Abstract
Spinal muscular atrophy (SMA) is a common autosomal recessive neurodegenerative disease caused by reduced survival motor neuron (SMN) levels. The assembly machinery containing SMN is implicated in the biogenesis of the spliceosomal small nuclear ribonucleoproteins (snRNPs). SMN is present in both the cytoplasm and nucleus, where it transiently accumulates in subnuclear domains named Cajal bodies (CBs) and functions in the maturation of snRNPs and small nucleolar (sno)RNPs. The impact of lowering SMN levels on the composition of CBs in SMA cells is still not completely understood. Here, we analyse the CB composition in immortalized and primary fibroblasts from SMA patients. We show that the U snRNA export factors PHAX and chromosome region maintenance 1 and the box C/D snoRNP core protein fibrillarin concentrate in CBs from SMA cells, whereas the box H/ACA core proteins GAR1 and NAP57/dyskerin show reduced CB localization. Remarkably, the functional deficiency in SMA cells is associated with decreased localization of the snoRNP chaperone Nopp140 in CBs that correlates with disease severity. Indeed, RNA interference knockdown experiments in control fibroblasts demonstrate that SMN is required for accumulation of Nopp140 in CBs. Conversely, overexpression of SMN in SMA cells restores the CB localization of Nopp140, whereas SMN mutants found in SMA patients are defective in promoting the association of Nopp140 with CBs. Taken together, we demonstrate that only a subset of CB functions (as indicated by the association of representative factors) are impaired in SMA cells and, importantly, we identify the decrease of Nopp140 localization in CBs as a phenotypic marker for SMA.
INTRODUCTION
Proximal spinal muscular atrophy (SMA) is a progressive degenerative disorder of lower (α) motor neurons (1). It is one of the most common genetic causes of mortality in childhood with an incidence of one in 6000–10 000 newborns. Based on the age of onset and severity, SMA has been classified into three main types that compose a continuum of severe (Type I), intermediate (Type II) and moderate (Type III) forms of disease. SMA is caused by mutations in the survival motor neuron 1 gene (SMN1) which result in reduced levels of the SMN protein (2–4). SMN1 possesses a near identical copy, SMN2. The copy number of SMN2 parallels the clinical severity (5). The functional difference between the two genes lies on a translational silent single nucleotide in exon 7 that generates the SMN2-specific alternative splicing of exon 7 (2,6–9). The removal of exon 7 from SMN2 transcript results in the replacement of the C-terminal 16 amino acid residues of SMN by 4-amino acid sequence (SMNΔex7) that might affect the stability of the truncated protein (6,10). SMN1 produces much more of the transcript encoding the full-length SMN than SMN2 gene, whereas the main transcript of SMN2 encodes SMNΔex7. Together these data identify SMN2 as a major modifier gene. Genetically engineered animal models of SMA confirm that SMN is a vital protein (11,12), and SMN2 gene products can partially compensate for the inactivation of SMN1 (13).
The mechanism by which the lower levels of SMN result in the selective degeneration of the α-motor neurons in SMA remains to be determined (14,15). The ubiquitously expressed SMN is a component of a large multiprotein complex involved in a number of cellular processes, including transcription, splicing, ribonucleoprotein (RNP) biogenesis, neurite and axon outgrowth, and the function of the neuromuscular junction (16–18). SMN interacts directly with various proteins, including the spliceosomal Sm core proteins (19–21). The SMN complex assists the cytoplasmic assembly of the Sm proteins onto the U snRNA (22,23) and together are imported to the nucleus (24). The newly imported small nuclear ribonucleoproteins (snRNPs) transiently accumulate in SMN-containing Cajal bodies (CBs) for their maturation into the fully assembled splicing snRNPs (25,26).
