Abstract
The RAD50/MRE11/NBS1 protein complex (RMN) plays an essential role during the early steps of DNA double-strand break (DSB) repair by homologous recombination. Previous data suggest that one important role for RMN in DSB repair is to provide a link between DNA ends. The striking architecture of the complex, a globular domain from which two extended coiled coils protrude, is essential for this function. Due to its DNA-binding activity, ability to form dimers and interact with both RAD50 and NBS1, MRE11 is considered to be crucial for formation and function of RMN. Here, we show the successful expression and purification of a stable complex containing only RAD50 and NBS1 (RN). The characteristic architecture of the complex was not affected by absence of MRE11. Although MRE11 is a DNA-binding protein it was not required for DNA binding per se or DNA-tethering activity of the complex. The stoichiometry of NBS1 in RMN and RN complexes was estimated by SFM-based volume analysis. These data show that in vitro, R, M and N form a variety of stable complexes with variable subunit composition and stoichiometry, which may be physiologically relevant in different aspects of RMN function.
INTRODUCTION
DNA double-strand breaks (DSBs) can be caused by endogenous or exogenous DNA-damaging agents. Unrepaired DSBs can be lethal, whereas misrepaired DSBs can cause chromosomal fragmentation, translocations and deletions. The resulting genome instability is a common precursor to carcinogenesis and therefore, effective repair of DSBs is of great importance (1). DSB repair involves several processes including recognition of DNA breaks, activation of cell-cycle checkpoints and eventual restoration of intact chromosomes. In eukaryotic cells DSB repair can occur by one of two distinct mechanisms: nonhomologous end-joining (NHEJ) and homologous recombination (HR) (2–4). The RAD50/MRE11/NBS1 (RMN) protein complex is a required component during the early steps of HR (5,6). The essential role of RMN is underscored by the fact that null mutations in genes encoding any of the three subunits are embryonic lethal in mice (7–9). Similarly, both budding and fission yeast cells deleted for RMN subunits have severe DNA-damage sensitivity phenotypes (10–12). In humans, hypomorphic mutations in the genes encoding RMN components cause genetic disorders characterized by marked cancer predisposition. The NBS1 gene gets its name from the human autosomal recessive disorder Nijmegen Breakage syndrome (NBS) (13). Mutations in MRE11 cause ataxia-telangiectasia-like disorder (ATLD), constituting a subset of the phenotype of patients with NBS (14).
The RMN complex has a striking architecture that is crucial for its function (15). The well-defined RM complex is a heterotetrameric assembly of two RAD50 and two MRE11 molecules (R2M2) (16). The RAD50 amino acid sequence has a long region predicted to form a coiled-coil domain separating the Walker A- and B-amino-acid motifs of an ATPase domain. The RAD50 polypeptide folds back on itself forming a 50-nm coiled coil, which juxtaposes the N- and C-termini to constitute a functional ATP binding and hydrolysing head (15–18). The role of ATP binding and hydrolysis in RM function has not been entirely defined. The distinct 50-nm-long coiled coil is a notably flexible structure (19). The coiled-coil apex consists of a pair of cysteine residues that can coordinate a Zn ion and pair with two analogous cysteines from another RAD50 apex (18,20,21). In the complex, MRE11 interacts with itself and with the RAD50 coiled coils near the connection to the ATPase domain, forming a stable heterotetramer (16,18). Several DNA-processing activities of MRE11 have been demonstrated in vitro including: DNA binding, DNA annealing, Mn2+-dependent 3′ to 5′ dsDNA exonuclease, ssDNA endonuclease and DNA duplex unwinding (11,22–29). In some in vitro situations these activities are influenced by association with RAD50 and NBS1 (11,22–25). Recently, MRE11 has been shown to be crucial for initiation and coordination of DNA end-processing during DSB repair (30–34). MRE11 is also expected to be crucial for NBS1 association with the complex, based on a reduced association of NBS1 in the presence of an MRE11 allele associated with ATLD (ATLD¾) (35), and the purification of a stable complex containing only MRE11 and NBS1 (14).
NBS1 is involved in signalling the presence of DNA damage to effect a cell-cycle checkpoint (2,11,36–38). DSB-repair-associated cell-cycle signalling occurs through NBS1 mediated activation of the ATM kinase (39,40). ATM activation is currently thought to involve interaction with NBS1 (41,42) in the RMN complex bound to DNA at the site of breaks. This interaction is proposed to cause dissociation of inactive ATM dimers, creating kinase active ATM monomers (38,40). Activated ATM effects on the cell cycle and DNA-damage response occur though phosphorylation of downstream target proteins (43). However, the architectural arrangement of protein components that contribute to these NBS1-specific functions has not been determined.
The diverse functions of RMN in DSB repair all involve interaction with DNA, and depend on the specific architecture of this protein complex. DNA is bound by the globular domains that include the RAD50 ATPase active site and MRE11, whereas the RAD50 coiled coils protrude away from DNA (15). On linear double-stranded (ds) DNA, this results in the accumulation of large RMN oligomers that tether DNA molecules via interaction of the RMN coiled coils (15,18,20,44). DNA is an allosteric effector of the RMN complex as binding DNA at the globular domain induces an ATP-independent reorientation of the RAD50 coiled coils to become parallel to one another (45). This latter orientation disables intracomplex association of the coiled-coil apexes, and thus stimulates the intercomplex interactions needed for DNA tethering. These observations all imply an important role for RMN in DSB repair organizing broken DNA strands. The above observations suggest that MRE11 has a crucial role in this process being a central element of the complex involved in protein architecture and of protein–DNA interaction (46). However, Mre11 was not present in Rad50 originally purified from Saccharomyces cerevisiae, which nevertheless appears to be a dimeric protein and also binds DNA (47). The reduced levels of NBS1 in purified RAD50/MRE11-(ATLD¾)/NBS1 preparations are presumably due to altered MRE11–NBS1 interaction. This observation does not exclude the presence of additional NBS1 interaction sites on RAD50.
