its known enzymatic functions. J. Virol. 823342-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pogany, J., K. A. White, and P. D. Nagy. 2005. Specific binding of tombusvirus replication protein p33 to an internal replication element in the viral RNA is essential for replication. J. Virol. 794859-4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prathiba, J., and R. Malathi. 2007. Group I introns and GNRA tetraloops: remnants of “the RNA world”? Mol. Biol. Rep. 3411-17. [DOI] [PubMed] [Google Scholar]
- 44.Priano, C., R. Arora, L. Jayant, and D. Mills. 1997. Translational activation in coliphage Qbeta: on a polycistronic messenger RNA, repression of one gene can activate translation of another. J. Mol. Biol. 271299-310. [DOI] [PubMed] [Google Scholar]
- 45.Price, B. D., R. R. Rueckert, and P. Ahlquist. 1996. Complete replication of an animal virus and maintenance of expression vectors derived from it in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 939465-9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Price, B. D., M. Roeder, and P. Ahlquist. 2000. DNA-Directed expression of functional flock house virus RNA1 derivatives in Saccharomyces cerevisiae, heterologous gene expression, and selective effects on subgenomic mRNA synthesis. J. Virol. 7411724-11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quinkert, D., R. Bartenschlager, and V. Lohmann. 2005. Quantitative analysis of the hepatitis C virus replication complex. J. Virol. 7913594-13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rajendran, K. S., and P. D. Nagy. 2003. Characterization of the RNA-binding domains in the replicase proteins of tomato bushy stunt virus. J. Virol. 779244-9258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rubino, L., and M. Russo. 1998. Membrane targeting sequences in tombusvirus infections. Virology 252431-437. [DOI] [PubMed] [Google Scholar]
- 50.Salonen, A., T. Ahola, and L. Kaariainen. 2005. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 285139-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 52.Schneemann, A., V. Reddy, and J. E. Johnson. 1998. The structure and function of nodavirus particles: a paradigm for understanding chemical biology. Adv. Virus. Res. 50381-446. [DOI] [PubMed] [Google Scholar]
- 53.Schwartz, M., J. Chen, M. Janda, M. Sullivan, J. den Boon, and P. Ahlquist. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9505-514. [DOI] [PubMed] [Google Scholar]
- 54.Scotti, P. D., S. Dearing, and D. W. Mossop. 1983. Flock house virus: a nodavirus isolated from Costelytra zealandica (White) (Coleoptera: Scarabaeidae). Arch. Virol. 75181-189. [DOI] [PubMed] [Google Scholar]
- 55.Selling, B. H., R. F. Allison, and P. Kaesberg. 1990. Genomic RNA of an insect virus directs synthesis of infectious virions in plants. Proc. Natl. Acad. Sci. USA 87434-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sullivan, M. L., and P. Ahlquist. 1999. A brome mosaic virus intergenic RNA3 replication signal functions with viral replication protein 1a to dramatically stabilize RNA in vivo. J. Virol. 732622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Wynsberghe, P. M., H. R. Chen, and P. Ahlquist. 2007. Nodavirus RNA replication protein a induces membrane association of genomic RNA. J. Virol. 814633-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, Y., C. L. Liu, J. D. Storey, R. J. Tibshirani, D. Herschlag, and P. O. Brown. 2002. Precision and functional specificity in mRNA decay. Proc. Natl. Acad. Sci. USA 995860-5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weiss, M. A., and N. Narayana. 2000. RNA recognition by arginine-rich peptide motifs. Biopolymers 48167-180. [DOI] [PubMed] [Google Scholar]
- 60.Winkles, J. A., and R. M. Grainger. 1985. Differential stability of Drosophila embryonic mRNAs during subsequent larval development. J. Cell Biol. 1011808-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 313406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zuker, M., and A. B. Jacobson. 1998. Using reliability information to annotate RNA secondary structures. RNA 4669-679. [DOI] [PMC free article] [PubMed] [Google Scholar]