Abstract
Methylation of specific histone lysine residues regulates gene expression and heterochromatin function, but little is known about its role in DNA repair. To examine how changes in conserved methylated residues of histone H3 affect nucleotide excision repair (NER), viable H3K4R and H3K79R mutants were generated in Saccharomyces cerevisiae. These mutants show decreased UV survival and impaired NER at the transcriptionally silent HML locus, while maintaining normal NER in the constitutively expressed RPB2 gene and transcriptionally repressed, nucleosome loaded GAL10 gene. Moreover, the HML chromatin in these mutants has reduced accessibility to Micrococcal nuclease (MNase). Importantly, chromatin immunoprecipitation analysis demonstrates there is enhanced recruitment of the Sir complex at the HML locus of these mutants, and deletion of the SIR2 or SIR3 genes restores the MNase accessibility and DNA repair efficiency at this locus. Furthermore, following UV irradiation expression of NER genes in these mutants remains at wild type levels, with the exception of RAD16 which decreases by more than 2-fold. These results indicate that impaired NER occurs in the silenced chromatin of H3K79R and H3K4,79R mutants as a result of increased binding of Sir complexes, which may reduce DNA lesion accessibility to repair enzymes.
INTRODUCTION
In eukaryotic cells, chromatin acts as the ‘platform’ for various nuclear processes including replication, recombination, repair and transcription. The dynamic structure of chromatin and its ability to adopt numerous conformations acts as a control mechanism for different processes that target DNA. The fundamental unit of chromatin is the nucleosome which is comprised of a nucleoprotein core of 147 bp of DNA wrapped 1.65 times around an octamer of the core histones H2A, H2B, H3 and H4 (1). These nucleosome core particles are further assembled into arrays of oligonucleosomes to give the increasingly compact structural hierarchy of chromatin, which is inhibitory to protein factors that interact with DNA. Based on the compaction and accessibility to nuclear machinery, the eukaryotic genome is organised into active regions known as euchromatin and inactive regions known as heterochromatin. Unlike higher eukaryotes, the majority of chromatin in budding yeast, Saccharomyces cerevisiae, is euchromatic in nature. Only a few regions, such as telomeres, the silent mating type loci (HM), and ribosomal RNA genes (rDNA), are packaged into transcriptionally silent heterochromatin-like structures (2). Formation of silent chromatin at HM loci and telomeres is governed by the binding of silencing proteins Sir2, Sir3 and Sir4 (which form the Sir complex) to specific DNA sequences (called silencers) (2,3)
Packaging of chromatin into silenced regions is also affected by post-translational modification of histone tails, including acetylation and methylation of the ε-amino groups of specific lysine residues (4). It has been observed that lysines in the tails of histone H3 and H4 are generally ‘hyperactelyated’ in active chromatin and ‘hypoacetylated’ in silenced chromatin, the latter facilitating binding of the Sir complex. Of particular importance is H4K16, which is the direct target of Sir2-mediated deacetylation (2,5). Apart from acetylation, Set1 and Dot1 (histone methyltransferases) methylate histone H3 at K4 and K79, respectively, and this ‘signature’ is important for preventing heterochromatin formation (6–9). Indeed, methylation of H3K4 and H3K79 is extremely low in heterochromatin, and it has been suggested that Sir proteins predominantly associate with nucleosomes that are hypomethylated at H3K79 (10). In addition, formation of the Sir complex inhibits methylation of K79 by Dot1 (9,10).
Nucleotide excision repair plays a key role in removing bulky, helix distorting DNA damage such as DNA photoproducts produced by UV-light (11–13). It appears that the rate-limiting step in nucleotide excision repair (NER) is recognition of DNA lesions in different chromatin ‘landscapes’. Several in vitro studies have demonstrated that chromatin is a barrier for efficient repair of various NER substrates including UV-induced DNA lesions (14–16). In vivo, however, such lesions are efficiently repaired despite being assembled into nucleosomes because of alterations in nucleosome structure during NER. A popular model for NER in chromatin is the ‘access-repair-restore’ model, which proposes that chromatin structure is altered during repair thereby exposing damage sites to repair factors (17,18). Following repair, the site is restored to its original state (17). Recent studies have indicated that modifications in histones assist in the recognition and accessibility of DNA repair sites (14,15,19). For example, histone H3 is hyperacetylated in vivo following UV irradiation, which may aid in damage recognition and NER (20). Furthermore, the human Gcn-5 containing HAT (histone acetyltransferase) complex TFTC has been shown to contain a DDB1 like protein, SAP130 (21). The DDB1 protein is a component of the UV-DDB heterodimer, which binds to UV lesions in vivo (17,21).
Unlike histone acetylation, histone methylation does not appear to be induced by DNA damage (22). However, methylation of histones appears to play a role in checkpoint control and methylated histones interact with checkpoint proteins following DNA damage (19,23). For example, the checkpoint protein in Schizosaccharomyces pombe Crb2, and its human homolog 53BP1, interacts with methylated histone H4K20 following ionizing radiation-induced DNA damage (22,24,25). Although p53BP1 also has an affinity for methylated H3K79 (22), the biological significance of this interaction remains to be determined. Similar observations were made in budding yeast where Dot1p, which methylates H3K79, is required for the activation of rad53 checkpoint control following UV damage (26). In addition, a recent study shows that Dot1 null mutants and H3K79 point mutants are sensitive to UV radiation (27). Finally, epitasis analysis between dot1 and various UV repair genes indicates that H3K79 methylation plays overlapping roles in NER, post replication repair, and Rad9-mediated checkpoint function (27).
Since histone proteins can undergo a number of different modifications at various residues, it is unclear how these modifications work in concert in the cell. Therefore, to examine the role of these modifications in DNA repair in intact cells, we have begun exploring site-specific mutation of histone modification sites in yeast chromatin that preserve cell viability. In the present study, we examined how histone methylation in both the core domain and tail region affect DNA repair of UV-induced cyclobutane pyrimidine dimers (CPDs). The following histone H3 methylation mutants were used: H3 K4R, H3K79R and H3K4,79R where lysines 4, 79 or 4 and 79, respectively, are mutated to an arginine. Our results indicate that change at the methylation site in the core domain of histone H3 alters cell survivability following UV irradiation and impairs the rate of CPD removal from transcriptionally silent loci by enhancing binding of the Sir complex. In contrast, change in the methylation site in the tail domain of histone H3 alone does not exert an effect of similar magnitude on the UV damage sensitivity of the cell.
