Abstract
Embryonic stem cells are immortalized cells whose proliferation rate is comparable to that of carcinogenic cells. To study the expression of embryonic stem cell genes in primary cells, genetic screening was performed by infecting mouse embryonic fibroblasts (MEFs) with a cDNA library from embryonic stem cells. Cold-inducible RNA-binding protein (CIRP) was identified due to its ability to bypass replicative senescence in primary cells. CIRP enhanced extracellular signal-regulated kinase 1 and 2 (ERK1/2) phosphorylation, and treatment with an MEK inhibitor decreased the proliferation caused by CIRP. In contrast to CIRP upregulation, CIRP downregulation decreased cell proliferation and resulted in inhibition of phosphorylated ERK1/2 inhibition. This is the first evidence that ERK1/2 activation, through the same mechanism as that described for a Val12 mutant K-ras to induce premature senescence, is able to bypass senescence in the absence of p16INK4a, p21WAF1, and p19ARF upregulation. Moreover, these results show that CIRP functions by stimulating general protein synthesis with the involvement of the S6 and 4E-BP1 proteins. The overall effect is an increase in kinase activity of the cyclin D1-CDK4 complex, which is in accordance with the proliferative capacity of CIRP MEFs. Interestingly, CIRP mRNA and protein were upregulated in a subgroup of cancer patients, a finding that may be of relevance for cancer research.
Embryonic stem (ES) cells can be propagated indefinitely in vitro and still maintain their capacity for differentiation into a wide variety of somatic and extraembryonic tissues (4). A common characteristic of ES cells and cancer cells is that both are immortal. When removed from optimal growth conditions, ES cells will spontaneously differentiate, and the ultimate consequence is a considerably decreased proliferation rate (46, 47).
The possibility that a defective stem cell may be the cause and origin of cancer is gaining credence since the recent discovery of tissue-specific stem cells (18, 27, 40). In the last decade, several studies have reported regulatory pathways that are common to both stem cells and cancer cells; the extent of this similarity is such that an epigenetic stem cell signature in cancer has been proposed (54, 59).
Primary mouse embryonic fibroblasts (MEFs) reach replicative senescence after a few passages in culture upon acquisition of genetic mutations that make them immortal (17). Replicative senescence is characterized by a phenotypic and genotypic change that results in a loss of proliferative potential (48, 49). Interestingly, senescent cells have recently been detected in human tumors, particularly in benign lesions (6, 15); senescence is thus an antitumorigenic mechanism developed by mammalian cells to avoid cell proliferation when any genetic alteration has occurred. Genes whose expression bypasses replicative potential are considered candidate oncogenes (13, 22).
The ultimate aim of this study is to identify genes in ES cells that are implicated in cellular proliferation and that may be involved in human cancer. If cancer cells mimic the behavior of ES cells to become immortal, then an examination of ES cell gene expression may help identify genes that have an important function in the process of tumorigenesis.
It has been demonstrated that several proteins detected in ES cells are overexpressed and promote proliferation in some types of human cancer (23, 37). This study investigates which ES cell genes induce proliferation when they are expressed in primary cells. To this end, a cDNA library of genes expressed in murine ES cells was overexpressed in primary MEFs, and then clones with a potentially high proliferative advantage were selected for further characterization. This led to the identification of cold-inducible RNA-binding protein (CIRP). A considerable number of RNA-binding proteins have been shown to be involved in various diseases, such as congenital dyskeratosis, cancer, and the genotoxic response (19).
Mammalian cells exposed to mild hypothermia show a general inhibition of protein synthesis that is accompanied by an increase in the expression of a small number of cold-shock mRNAs and proteins (56). Cold-inducible proteins have been characterized as sensor proteins whose expression increases in response to the mild cold stress that is induced by a decrease in the physiological temperature (37°C) (11). In clinics, hypothermia is currently employed in both heart and brain surgery (2, 50); however, the molecular mechanisms regulating the response to cold stress in mammalian cells is unknown. Apart from its activation with mild cold induction, CIRP is induced by hypoxia (57) and has been associated with diurnal changes in the mouse brain, suggesting a relationship with the regulation of circadian rhythms (36). Therefore, CIRP seems to be a multifunctional protein that may contribute to the maintenance of normal cell function, apart from the stress response; however, the precise function of CIRP and its biological role in cell proliferation are not yet understood.
MATERIALS AND METHODS
Construction of the cDNA library and genome screen.
RNA was extracted from CGR8 and R1 ES cells using TRIzol reagent (Invitrogen SA, Barcelona, Spain) according to the manufacturer's instructions. Construction of the library was performed using a ZAP express cDNA synthesis and cloning kit (Stratagene, La Jolla, CA). The synthesized cDNAs from CGR8 and R1 ES cells were cloned into the EcoRI and XhoI sites of the pHygroMaRX vector, as previously described (16). The resulting cDNA library contained 6 × 106 different clones, and more than 95% of fragments contained a cDNA insert. The MaRX vector contains two loxP sites flanking the multiple cloning site, which enables gene excision upon infection with the Cre recombinase. Proliferation screen conditions were based on the clear proliferative effect of a positive control (p53DN) versus a negative control (green fluorescent protein [GFP]) of proliferation in primary MEFs, as previously described (22).
Cell culture and infections.
CGR8 and E14 ES cell lines were grown in Dulbecco's modified Eagle's medium (DMEM) with 20% serum (HyClone) and the presence of leukemia inhibitory factor (LGC Promochen). Medium was changed daily, and cells were not allowed to grow to more than 70% confluence to avoid spontaneous differentiation. MEFs and HeLa cells were cultured in DMEM (Lonza, Basel, Switzerland) supplemented with 10% fetal calf serum (FCS; Lonza) plus antibiotics. Human mammalian epithelial cells (HMECs) from breast tissue were grown in mammary epithelial cell basal medium plus supplements (Lonza). TERA cells (human embryonal carcinoma-derived cell line TERA2) were cultured in DMEM (Gibco) plus 10% FCS, and they were grown at 10% CO2. MEFs were generated from pregnant mice as previously described (5). The packaging cell line LinXE was transfected in 10-cm dishes using the calcium phosphate method with 30 μg of library each. Two plates of negative GFP and positive p53DN controls of proliferation were transfected in parallel as previously described (16). At 48 h posttransfection, viral supernatants were collected, filtered, supplemented with 8 μg/ml of Polybrene, and used to infect 10-cm dishes of MEFs at early passage (passage 4 [P4]). After infection, cells were selected with hygromycin at 50 μg/ml (Invitrogen) for 10 days. After selection, MEFs were seeded at a density of 105 cells per 10-cm plate to allow proliferative clones to arise. For a scheme of the protocol for performing the genetic screen and the number of clones obtained, see Fig. S1 in the supplemental material. Every 2 to 3 days, the medium was changed, allowing proliferative clones to grow and expand in culture. Finally, two clones were obtained; one clone was identified as CIRP. The clones that bypassed senescence after being infected with the library were selected for RNA extraction, cDNA synthesis, and PCR amplification with specific primers for the MaRx vector. For an example of the PCR amplification of such clones, see Fig. S2 in the supplemental material. Sequencing of each PCR product was performed to identify the inserted cDNA (data not shown). PCR products from the clones that contained a full-length cDNA were purified, digested with EcoRI and XhoI, and inserted into retroviral MaRx vectors containing the hygromycin or puromycin resistance genes. In all the infections, a heterogeneous population resulted after selection of MEFs with each gene (a GFP, CIRP, p53175H, or Val12 mutant K-ras [K-rasVal12] gene). All experiments were repeated independently at least three times, each in triplicate.
Growth curves.
CGR8, HeLa, and primary MEFs were seeded at 1 × 106 cells per 10-cm plate. Cells were grown simultaneously, and every 3 days, cells from each cell line were counted and seeded at a density of 106 cells per dish of the 10-cm plates, as indicated by a 3T3 cell protocol. MEFs at P4 were infected with GFP-, CIRP-, and p53DN (p53175H)-pII-hygroMaRx and selected over 2 weeks with hygromycin (50 μg/ml; Invitrogen, Carlsbad, CA). After selection, cells were seeded according to a 3T3 protocol. The relative number of cells is considered as a measure of the number of cells per passage related to the initial number of cells seeded per plate.
For the colony formation assay, MEFs were infected with pII-hygro-MaRx-CIRP or pII-hygro-MaRx-GFP and selected with the appropriate antibiotic. After selection, 1 × 105 cells were seeded in triplicate in 10-cm plates and maintained for 20 days. Finally, cells were fixed and stained with crystal violet for colony quantification. Senescence-associated β-galactosidase activity was analyzed as previously described (8). Briefly, cells were fixed with 0.5% glutaraldehyde, washed in phosphate-buffered saline (PBS; pH 6.0) supplemented with 1 mM MgCl2, and stained in X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining solution overnight at 37°C, as specified by the manufacturer (Cell Signaling, Danvers, MA). The MEK inhibitor PD98058 (Invitrogen) was used at a concentration of 50 μM, and control plates were treated with dimethyl sulfoxide.
Growth in soft agar.
To measure the anchorage-independent growth, 1 × 104 cells were suspended in 1.4% agarose D-1 low electroendosmosis (Pronadisa, Madrid, Spain) growth medium containing 10% FCS, deposited onto a solidified base of growth medium containing 2.8% agar (agarose D-1, low electroendosmosis; Pronadisa), and overlaid with 1 ml of growth medium. After 24 h, medium containing 10% FCS was added to each dish and renewed every 2 to 3 days. Colonies were photographed 3 weeks later. The results were verified in three independent experiments.
Protein synthesis measurement.
Established cell lines of MEFs expressing CIRP and GFP at P4 were counted and seeded in triplicate wells in 24-well plates at a density of 105 cells. On the next day, [35S]methionine (Amersham, Buckinghamshire, United Kingdom) was added to the culture medium for 2 h at a final concentration of 5 μCi/ml, as previously described (34). Then, cells were washed five times with ice-cold PBS and lysed by adding 400 μ1 of sodium dodecyl sulfate (SDS) 0.2%. A total of 100 μl of ice-cold trichloroacetic acid ([TCA] wt/vol) was added to each well, and the cells were incubated on ice for 30 min. Then, 50 μl of lysate was applied to GF/C filters to measure 35S incorporation by using liquid scintillation spectroscopy. Filters were allowed to dry and were then washed and boiled in 5% TCA for 2 min; 5% TCA was replaced, and filters were washed further by boiling them in 5% TCA for an 2 additional min. Finally, they were rinsed in 100% ethanol and allowed to dry before counting in 3 ml of scintillation liquid. Replicative wells from the same experiment were harvested simultaneously for both protein determination by quantifying proteins by Bradford reagent and/or cell number by counting cells with an inverted microscope using a hemocytometer (viability was determined by trypan blue exclusion).