Here, we show the successful expression and purification of a complex containing only RAD50 and NBS1. Scanning force microscopy (SFM) analysis of purified RN preparations showed that RN, like RM and RMN, formed dimers as well as higher-order multimers. SFM-based volume analysis of different (RAD50)2 complexes further identified RN and RMN to be mixtures of at least two different species with different stoichiometry, the main fractions nicely fitting an R2N2 (RN) and R2M2N2 (RMN) stoichiometry. In addition, we observed that RN is more active in DNA-binding and -tethering assays than RM and RMN.
MATERIALS AND METHODS
Protein expression and purification
Human RM, RMN and RN preparations were produced by co-infection of Sf21 cells (7500 cm2) in adherent culture with baculoviruses expressing C-terminally 6-histidine tagged RAD50, untagged MRE11 (RM and RMN) and untagged NBS1 (RMN and RN) at an MOI of ∼10 (constructs for viruses were a generous gift from T. Paull and M. Gellert). Cells were harvested after 72 h. The purification procedure was based on a method described previously (48). Briefly, infected cells were collected, washed three times in PBS and frozen in liquid nitrogen. Cells were thawed and re-suspended in 40-ml cold buffer A (50 mM KH2PO4, pH 7.0, 0.5 M NaCl, 0.5% Tween 20, 10% glycerol, 20 mM β-mercaptoethanol, 10% glycerol) containing 5 mM imidazole and 1 mM Pefablock (Merck). Then, the cells were disrupted by 30 strokes of a type B pestle in a Dounce homogenizer. After 1 h of centrifugation at 100 000 g, the soluble fraction was loaded on a 3-ml Ni2+-NTA agarose column (Qiagen), equilibrated in buffer A containing 5 mM imidazole. The column was washed with 10 vol. of the same buffer and then with 10 vol. of buffer A containing 40 mM imidazole. Bound proteins were eluted in buffer A containing 125 mM imidazole. RAD50 containing fractions were pooled and dialysed against buffer B (20 mM Tris–HCl, pH 8.0, 100 mM NaCl, 10% glycerol, 1 mM DTT). This preparation was loaded on a 1-ml Resource Q column (GE Healthcare) equilibrated in buffer B. After washing the column with 10 column volumes, the proteins were eluted by a 10-ml linear salt gradient from 100 mM to 800 mM NaCl. After addition of Tween 20 (0.1% final concentration), the pooled fractions were loaded on a Superdex 200 size-exclusion column (GE Healthcare) that was equilibrated in buffer B containing 0.1% Tween 20. (For purification of RMN and RN, buffer B containing 500 mM NaCl was used.) RAD50 complex containing fractions were pooled, aliquoted and frozen in liquid nitrogen. Immuno-blotting of RAD50, MRE11 and NBS1 was performed with antibodies MS-RAD10 (GeneTex), PC388 (Oncogene) and sc-8580 (Santa Cruz) respectively, on nitrocellulose transfer membrane (Whatman) using standard immuno-blotting techniques. Protein concentrations were determined by the Bradford method, using BSA as a standard (49).
SFM analysis
RAD50 stoichiometry
Protein preparations were diluted in protein buffer (20 mM Tris–HCl, pH 8.0, 100 mM NaCl, 10% glycerol, 1 mM DTT, 0.1% Tween 20). Five to 15 ng of protein was deposited on freshly cleaved mica. After ∼1 min the mica was rinsed with glass-distilled water (Sigma) and dried with filtered air. Samples were imaged in air at room temperature and humidity by tapping mode SFM using a Nanoscope IV (Digital Instruments). Silicon Nanotips were from Digital Instruments (Nanoprobes). Images were collected at 1 μm × 1 μm, and processed only by flattening to remove background slope. RAD50 stoichiometry was determined by counting the number of 50-nm long coiled coils of individual RAD50 complexes. Relative occurrence of different conformations was determined for all three protein preparations (RM, RMN and RN) from 400–600 individual complexes each.
SFM-based volume analysis
The volume of the globular part of complexes from the three protein preparations was derived from images obtained as described above, but here, only dimeric RAD50 complexes (two coiled coils) were selected. Such dimeric complexes were used for volume analysis of their globular part. Volume analysis was also performed on whole dimeric RM complexes (globular part + coiled coils). Volume determination was performed on images imported into IMAGE SXM 1.69 (National Institutes of Health IMAGE version modified by Steve Barrett, Surface Science Research Centre, University of Liverpool, Liverpool, UK). The average height and area of a manually defined object was used to calculate a volume in arbitrary pixel units. The average volume of three adjacent background regions was then subtracted to determine the volume (50). As RM has a known R2M2 stoichiometry (16,35), the volume of this complex was assumed to represent R2M2, which was used as a basis for the calculation of mass and stoichiometry of the other RAD50 complexes. For each protein preparation between 258 and 318 complexes were analysed, and from these data, a Gaussian distribution was calculated using Origin software (OriginLab Corporation, USA). As an internal control, the mass of RM was also determined as described before (51) via comparison of the measured volume for RM with the volume that was determined for the Ku70/80 heterodimer, which has a known mass of 155 kDa.