MATERIALS AND METHODS
Cell growth and UV irradiation
For DNA repair studies, yeast cells were grown in YPD medium at 30°C until early log phase (A600 ∼0.6), harvested, washed and resuspended in ice-cold PBS (phosphate buffered saline). The resuspended cells were irradiated at 100 J/m2 UV light (254 nm), measured with a Spectroline DM-254N short wave ultraviolet meter (Spectronics Corp.), and incubated in pre-warmed YPD medium in the dark at 30°C. After various time periods, cells were harvested and the DNA isolated using the glass-bead method described previously (28). For UV sensitivity assays, cells were diluted to different concentrations, spread onto YPD-containing agarose plates and irradiated with different UV doses, measured as described above. Colonies were counted after 48 h of incubation in the dark at 30°C.
Locus-specific repair
Gene-specific repair analyses at the HML, RPB2 and GAL10 chromatin loci were performed as described in Nag et al. (29). Briefly, equal amounts of restriction-digested DNA were treated with or without a saturating concentration of T4 endonuclease V for 120 min at 37°C. The samples were then electrophoresed on 1% alkaline agaorse gels, transferred to Hybond N+ membranes (Amersham), and hybridized with gene specific (α32P)dATP-labeled DNA probes. The number of CPDs in these restriction fragments was determined by methods using Poisson analysis, as described in detail elsewhere (30,31). The level of repair was then calculated as a function of the number of CPDs remaining per fragment (32).
RT–PCR and western blot
Cells were grown to early log phase (A600 ∼0.6) under the same conditions as for the repair experiments. Total RNA was isolated from each sample as described previously (28), and 5 μg of RNA was reverse transcribed using Superscript III RT enzyme (Invitrogen), as per manufacturer's instructions. The resultant cDNA was PCR amplified for 30 cycle using primers specific for the HMLα1, RPB2, GAL10 gene and the NER genes listed in Supplementary Table 1. The mRNA levels of the ACT1 gene were used as a loading control.
For western blot, total cellular protein was isolated using a trichloroacetic acid precipitation method as described previously (33). Both anti-Sir2 (Santa Cruz Biotechnology, sc-6666) and anti-tubulin antibodies (Abcam, ab-6160) were used.
Chromatin immunoprecipitation (ChIP)
ChIP was performed as described by Kuo and Allis (34). Mid-log phase yeast cells were crosslinked by 1% formaldehyde, lysed in buffer (50 mM Hepes–KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 1 mM PMSF, 1 μg/ml leupeptin, 1 μg/ml pepstatin A) using glass beads (425–600 µm, Sigma), followed by sonication. Aliquots of 500 μg of protein from each sample were immunoprecipitated with 10 μl of anti-Sir2 antibody (Santa Cruz: sc-6666) overnight at 4°C. The immunoprecipitated complex was purified using Protein A sepharose beads (50% slurry). Chromatin was then eluted from the beads with elution buffer (1% SDS, 0.1 M NaHCO3) and crosslinks reversed by incubation at 65°C overnight. The HML locus was amplified using the primer sets 5′-AGTTTTCGGCACGGACTTATTTGG-3′ and 5′-TCGTCTAATACAAGTTTGAATGACG-3′ for the HML-E silencer region and 5′-GATGCAATTTATTGCTTCCC-3′ and 5′-CATATTGTGAATGTCGTC-3′ for the fragment adjacent to the HMLα2 transcription start site.
Chromatin accessibility assay
Micrococcal nuclease digestion was performed using methods described previously (35,36) with modifications described in Nag et al. (29). Briefly, spheroplast were isolated from mid-log phase (∼1.0 × 107 to 2.0 × 107 cells/ml) cells using Zymolyase. The spheroplasts were suspended in spheroplast digestion buffer [1 M Sorbitol, 50 mM NaCl, 10 mM Tris–HCl, pH 7.5, 5 mM MgCl2, 1 mM CaCl2, 1 mM β-mercaptoethanol, 0.5 mM spermidine and 0.075% (v/v) NP-40]. 200 μl aliquots of spheroplasts were digested with varying concentrations of micrococcal nuclease (MNase) (Worthington) for 10 min at 37°C. The reactions were terminated with 0.1 X vol of stop solution (5% SDS, 250 mM EDTA) followed by Proteinase-K (50 μg/ml) treatment for 2 h at 55°C. After isolation, the DNA was electrophoresed on 1.2% agarose gels, transferred to Hybond N+ membranes via the alkaline transfer method and hybridized to probes specific for either the HML locus (i.e. 518 bp probe to the HMLα1 ORF, and 900 bp probe to the HMLα1 promoter and HMLα2 ORF region) or RPB2 gene. The blots were quantified using ImageQuant 5.2 software and expressed as the ratio of mono- to tri-nucleosome as a function of MNase concentration.
RESULTS
Histone H3K79 mutants are more sensitive to UV irradiation
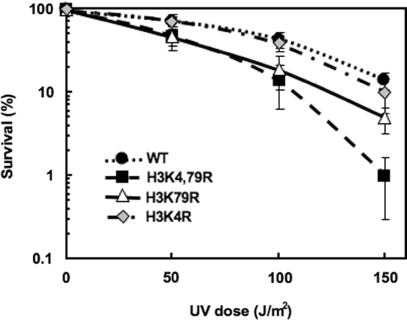
Histone methylation mutants H3K4R, H3K79R and H3K4,79R were generated by plasmid shuffling into the wild-type (wt) yeast strain WY121 (37,38). Both H3K79R and H3K4,79R mutants show enhanced sensitivity to UV-irradiation, compared to wt cells (Figure 1), with the double mutant H3K4,79R being the most sensitive. For example, following a UV dose of 150 J/m2, H3K4,79R cells were over 10 times more sensitive than wt cells, while H3K79R cells were about 3 times more sensitive and H3K4R cells showed almost no sensitivity (Figure 1). As increased UV sensitivity can reflect a deficiency in DNA repair, we examined NER of UV damage to the DNA of H3K79R and H3K4,79R mutants.
Figure 1.