Immunoblotting.
Total cell lysates were prepared from a confluent 10-cm dish. Cells were washed in 1× PBS and lysed in 1 ml of lysis buffer (50 mM Tris-HCl, pH 7.5, 1% NP-40, 10% glycerol, 150 mM NaCl, 2 mM complete protease inhibitor cocktail; Roche Diagnostics, Barcelona, Spain). At this time, 50 μg of protein from each sample, which had been previously quantified with a Bio-Rad protein assay (Bio-Rad, Hercules, CA), was electrophoresed on a 10 to 12% SDS-polyacrylamide gel electrophoresis gel and transferred to a nitrocellulose membrane. Western analysis was performed as previously described (5) using antibodies to the following proteins: β-actin (Sigma-Aldrich, St. Louis, MO); cyclin D1 (Neomarkers, Fremont, CA); p53, p21WAF1, p16INK4a, cyclin A2, c-fos, and total extracellular signal-regulated kinases 1 and 2 (ERK1/2; Santa Cruz Biotechnology Inc., Santa Cruz, CA); CIRP (Proteintech, Chicago, IL); p19ARF (Abcam, Cambridge, United Kingdom); and phosphorylated ERK1/2 (P-ERK1/2), P-MEK1/2, total MEK1/2, S6, S6 phosphorylated at positions 235 and 236 [P-S6 (235/236)], P-S6 (240/244), 4E-BP1 phosphorylated at Thr 37 and 46 [P-4E-BP1 (Thr 37/46)], and 4E-BP1 (Cell Signaling). A rabbit polyclonal antibody against CIRP was also generated in our laboratory by immunizing rabbits with a carboxyl-terminal oligopeptide (DSYDSYATHNE) as previously described (35).
Kinase assay.
CIRP and GFP MEFs were resuspended in immunoprecipitation buffer (50 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid [HEPES; pH 7.5], 150 mM NaCl, 1 mM EDTA, 2.5 mM ethylene glycol-bis(β-aminoethylether)-N,N,N′,N′-tetraacetic acid [EGTA], 1 mM dithiothreitol [DTT], 0.1% Tween 20) containing 10% glycerol, 0.1 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin per ml, 20 U of aprotinin per ml, 10 mM β-glycerophosphate, 1 mM NaF, and 0.1 mM sodium orthovanadate (Sigma-Aldrich) and sonicated at 4°C. Lysates were clarified by centrifugation, and the supernatants were precipitated for 2 h at 4°C with protein G-Sepharose beads precoated with saturating amounts of cyclin D1 antibody (Santa Cruz). Immunoprecipitated proteins on beads were washed four times with 1 ml of immunoprecipitation buffer and twice with 50 mM HEPES (pH 7.5) containing 1 mM DTT. At this stage, the supernatant was collected for analysis of β-actin by Western blotting as a loading control for the kinase assay. The beads were resuspended in kinase buffer (50 mM HEPES, 10 mM MgCl2, 1 mM DTT) containing a substrate pRb fusion protein (Santa Cruz), 1 μg of histone H1 (Roche, Basel, Switzerland), 20 μM ATP, and 10 μCi of [γ-32P]ATP. After incubation for 30 min at 30°C with occasional mixing, the samples were boiled in polyacrylamide gel sample buffer containing SDS and separated by electrophoresis. Phosphorylated proteins were visualized by autoradiography of the dried gels.
siRNA experiments.
MEFs, HMECs, HeLa, and TERA cells were reverse transfected with 4.4 to 10 μl of Hyperfect reagent (Qiagen, Hilden, Germany) per well of 24-well plates according to maximal transfection efficiency and absence of toxicity for each cell line. Optimal cell number for transfection was chosen specifically for each cell line. Transfection optimization was done by using a final concentration of each small interfering RNA (siRNA) at 30 nM. The following siRNAs were used: murine CIRP (ID 60854 and ID 160217), human CIRP (ID 96936 and ID 145723) (Ambion, Austin, TX), a Cy5-labeled siRNA targeting cyclophilin B (siGLO; Cultek, Landgraaf, The Netherlands) for human cells and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (AM649; Ambion) for murine cells as labeled controls to determine transfection efficiency. SiGLO and GAPDH controls were chosen because they behaved neutrally in proliferation after several negative controls for both murine and human cells were tested. Two independent siRNAs per gene were used. Two days after transfection, the medium was changed, and cells were maintained for three additional days. Results were done in triplicate for each siRNA in at least two independent experiments. Finally, after 5 days of transfection, cell number was determined by counting cells with an inverted microscope using a hemocytometer (viability was determined by trypan blue exclusion). The absence of apoptotic cells was verified by analyzing the cell cycle profiles of CIRP siRNA-transfected cells and comparing them to the profile of our control siRNA (data not shown).
Real-time PCR.
Quantitative real-time TaqMan reverse transcription-PCR was used to determine the differential expression of CIRP mRNA and the housekeeping controls: polymerase 2A (POL2A), β-actin, and/or 18S RNA. Such negative controls, which showed no differences between normal and tumor tissue, were specifically chosen after an endogenous control array was tested by using microfluidic cards (data not shown) (Applied Biosystems, Foster City, CA). Relative quantification analysis was performed with an ABI Prism 7700 instrument. The PCR program consisted of denaturing at 95°C for 10 min and 50 cycles at 95°C for 15 s with annealing and elongation at 60°C for 1 min. Real-time PCRs were performed in triplicate using Assays-on-Demand TaqMan Gene Expression Assays (Applied Biosystems) with probes for murine CIRP (Mm01144803_g1) or human CIRP (Hs00992239_g1). The following probes were used for endogenous human genes: β-actin (Hs99999903_m1), POL2A (Hs00172187_m1), β-actin (Mn00607939_s1) for mouse, and 18S RNA (4310893E) for both species. All results were validated with at least two different endogenous genes to normalize variations in cDNA quantities from different samples.
Patient samples.
Normal tissue and tumor tissue from 31 patients with colon carcinoma and 19 patients with prostate carcinoma were randomly chosen from the tumor bank at the Pathology Department of Vall d'Hebron Hospital (Barcelona, Spain). Biopsy samples are routinely collected, quickly frozen, and stored at −80°C immediately after surgery. All tumors were histologically examined to confirm the diagnosis of carcinoma. All procedures of the present study have been approved by the Ethics Committee of Hospital Vall d'Hebron.
Total RNA was extracted from normal and tumor tissue with an RNAeasy minikit (Qiagen). An RNA Nano Lab Chip kit (Agilent, Palo Alto, CA) was used to quantify and determine the integrity of the isolated total RNA. cDNA synthesis was performed as previously described (26). Quantitative real-time TaqMan reverse transcriptase PCR technology (Applied Biosystems) was used to determine the differential expression of CIRP, POL2A, and 18S RNA. Relative quantification analysis was performed with the ABI Prism 7700 instrument, and data were analyzed with sequence detection software (Applied Biosystems). For protein extracts, to facilitate complete homogenization of the samples, normal and tumor tissue from each patient was quickly disrupted by using a homogenizer in protein extraction buffer, following the same protocol as previously described for cells. Proteins were detected by antibodies for the following: CIRP, β-actin, P-ERK1/2, and ERK1/2. Two different CIRP antibodies were used; one was from Proteintech, and the other was generated in our laboratory.
TMA.
To corroborate the in vitro results, immunohistochemical studies were performed in the following cancer tissues using a tissue microarray (TMA): breast (26 infiltrating carcinomas and 7 in situ carcinomas), lung (22 patients), prostate (9 patients), ovarian (25 patients), colon (17 patients), gastric (19 patients), skin (13 patients), rhabdomyosarcomas (12 patients), liver and pancreas (17 patients), nervous system (11 patients), and kidney (15 patients) cancers. The TMA was constructed by a standard method (41) using cases kindly provided by the Tumor Bank of the Pathology Department, Hospital Vall d'Hebron. The cases contained representative samples of the most common tumors. Three-micrometer tissue sections from the TMA block were sectioned and applied to special immunohistochemistry coated slides (Dako, Glostrup, Denmark). The slides were baked overnight in a 56°C oven, deparaffinized in xylene for 20 min, rehydrated through a graded ethanol series, and washed with PBS. A heat-induced epitope retrieval step was performed in a solution of sodium citrate buffer with a pH of 6.5. The slide was then heated for 2 min in a conventional pressure cooker. After being heated, the slide was incubated with proteinase K for 10 min and rinsed in cool running water for 5 min. Endogenous peroxide activity was quenched with 1.5% hydrogen peroxide (Dako) in methanol for 10 min. Incubation with the primary antibodies to CIRP at 1:200 (our antibody) and P-ERK1/2 (Cell Signaling) at 1:100 was performed. After incubation, immunodetection was carried out with the EnVision (Dako) visualization system using diaminobenzidine chromogen as the substrate, according to manufacturer's instructions. Incubations omitting the primary antibody were used as a control for this technique (data not shown). All immunostaining was performed in an BenchMark XT IHQ/ISF Ventana automatic immunostaining device (Roche). To evaluate immunohistochemical staining, we scored the percentage of positive cells and intensity of the staining, which was assessed semiquantitatively. Samples that showed no immunostaining were considered negative, and samples that showed any positivity were grouped together for statistical purposes. P-ERK1/2 showed three types of positive expression: weak, moderate, or intense. Moderate to strong positivity was identified easily at low magnification (×40), whereas weak positivity was visible at high magnification (×400). Next, CIRP staining was evaluated. Tumors were considered positive if more than 10% of the cells showed moderate to strong staining with CIRP antibody.
Statistical analysis.
Statistical analysis was performed with the Statistical Package from Social Science, version 11.5 (SPSS, Inc., Chicago, IL). A Wilcoxon test was used to compare normal and tumor tissue of all patients with prostate and colon cancer (colon cancer, P = 0.02; prostate cancer, P = 0.33). In the growth curves shown in Fig. 1A and C, 3C, and 6A, a t test has been performed in order to validate the significance of the data. In addition, a Mann Whitney U analysis was performed to correlate the expression of CIRP and P-ERK1/2 proteins in each kind of cancer from a total of 193 patients.