DNA tethering reactions
EcoRV linearized pBluescript plasmid was incubated at 1.5 nM with 36 nM RAD50 complex (expressed with respect to RAD50) in binding buffer (20 mM Tris–HCl, pH 8.0, 100 mM NaCl, 10% glycerol, 0.1% Tween 20) for 15 min at 25°C in a volume of 80 μl. Reactions were diluted 8-fold in deposition buffer (10 mM HEPES-KOH, pH 8.0, 10 mM MgCl2), deposited and imaged by SFM as described above. DNA tethering was quantified by determining the number of DNA tethers, the volume of each tether, the number of free DNA molecules and the number of DNA molecules involved in tethering from 32 images (8 μm × 8 μm) for each protein preparation. To determine the number of DNA molecules that were tethered, each DNA molecule coming out of a tether that was visible for more than 50% was counted as one whereas shorter pieces were counted as 0.5.
Electrophoretic mobility shift assays (EMSAs)
Aliquots of dsDNA66 (5′-AF 532-AGA AAC TGG GCA TGT GGA GAC AGA GAA GAC TCT TGG GTT TCT GAT AGG CAC TGA CTC TCT CTG CCT-3′ annealed to its complementary oligo 5′-AGG CAG AGA GAG TCA GTG CCT ATC AGA AAC CCA AGA GTC TTC TCT GTC TCC ACA TGC CCA GTT TCT-3′ from Eurogentec S.A.) labelled with Alexa Fluor at the 5′ end of the upper strand (1 nM) were incubated with the indicated amounts of protein complex in binding buffer (5% glycerol, 25 mM Tris–HCl, pH 7.5, 100 mM KCl, 5 mM MgCl2, 1 mM DTT and 2% PEG-6000) for 20 min at 25°C, in a final volume of 20 μl. The reactions products were separated on a 5% non-denaturing polyacrylamide gel running in 0.5× TBE buffer at 4°C. The labelled DNA was visualized by direct scanning of the wet gel with a 532-nm laser using an image analyser (Typhoon 9200). The emission signal was sorted with a 555BP20-nm filter. Quantification of the data was performed with ImageQuant 5.2 software. To check the protein preparations for contaminant nuclease activity, protein (125 ng) was also incubated with DNA (1 nM) as described above and then subjected to an additional incubation at 37°C in the presence or absence of proteinase K (1 mg/ml). Separation and analysis of complexes formed was performed as described above.
RESULTS
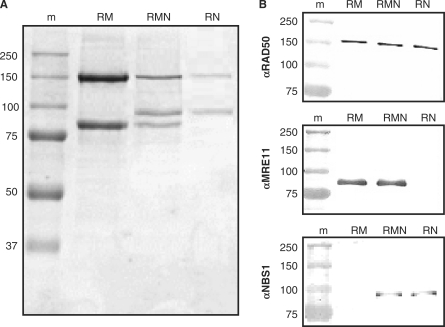
During purification of RMN, some preparations included fractions containing mainly RAD50 and NBS1 (Supplementary Figure S1A). Thus, we pursued the possibility that RAD50 and NBS1 might form a stable complex. Native polyacrylamide gel electrophoresis (PAGE) analysis of such RAD50 and NBS1 containing fractions confirmed that both proteins were indeed part of the same RAD50/NBS1 complex (Supplementary Figure S1B). To see if an RN complex could also be formed in the absence of MRE11, we expressed RN in Sf21 cells that were co-infected with only the baculoviruses expressing C-terminally 6-histidine tagged RAD50, and untagged NBS1. Indeed, we successfully purified RN, showing that in addition to RM and RMN, RN is also a stable protein complex that can be individually expressed and purified (Figure 1A). Immuno-blotting analysis confirmed the unambiguous identity of RAD50, MRE11 and NBS1, as expected in the three purified complexes (Figure 1B).
Figure 1.
(A) Coomassie stained SDS-PAGE gel of purified protein preparations. Lane 1, molecular size marker (m; molecular mass indicated in kilo Dalton); lane 2, RAD50/MRE11 complex (RM); lane 3, RAD50/MRE11/NBS1 complex (RMN); lane 4, RAD50/NBS1 complex (RN). (B) Immuno-blotting analysis of purified proteins. Blots containing purified RM, RMN and RN preparations were probed with antibodies directed against RAD50, MRE11 and NBS1, as indicated.