H3K79R and H3K4,79R cells are more sensitive to UV radiation than wt cells. Cells were diluted to appropriate concentrations, spread on YPD plates and irradiated at different UV doses. Colony forming ability following UV radiation was monitored and expressed as percent survival relative to unirradiated cells.
Mutation of Histone H3K79 leads to impaired NER in the HML locus
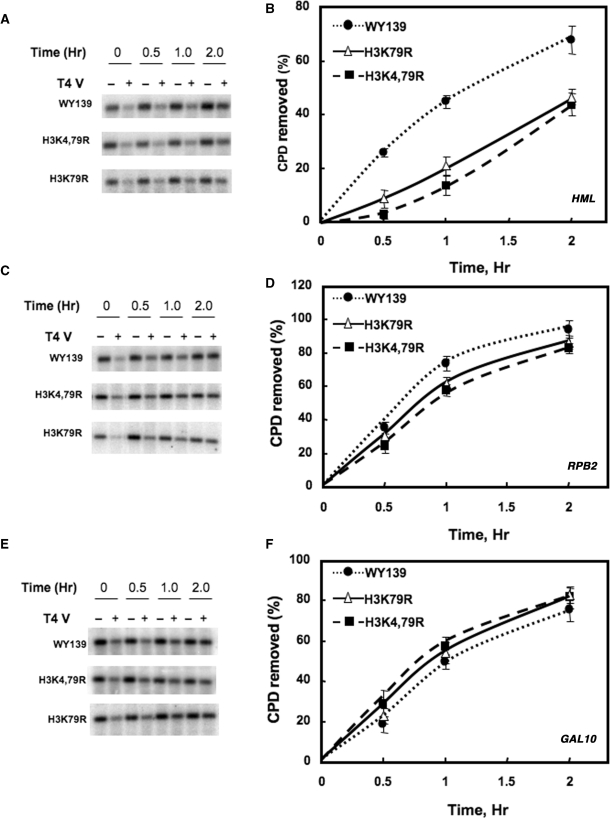
Repair of CPDs was examined in three different chromatin loci in yeast: (i) HML, a nucleosome-loaded, transcriptionally silent mating-type locus; (ii) RPB2, a constitutively expressed gene encoding the second largest subunit of RNA Pol II and (iii) GAL10, a nucleosome loaded inducible gene under transcriptionally repressed conditions. For repair experiments, cells were irradiated with 100 J/m2 UV light (predominately 254 nm) and incubated for different times following irradiation. As shown in Figure 2, both of the methylated lysine mutants showed a significant decrease in the rate of CPD removal from a 2.3 kb Bsp1286I fragment containing the HML locus compared to wt cells (Figure 2A and B). On the other hand, only a small difference was observed when CPD removal was examined in a 3.4 kb NruI fragment containing the RPB2 locus (Figure 2C and D). Furthermore, in a 2.2 kb EcoR1-EcoRV fragment containing the transcriptionally repressed GAL10 gene, the rate of CPD removal was again almost identical between wt and mutant cells (Figure 2E and F). In addition, RT-PCR analysis of the expression of HMLα, GAL10 and RPB2 genes in the presence of glucose indicates there is no leaky expression of either the HMLα or GAL10 genes of these mutants, while expression of the RPB2 gene remains high in both wt and mutant cells (Figure S1A). This indicates that neither repair nor transcription is altered at the RPB2 and GAL10 genes of H3K79R and H3K4,79R cells. Moreover, the enhanced repair of the HML locus in wt cells, compared to the mutants, is not due to the additional participation of transcription coupled repair (TCR) at these loci in wt cells.
Figure 2.
NER of the HML, RPB2 and GAL10 loci in H3 methylation mutants. Cells were irradiated with 100 J/m2 UV light and allowed to repair in the dark at 30°C for various times. Genomic DNA was isolated, digested with appropriate restriction enzyme(s) and subjected to T4 endonuclease V digestion. Southern analysis was performed to determine CPD removal using radioactive probes to a 2.3 kb HML fragment (A), a 3.4 kb RPB2 fragment (C) and a 2.2 kb GAL10 fragment (E). The time course of CPD removal for each strain was plotted for HML (B), RPB2 (D) and GAL10 (F) as the mean ± 1 SD for three independent experiments.
Mutation of Histone H3K79 renders chromatin less accessible to micrococcal nuclease
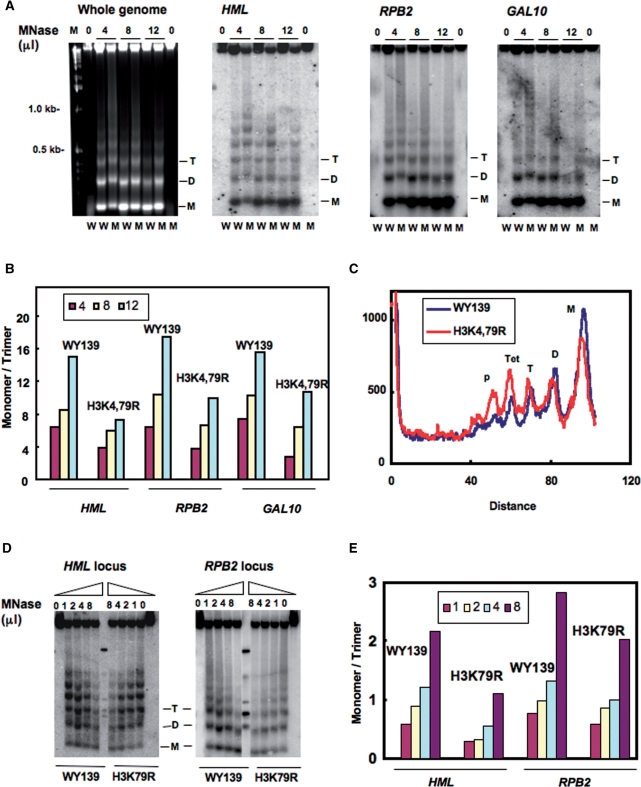
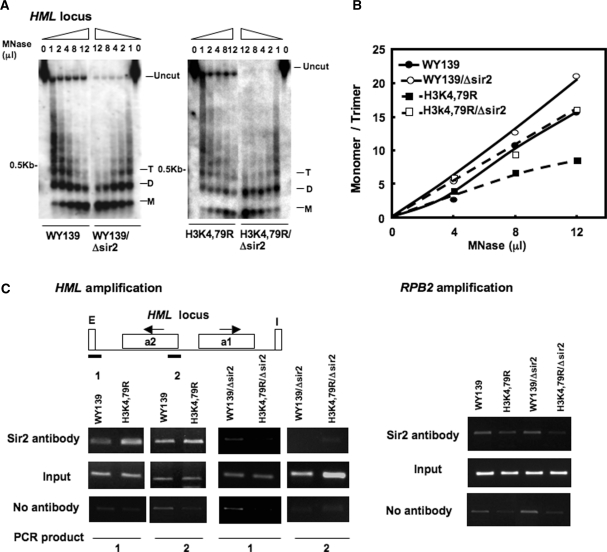
An explanation for the reduced NER efficiency in the HML locus in H3K79R cells is that the H3K79 mutation affects the local chromatin structure and reduces the accessibility of nucleosome DNA to repair enzymes. Therefore, we examined the MNase accessibility of both bulk chromatin and chromatin at the HML, RPB2 and GAL10 loci of these mutants. Spheroplasts isolated from wt and mutant strains were treated with increasing concentrations of MNase and the resulting DNA fragments separated on agarose gels (Figure 3). As shown in Figure 3A and D, chromatin from mutant cells is more resistant to MNase digestion compared to wt. However, the bulk DNA banding patterns are similar (Figure 3A, left panel), indicating there is no significant difference in overall nucleosome repeat length between wt and mutant cells. Furthermore, Southern blot analysis indicates that the HML chromatin of H3K4,79R mutant is also less accessible to MNase than that of wt cells (Figure 3A, second panel from left).