CBs are defined as subnuclear bodies that concentrate the CB marker protein coilin (27). They contain several functionally distinct categories of components that are involved in RNA metabolism, such as factors implicated in the maturation of the telomerase RNP and in pre-ribosomal RNA processing, as well as snRNPs and small nucleolar (sno)RNPs. CBs are also involved in the complex intranuclear trafficking of those RNPs (28,29). Interestingly, SMN interacts directly with the core snoRNP components fibrillarin and GAR1, raising the possibility that SMN might also be involved in the late snoRNP assembly step in CBs (30,31). Similar to snRNPs, box C/D snoRNPs transiently localize in CBs before ending up in the nucleolus (32,33). It has been proposed that PHAX (phosphorylated adaptor for RNA export) recruits the snoRNP precursors to CBs, and chromosome region maintenance 1 (CRM1) targets the mature snoRNPs to the nucleolus (34). The presence of CRM1 in CBs might also be related to the retention of newly imported snRNPs into CBs (35). We have previously reported that snRNPs are not detected in CBs of fibroblast cells from all three types of SMA (36), suggesting that other CB constituents could also be absent.
In this study, we analysed the subnuclear localization of other CB components to address the consequences of reduced SMN levels in fibroblast cells derived from SMA patients. Interestingly, the presence of the snoRNP chaperone Nopp140 in CBs from fibroblast cells was correlated with the SMA disease severity. The relationship between SMN and Nopp140 was further assayed using RNAi to deplete immortalized control fibroblast cells of SMN or Nopp140. The immunofluorescence (IF) analyses showed that no localization of Nopp140 is observed in residual CBs of SMN-depleted fibroblast cells. The depletion of Nopp140 slightly reduced the localization of SMN in CBs. In SMA Type I-derived fibroblast cells, overexpression of SMN restored the CB localization of Nopp140. Moreover, SMN overexpression in COS cells enhanced the localization of Nopp140 in SMN-containing CBs. In contrast, cells transfected with SMA mutant SMN failed to enhance the CB localization of Nopp140. Altogether, our results show that SMN levels can modulate the composition of CBs and, therefore, influence the intranuclear organization and regulation of gene expression. Furthermore, our data reveal that Nopp140 localization in CBs is correlated with the phenotypic expression of SMA.
RESULTS
Nopp140 is barely detectable in CBs of SMA-derived fibroblast cells
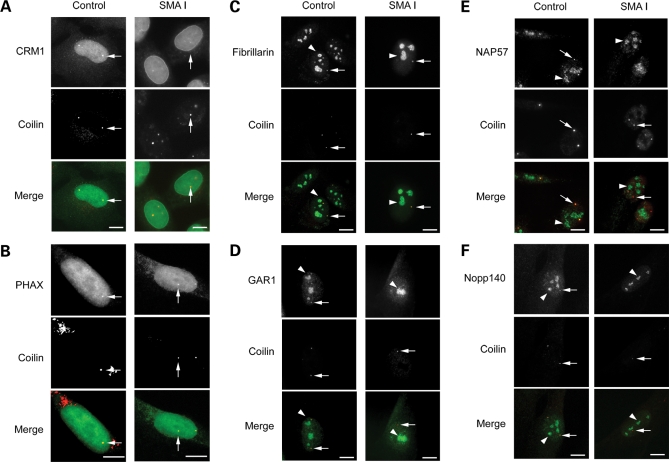
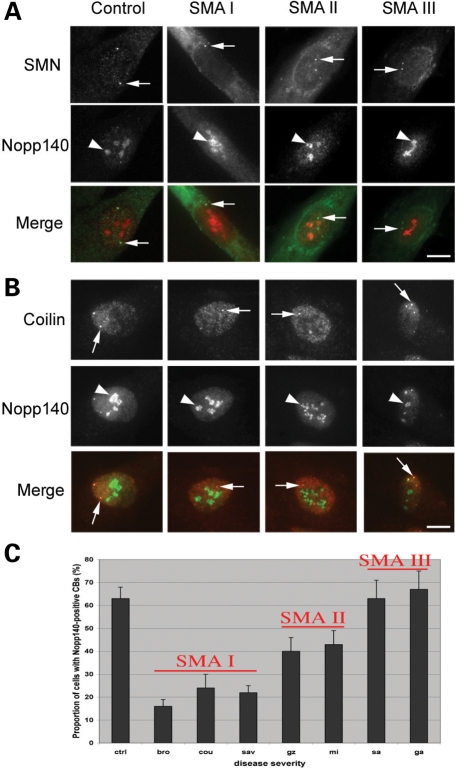