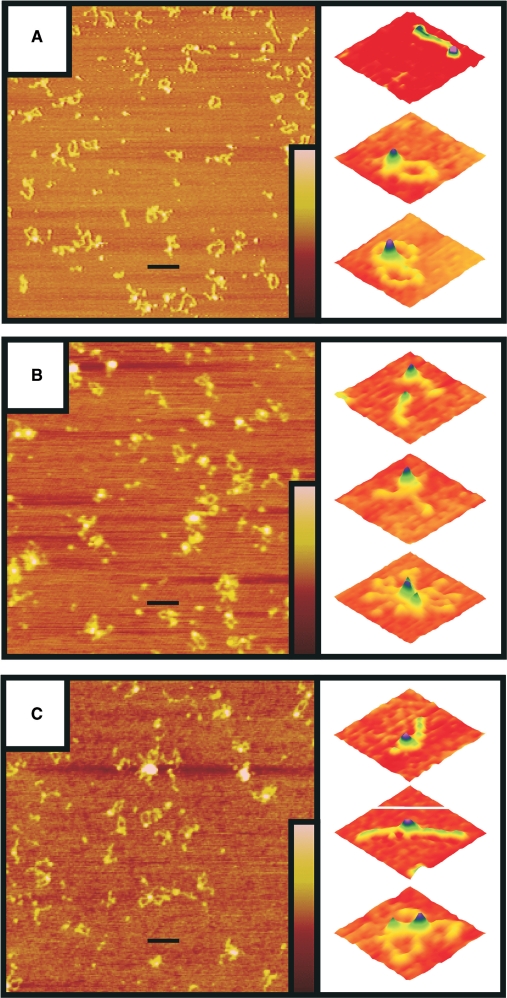
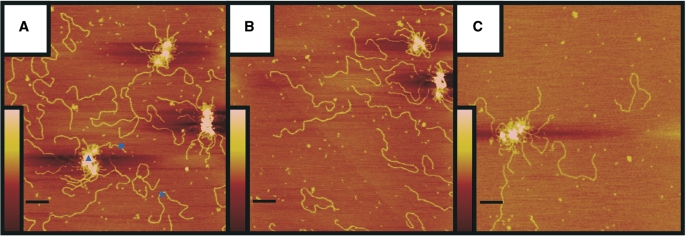
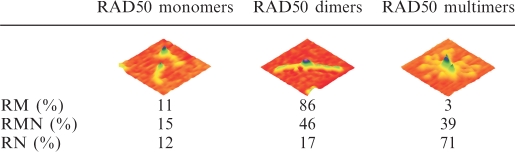
SFM imaging showed that RN looks very similar to RM and RMN with a clear globular domain from which about 50-nm-long coiled coils protrude (Figure 2). Furthermore, RN as well as RM and RMN were all inhomogeneous mixtures of complexes with different amounts of RAD50. By counting the number of coiled coils of individual complexes, they were classified as monomeric, dimeric or multimeric with respect to RAD50. The distribution of complexes with these RAD50 stoichiometries differed among the RMN, RM and RN protein preparations (Table 1). For all three preparations, between 11 and 15% of the complexes were monomeric. For RM, the majority of remaining complexes was dimeric (86%). For RMN, on the other hand, dimeric and multimeric complexes were almost equally prevalent, while for RN, the multimeric form was most common. More than 95% of the multimers observed for all preparations was formed via interaction of the globular domains. The above data suggest that the presence of NBS1 as a part of the complex stimulates multimerization via interaction of the globular domains, whereas the presence of MRE11 reduces multimerization.
Figure 2.
SFM analysis of RM (A), RMN (B) and RN preparations (C). Purified protein was deposited on mica and imaged by tapping mode SFM at 1 μm × 1 μm scale in air (left panels). The scale bars are 100 nm. The colour bars represent the height from 0 to 3 nm (brown to pink). The right panels are examples of individual complexes enlarged and presented as surface plots (0.105 μm × 0.105 μm scale) in which RAD50 has a monomeric (I), dimeric (II) or multimeric (III) stoichiometry.
Table 1.
Distribution of RAD50 stoichiometry on the different purified protein preparations
 |
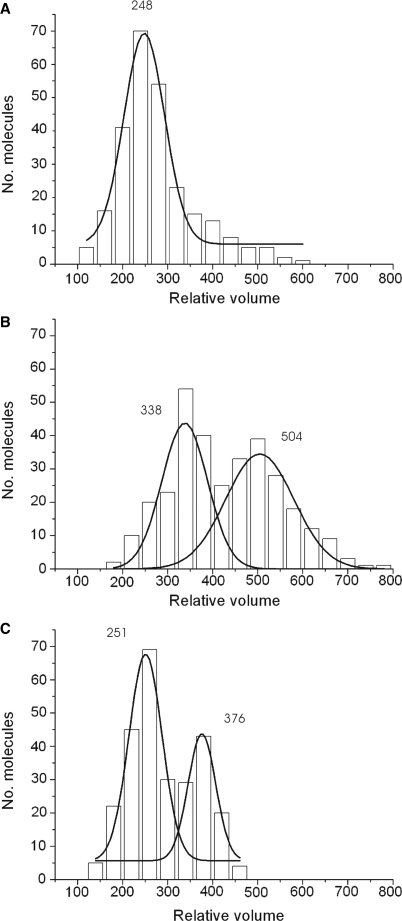
A heterotetrameric (R2M2) complex is formed by the archaeal homolog and this stoichiometry is assumed to be general (16). Although the above data clearly show the presence of monomeric and multimeric RM complexes, the dimeric RAD50 complex is indeed the most abundant form. In contrast to RM, the stoichiometry of RMN and the newly purified RN complex are not well characterized. We estimated the stoichiometry of RN and RMN, by SFM-based volume analysis on the different protein preparations. Dimeric RAD50 complexes were selected from SFM images of the different purified protein preparations. Such dimeric complexes were used to determine the volume of their globular part. The volume distributions were plotted in histograms and Gaussian distributions were calculated (Figure 3). Volume analysis was also performed on whole dimeric complexes (globular part + coiled coils) to determine the volume of the coiled coils separately, and thus also the volume of the dimeric complexes as a whole. As RM has a known R2M2 stoichiometry (16,35), the measured volume for RM (globular part + coiled coils) was assumed to represent R2M2 and was used as a basis for calculating the mass and stoichiometry of the other RAD50 complexes (Table 2). To check this assumption, the molecular mass of RM was also calculated by performing a volume determination on the Ku70/80 heterodimer with a known mass of 155 kDa (Supplementary Figure S2). By multiplying the measured number of Ku70/80 equivalents for RM with the size of the Ku70/80 protein, the molecular weight of RM was determined to be 444 kDa, which is close to the 473 kDa as determined by its amino acid sequence, assuming an R2M2 stoichiometry.