Figure 3.
Nucleosome DNA is less accessible to MNase in H3 methylation mutants. Spheroplasts were isolated from WY139 (wt) and methylation mutants, treated with different concentrations of MNase (10 U/μl stock solution) and genomic DNA was isolated, electrophoresed on agarose gels, stained with ethidium bromide, blotted and hybridized with a probe specific for the HMLα1, RPB2 and GAL10 ORF. (A) MNase digestion patterns are shown for both bulk chromatin, HML, RPB2 and GAL10 chromatin. W: WY139 and M: H3K4,79R. (B) Quantitative analysis of MNase accessibility at the three loci. Data is expressed as the ratio of mono- to tri-nucleosome signal at different concentrations of MNase. (C) Comparative scans of the 4 μl MNase lanes for the HML locus in WY139 and mutant H3K4,79R cells. (D) MNase digestion patterns for HML and RPB2 chromatin of WY139 and H3K79R. (E) Quantitative analysis of MNase accessibility at HML and RPB2. Data is expressed as the ratio of mono- to tri-nucleosome formation at different concentration of MNase.
Quantitative analysis of the blots indicate that MNase digestion is reduced at the HML chromatin locus in the mutant as there is less of an increase in the ratio of mono- to tri-nucleosome formation with increased digestion, compared to wt (Figure 3B). In support of this, scans of lanes from equivalent digestion conditions show a relatively high yield of di- and mono-nucleosomes from wt chromatin, while higher order nucleosome persist for the HML chromatin of the mutant cells (Figure 3C). MNase accessibility at the RPB2 and GAL10 loci also shows a difference between wt and H3K4,79R mutants, but this difference was less pronounced compared to silenced HML chromatin (Figure 3B). Like H3K4,79R, the H3K79R mutant also showed a lower digestion level with MNase and relatively low yield of di- and mono-nucleosome populations at the HML locus under different enzyme concentrations compared to wt (Figure 3D and E). Again, the difference in MNase accessibility at the RPB2 and GAL10 loci of H3K79R is less significant compared to wt cells (Figures 3D and S2). This suggests that lysine to arginine mutations of histone H3K4,79 predominantly affects silenced chromatin.
Deletion of Sir2 or Sir3 increases repair of the HML locus
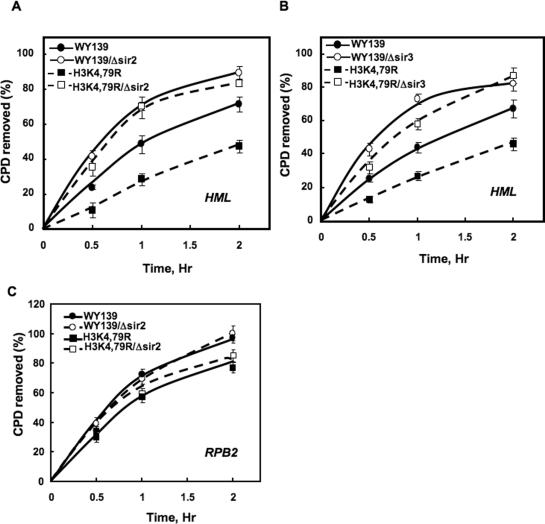
From the above results, it seems likely that nucleosome DNA in the HML locus is less accessible to exogenous enzymes in H3K79 mutants. To examine if this is due to enhanced binding of the Sir complex to HML nucleosomes, the effect of Sir2 and Sir3 deletion on NER was investigated. As shown in Figure 4A and B, deletion of these genes from wt and mutant H3K4,79R cells significantly increases the rate of CPD removal at the HML locus. Furthermore, the rate of repair in Sir deleted H3K4,79R cells is comparable to wt cells having Sir2 or Sir3 deleted (Figure 4, compare open symbols in panels A and B). The CPD removal was also measured at the RPB2 locus in Sir2 deleted strains. The results show there is no significant difference in the repair rate between wt and H3K4,79R, and their corresponding Sir2 deletion strains (Figure 4C). This suggests that decreased repair at the HML locus of H3K4,79R mutant cells is primarily due to enhanced binding of Sir proteins.
Figure 4.
Deletion of SIR genes increases repair in the H3K4,79R mutant. The Sir2 and Sir3 genes were deleted from WY139 (wt) and H3K4,79R mutants using gene replacement, and CPD removal was measured at the HML and RPB2 loci after exposure to 100 J/m2 UV light. The time courses of CPD removal are shown for the 2.3 kb HML fragment for Sir2 and Sir3 deletion mutants in (A) and (B), respectively, and the time course of CPD removal from the 3.4 kb RPB2 fragment for the Sir2 deletion mutant is shown in (C). Data represent the mean ± 1 SD for three independent experiments.