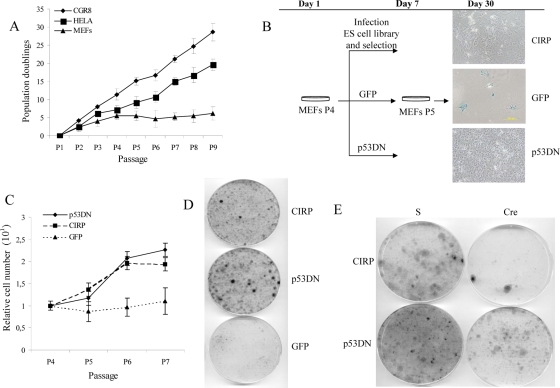
FIG. 1.
CIRP bypasses replicative senescence in primary MEFs. (A) Growth curve of CGR8 ES cells versus HeLa cancer cells or primary MEFs following a 3T3 protocol. Passage number versus population doublings are represented in the graph. (B) Design of the experimental protocol performed in MEFs to carry on the genetic screen. β-Galactosidase staining was performed in GFP-, CIRP-, and p53DN-expressing MEFs at P6. Note the presence of β-galactosidase-positive cells (blue) only in GFP MEFs and their correlation with the characteristic cell morphology of senescent cells. (C) Growth curve of MEF cells after infection and selection of the indicated genes after performing a 3T3 protocol. The relative number of cells is considered a measure of the number of cells per passage relative to the initial number of cells seeded per plate. Negative and positive controls of proliferation, the GFP and p53DN genes, respectively, were included. (D) Colony formation assay. MEF cells after infection and selection of the indicated genes were seeded at a density of 105 cells per 10-cm plate and allowed to grow for 15 to 20 days. Finally, they were stained with crystal violet to observe the colonies formed from individual cells. (E) MEF cells were infected with a retroviral vector carrying CIRP plus the Cre cDNA or with the empty vector designated S. In parallel, a positive control of proliferation is shown (p53DN).
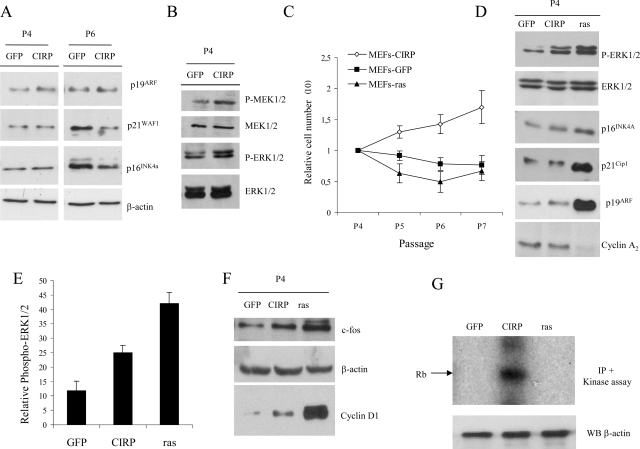
FIG. 3.
Cell cycle proteins during senescence. (A) MEF cells were infected with the indicated genes, selected with appropriate antibiotics, and, after selection, were split following a 3T3 protocol. Immunoblot of CIRP MEFs versus GFP MEFs with antibodies to p16INK4a, p19ARF, and p21WAF1 at P4 and in later passage when GFP vector-infected cells reached a clear senescence morphology (P6). (B) Immunoblot of CIRP and GFP MEFs with P-ERK1/2 and P-MEK1/2 antibodies; total ERK1/2 and MEK1/2 are shown as loading controls. (C) Growth curve of GFP, CIRP, and K-rasVal12 MEFs after following a 3T3 protocol. Cells were counted at different passages, and relative cell numbers are shown. The relative number of cells is considered a measure of the number of cells per passage relative to the initial number of cells seeded per plate. (D) Immunoblot of CIRP, GFP, and K-rasVal12 MEFs with antibodies to p21WAF1, p19ARF, p16INK4a, and cyclin A2. Total ERK1/2 is shown as a loading control. (E) P-ERK1/2 quantification versus total ERK1/2 in CIRP and K-rasVal12 MEFs. Results represent the mean of three independent experiments. (F) Immunoblot of CIRP, GFP, and K-rasVal12 MEFs with antibodies to c-fos and cyclin D1. β-Actin is shown as loading control. (G) Kinase assay in CIRP, K-rasVal12, and GFP MEFs. CIRP MEFs exhibit high kinase activity compared to senescent cells, either by replicative senescence (GFP) or by oncogene-induced senescence (K-rasVal12). β-Actin is shown as a loading control from the same supernatants of the kinase assay samples. IP, immunoprecipitation; WB, Western blotting.
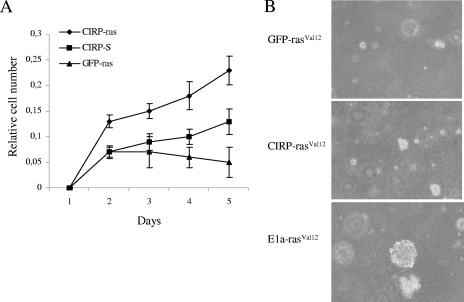
FIG. 6.
CIRP is not involved in transformation. (A) MEF cells were infected with a retroviral vector carrying CIRP and compared to those with GFP cDNAs. The cells were selected with appropriate antibiotics and coinfected with retroviruses expressing an empty vector (S) or K-rasVal12. Cells were selected again with appropriate antibiotics. Then, 105 cells were seeded in the wells of 24-well plates and fixed daily for 5 days. Finally, they were stained with crystal violet and destained with 10% acetic acid. Absorbance measurement at 595 nm was used as an indicator of the relative number of cells. (B) Soft-agar assay using GFP-rasVal12 and E1a-rasVal12 as negative and positive controls, respectively, of proliferation. K-rasVal12 does not contribute to transformation in CIRP MEF cells as no clear colonies were observed in medium containing agar.
RESULTS
CIRP bypasses replicative senescence in MEFs.
In order to compare the proliferation pattern of CGR8, HeLa, and MEFs cells, a 3T3 protocol was performed (Fig. 1A). Differences between CGR8 cells and MEFs were statistically significant (t test, P = 0.006). Differences between HeLa cells and MEFs were also statistically significant (t test, P = 0.03). However, CGR8 grew in a similar fashion to HeLa cells (t test, P = 0.15). These results led to the conclusion that the proliferative capacity of ES cells and cancer cells is quite similar but markedly different from that of primary cells (Fig. 1A). Based on this observation, wild-type MEFs were infected with a mouse embryonic cDNA library in the MaRX retroviral vector to identify genes involved in proliferation and immortalization. This led to the finding that the CIRP gene is an immortalizing gene. A retrovirus-encoded CIRP cDNA conferred unlimited proliferation capacity on primary MEFs with an efficacy comparable to that of p53175H (Fig. 1B). While control cells (GFP MEFs) entered senescence, verification that CIRP bypassed replicative senescence was obtained by the absence of β-galactosidase-positive cells in CIRP-expressing MEFs (Fig. 1B). After infection and subsequent selection, the proliferative function of CIRP in MEFs was assessed by performing growth curves following a 3T3 protocol. While GFP MEFs stop growing, CIRP-expressing cells continue to proliferate (Fig. 1C). Differences between CIRP and GFP expression levels were statistically significant (t test, P = 0.04). Moreover, no statistical differences were obtained in a comparison of CIRP to our positive control of proliferative p53DN (t test, P = 0.85), indicating that CIRP behaves as a true positive control. A characteristic of naïve cells is their low doubling number. Since the formation of a colony requires several doublings, naïve cells will form no colonies or very few colonies. In contrast, immortal cells do not have this limitation. In order to verify that CIRP MEFs were able to proliferate and form colonies after being seeded at a low density, a colony formation assay was performed comparing CIRP and GFP MEFs in parallel to a positive control of proliferation (p53DN). Interestingly, CIRP overexpression contributed to the formation of colonies in MEFs, whereas the control cells (GFP MEFs) did not (Fig. 1D). A quantification of the colony number from two independent experiments is shown in Fig. S3 in the supplemental material. Finally, to verify that the phenotypic effect of CIRP MEFs was the result of overexpression of CIRP cDNA alone, coinfection of MEFs was done with CIRP plus an empty vector and CIRP plus a Cre recombinase cDNA. Proliferation ceased only when the Cre cDNA was overexpressed (Fig. 1E).
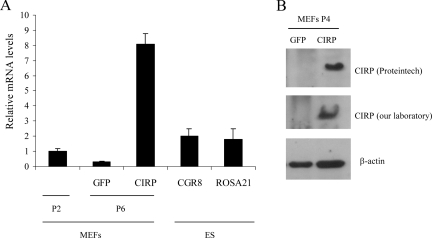
In order to verify CIRP overexpression in our model, CIRP mRNA was measured by quantitative real-time-PCR (Fig. 2A). CIRP mRNA was studied in primary MEFs at P2, in senescent MEFs at P6, and in ES cells. CIRP protein expression was verified by Western blotting (Fig. 2B) by using two different antibodies against CIRP. One antibody used was from Proteintech, and the other was generated in our laboratory. CIRP expression was verified in all our experiments at the mRNA and/or protein level.
FIG. 2.
CIRP expression in various cell lines. (A) MEF cells were infected with a retroviral vector carrying the indicated genes, selected with appropriate antibiotics, and after selection were split following a 3T3 protocol. At P6 when clear phenotypic characteristics distinguish GFP- from CIRP-expressing cells, RNA was extracted to perform quantitative real-time PCR. Primary MEFs at early passage (P2) were also included, and ES cell lines CGR8 and ROSA 21 are shown as positive controls of CIRP expression as the original cDNA library was from both ES cell lines. (B) Duplicate dishes from CIRP- and GFP-expressing MEFs were kept for protein extraction. An immunoblot with anti-CIRP antibody is shown with two independent CIRP antibodies (one from Proteintech and the other generated in our laboratory). β-Actin is shown as loading control.
CIRP stimulates P-ERK1/2.