Figure 3.
The volume distributions of the globular part for RM (A), RMN (B) and RN (C) are presented in histograms. Purified protein was deposited on mica and imaged by tapping mode SFM at 1 μm × 1 μm scale in air. From such images isolated dimeric RAD50 complexes were selected for volume determination of the globular part. The x-axis is the relative molecular volume obtained from the SFM data, the y-axis is the number of protein molecules in each peak. A Gaussian distribution was calculated for the data and is displayed as a solid black line. The average volumes estimated from the Gaussian distributions are shown above each peak.
Table 2.
Estimated mass of different RAD50 complexes measured by SFM-based volume analysis
| Protein/Fraction | SFM-based volume | Mass | Stoichiometrya |
|---|---|---|---|
| RM | 379 ± 9b | 473a | R2M2a |
| RMN (low MW fraction) | 469 ± 18 | 586 ± 22 | R2(M+N)n, n = 1.65 ± 0.14 |
| RMN (high MW fraction) | 635 ± 28 | 794 ± 34 | R2(M+N)n, n = 2.91 ± 0.21 |
| RN (low MW fraction) | 382 ± 10 | 477 ± 13 | R2Nn, n = 1.95 ± 0.16 |
| RN (high MW fraction) | 507 ± 13 | 633 ± 17 | R2Nn, n = 3.79 ± 0.20 |
aRM with its known R2M2 stoichiometry is used as a basis for the calculation of mass and stoichiomctry of the other RAD50 complexes. Molecular weight of each polypeptide including C-terminal histidine tag on RAD50 (kDa): RAD50, 155.70; MRE11, 81.03; NBS1, 84.91.
bDetermined volumes and masses are represented by the mean ± SE.
The distribution of volumes for RMN and RN were best fit by two Gaussians, indicating that both preparations contain two components, low and high molecular weight (Figure 3B and C). For RN, the more prevalent, low-molecular-weight complex correlated with an R2N2 stoichiometry (R2Nn; n = 1.95 ± 0.16) while the less abundant high-molecular-weight complex nicely fits a R2N4 stoichiometry (R2Nn; n = 3.79 ± 0.20). For RMN, the determined volume of the low-molecular-weight species corresponds to a molecular mass of 586 ± 22 kDa. This fits a stoichiometry of R2(M+N)n with n being 1.65 ± 0.14. For the high-molecular-weight species this mass is 794 ± 34 kDa correlating with a stoichiometry of R2(M+N)n with n being 2.91 ± 0.21. Within this method, we are not able to discriminate between MRE11 and NBS1. However, if we assume MRE11 to be present as a dimer in both complexes, the calculated NBS1 stoichiometry in both complexes becomes 1.32 ± 0.26 for the low-molecular-weight complex and 3.77 ± 0.41 for the high-molecular-weight complex.
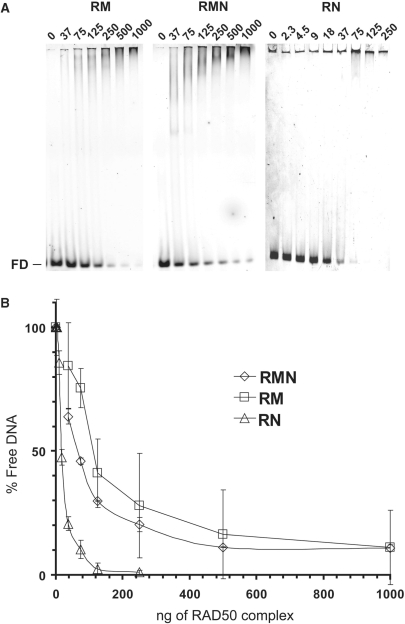
Due to its ability to form dimers and to interact with both RAD50 and NBS1, MRE11 is considered to be crucial in the formation of stable RMN complexes (46). Here, we see that RN also forms a stable dimeric protein complex with an architecture similar to RM and RMN. As MRE11 is able to bind DNA by itself, it is also considered to be an important factor in the DNA-binding and -tethering activities of RMN. To address the role of MRE11 in DNA binding, we performed EMSAs for the different purified preparations in the presence of linear dsDNA using non-denaturing PAGE (Figure 4). The DNA-binding affinity for each complex was determined via quantification of the remaining free DNA after incubation with increasing concentrations of protein complex. Disappearance of the free DNA was not due to contaminant nuclease activity, as for all preparations, treatment with proteinase K led to reappearance of the unbound DNA (Supplementary Figure S3). Surprisingly, we observed that RN is more active in DNA binding than RM and RMN. The amount of RAD50 protein complex needed for binding 50% of the DNA was 58, 32 and 8 ng, corresponding to 6.1, 2.5 and 0.8 nM for RM (R2M2), RMN (R2M2N2) and RN (R2N2), respectively, at 1 nM of DNA. This means that the presence of MRE11 modulates DNA-binding activity of RAD50 containing complexes negatively, whereas NBS1 stimulates this activity.