The accessibility of nucleosome DNA was also mapped in the HML locus of Sir2Δ mutants. As shown in Figure 5A, we found that deletion of Sir2 from both wt and H3K4,79R renders HML chromatin more accessible to MNase. While a nucleosome ladder consisting of six to seven repeats could be easily detected in wt and H3K4,79R chromatin, formation of dinucleosomes is more prominent in the respective Sir2 deletion strain. Moreover, quantitative analysis of the MNase digestion pattern clearly shows the increase of mono- to tri-nucleosome DNA ratio with increasing concentration of MNase in the Sir2 deletion strains of both wt and H3K4,79R compared to the respective parental strains (Figure 5B). When tested for MNase accessibility in the RPB2 locus, we observed little change in chromatin structure between wt and H3K4,79R, and their corresponding Sir2 deletion strain (Figure S3B). Moreover, the Sir2 protein level in both wt and H3K4,79R cells is comparable (Figure S3A), indicating that the difference in nucleosome organization between these two strains is not due to differences in SIR2 protein levels.
Figure 5.
MNase accessibility increases in Sir2 deletion strains. Spheroplasts were isolated from the Sir2 deletion strains of WY139 (wt) and H3K4,79R, treated with different concentrations of MNase, blotted and hybridized with a HMLα1 ORF-specific probe, as described in legend to Figure 3. (B) Quantitative analysis of MNase accessibility at the HML locus. Data is expressed as the ratio of mono- to tri-nucleosome formation at different concentration of MNase. (C) ChIP analysis, using Sir2 antibody and HMLα1 ORF primers, showing recruitment of Sir proteins at the HML locus. Chromatin immunoprecipitated from Sir2 deleted strains was used as control (right-hand panels). Each experiment was repeated three times and the data shown are for a single representative experiment. Right panel shows amplification of RPB2 gene from the ChIP DNA, which served as a control.
The results discussed above indicate that the suppression of NER at the HML locus in H3K4,79R mutants is due to the presence of Sir complexes. To directly explore this possibility, ChIP analysis was performed. As shown in Figure 5C, the amount of Sir2 bound at the HML-E silencer is greater in H3K4,79R mutant cells compared to wt cells. A small increase in Sir proteins is also observed at the distal HML α2 gene suggesting that there may be an increase in spreading of Sir complex at the HML locus in the mutant compared to wt cells. Chromatin from Sir2 deletion strains were used as control in the ChIP analysis (Figure 5C, right-hand panels).
Loss of H3K4 or H3K79 methylation affects expression of repair genes
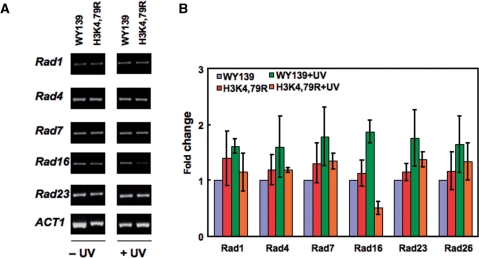
Another explanation for the reduced repair efficiency in H3K79 mutants compared to wt is there may be a change in the expression of repair genes. To test this possibility, we measured the expression levels of a number of NER genes between H3K4,79R mutant and wt cells under normal growth conditions using RT–PCR. As shown in Supplementary Table 1 and Figure 6A, the H3K4,79 mutations do not affect expression of the repair genes tested. However, when these cells were subjected to UV irradiation, we found that among these NER genes, there is an ∼50% decrease in the expression of RAD16 in H3K4,79R cells rather than the ∼2-fold increase observed in wt cells (Figure 6B).
Figure 6.
Expression of NER genes in H3 methylation mutants after UV irradiation. Both wt and H3K4,79R cells were allowed to repair for 20 min after exposure to 100 J/m2 UV radiation. Total RNA was isolated and RT-PCR was performed using gene-specific primers. Cells not treated with UV radiation served as controls. (A) Gel showing the RT-PCR products. (B) Fold change in RT-PCR product for each of the NER genes shown in (A). Data represent the mean ± 1 SD for three independent experiments.
DISCUSSION
In eukaryotes, the packaging of DNA in chromatin not only governs DNA damage formation but also restricts DNA repair machinery from accessing these damage sites (12,13). The accessibility to damaged DNA can be achieved by altering histone–histone or histone–DNA interactions in chromatin through the action of chromatin remodelers or histone modification enzymes (39). In the present study, we have focused on methylation of two conserved lysine residues, K4 and K79, which are located in the tail and histone fold domain, respectively, of histone H3. These residues were mutated to arginine and the mutants were tested for both UV sensitivity and NER efficiency. The H3K79 mutants were found to be more sensitive to UV irradiation compared to wt cells (Figure 1), while H3K4R mutants showed nearly the same sensitivity as wt (at least for UV doses ≤150 J/m2). This suggests that H3K79 methylation plays a major role in cell survival following UV irradiation, in agreement with Thompson and colleagues (27,40). Interestingly, in our case, the double mutant H3K4,79R is more sensitive to UV radiation than either of the single mutants. It is possible that H3K79 methylation may (at least partially) compensate for the effect caused by the loss of H3K4 methylation, or vice versa. Alternatively, the combined methylation states of H3K4 and H3K79 may have a cumulatively greater effect on UV sensitivity than either residue individually. It has also been shown previously that H3K79 methylation not only plays a role in UV induced RAD9-mediated checkpoint function but also affects nucleotide excision repair and RAD5 post-replication repair pathways (27). Therefore, collectively these effects in combination can cause hypersensitivity to UV radiation. Moreover, lysine 79 methylation can also affect the structure of chromatin (7,9,10,41), but how it can influence the repair dynamics especially during NER is not known. In this study, we have addressed this aspect of histone modification by studying the efficiency of removal of CPDs in H3K79 methylation mutants.
We tested NER efficiency of the different mutants in three different, well-characterized chromatin loci of yeast. Compared to wt cells, all three methylation mutants exhibited reduced NER of UV-induced CPDs at the transcriptionally silenced HML locus, with H3K4,79R being the least efficient at carrying out CPD removal. On the other hand, there was no significant difference between these mutants and wt cells in repair of the transcriptionally active RPB2 locus and the transcriptionally repressed GAL10 locus (Figure 2). Importantly, the amount of CPDs formed in each of the loci tested was similar for each strain (data not shown), indicating that these mutations do not affect CPD yields in these different loci.