We investigated the molecular mechanisms in CIRP MEFs that resulted in maintained growth under conditions in which senescence occurred in control cells. This investigation was accomplished by analysis of the status of the p16INK4a, p19ARF, and p21WAF1 proteins, which have a known role in the Rb and p53 pathways (51). No differences were observed in the levels of p16INK4a, p19ARF, or p21WAF1 proteins between GFP and CIRP MEFs after infection and selection with the appropriate antibiotics at P4 (Fig. 3A). To verify that p16INK4a, p19ARF, or p21WAF1 was not accumulating in CIRP MEFs (as they did in GFP MEFs), cells were analyzed after selection (P4) and during senescence (P6) when clear phenotypic, β-galactosidase-positive cells were present in GFP MEFs (Fig. 1B). p16INK4a and p21WAF1 clearly accumulated in GFP MEFs at P6 in the presence of β-galactosidase-positive cells, indicating their senescent status (Fig. 1B). However, this accumulation was not observed in CIRP MEFs at P6 (Fig. 3A). β-Actin protein was included as a loading control. Absence of the above-mentioned accumulated cell cycle-inhibitory proteins confirmed the finding that the CIRP gene is an immortalizing gene. ERK1/2 activation was measured by levels of dually phosphorylated ERK1/2 (P-ERK1/2). In contrast, ERK1/2 activation was observed upon treatment with apoptotic inducers in CIRP-deficient cells when CIRP was overexpressed (44). Hence, the phosphorylation status of the ERK1/2 protein was analyzed in this model. Surprisingly, clear differences were observed between P-ERK1/2 and total ERK1/2 in CIRP MEFs and GFP MEFs at P4 (Fig. 3B). The upstream protein MEK1/2 was also affected (Fig. 3B). Total ERK1/2 and MEK1/2 were included as protein loading standards for their respective phosphorylated forms.
Since K-rasVal12 provokes premature entry into senescence through the activation of P-ERK1/2 (24, 61), verification that K-rasVal12 MEFs entered premature senescence was performed (Fig. 3C; see also Fig. S4 in the supplemental material). A t test was performed to validate the results from Fig. 3C. CIRP MEFs differed significantly from GFP (P = 0.01) and K-rasVal12 (P = 0.01) MEFs. In contrast, no statistical differences were found between GFP and K-rasVal12 MEFs (P = 0.21). It was also verified that K-rasVal12 induced an increase in P-ERK1/2 at P4 (Fig. 3D). Senescence was also verified by studying several cell cycle-regulatory proteins in different cell lines, including CIRP, K-rasVal12, and GFP MEFs. In agreement with previous reports (24, 25, 49), there was a great increase in p19ARF and p21WAF1 levels in K-rasVal12 MEFs at the same time that ERK1/2 phosphorylation occurred (Fig. 3D). However, this strong increase did not occur in CIRP or GFP MEFs at the same passage. Cyclin A2 was also studied as an indicator of decreased proliferation in primary cells. Its role as an indicator was previously reported in K-rasVal12 overexpression (49). To determine whether the opposing cell responses mediated by P-ERK1/2 are related to the amount of active ERK1/2, P-ERK1/2 was quantified and compared in CIRP, K-rasVal12, and GFP MEFs at P4 (Fig. 3E). ERK1/2 activation was stronger in K-rasVal12 MEFs than in CIRP MEFs. Based on these observations, it is speculated that both the intensity and duration of the P-ERK1/2 signal might be involved in the determination of whether a cell enters into senescence. Therefore, P-ERK1/2 in CIRP and K-rasVal12 MEFs was analyzed in a later passage than P4 (e.g., P7). As predicted, P-ERK1/2 activation was maintained in K-rasVal12 MEFs but not in CIRP MEFs (see Fig. S5 in the supplemental material).
In order to determine how CIRP bypasses replicative senescence, several ERK1/2 substrates have been studied (55). Although no differences in c-myc or p53 status were detected between CIRP and GFP MEFs (data not shown), c-fos was upregulated in CIRP MEFs (Fig. 3F). Fos family members induce cell cycle entry by activating cyclin D1 (3). Therefore, cyclin D1 was analyzed in CIRP, GFP and K-rasVal12 MEFs. Interestingly, cyclin D1 was clearly upregulated in CIRP MEFs compared to GFP MEFs (Fig. 3F). Unexpectedly, cyclin D1 protein was also upregulated in K-rasVal12 MEFs compared to GFP MEFs at P4 in the concomitant presence of β-galactosidase-positive cells (Fig. 3F; see also Fig. S4 in the supplemental material). A key step required for passage through the G1/S transition is induction of cyclin D1 and activation of cyclin D1-CDK4/6 complexes (31). Consistent with the observation that cyclin D1 was upregulated in CIRP MEFs, kinase activity assays of cyclin D1-CDK4 complexes demonstrated that the kinase activity was present in CIRP MEFs while absent in GFP and K-rasVal12 MEFs. β-Actin, which was used as a loading control, did not vary (Fig. 3G). Thus, cyclin D1-CDK4-associated kinase activity in CIRP MEFs stimulates progression through the cell cycle and ultimately activates proliferation. In order to determine if CIRP overexpression causes a proliferative effect in cell lines other than MEFs, CIRP was overexpressed in the murine immortalized cell line NIH 3T3, the human cancer cell line HeLa, the human primary cell line IMR90, and the human primary cell line HMEC. In contrast to MEF cells, only slight differences were observed in proliferation when these cells were tested with CIRP overexpression (data not shown). No differences were observed in the P-ERK1/2 status of CIRP or GFP cells in any human cell lines or in NIH 3T3 cells (data not shown).
CIRP increases protein synthesis.
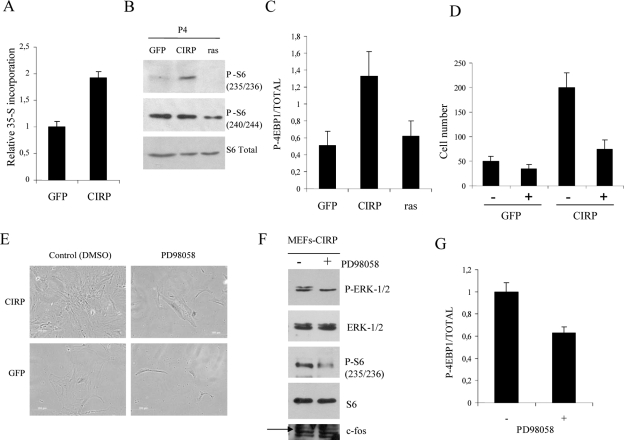
The cold-inducible protein Rbm3, which belongs to the same family as CIRP, has been shown to increase global protein synthesis by binding to 60S ribosomal subunits (9). Given that a proliferative effect caused by CIRP was clearly observed in these experiments, the next study addressed whether general protein synthesis was also affected. Total protein synthesis production was investigated by measuring 35S methionine incorporation in CIRP, GFP and K-rasVal12 MEFs (Fig. 4A). The increase in general protein synthesis production observed in CIRP MEFs led to an investigation of the proteins involved in controlling the translational machinery. This included S6 and proteins that regulate the initiation and elongation phases of translation, such as 4E-BP1. Interestingly, the phosphorylation status of S6 at positions 235 and 236 was increased in CIRP MEFs compared to GFP or K-rasVal12 MEFs (Fig. 4B). Moreover, the phosphorylation status of S6 at positions 240 and 244 decreased in K-rasVal12 MEFs, correlating with their entry into senescence (Fig. 3C and 4B). Total 4E-BP1 and P-4E-BP1 (Thr 37/46) were both upregulated in CIRP MEFs compared to GFP and K-rasVal12 MEFs (Fig. 4C). This rendered the 4E-BP1 protein inactive and facilitated initiation of translation. eIF-4E protein did not vary between CIRP MEFs and GFP MEFs (data not shown). Akt is an upstream protein of S6 which functions in the phosphatidylinositol 3-kinase pathway but not the mitogen-activated protein kinase (MAPK) pathway (30). Akt did not vary between CIRP and GFP (data not shown). This finding ruled out the possibility that the phosphatidylinositol 3-kinase pathway could affect the phosphorylation status of S6.
FIG. 4.
CIRP increases protein synthesis. (A) MEF cells were infected with the indicated genes and selected with appropriate antibiotics. After selection they were seeded at 105 cells per well in a 24-well plate. The following day, [35S]methionine was added to the culture medium and incubated for 2 h. The graph shows [35-S]methionine incorporation in CIRP MEFs relative to GFP MEFs as a measurement of general protein synthesis. (B) MEF cells were infected with the indicated genes and selected with appropriate antibiotics. After selection, protein extracts were collected for immunoblotting with S6 ribosomal protein and an antibody that recognizes its phosphorylation (P-S6) at positions 235/236 and 240/244. (C) Relative quantification of P-4E-BP1 protein at positions Thr 37/46, total 4E-BP1 in CIRP MEFs, and two negative controls of proliferative GFP and K-rasVal12 MEFs. Results represent the mean of three independent experiments. The bands of the immunoblot were quantified by specific software (Quantity One). (D) MEF cells were infected with the indicated genes and selected with appropriate antibiotics. After selection, they were seeded at 105 cells per well in a 24-well plate. Cell number after treatment of CIRP MEFs compared to GFP MEFs at P4 with the MEK inhibitor PD98058 for 1 week is shown. (E) The MEK inhibitor PD98058 is able to reverse the proliferative phenotype in CIRP MEFs, but it does not affect GFP MEFs. (F) MEF cells were infected with CIRP, selected with appropriate antibiotics, and further seeded at a density of 0.5 × 106 cells per 10-cm plate. Two plates were treated with PD98058 and compared to control plates (dimethyl sulfoxide [DMSO]). Protein extracts were collected for immunoblotting of P-ERK1/2, ERK1/2, P-S6 235/236, S6, and c-fos. (G) Relative quantification of P-4E-BP1 protein at positions Thr 37/46 compared to total 4E-BP1 in CIRP MEFs treated with the MEK inhibitor PD98058 from the same experiment as in panel F. All experiments were performed independently at least two times.
In order to test if ERK1/2 activation was responsible for the proliferative phenotype of CIRP, CIRP and GFP MEFs were treated daily with the MEK inhibitor PD98058. While PD98058 had no significant effect on GFP MEFs, it did significantly decrease the proliferative capacity of CIRP MEFs (Fig. 4D and E). A decrease in P-S6, a decrease in P-4E-BP1, and no change in c-fos accompanied the decrease in proliferation (Fig. 4F and G).