Figure 4.
DNA binding by RM, RMN and RN. (A) Alexa Fluor 532 labeled dsDNA66 (1 nM), was incubated with RM (37, 75, 125, 250, 500 and 1000 ng), RMN (idem) or RN (2.3, 4.5, 9, 18, 37, 75, 125 and 250 ng) for 20 min at 25°C in a final volume of 20 μl. Complexes formed were separated by 5% non-denaturing PAGE and visualized by fluorescence scanning. FD, free DNA. (B) Free DNA was quantified and plotted against the amounts of protein added.
We also tested the importance of MRE11 for the DNA-tethering activity of RAD50 complexes by SFM imaging. We incubated the protein with 3-kb linear DNA and deposited it for SFM imaging, where tethering is defined as association of DNA molecules via interaction between bound RAD50 complex multimers. RM and RMN have previously been shown to possess DNA-tethering activity (15,18,20,44), which we also observe here (Figure 5A and B). In addition, we also observed that RN is fully functional in DNA-tethering activity (Figure 5C). The RN preparation appeared more active in DNA tethering based on three different quantifications from the SFM images: (i) a higher ratio of tethered complexes to free DNA (RN = 1:9.4, RM = 1:21.7 and RMN = 1:17.2), (ii) larger average volume of tethered complexes (RN = 254 000 nm3, RM = 133 000 nm3 and RMN = 157 000 nm3) and (iii) higher percentage of DNA in tethered complexes (RN = 30%, RM = 16% and RMN = 20%). Together, this shows that RAD50 and NBS1 can form a stable complex that maintains its characteristic architecture and that is fully functional in the DNA-binding and the DNA-tethering assays.
Figure 5.
DNA tethering by RM (A), RMN (B) and RN (C). Reaction mixtures containing 15 nM of 3.0-kb linear DNA fragment and 25 nM of purified protein were deposited on mica and imaged by tapping mode SFM. The scale bars are 200 nm. Colour represents height from 0 to 3 nm (brown to pink), as shown by the inserts. DNA tethers (triangle), free DNA molecules (star) and DNA molecules involved in DNA tethering (circle) were all quantified as described in ‘Materials and Methods’ section.
DISCUSSION
It is generally accepted that RMN plays an essential role in the early steps of DSB repair. The multiple roles that RMN may have during this process are, however, not yet all clearly defined. Several lines of evidence indicate that one important role for RMN in DSB repair is to link broken DNA ends. Due to its DNA-binding activity and ability to bind RAD50 and NBS1, MRE11 is considered as the central element of the complex involved in protein–protein and protein–DNA interaction. The ability of MRE11 to homodimerize is considered crucial for formation of a functional dimeric RMN complex (30).
Here, we show that, in addition to RM and RMN, RN is also a stable protein complex with a similar molecular architecture. This shows that RAD50 is able to directly interact with NBS1. Previous data suggested that NBS1 binding to the complex is linked to MRE11 (14,35). However, the presence of NBS1 interaction sites on RAD50 cannot be excluded. It is even possible that MRE11 and NBS1 compete for binding in the complex, or that RAD50 and MRE11 both contribute to NBS1 interaction. The latter would also explain the reduced yield of protein upon purifying RN compared to MRE11-containing complexes (Figure 1A).
We show that a protein complex with dimeric RAD50 does not depend on the presence of MRE11. This is in line with the observation that purified Rad50 from S. cerevisiae was a dimeric protein by itself (47). In our SFM assay mulitimerization of the protein complex is an important aspect of DNA tethering. In that respect, it is interesting to see that the presence of NBS1 stimulates multimerization, whereas the presence of MRE11 seems to inhibit such globular domain interactions.
A previous sedimentation equilibrium analysis of RMN failed to assign a clear stoichiometry for the components and suggested that this complex may exist as a mixture with different stoichiometries (35). Indeed, our SFM-based single-particle volume analysis shows that both RMN and RN are mixtures of at least two complexes with different stoichiometry. For RN, the low- and high-molecular-weight complexes nicely correlate with an R2N2 and R2N4 stoichiometry. For RMN, we cannot discriminate between MRE11 and NBS1, but assuming that MRE11 is present in both complexes as a dimer, the calculated stoichiometry for the two RMN species is R2M2Nn with n being 1.32 for the low-molecular-weight species, and n being 3.77 for the high-molecular-weight species. The existence of the latter complex is in line with the relative intensity of Coomassie staining bands on denaturing polyacrylamide gels of purified RMN preparations, where the NBS1 band is up to 1.5 times more intense than the MRE11 band (Figure1A).
Because MRE11 binds DNA by itself, it is also considered centrally important for the DNA-binding and -tethering activity of RMN. Here we see that this is not the case. The RN complex is more active than RMN in both DNA-binding and -tethering activity. Surprisingly, RM appeared to be the least active in DNA binding and tethering. The increased activity for RN in DNA tethering could reflect the increased affinity to bind DNA as well as the preference of RN to form the oligomeric protein complexes required for DNA tethering (15).