Mutation of H3 K4,79 affects chromatin accessibility at a silenced locus
The crystal structure of the nucleosome indicates that Lys79 of the two histone H3 proteins are located at the top and bottom surfaces of the nucleosome disk and most likely regulate interactions with exogenous proteins, like Dot1p (9,42). Therefore, it is likely that any change in this residue can affect the accessibility of nucleosome DNA to nuclear proteins. In agreement with this hypothesis, we found that DNA in the HML locus of H3K79R mutants was distinctly less accessible to MNase, compared to wt (Figure 3). The HML locus is distinct from the other two loci examined in our study in that it is a transcriptionally silent locus that contains positioned nucleosomes along with bound Sir complexes. Although the exact mechanism of transcriptional silencing in the HML locus is not clear, various studies have proposed the ‘steric hindrance model’ of reduced chromatin accessibility to DNA binding proteins in the silenced chromatin region (2). Our observation that deletion of Sir2 increases accessibility of MNase to nucleosome DNA and increases NER efficiency at the HML locus (Figures 4 and 5) supports this model. It is worth noting that deletion of Sir2 causes increased transcription of the HML gene (Figure S1B), indicating TCR could also play a role in repair of this locus, in addition to GGR, in Sir2 deletion cells. This situation, however, should be present in both wt and H3K4,79R cells, and the addition of TCR to the overall repair of HML would not be expected to change the NER response between the two cell types, as observed.
Dot1-mediated methylation of Lys79 plays an important role in establishing the transcriptional silencing and Sir protein association with heterochromatin (7,9,10). It has been found that ∼10% of the yeast genome is heterochromatic and is hypomethylated at H3K79, whereas the remaining ∼90% of the genome is hypermethylated at H3K79 (9). It has been proposed that Sir proteins prefer to bind to the core nucleosome surface that have hypo-methylated Lys79 and maintain a stable heterochromatin condition by further preventing Dot1 mediated methylation of this residue (10). Indeed, our ChIP analysis shows that in the H3K4,79R mutant more Sir2 protein is recruited to the HML locus (Figure 5C) than in wt cells. This is also in agreement with previous studies that have shown that the LRS mutant H3K79R has enhanced silencing at telomeres and mating-type loci (41) due to increased recruitment of Sir proteins (43). Furthermore, it has been suggested that the Lys79 to Arg79 mutation mimics the hypomethylation state of H3K79 (37), which would imply that nucleosome cores of H3K4,79R cells have a permanent modification state that results in stronger Sir binding compared to wt cells and hinders accessibility of repair factors. Our results support this hypothesis, and was further strengthened by testing NER in H3 K4,79G and Dot1 mutant cells. The H3K79G mutation disrupts the interaction between Sir protein and the nucleosome core, leading to loss of Sir-mediated silencing, while deletion of Dot1 leads to random binding of Sir proteins throughout the genome (9). In both mutant types, we found that NER at the HML locus was similar to wt cells (Figure S4A and S4C). Indeed, a small but consistent increase was observed in the overall repair rate of these mutants compared to wt cells. Furthermore, MNase digestion patterns show little difference in the chromatin accessibility of the HML locus between these mutants and wt cells (Figure S4B and S4D).
To date, only a few reports have appeared on the effect of Sir proteins on NER in vivo. Initially, it was reported that the silenced HML locus is repaired more slowly by NER than the active MATα locus, which could be explained by TCR in the active MAT locus (44,45). Another study showed that Sir3 protein interacts with NER protein Rad7 in vitro and that deletion of the SIR3 gene rescues ∼25% of the UV sensitivity caused by deletion of Rad7 (46). It was suggested that this interaction may play a role in damage recognition during NER in transcriptionally silent chromatin (46). More recently, Thoma and colleagues examined repair of CPDs by UV photolyase and NER in yeast strains containing the URA3 gene inserted near a telomere end of chromosome V, where URA3 was transcriptionally active in sir3 mutants, partially silenced in SIR3 cells, or completely silenced by overexpression of SIR3 (47). These authors observed (i) efficient repair of active URA3 by both pathways, (ii) reduced NER in partially silenced URA3 and (iii) considerably reduced repair by both pathways in completely silenced URA3. Our results are in agreement with these studies, showing that increased binding of Sir proteins at the HML locus decreases UV survivability and reduces NER of the HML locus, while having no effect on NER in the active RPB2 and repressed GAL10 genes, in yeast cells.
An interesting observation in this report is that expression of RAD16 decreases after UV irradiation in H3K4,79R cells compared to wt (Figure 6). It has been shown that Rad16 and Rad7 form an ATP-dependent DNA damage sensing complex that is needed for efficient NER of silenced chromatin (48,49). Furthermore, it was reported that these proteins can interact with autonomously replicating sequence binding factor Abf1 to generate superhelicity in DNA, possibly required for nucleosome reloading after NER (50). In addition, it was recently reported that Rad16 mediates UV-induced acetylation of histone H3 in yeast, necessary for efficient NER of non-transcribed DNA (51). Therefore, Rad7 and Rad16 proteins may play a role in both early and late steps of NER (52), and the increased expression of RAD16 after UV irradiation (53) is required for the later stages of NER. In our case, whether the decreased expression of Rad16 further retards the NER efficiency at the HML locus in H3K4,79R cells, is not clear and requires further investigation. It seems likely, however, that the presumed ATP-dependent chromatin remodelling activity of RAD16 is involved in NER of nucleosome bound regions, suggesting that low RAD16 expression may indeed reduce the NER rate of silenced loci in H3K4,79R mutants.