CIRP downregulation inhibits proliferation.
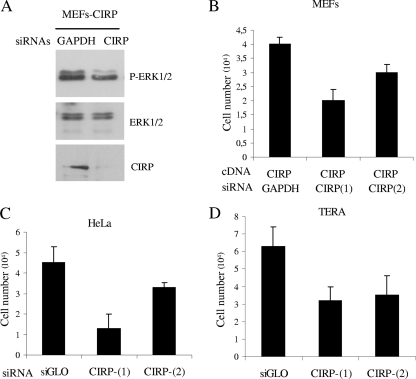
In order to study if CIRP inhibition provoked an effect opposite to CIRP overexpression, an siRNA strategy was used. For this purpose, two different CIRP siRNAs were tested in different cell lines. Murine and human siRNAs against CIRP were able to downregulate CIRP mRNA and protein (Fig. 5A and data not shown). Protein extraction of CIRP MEFs transfected with siRNA against GAPDH was compared to cells transfected with siRNAs against CIRP using an analysis of P-ERK1/2 status (Fig. 5A). Interestingly, CIRP inhibition provoked a decrease in cell number in CIRP MEFs compared to cells with siRNA against GAPDH (Fig. 5B). The cancer cell line HeLa was also studied. In this study, CIRP downregulation provoked a decrease in cell numbers at different levels (Fig. 5C).
FIG. 5.
CIRP downregulation in various cell lines. (A) Immunoblot of P-ERK1/2, ERK1/2, and CIRP (our laboratory) from MEF cells transfected with the CIRP(1) siRNA. (B) MEFs cells at P4 were infected with a vector carrying CIRP cDNA and selected with appropriate antibiotics. Then, cells were counted and inversely transfected with Hyperfect reagent at a density of 10 × 103 to 25 × 103 cells per well in a 24-well plate. Two different siRNAs against CIRP were used and compared to a neutral siRNA against GAPDH. Cells were counted 5 days after transfection with an inverted microscope using a hemocytometer (viability determined by trypan blue exclusion). (C) Cell number of HeLa cells after transient transfection with two different CIRP siRNAs compared to neutral siRNA (siGLO). (D) Cell number of TERA cells after transient transfection of two different CIRP siRNAs compared to neutral siRNA (siGLO). siRNA results from two independent experiments are shown for all cell lines.
Finally, CIRP was identified from a cDNA library of ES cells. Using this information, the effect of CIRP on other cells with ES cell-like properties (i.e., TERA cells derived from teratocarcinoma) was determined. Interestingly, CIRP downregulation decreased the cell number of TERA cells. Figure 5D contains graphs representing counted cell numbers 5 days after transient transfection of the indicated siRNAs.
Although CIRP decreased cell numbers in HeLa and TERA cells compared to a negative control of proliferation (siGLO), their P-ERK1/2 status was not affected (data not shown).
CIRP is not involved in transformation.
The ability of CIRP to protect cells against premature senescence induced by K-rasVal12 was also examined. Wild-type MEFs rapidly arrested upon infection with a retrovirus-encoded K-rasVal12. Conversely, MEFs expressing CIRP (Fig. 6A) in the presence of K-rasVal12 continued to proliferate. Statistical comparisons showed that coinfected CIRP and rasVal12 MEFs differ from coinfected GFP and rasVal12 MEFs (t test, P = 0.03). A previous study proposed that another cold-inducible protein (Rbm3) belonging to the same family of CIRP should be considered an oncogenic protein (53). In order to study whether CIRP could be considered an oncogenic protein, the ability of CIRP to induce transformation was studied. In this study, the ability of CIRP to transform cells in cooperation with mutant K-rasVal12 was examined. K-rasVal12 MEFs are unable to form colonies in soft agar due to the ability of K-rasVal12 to induce premature senescence in primary cells. Therefore, the cooperation of oncogenic proteins may provide their ability to form colonies in the presence of K-rasVal12. These experiments demonstrated that while K-rasVal12 caused neoplastic transformation of E1a vector-infected cells, K-rasVal12 did not acquire the ability to form foci or colonies in soft-agar assays in CIRP-expressing MEFs. Therefore, coexpression of K-rasVal12 and CIRP did not contribute to transformation in either primary MEFs or NIH 3T3 immortalized cells (Fig. 6B and data not shown).
CIRP in human tumors.
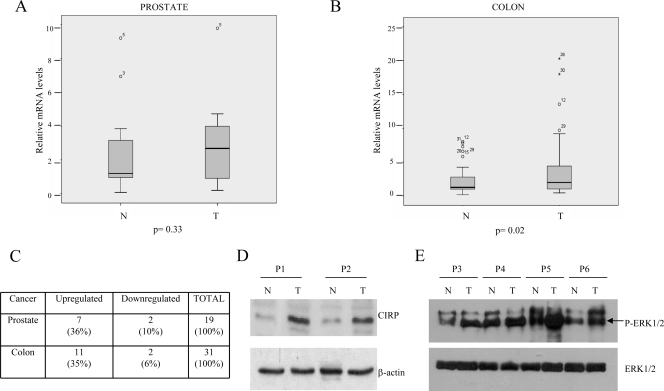
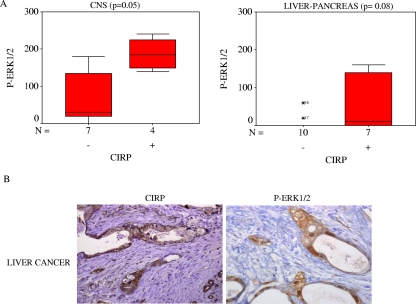
The next study investigated whether the CIRP gene had any effect in human tumors by comparing CIRP mRNA expression in normal and mutant tissue from the same patient. CIRP mRNA was analyzed by quantitative real-time PCR in 19 prostate and 31 colon carcinomas. Mean values from all patients analyzed are shown in Fig. 7A and B. CIRP mRNA upregulation was observed in 7 of 19 (36%) prostate and 11 of 31 (35%) colon cancer patients (Fig. 7C). To confirm CIRP protein upregulation, protein analysis was performed in four cases in which mRNA was found to be upregulated. An example of two patients is shown (Fig. 7D). Statistical analysis showed that in colon carcinomas our results were significant in comparisons of normal and mutant tissues (Wilcoxon test, P = 0.02). In contrast, our results were not significant in prostate carcinomas. In order to determine if CIRP overexpression might be associated with an increase in P-ERK1/2 status in human tumors, P-ERK1/2 was analyzed in six patients with colon cancer and two patients with prostate cancer. P-ERK1/2 was upregulated in three of six samples of colon cancer and one of two cases of prostate cancer. An example of four patients is shown in Fig. 7E. In order to determine if this association was truly significant, we assayed a larger series of tumors (193 patients). The patients were analyzed according to cancer type using immunohistochemistry for CIRP and P-ERK1/2 antibodies. Positive and negative controls were included for each antibody. P-ERK1/2 was found in both the nucleus and the cytoplasm, and both types of staining were considered positive. In contrast, CIRP was expressed mainly in the cytoplasm. The number of patients staining positive for either P-ERK1/2 or CIRP in several types of cancer is represented in Tables 1 and 2, respectively. Interestingly, central nervous system-related tumors showed a significant correlation (P = 0.05) between P-ERK1/2 and CIRP (Fig. 8A). Results approached significance in the liver-pancreas carcinomas (Fig. 8A and B).
FIG. 7.
CIRP expression in prostate and colon tumors. (A) Average of mRNA levels quantified by real-time PCR in 19 prostate carcinomas. N, normal; T, tumor. (B) Average of mRNA levels quantified by real-time PCR in 31 colon carcinomas. (C) Summary of all patients analyzed by quantitative real-time PCR. (D) Immunoblot of the CIRP protein in two patients with colon carcinoma. A total of 250 μg of total protein extract was used. (E) Example of a P-ERK1/2 immunoblot in four patients where CIRP was upregulated (P3, P4, and P5, patients with colon cancer; P6, patient with prostate cancer).
TABLE 1.
P-ERK1/2 protein expression in human cancer
| Cancer type | No. of P-ERK1/2-positive patients | Total no. of patients |
|---|---|---|
| Liver-pancreas | 5 | 17 |
| Gastric | 4 | 19 |
| Nervous system | 11 | 11 |
| Skin | 4 | 13 |
| Lung | 1 | 22 |
| Kidney | 2 | 15 |
| Ovarian | 7 | 25 |
| Rhabdomyosarcomas | 5 | 12 |
| Breast (infiltrating) | 7 | 26 |
| Breast (in situ) | 1 | 7 |
| Colon | 4 | 17 |
| Prostate | 0 | 9 |
| Total | 46 | 193 |
TABLE 2.
CIRP protein expression in human cancer
| Cancer type | No. of CIRP-positive patients | Total no. of patients |
|---|---|---|
| Liver-pancreas | 6 | 17 |
| Gastric | 9 | 19 |
| Nervous system | 4 | 11 |
| Skin | 0 | 13 |
| Lung | 6 | 22 |
| Kidney | 5 | 15 |
| Ovarian | 3 | 25 |
| Rhabdomyosarcomas | 2 | 12 |
| Breast (infiltrating) | 8 | 26 |
| Breast (in situ) | 3 | 7 |
| Colon | 7 | 17 |
| Prostate | 2 | 9 |
| Total | 55 | 193 |
FIG. 8.
Concomitant CIRP and P-ERK expression in human tumors. (A) A Mann-Whitney U test was used to validate the correlation of CIRP and P-ERK1/2 proteins detected by immunohistochemistry from patients with cancer of the central nervous system (n = 11 patients) and liver-pancreas (n = 17 patients). (B) Example of immunohistochemistry of CIRP and P-ERK1/2 proteins in the tumor from a patient with liver cancer.
DISCUSSION
We identified the CIRP gene by exploiting the entry of MEFs into stress-induced senescence in culture. CIRP overexpression bypasses replicative senescence in primary MEFs. CIRP is a cold-inducible member of the glycine-rich RNA-binding protein family. Although the function of CIRP has not yet been determined, it has been suggested that this protein may affect gene expression by facilitating translation at mildly cold temperatures, such as 32°C (11). We have shown in this study that CIRP-expressing MEFs at physiological temperature, in contrast to GFP MEFs, did not overexpress the senescence-associated protein p16INK4a or p21WAF1. Furthermore, they did not accumulate in β-galactosidase-positive cells, which would indicate senescence.