These data present a new picture of the RMN complex. Biochemically functional complexes exist with a variety of subunit stoichiometries but all with the same striking architecture. The different components appear to modulate protein oligomerization. This raises the possibility that RAD50 participates in a variety of complexes where dynamic interchange of component parts may modulate biological function. An example of such behaviour is observed at human telomeres where RAD50 and MRE11 are present during interphase, whereas NBS1 is only present during S-phase, and not in G1 or G2 (52). Here, the transient recruitment of NBS1 in S-phase is hypothesized to play a role in telomere replication as addition of NBS1 to RM potentiates activities that could play an important role in the opening of the t-loop facilitating progression of DNA replication to the end of the chromosome. Although MRE11 is clearly an essential protein, it may not contribute to all functions attributed to RMN. For instance, during HR-mediated DSB repair, there might be times where DNA tethering is required but where the nuclease activity contributed by MRE11 is not. The specific role of the different RMN components in biochemical functions can now be addressed.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The European Commission (IP 512113 and IP 503259); the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research (NWO); NCI (USA) (SBDR 5PO1CA092584); NWO-Chemical Sciences TOP; and the Dutch Cancer Society (H.S. is a Marie Currie fellow and C.W. is the recipient of an NWO Vici award). Funding for open access charge: NWO-Chemical Sciences Vici award.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Su TT. Cellular responses to DNA damage: one signal, multiple choices. Ann. Rev. Genetics. 2006;40:187–208. doi: 10.1146/annurev.genet.40.110405.090428. [DOI] [PubMed] [Google Scholar]
- 2.Khanna KK, Jackson SP. DNA double-strand breaks: signalling, repair and the cancer connection. Nat. Gen. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 3.Valerie K, Povirk LF. Regulation and mechanisms of mammalian double-strand break repair. Oncogene. 2003;22:5792–5812. doi: 10.1038/sj.onc.1206679. [DOI] [PubMed] [Google Scholar]
- 4.Van Gent DC, Hoeijmakers JHJ, Kanaar R. Chromosomal stability and the DNA double-stranded break connection. Nat. Rev. Genet. 2001;2:196–206. doi: 10.1038/35056049. [DOI] [PubMed] [Google Scholar]
- 5.Borde V, Lin W, Novikov E, Petrini JH, Lichten M, Nicolas A. Association of Mre11p with double-strand break sites during yeast meiosis. Mol. Cell. 2004;13:389–401. doi: 10.1016/s1097-2765(04)00034-6. [DOI] [PubMed] [Google Scholar]
- 6.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:666–668. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 7.Luo G, Yao MS, Bender CF, Mills M, Bladl AR, Bradley A, Petrini JHJ. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc. Natl Acad. Sci. USA. 1999;96:7376–7381. doi: 10.1073/pnas.96.13.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao Y, Weaver DT. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res. 1997;25:2985–2991. doi: 10.1093/nar/25.15.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu J, Petersen S, Tessarollo L, Nussenzweig A. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr. Biol. 2001;11:105–109. doi: 10.1016/s0960-9822(01)00019-7. [DOI] [PubMed] [Google Scholar]
- 10.Chahwan C, Nakamura TM, Sivakumar S, Russell P, Rhind N. The fission yeast Rad32 (Mre11)-Rad50-Nbs1 complex is required for the S-phase DNA damage checkpoint. Mol. Cell. Biol. 2003;23:6564–6573. doi: 10.1128/MCB.23.18.6564-6573.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D'Amours D, Jackson SP. The MRE11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell Biol. 2002;3:317–327. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- 12.Pâques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Varon R, Viussinga C, Platzer M, Cerosaletti KM, Chrzanowska KH, Saar K, Beckmann G, Seemanová E, Cooper PR, Nowak NJ, et al. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 1998;93:467–476. doi: 10.1016/s0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- 14.Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NGJ, Raams A, Byrd PJ, Petrini JHJ, Taylor AM. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an Ataxia-Telangiectasia-like disorder. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- 15.de Jager M, van Noort J, van Gent D, Dekker C, Kanaar R, Wyman C. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol. Cell. 2001;8:1129–1135. doi: 10.1016/s1097-2765(01)00381-1. [DOI] [PubMed] [Google Scholar]
- 16.Hopfner KP, Karcher A, Craig L, Woo TT, Carney JP, Tainer JA. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell. 2001;105:473–485. doi: 10.1016/s0092-8674(01)00335-x. [DOI] [PubMed] [Google Scholar]
- 17.Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, Carney JP, Tainer JA. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 18.Hopfner KP, Craig L, Moncalian G, Zinkel RA, Usui T, Owen BAL, Karcher A, Henderson B, Bodmer JL, McMurray CT, et al. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- 19.Van Noort J, Van der Heijden T, De Jager M, Wyman C, Kanaar R, Dekker C. The coiled-coil of the human Rad50 DNA repair protein contains specific segments of increased flexibility. Proc. Natl Acad. Sci. USA. 2003;100:7581–7586. doi: 10.1073/pnas.1330706100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiltzius JJW, Hohl M, Fleming JC, Petrini JHJ. The Rad50 hook domain is a critical determinant of Mre11 complex functions. Nat. Struct. Mol. Biol. 2005;12:403–407. doi: 10.1038/nsmb928. [DOI] [PubMed] [Google Scholar]
- 21.Cahill D, Carney JP. Dimerization of the Rad50 protein is independent of the conserved hook domain. Mutagenesis. 2007;22:269–274. doi: 10.1093/mutage/gem011. [DOI] [PubMed] [Google Scholar]