In summary, we have shown that H3K79R methylation mutants have a constitutively altered nucleosome structure in the transcriptionally silent HML region. This altered chromatin structure arises from enhanced recruitment of Sir proteins to this locus, even though there is no apparent change in the overall level of Sir protein in these cells. These results indicate that increased recruitment of Sir proteins to the HML locus results in a ‘less flexible’ chromatin structure, one in which NER is significantly impaired. Although subtle changes in chromatin structure of Sir independent (devoid/depleted) loci, like RPB2 and GAL10, were also observed the exact reason for these changes is not clear at present. However, it is possible that exchange of lysine to arginine in H3 may cause some irregular binding of Sir proteins in these loci, which could, at least slightly, impede the access to nucleosome DNA. Nevertheless, these differences are minor compared to those observed for the HML locus. Thus, changes in the histone methylation pattern can promote abnormal silencing in chromatin and impair removal of DNA lesions in chromatin, which can enhance rates of mutagenesis in intact cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (ES002614 to M.J.S.); American Cancer Society (RSG-03-181-01-GMC to J.J.W.). Funding for open access charge: National Institutes of Health (ES002614).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Yi Jin for his helpful advice regarding many of the yeast genetic techniques and microarray data analyses used in this study, and Drs John Hinz and Anamaria Zavala for helpful comments on the manuscript. The contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Environmental Health Sciences (NIEHS, NIH).
REFERENCES
- 1.Luger K. Structure and dynamic behavior of nucleosomes. Curr. Opin. Genet. Dev. 2003;13:127–135. doi: 10.1016/s0959-437x(03)00026-1. [DOI] [PubMed] [Google Scholar]
- 2.Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- 3.Moazed D. Common themes in mechanisms of gene silencing. Mol. Cell. 2001;8:489–498. doi: 10.1016/s1097-2765(01)00340-9. [DOI] [PubMed] [Google Scholar]
- 4.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 5.Suka N, Suka Y, Carmen AA, Wu J, Grunstein M. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol. Cell. 2001;8:473–479. doi: 10.1016/s1097-2765(01)00301-x. [DOI] [PubMed] [Google Scholar]
- 6.Krogan NJ, Dover J, Khorrami S, Greenblatt JF, Schneider J, Johnston M, Shilatifard A. COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression. J. Biol. Chem. 2002;277:10753–10755. doi: 10.1074/jbc.C200023200. [DOI] [PubMed] [Google Scholar]
- 7.Ng HH, Feng Q, Wang H, Erdjument-Bromage H, Tempst P, Zhang Y, Struhl K. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 2002;16:1518–1527. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santos-Rosa H, Bannister AJ, Dehe PM, Geli V, Kouzarides T. Methylation of H3 lysine 4 at euchromatin promotes Sir3p association with heterochromatin. J. Biol. Chem. 2004;279:47506–47512. doi: 10.1074/jbc.M407949200. [DOI] [PubMed] [Google Scholar]
- 9.van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 10.Ng HH, Ciccone DN, Morshead KB, Oettinger MA, Struhl K. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proc. Natl Acad. Sci. USA. 2003;100:1820–1825. doi: 10.1073/pnas.0437846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gillet LC, Scharer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem. Rev. 2006;106:253–276. doi: 10.1021/cr040483f. [DOI] [PubMed] [Google Scholar]
- 12.Prakash S, Prakash L. Nucleotide excision repair in yeast. Mutat. Res. 2000;451:13–24. doi: 10.1016/s0027-5107(00)00037-3. [DOI] [PubMed] [Google Scholar]
- 13.Thoma F. Light and dark in chromatin repair: repair of UV-induced DNA lesions by photolyase and nucleotide excision repair. EMBO J. 1999;18:6585–6598. doi: 10.1093/emboj/18.23.6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong F, Kwon Y, Smerdon MJ. Nucleotide excision repair in chromatin and the right of entry. DNA Repair. 2005;4:884–896. doi: 10.1016/j.dnarep.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 15.Peterson CL, Cote J. Cellular machineries for chromosomal DNA repair. Genes Dev. 2004;18:602–616. doi: 10.1101/gad.1182704. [DOI] [PubMed] [Google Scholar]
- 16.Thoma F. Repair of UV lesions in nucleosomes–intrinsic properties and remodeling. DNA Repair. 2005;4:855–869. doi: 10.1016/j.dnarep.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 17.Green CM, Almouzni G. When repair meets chromatin. First in series on chromatin dynamics. EMBO Rep. 2002;3:28–33. doi: 10.1093/embo-reports/kvf005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smerdon MJ. DNA repair and the role of chromatin structure. Curr. Opin. Cell Biol. 1991;3:422–428. doi: 10.1016/0955-0674(91)90069-b. [DOI] [PubMed] [Google Scholar]
- 19.Altaf M, Saksouk N, Cote J. Histone modifications in response to DNA damage. Mutat. Res. 2007;618:81–90. doi: 10.1016/j.mrfmmm.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 20.Yu Y, Teng Y, Liu H, Reed SH, Waters R. UV irradiation stimulates histone acetylation and chromatin remodeling at a repressed yeast locus. Proc. Natl Acad. Sci. USA. 2005;102:8650–8655. doi: 10.1073/pnas.0501458102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brand M, Moggs JG, Oulad-Abdelghani M, Lejeune F, Dilworth FJ, Stevenin J, Almouzni G, Tora L. UV-damaged DNA-binding protein in the TFTC complex links DNA damage recognition to nucleosome acetylation. EMBO J. 2001;20:3187–3196. doi: 10.1093/emboj/20.12.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huyen Y, Zgheib O, Ditullio RA, Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 23.Vidanes GM, Bonilla CY, Toczyski DP. Complicated tails: histone modifications and the DNA damage response. Cell. 2005;121:973–976. doi: 10.1016/j.cell.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 24.Sanders SL, Portoso M, Mata J, Bahler J, Allshire RC, Kouzarides T. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 2004;119:603–614. doi: 10.1016/j.cell.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 25.Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giannattasio M, Lazzaro F, Plevani P, Muzi-Falconi M. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J. Biol. Chem. 2005;280:9879–9886. doi: 10.1074/jbc.M414453200. [DOI] [PubMed] [Google Scholar]