Interestingly, CIRP expression led to an increase in the phosphorylation state of MEK1/2 and ERK1/2 kinases. MAPKs are important mediators of signal transduction from the cell membrane to the nucleus, and ERK1/2 plays a central role in cell proliferation control (32). Activation of the ERK pathway may critically contribute to tumorigenesis or cancer growth, as seen by the presence of activated ERK1/2 in a variety of human tumor cell lines and cancer tissues.
The elevated phosphorylation level of ERK1/2 kinase in CIRP MEFs compared to GFP MEFs is an interesting discovery. ERK1/2 activation has been described in K-rasVal12 cells as the main cause of premature entry into senescence when it is overexpressed in primary MEFs (24, 61). Therefore, K-rasVal12-expressing MEFs were studied in parallel to CIRP and GFP. In order to unravel the intricate mechanism occurring downstream of P-ERK1/2 in K-rasVal12 and CIRP MEFs, several cell cycle regulatory proteins were studied. These proteins would explain why some cells entered senescence while other cells bypassed it. Interestingly, the senescence-associated proteins p16INK4a, p19ARF, and p21WAF1 were highly upregulated in K-rasVal12 MEFs but not in CIRP MEFs. Although cyclin D1 was also upregulated in K-rasVal12 MEFs, these data suggest that p19ARF and p21WAF1 stimulation plays a crucial role in inducing cell arrest and premature senescence. There are three explanations for these findings: (i) upregulation of p19ARF and p21WAF1 occurs as early as ERK1/2 activation; (ii) p19ARF and p21WAF1 activation is about 10-fold stronger than GFP; (iii) p19ARF and p21WAF1 upregulation correlates with the presence of β-galactosidase-positive cells at P4 (see Fig. S4 in the supplemental material).
Interestingly, our results suggest that although both CIRP and K-rasVal12 activate the same protein (P-ERK1/2), each of them shuts down divergent downstream pathways that evolve into different cellular responses. p19ARF, p21WAF1, and p16INK4a prevent cell proliferation in K-rasVal12 MEFs while immortalization occurs in CIRP MEFs.
In MEFs, CIRP overexpression induces ERK1/2 phosphorylation and proliferation in a manner that seems to be specific for MEFs. The phosphorylation does not occur in immortalized murine cells or primary and immortalized human cells (data not shown). This could be explained by the complexity of primary human cells compared to murine cells. Human cells may need more than a single event to bypass senescence and become completely immortalized. Therefore, the molecular pathways that immortalize human and murine cells do not necessarily focus on the same target proteins. Overall, it is not surprising that CIRP overexpression bypasses senescence in primary murine cells but has no effect on their human counterparts. The fact that CIRP overexpression does not increase proliferation of NIH 3T3 cells could be explained by the immortalized nature of NIH 3T3 cells. The ability of primary cells to modulate the expression of certain genes is lost with immortalization (28). This occurs because a mutation is responsible for the immortal phenotype (42). How such mutation(s) affect CIRP downstream pathways should be deeply investigated for each specific immortalized cell line. In addition, NIH 3T3 cells proliferate in response to the activation of the ERK pathway (21). Thus, the fact that P-ERK1/2 is not increased in NIH 3T3 cells upon CIRP overexpression could explain at least in part their lack of proliferation.
P-ERK1/2 has been proposed to be a master regulator of the G1/S transition (32). It is puzzling how P-ERK1/2 could function as a master regulator of cell cycle progression in our model. It was found that the level of P-ERK1/2 in CIRP MEFs was approximately half that in K-rasVal12 MEFs. It is hypothesized that the intensity of P-ERK1/2 activation might be important when the cell shuts down a downstream pathway of P-ERK1/2 that ends in a different cell response. Moreover, the fact that P-ERK1/2 is maintained in K-rasVal12 MEFs at P7 suggests that the duration of ERK1/2 signaling might be important in the maintenance of a senescence phenotype in primary MEFs (see Fig. S5 in the supplemental material). Our results support previous studies performed in vivo and in vitro that describe a dose-dependent oncogene-induced senescence in different contexts (45).
The presence of proliferative signals downstream of ERK1/2 that could differentiate CIRP from GFP and K-rasVal12 MEFs was investigated. This investigation was based on the presumption that there is no lack of inhibitory signals in CIRP MEFs that would explain why they do not enter into senescence. Several ERK1/2 substrates have been described (55). Among them, we found the c-fos protein to be upregulated in CIRP MEFs relative to GFP MEFs. Given the fact that c-fos is a component of the AP-1 complex and that the cyclin D1 promoter contains a functional AP-1 binding site (52), one possible mechanism by which the ERK pathway might stimulate proliferation could be by activating the cyclin D1 protein. Interestingly, both the levels of cyclin D1 and its kinase activity were highly upregulated in CIRP MEFs compared to GFP MEFs. Although cyclin D1 was also upregulated in K-rasVal12 MEFs, some studies have shown that expression of activated ras is sufficient to induce an accumulation of cyclin D1 in several cell types (10, 60). For example, Quereda et al. linked an increase in cyclin D1 to p21WAF1 in MEFs since cyclin D1 is absent in p21WAF1-deficient MEFs (38).
Our data suggest that a myriad of molecular events take place upon K-rasVal12 overexpression. Furthermore, the final cell response is generated by a balance between proliferative and inhibitory signals. However, the high cyclin D1-CDK4 activity observed in CIRP MEFs suggests that another downstream ERK1/2 protein might be separately involved from c-fos activation.
A major role has been described for the ERK1/2 pathway in neuronal activity-induced phosphorylation of S6, eIF-4E, and 4E-BP1. This phosphorylation occurs in long-lasting forms of synaptic plasticity and memory, linking the ERK pathway to proteins that regulate translational machinery (20). The increase in total protein synthesis observed in CIRP MEFs compared to GFP MEFs prompted a study of translation-related proteins. Our results demonstrated an association between CIRP and an increase in total protein synthesis. These findings agree with a previous study that shows that Rbm3, another RNA-binding protein, increases protein synthesis in a different model of immortalized N2a cells (9). Further studies have shown the ability of Rbm3 to act as an oncogene (53). Therefore, we hypothesize that any of these proteins (S6, 4E-BP1, or eIF-4E) could be considered a candidate target for cyclin D1 induction of associated kinase activity in CIRP MEFs.
S6, 4E-BP1, and eIF-4s have a fundamental role in translational control and the ultimate stimulation of protein synthesis. This is a result of their involvement in cap-dependent mRNA translation, which accounts for the synthesis of the vast majority of cellular proteins (all mRNAs transcribed in the nucleus bear a 5′ cap). S6 phosphorylation has been linked to an increase in the initiation of translation. In contrast to the activation caused by S6 phosphorylation, 4E-BP1 phosphorylation produces an inactive protein that releases other transcription factors responsible for initiating translation (29).
Interestingly, we found that the phosphorylation levels of both S6 ribosomal and 4E-BP1 proteins were increased in CIRP MEFs compared to GFP MEFs. Based on these data, we propose that the principal mechanism used by CIRP to immortalize cells is the activation of general protein synthesis. This is achieved through the phosphorylation of S6 ribosomal protein, which increases the ratio of P-4E-BP1 to total 4E-BP1 through ERK1/2. Specific phosphorylation of ribosomal protein S6 and 4E-BP1 has been linked mechanistically to increases in translational efficiency in response to a variety of growth-inducing stimuli (39). Specific phosphorylation at positions 235 and 236 in S6 and positions 37 and 46 in 4E-BP1 would explain the general increase in protein synthesis observed in CIRP MEFs. As expected in the senescent status of K-rasVal12 MEFs, we found a downregulation of S6 phosphorylation at positions 240 and 244 and a downregulation of P-4E-BP1. From these studies, we could not rule out the possibility of a direct/indirect interaction of CIRP with proteins involved in the transcriptional machinery. However, S6 and 4E-BP1 activation would seem to be a more important mechanism than c-fos activation in the mediation of CIRP immortalization. The fact that c-fos protein, unlike P-S6 and P-4E-BP1, does not decrease upon treatment of CIRP-expressing MEFs with the MEK inhibitor PD98058 supports this proposal.
In agreement with our previous data, it is important to note that CIRP inhibition decreases proliferation in not only CIRP MEFs but also HeLa cancer cells and the ES cell-derived TERA cell line. The P-ERK1/2 status is specifically compromised in MEFs but not in the other cancer cell types tested (Fig. 3B and D and 5B; also data not shown). Although CIRP siRNAs decrease cell number in these cancer cell lines, CIRP inhibition might affect different target proteins. The fact that P-ERK1/2 status is not compromised in HeLa or TERA cells when CIRP is inhibited could be due to the ability of cancer cells to activate simultaneously multiple growth-promoting pathways in order to fully transform (12). Under these circumstances, inhibition of one proliferation-promoting pathway would not compromise proliferation since the other pathways could maintain the proliferative status. For example, human colorectal cancer cells harboring a BRAF(V600E) mutation are growth factor independent for the activation of ERK1/2 and survival (58). Given the fact that CIRP is an RNA-binding protein involved in several biological processes (7), its ability to act on different targets might differ in a cell context-dependent manner.
Three concepts led us to hypothesize that CIRP may be relevant in human cancer. (i) ERK1/2 activity potentially has a prognostic value in patient outcome (33). (ii) Total 4E-BP1 is potentially overexpressed in high-grade (compared to low-grade) human tumors, and nearly all the 4E-BP1 proteins are phosphorylated (1, 43). (iii) In accordance with these two facts, we observed that the phosphorylation status of ERK1/2 and 4E-BP1 is upregulated in CIRP MEFs.
A subgroup of colon (33%) and prostate (36%) cancers showed upregulated CIRP mRNA in tumor cells compared to normal tissue. This finding suggests that CIRP upregulation may have biological significance in human tumors. Our results disagree with a previous study by Hamid et al. in which CIRP expression was reported to decrease in tumor tissue (14). Differences between the results of that study and ours may be related to the type of cancer analyzed and to the methodology used in our series of colon and prostate cancers. In order to validate CIRP expression in a myriad of samples, a comprehensive study of the CIRP protein in several tumor types was performed in a total of 193 patients. CIRP and P-ERK1/2 proteins were found to be expressed in 28% and 23%, respectively, of the patients analyzed. Interestingly, CIRP expression was associated with P-ERK1/2 in a group of patients with tumors derived from the central nervous system (P = 0.05). Moreover, a tendency toward significance was observed in some patients with liver-pancreas carcinomas (P = 0.08).