- 22.Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR, III, Hays L, Morgan WF, Petrini JHJ. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 23.Moncalian G, Lengsfeld B, Bhaskara V, Hopfner KP, Karcher A, Alden E, Tainer JA, Paull TT. The Rad50 signature motif: Essential to ATP binding and biological function. J. Mol. Biol. 2004;335:937–951. doi: 10.1016/j.jmb.2003.11.026. [DOI] [PubMed] [Google Scholar]
- 24.Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre11 facilitates repair of DNA double-strand breaks. Mol. Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 25.Paull TT, Gellert M. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 1999;13:1276–1288. doi: 10.1101/gad.13.10.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Jager M, Dronkert MLG, Modesti M, Beerens CEMT, Kanaar R, van Gent DC. DNA-binding and strand-annealing activities of human Mre11: implications for its roles in DNA double-strand break repair pathways. Nucleic Acids Res. 2001;29:1317–1325. doi: 10.1093/nar/29.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hopfner KP, Karcher A, Shin DS, Fairley C, Tainer JA, Carney JP. Mre11 and Rad50 from Pyrococcus furiosus: Cloning and biochemical characterization reveal an evolutionarily conserved multiprotein machine. J. Bacteriol. 2000;182:6036–6041. doi: 10.1128/jb.182.21.6036-6041.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Furuse M, Nagase Y, Tsubouchi H, Murakami-Murofushi K, Shibata T, Ohta K. Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J. 1998;17:6412–6425. doi: 10.1093/emboj/17.21.6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trujillo KM, Yuan SF, Lee EYHP, Sung P. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J. Biol. Chem. 1998;273:21447–21450. doi: 10.1074/jbc.273.34.21447. [DOI] [PubMed] [Google Scholar]
- 30.Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LN, Cahill D, Hitomi C, Guenther G, et al. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buis J, Wu Y, Deng Y, Leddon J, Westfield G, Eckersdorff M, Sekiguchi JM, Chang S, Ferguson DO. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008;135:85–96. doi: 10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hopkins BB, Paull TT. The P. furiosus Mre11/Rad50 complex promotes 5' strand resection at a DNA double-strand break. Cell. 2008;135:250–260. doi: 10.1016/j.cell.2008.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu Z, Chung W, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee JH, Ghirlando R, Bhaskara V, Hoffmeyer MR, Gu J, Paull TT. Regulation of Mre11/Rad50 by Nbs1. J. Biol. Chem. 2003;278:45171–45181. doi: 10.1074/jbc.M308705200. [DOI] [PubMed] [Google Scholar]
- 36.Assenmacher N, Hopfner KP. MRE11/RAD50/NBS1: complex activities. Chromosoma. 2004;113:157–166. doi: 10.1007/s00412-004-0306-4. [DOI] [PubMed] [Google Scholar]
- 37.Van den Bosch M, Bree RT, Lowndes NF. The MRN complex: coordinating and mediating the response to broken chromosomes. EMBO Rep. 2003;4:844–849. doi: 10.1038/sj.embor.embor925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.You Z, Chahwan C, Bailis J, Hunter T, Russell P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol. Cell. Biol. 2005;25:5363–5379. doi: 10.1128/MCB.25.13.5363-5379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee JL, Paull TT. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004;304:93–96. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- 40.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 41.Berkovich E, Monnat RJ, Jr, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat. Cell Biol. 2007;9:683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- 42.Stracker TH, Morales M, Couto SS, Hussein H, Petrini JHJ. The carboxy terminus of NBS1 is required for induction of apoptosis by the MRE11 complex. Nature. 2007;447:218–221. doi: 10.1038/nature05740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, III, Hurov KE, Corey JL, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to the DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 44.Wyman C, Kanaar R. Chromosome orginization: reaching out to embrace new models. Curr. Biol. 2002;12:R446–R448. doi: 10.1016/s0960-9822(02)00941-7. [DOI] [PubMed] [Google Scholar]
- 45.Moreno-Herrero F, de Jager M, Dekker NH, Kanaar R, Wyman C, Dekker C. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature. 2005;437:440–443. doi: 10.1038/nature03927. [DOI] [PubMed] [Google Scholar]
- 46.Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem. Cell. Biol. 2007;85:509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- 47.Raymond WE, Kleckner N. RAD50 protein of S. cerevisiae exhibits ATP-dependent DNA binding. Nucleic Acids Res. 1993;21:3851–3856. doi: 10.1093/nar/21.16.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee JH, Paull TT. Purification and biochemical characterization of ataxia-telangiectasia mutated and Mre11/Rad50/Nbs1. Methods Enzymol. 2006;408:529–539. doi: 10.1016/S0076-6879(06)08033-5. [DOI] [PubMed] [Google Scholar]
- 49.Bradford MM. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principles of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 50.Wyman C, Rombel I, North AK, Bustamente C, Kustu S. Unusual oligomerization required for activity of NtrC, a bacterial enhancer-binding protein. Science. 1997;275:1658–1661. doi: 10.1126/science.275.5306.1658. [DOI] [PubMed] [Google Scholar]
- 51.Verhoeven EEA, Wyman C, Moolenaar GF, Goosen N. The presence of two UvrB subunits in the UvrAB complex ensures damage detection in both DNA strands. EMBO J. 2002;21:4196–4205. doi: 10.1093/emboj/cdf396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu X-D, Küster B, Mann M, Petrini JHJ, de Lange T. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat. Genet. 2000;25:347–352. doi: 10.1038/77139. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.