- 27.Bostelman LJ, Keller AM, Albrecht AM, Arat A, Thompson JS. Methylation of histone H3 lysine-79 by Dot1p plays multiple roles in the response to UV damage in Saccharomyces cerevisiae. DNA Repair. 2007;6:383–395. doi: 10.1016/j.dnarep.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 28.Li S, Smerdon MJ. Rpb4 and Rpb9 mediate subpathways of transcription-coupled DNA repair in Saccharomyces cerevisiae. EMBO J. 2002;21:5921–5929. doi: 10.1093/emboj/cdf589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nag R, Gong F, Fahy D, Smerdon MJ. A single amino acid change in histone H4 enhances UV survival and DNA repair in yeast. Nucleic Acids Res. 2008;36:3857–3866. doi: 10.1093/nar/gkn311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 31.Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 32.Sweder KS, Hanawalt PC. Preferential repair of cyclobutane pyrimidine dimers in the transcribed strand of a gene in yeast chromosomes and plasmids is dependent on transcription. Proc. Natl Acad. Sci. USA. 1992;89:10696–10700. doi: 10.1073/pnas.89.22.10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pellicioli A, Lucca C, Liberi G, Marini F, Lopes M, Plevani P, Romano A, Di Fiore PP, Foiani M. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 1999;18:6561–6572. doi: 10.1093/emboj/18.22.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuo MH, Allis CD. In vivo cross-linking and immunoprecipitation for studying dynamic Protein:DNA associations in a chromatin environment. Methods. 1999;19:425–433. doi: 10.1006/meth.1999.0879. [DOI] [PubMed] [Google Scholar]
- 35.Kent NA, Bird LE, Mellor J. Chromatin analysis in yeast using NP-40 permeabilised sphaeroplasts. Nucleic Acids Res. 1993;21:4653–4654. doi: 10.1093/nar/21.19.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kent NA, Mellor J. Chromatin structure snap-shots: rapid nuclease digestion of chromatin in yeast. Nucleic Acids Res. 1995;23:3786–3787. doi: 10.1093/nar/23.18.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin Y, Rodriguez AM, Stanton JD, Kitazono AA, Wyrick JJ. Simultaneous mutation of methylated lysine residues in histone H3 causes enhanced gene silencing, cell cycle defects, and cell lethality in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007;27:6832–6841. doi: 10.1128/MCB.00745-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin AM, Pouchnik DJ, Walker JL, Wyrick JJ. Redundant roles for histone H3 N-terminal lysine residues in subtelomeric gene repression in Saccharomyces cerevisiae. Genetics. 2004;167:1123–1132. doi: 10.1534/genetics.104.026674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cosgrove MS, Boeke JD, Wolberger C. Regulated nucleosome mobility and the histone code. Nat. Struct. Mol. Biol. 2004;11:1037–1043. doi: 10.1038/nsmb851. [DOI] [PubMed] [Google Scholar]
- 40.Evans ML, Bostelman LJ, Albrecht AM, Keller AM, Strande NT, Thompson JS. UV sensitive mutations in histone H3 in Saccharomyces cerevisiae that alter specific K79 methylation states genetically act through distinct DNA repair pathways. Curr. Genet. 2008;53:259–274. doi: 10.1007/s00294-008-0182-1. [DOI] [PubMed] [Google Scholar]
- 41.Park JH, Cosgrove MS, Youngman E, Wolberger C, Boeke JD. A core nucleosome surface crucial for transcriptional silencing. Nat. Genet. 2002;32:273–279. doi: 10.1038/ng982. [DOI] [PubMed] [Google Scholar]
- 42.White CL, Suto RK, Luger K. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 2001;20:5207–5218. doi: 10.1093/emboj/20.18.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fry CJ, Norris A, Cosgrove M, Boeke JD, Peterson CL. The LRS and SIN domains: two structurally equivalent but functionally distinct nucleosomal surfaces required for transcriptional silencing. Mol. Cell. Biol. 2006;26:9045–9059. doi: 10.1128/MCB.00248-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Terleth C, van Sluis CA, van de PP. Differential repair of UV damage in Saccharomyces cerevisiae. Nucleic Acids Res. 1989;17:4433–4439. doi: 10.1093/nar/17.12.4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Terleth C, Schenk P, Poot R, Brouwer J, van de PP. Differential repair of UV damage in rad mutants of Saccharomyces cerevisiae: a possible function of G2 arrest upon UV irradiation. Mol. Cell. Biol. 1990;10:4678–4684. doi: 10.1128/mcb.10.9.4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paetkau DW, Riese JA, MacMorran WS, Woods RA, Gietz RD. Interaction of the yeast RAD7 and SIR3 proteins: implications for DNA repair and chromatin structure. Genes Dev. 1994;8:2035–2045. doi: 10.1101/gad.8.17.2035. [DOI] [PubMed] [Google Scholar]
- 47.Livingstone-Zatchej M, Marcionelli R, Moller K, de PR, Thoma F. Repair of UV lesions in silenced chromatin provides in vivo evidence for a compact chromatin structure. J. Biol. Chem. 2003;278:37471–37479. doi: 10.1074/jbc.M306335200. [DOI] [PubMed] [Google Scholar]
- 48.Guzder SN, Sung P, Prakash L, Prakash S. Yeast Rad7-Rad16 complex, specific for the nucleotide excision repair of the nontranscribed DNA strand, is an ATP-dependent DNA damage sensor. J. Biol. Chem. 1997;272:21665–21668. doi: 10.1074/jbc.272.35.21665. [DOI] [PubMed] [Google Scholar]
- 49.Verhage R, Zeeman AM, de GN, Gleig F, Bang DD, van de PP, Brouwer J. The RAD7 and RAD16 genes, which are essential for pyrimidine dimer removal from the silent mating type loci, are also required for repair of the nontranscribed strand of an active gene in Saccharomyces cerevisiae. Mol. Cell. Biol. 1994;14:6135–6142. doi: 10.1128/mcb.14.9.6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reed SH, Akiyama M, Stillman B, Friedberg EC. Yeast autonomously replicating sequence binding factor is involved in nucleotide excision repair. Genes Dev. 1999;13:3052–3058. doi: 10.1101/gad.13.23.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teng Y, Liu H, Gill HW, Yu Y, Waters R, Reed SH. Saccharomyces cerevisiae Rad16 mediates ultraviolet-dependent histone H3 acetylation required for efficient global genome nucleotide-excision repair. EMBO Rep. 2008;9:97–102. doi: 10.1038/sj.embor.7401112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reed SH. Nucleotide excision repair in chromatin: the shape of things to come. DNA Repair. 2005;4:909–918. doi: 10.1016/j.dnarep.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 53.Bang DD, Timmermans V, Verhage R, Zeeman AM, van de PP, Brouwer J. Regulation of the Saccharomyces cerevisiae DNA repair gene RAD16. Nucleic Acids Res. 1995;23:1679–1685. doi: 10.1093/nar/23.10.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.