The decrease in growth rate found in mammalian cells treated with mild cold stress is not entirely attributable to arrested metabolism. This decrease may also involve an active process in which CIRP and other stress-responsive proteins (e.g., Rbm3 [9]) play a fundamental role in stimulating proliferation. We hypothesize that although most cell proteins are downregulated or inhibited with cold stress, CIRP is activated. CIRP shuts down a signal, thereby maintaining cells in an active proliferative status. Although the external temperature under mild cold stress may not be the most appropriate, it is not so unbearable that cell growth would be stopped. In this report, we describe for the first time that CIRP bypasses replicative senescence when overexpressed at a physiological temperature (37°C). This bypass is achieved through ERK1/2 activation in MEFs. The mechanisms are similar, but not identical, to those occurring in K-rasVal12 overexpression. It should be noted that these results indicate that CIRP may operate in a different manner than K-rasVal12 (which may be ERK1/2 independent).
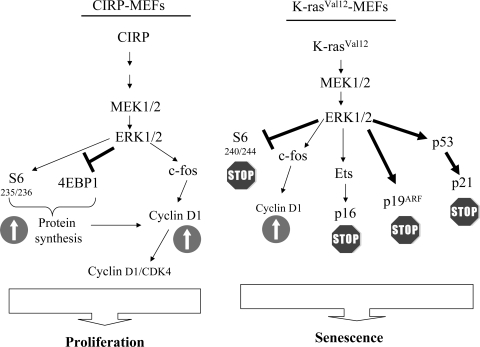
Senescence is an integrated response to diverse signals. In this study, we showed that ERK1/2 activation in primary cells does not necessarily involve premature entry into senescence—it can also promote proliferation. Premature entry into senescence has been previously described, but proliferation has not (24, 61). From our results, we conclude that CIRP bypasses senescence by ultimately upregulating cyclin D1. Cyclin D1 stimulates an increase in cyclin D1-CDK4 kinase activity through the MAPK pathway and proteins involved in the initiation of translation. We propose a comparative model between CIRP and K-rasVal12 explaining why CIRP MEFs bypass senescence and K-rasVal12 MEFs enter it (Fig. 9). Our data suggest that different cell responses are shut down depending on P-ERK1/2 levels. Therefore, ERK1/2 activation must be finely tuned by the cell to allow for the most appropriate response.
FIG. 9.
Proposed model to explain why CIRP and K-rasVal12 genes evolve into different cell responses upon activation with the same MAPK cascade. In CIRP-expressing MEFs, the proliferative signals driven mainly by P-S6, P-4E-BP1, and c-fos activate cyclin D1 and its associated kinase activity. Although the same MAPK pathway is activated in K-rasVal12 MEFs, major inhibitory signals are present that are driven by the proteins p19ARF, p21WAF1, and p16INK4a. These proteins dictate a different cell fate.
Insight into the molecular mechanisms that control ES cell growth will help us understand how cancer cells become immortalized. CIRP and other RNA-binding proteins should be further studied to elucidate their interaction with mRNAs in cell signaling, senescence, and human tumors.
Supplementary Material
Acknowledgments
We thank T. Moliner, R. Somoza, and T. Osteso for their excellent technical assistance. We thank the Tumor Bank from the Pathology Department (Hospital Vall d'Hebron) for providing the tumor samples analyzed here. We are very grateful to J. A. Leal for preparing the manuscript and C. Cavallo for English language support.
This study was supported by Fondo de Investigación Sanitario (FIS) project 04/0530 and Marato TV3 project 052130. M.E.L. is an FIS investigator (CP03/00101).
Footnotes
Published ahead of print on 21 January 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Armengol, G., F. Rojo, J. Castellvi, C. Iglesias, M. Cuatrecasas, B. Pons, J. Baselga, and S. Ramon y Cajal. 2007. 4E-binding protein 1: a key molecular “funnel factor” in human cancer with clinical implications. Cancer Res. 677551-7555. [DOI] [PubMed] [Google Scholar]
- 2.Bernard, S. A., T. W. Gray, M. D. Buist, B. M. Jones, W. Silvester, G. Gutteridge, and K. Smith. 2002. Treatment of comatose survivors of out-of-hospital cardiac arrest with induced hypothermia. N. Engl. J. Med. 346557-563. [DOI] [PubMed] [Google Scholar]
- 3.Brown, J. R., E. Nigh, R. J. Lee, H. Ye, M. A. Thompson, F. Saudou, R. G. Pestell, and M. E. Greenberg. 1998. Fos family members induce cell cycle entry by activating cyclin D1. Mol. Cell. Biol. 185609-5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burdon, T., A. Smith, and P. Savatier. 2002. Signalling, cell cycle and pluripotency in embryonic stem cells. Trends Cell. Biol. 12432-438. [DOI] [PubMed] [Google Scholar]
- 5.Carnero, A., J. D. Hudson, C. M. Price, and D. H. Beach. 2000. p16INK4A and p19ARF act in overlapping pathways in cellular immortalization. Nat. Cell Biol. 2148-155. [DOI] [PubMed] [Google Scholar]
- 6.Collado, M., J. Gil, A. Efeyan, C. Guerra, A. J. Schuhmacher, M. Barradas, A. Benguria, A. Zaballos, J. M. Flores, M. Barbacid, D. Beach, and M. Serrano. 2005. Tumour biology: senescence in premalignant tumours. Nature 436642. [DOI] [PubMed] [Google Scholar]
- 7.De Leeuw, F., T. Zhang, C. Wauquier, G. Huez, V. Kruys, and C. Gueydan. 2007. The cold-inducible RNA-binding protein migrates from the nucleus to cytoplasmic stress granules by a methylation-dependent mechanism and acts as a translational repressor. Exp. Cell. Res. 3134130-4144. [DOI] [PubMed] [Google Scholar]
- 8.Dimri, G. P., X. Lee, G. Basile, M. Acosta, G. Scott, C. Roskelley, E. E. Medrano, M. Linskens, I. Rubelj, O. Pereira-Smith, et al. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA 929363-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dresios, J., A. Aschrafi, G. C. Owens, P. W. Vanderklish, G. M. Edelman, and V. P. Mauro. 2005. Cold stress-induced protein Rbm3 binds 60S ribosomal subunits, alters microRNA levels, and enhances global protein synthesis. Proc. Natl. Acad. Sci. USA 1021865-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filmus, J., A. I. Robles, W. Shi, M. J. Wong, L. L. Colombo, and C. J. Conti. 1994. Induction of cyclin D1 overexpression by activated ras. Oncogene 93627-3633. [PubMed] [Google Scholar]
- 11.Fujita, J. 1999. Cold shock response in mammalian cells. J. Mol. Microbiol. Biotechnol. 1243-255. [PubMed] [Google Scholar]
- 12.Garraway, L. A., and W. R. Sellers. 2006. Lineage dependency and lineage-survival oncogenes in human cancer. Nat. Rev. Cancer. 6593-602. [DOI] [PubMed] [Google Scholar]
- 13.Gil, J., P. Kerai, M. Lleonart, D. Bernard, J. C. Cigudosa, G. Peters, A. Carnero, and D. Beach. 2005. Immortalization of primary human prostate epithelial cells by c-Myc. Cancer Res. 652179-2185. [DOI] [PubMed] [Google Scholar]
- 14.Hamid, A. A., M. Mandai, J. Fujita, K. Nanbu, M. Kariya, T. Kusakari, K. Fukuhara, and S. Fujii. 2003. Expression of cold-inducible RNA-binding protein in the normal endometrium, endometrial hyperplasia, and endometrial carcinoma. Int. J. Gynecol. Pathol. 22240-247. [DOI] [PubMed] [Google Scholar]
- 15.Hanahan, D., and R. A. Weinberg. 2000. The hallmarks of cancer. Cell 10057-70. [DOI] [PubMed] [Google Scholar]
- 16.Hannon, G. J., P. Sun, A. Carnero, L. Y. Xie, R. Maestro, D. S. Conklin, and D. Beach. 1999. MaRX: an approach to genetics in mammalian cells. Science 2831129-1130. [DOI] [PubMed] [Google Scholar]
- 17.Hayflick, L., and P. S. Moorhead. 1961. The serial cultivation of human diploid cell strains. Exp. Cell. Res. 25585-621. [DOI] [PubMed] [Google Scholar]
- 18.Jordan, C. T., M. L. Guzman, and M. Noble. 2006. Cancer stem cells. N. Engl. J. Med. 3551253-1261. [DOI] [PubMed] [Google Scholar]
- 19.Keene, J. D. 2007. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 8533-543. [DOI] [PubMed] [Google Scholar]
- 20.Kelleher, R. J., 3rd, A. Govindarajan, H. Y. Jung, H. Kang, and S. Tonegawa. 2004. Translational control by MAPK signaling in long-term synaptic plasticity and memory. Cell 116467-479. [DOI] [PubMed] [Google Scholar]
- 21.Kim, S. E., and K. Y. Choi. 2007. EGF receptor is involved in WNT3a-mediated proliferation and motility of NIH 3T3 cells via ERK pathway activation. Cell Signal. 191554-1564. [DOI] [PubMed] [Google Scholar]
- 22.Kondoh, H., M. E. Lleonart, J. Gil, J. Wang, P. Degan, G. Peters, D. Martinez, A. Carnero, and D. Beach. 2005. Glycolytic enzymes can modulate cellular life span. Cancer Res. 65177-185. [PubMed] [Google Scholar]
- 23.Lawson, D. A., and O. N. Witte. 2007. Stem cells in prostate cancer initiation and progression. J. Clin. Investig. 1172044-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin, A. W., M. Barradas, J. C. Stone, L. van Aelst, M. Serrano, and S. W. Lowe. 1998. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev. 123008-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin, A. W., and S. W. Lowe. 2001. Oncogenic ras activates the ARF-p53 pathway to suppress epithelial cell transformation. Proc. Natl. Acad. Sci. USA 985025-5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lleonart, M. E., F. Vidal, D. Gallardo, M. Diaz-Fuertes, F. Rojo, M. Cuatrecasas, L. Lopez-Vicente, H. Kondoh, C. Blanco, A. Carnero, and S. Ramon y Cajal. 2006. New p53 related genes in human tumors: significant downregulation in colon and lung carcinomas. Oncol. Rep. 16603-608. [DOI] [PubMed] [Google Scholar]
- 27.Ma, S., K. W. Chan, L. Hu, T. K. Lee, J. Y. Wo, I. O. Ng, B. J. Zheng, and X. Y. Guan. 2007. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology 1322542-2556. [DOI] [PubMed] [Google Scholar]
- 28.Malyankar, U. M., S. R. Rittling, A. Connor, and D. T. Denhardt. 1994. The mitogen-regulated protein/proliferin transcript is degraded in primary mouse embryo fibroblast but not 3T3 nuclei: altered RNA processing correlates with immortalization. Proc. Natl. Acad. Sci. USA 91335-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mamane, Y., E. Petroulakis, O. LeBacquer, and N. Sonenberg. 2006. mTOR, translation initiation and cancer. Oncogene 256416-6422. [DOI] [PubMed] [Google Scholar]
- 30.Manning, B. D. 2004. Balancing Akt with S6K: implications for both metabolic diseases and tumorigenesis. J. Cell Biol. 167399-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsushime, H., D. E. Quelle, S. A. Shurtleff, M. Shibuya, C. J. Sherr, and J. Y. Kato. 1994. D-type cyclin-dependent kinase activity in mammalian cells. Mol. Cell. Biol. 142066-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meloche, S., and J. Pouyssegur. 2007. The ERK1/2 mitogen-activated protein kinase pathway as a master regulator of the G1- to S-phase transition. Oncogene 263227-3239. [DOI] [PubMed] [Google Scholar]
- 33.Milde-Langosch, K., A. M. Bamberger, G. Rieck, D. Grund, G. Hemminger, V. Muller, and T. Loning. 2005. Expression and prognostic relevance of activated extracellular-regulated kinases (ERK1/2) in breast cancer. Br. J. Cancer 922206-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mishra-Gorur, K., H. A. Singer, and J. J. Castellot, Jr. 2002. The S18 ribosomal protein is a putative substrate for Ca2+/calmodulin-activated protein kinase II. J. Biol. Chem. 27733537-33540. [DOI] [PubMed] [Google Scholar]
- 35.Nishiyama, H., K. Itoh, Y. Kaneko, M. Kishishita, O. Yoshida, and J. Fujita. 1997. A glycine-rich RNA-binding protein mediating cold-inducible suppression of mammalian cell growth. J. Cell Biol. 137899-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishiyama, H., J. H. Xue, T. Sato, H. Fukuyama, N. Mizuno, T. Houtani, T. Sugimoto, and J. Fujita. 1998. Diurnal change of the cold-inducible RNA-binding protein (Cirp) expression in mouse brain. Biochem. Biophys. Res. Commun. 245534-538. [DOI] [PubMed] [Google Scholar]
- 37.Pardal, R., A. V. Molofsky, S. He, and S. J. Morrison. 2005. Stem cell self-renewal and cancer cell proliferation are regulated by common networks that balance the activation of proto-oncogenes and tumor suppressors. Cold Spring Harb. Symp. Quant. Biol. 70177-185. [DOI] [PubMed] [Google Scholar]
- 38.Quereda, V., J. Martinalbo, P. Dubus, A. Carnero, and M. Malumbres. 2007. Genetic cooperation between p21Cip1 and INK4 inhibitors in cellular senescence and tumor suppression. Oncogene 267665-7674. [DOI] [PubMed] [Google Scholar]
- 39.Raught, B., A. C. Gingras, S. P. Gygi, H. Imataka, S. Morino, A. Gradi, R. Aebersold, and N. Sonenberg. 2000. Serum-stimulated, rapamycin-sensitive phosphorylation sites in the eukaryotic translation initiation factor 4GI. EMBO J. 19434-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ricci-Vitiani, L., D. G. Lombardi, E. Pilozzi, M. Biffoni, M. Todaro, C. Peschle, and R. De Maria. 2007. Identification and expansion of human colon-cancer-initiating cells. Nature 445111-115. [DOI] [PubMed] [Google Scholar]
- 41.Rimm, D. L., R. L. Camp, L. A. Charette, D. A. Olsen, and E. Provost. 2001. Amplification of tissue by construction of tissue microarrays. Exp. Mol. Pathol. 70255-264. [DOI] [PubMed] [Google Scholar]
- 42.Rittling, S. R. 1996. Clonal nature of spontaneously immortalized 3T3 cells. Exp. Cell. Res. 2297-13. [DOI] [PubMed] [Google Scholar]
- 43.Rojo, F., L. Najera, J. Lirola, J. Jimenez, M. Guzman, M. D. Sabadell, J. Baselga, and S. Ramon y Cajal. 2007. 4E-binding protein 1, a cell signaling hallmark in breast cancer that correlates with pathologic grade and prognosis. Clin. Cancer Res. 1381-89. [DOI] [PubMed] [Google Scholar]
- 44.Sakurai, T., K. Itoh, H. Higashitsuji, K. Nonoguchi, Y. Liu, H. Watanabe, T. Nakano, M. Fukumoto, T. Chiba, and J. Fujita. 2006. Cirp protects against tumor necrosis factor-alpha-induced apoptosis via activation of extracellular signal-regulated kinase. Biochim. Biophys. Acta 1763290-295. [DOI] [PubMed] [Google Scholar]
- 45.Sarkisian, C. J., B. A. Keister, D. B. Stairs, R. B. Boxer, S. E. Moody, and L. A. Chodosh. 2007. Dose-dependent oncogene-induced senescence in vivo and its evasion during mammary tumorigenesis. Nat. Cell Biol. 9493-505. [DOI] [PubMed] [Google Scholar]
- 46.Savatier, P., H. Lapillonne, L. Jirmanova, L. Vitelli, and J. Samarut. 2002. Analysis of the cell cycle in mouse embryonic stem cells. Methods Mol. Biol. 18527-33. [DOI] [PubMed] [Google Scholar]
- 47.Savatier, P., H. Lapillonne, L. A. van Grunsven, B. B. Rudkin, and J. Samarut. 1996. Withdrawal of differentiation inhibitory activity/leukemia inhibitory factor up-regulates D-type cyclins and cyclin-dependent kinase inhibitors in mouse embryonic stem cells. Oncogene 12309-322. [PubMed] [Google Scholar]
- 48.Schmitt, C. A., J. S. Fridman, M. Yang, S. Lee, E. Baranov, R. M. Hoffman, and S. W. Lowe. 2002. A senescence program controlled by p53 and p16INK4a contributes to the outcome of cancer therapy. Cell 109335-346. [DOI] [PubMed] [Google Scholar]
- 49.Serrano, M., A. W. Lin, M. E. McCurrach, D. Beach, and S. W. Lowe. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88593-602. [DOI] [PubMed] [Google Scholar]
- 50.Shann, F. 2003. Hypothermia for traumatic brain injury: how soon, how cold, and how long? Lancet 3621950-1951. [DOI] [PubMed] [Google Scholar]
- 51.Sherr, C. J., and F. McCormick. 2002. The RB and p53 pathways in cancer. Cancer Cell 2103-112. [DOI] [PubMed] [Google Scholar]
- 52.Shiozawa, T., T. Miyamoto, H. Kashima, K. Nakayama, T. Nikaido, and I. Konishi. 2004. Estrogen-induced proliferation of normal endometrial glandular cells is initiated by transcriptional activation of cyclin D1 via binding of c-Jun to an AP-1 sequence. Oncogene 238603-8610. [DOI] [PubMed] [Google Scholar]
- 53.Sureban, S. M., S. Ramalingam, G. Natarajan, R. May, D. Subramaniam, K. S. Bishnupuri, A. R. Morrison, B. K. Dieckgraefe, D. J. Brackett, R. G. Postier, C. W. Houchen, and S. Anant. 2008. Translation regulatory factor RBM3 is a proto-oncogene that prevents mitotic catastrophe. Oncogene 274544-4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taipale, J., and P. A. Beachy. 2001. The Hedgehog and Wnt signalling pathways in cancer. Nature 411349-354. [DOI] [PubMed] [Google Scholar]
- 55.Turjanski, A. G., J. P. Vaque, and J. S. Gutkind. 2007. MAP kinases and the control of nuclear events. Oncogene 263240-3253. [DOI] [PubMed] [Google Scholar]
- 56.Wassmann, H., C. Greiner, S. Hulsmann, D. Moskopp, E. J. Speckmann, J. Meyer, and H. Van Aken. 1998. Hypothermia as cerebroprotective measure. Experimental hypoxic exposure of brain slices and clinical application in critically reduced cerebral perfusion pressure. Neurol. Res. 20S61-65. [DOI] [PubMed] [Google Scholar]
- 57.Wellmann, S., C. Buhrer, E. Moderegger, A. Zelmer, R. Kirschner, P. Koehne, J. Fujita, and K. Seeger. 2004. Oxygen-regulated expression of the RNA-binding proteins RBM3 and CIRP by a HIF-1-independent mechanism. J. Cell Sci. 1171785-1794. [DOI] [PubMed] [Google Scholar]
- 58.Wickenden, J. A., H. Jin, M. Johnson, A. S. Gillings, C. Newson, M. Austin, S. D. Chell, K. Balmanno, C. A. Pritchard, and S. J. Cook. 2008. Colorectal cancer cells with the BRAF(V600E) mutation are addicted to the ERK1/2 pathway for growth factor-independent survival and repression of BIM. Oncogene 277150-7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Widschwendter, M., H. Fiegl, D. Egle, E. Mueller-Holzner, G. Spizzo, C. Marth, D. J. Weisenberger, M. Campan, J. Young, I. Jacobs, and P. W. Laird. 2007. Epigenetic stem cell signature in cancer. Nat. Genet. 39157-158. [DOI] [PubMed] [Google Scholar]
- 60.Winston, J. T., S. R. Coats, Y. Z. Wang, and W. J. Pledger. 1996. Regulation of the cell cycle machinery by oncogenic ras. Oncogene 12127-134. [PubMed] [Google Scholar]
- 61.Zhu, J., D. Woods, M. McMahon, and J. M. Bishop. 1998. Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev. 122997-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.