To assess the composition of CBs in SMA cells, immortalized Type I SMA-derived fibroblasts (from here on simply referred to as SMA cells) were further subjected to IF analyses with antibodies against proteins that transiently accumulate in CBs, subnuclear structures involved in non-coding RNA maturation. Antibodies specific for the CB marker coilin were used to visualize CBs. We first examined the distribution of CRM1 and PHAX that are part of the U snRNA export complex (29) and that are also intranuclear transport factors for snoRNAs (34,37). CRM1 and PHAX were shown here to distribute in the nucleoplasm and colocalized with the CB marker coilin in CBs in both SMA and control cells (Fig. 1). The statistical analyses of the double immunolabelling studies revealed that CBs accumulated CRM1 in 23 and 11% of SMA and control cells, respectively (Fig. 1A and Table 1). PHAX foci showed complete localization in CBs in 14% of SMA and in 21% of control cells (Fig. 1B and Table 1). We previously showed that SMN accumulates in CBs in ∼6% of SMA compared with 64% of control cells (36). Together these results indicated that the retention of CRM1 and PHAX in CBs appears independent of the presence of SMN.
Figure 1.
Subnuclear localization of CRM1, PHAX, C/D box snoRNP component fibrillarin, H/ACA box snoRNP proteins GAR1 and NAP57/dyskerin, snoRNP chaperone Nopp140 and coilin in SMA-derived fibroblast cells. Immortalized control and Type I SMA patient fibroblasts were costained for CRM1 (A), PHAX (B), fibrillarin (C), GAR 1 (D), NAP57/dyskerin (E) or Nopp140 (F) in green and the CB marker coilin in red. CRM1 and PHAX nuclear foci in both control and SMA cells were localized in CBs, as shown in yellow (merge). The arrow points to one of the CBs. It should be noted for both control and SMA cells that most of the immunolabelling for fibrillarin, GAR1, NAP57/dyskerin or Nopp140 concentrates in nucleoli (arrowhead) as expected. Fibrillarin nuclear foci (arrow) in both control and SMA cells were localized with coilin in CBs, as shown in yellow. GAR1, NAP57/dyskerin and Nopp140 nuclear foci (arrow) were found colocalized with coilin in control cells. In contrast, only a few Nopp140 foci (arrow) were detected in SMA cells and were not colocalized with coilin. The arrowhead and arrow point to one of the nucleoli and one of the CBs, respectively. Scale bars, 3 µm.
Table 1.
Analyses of nuclear foci in immortalized Type I SMA-derived fibroblast cells
| Immortalized fibroblasts | Proportion of cells witha |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SMN foci (%) | CBs with SMN (%) | CRM1 foci (%) | CBs with CRM1 (%) | PHAX foci (%) | CBs with PHAX (%) | Fib foci (%) | CBs with Fib (%) | CBs with GAR1 (%) | CBs with NAP57 (%) | Nopp140 foci (%) | CBs with Nopp140 (%) | |
| Control | 59 (n = 600) | 64 (n = 200) | 11 (n = 200) | 11 (n = 200) | 21 (n = 200) | 21 (n = 200) | 43 (n = 300) | 37 (n = 60b) | 23 (n = 505) | 36 (n = 438) | 44 (n = 200) | 41 (n = 200) |
| SMAI | 6 (n = 870) | 6 (n = 870) | 23 (n = 100) | 23 (n = 100) | 14 (n = 200) | 14 (n = 200) | 16 (n = 386) | 16 (n = 386) | 6 (n = 425) | 5 (n = 559) | 10 (n = 400) | 1 (n = 100) |
n corresponds to the number of cells that were counted.
afoci were identified with antibodies to one protein only. CBs were identified with antibodies to coilin.
bOnly the cells with fibrillarin (Fib) foci were considered for the statistical analyses.
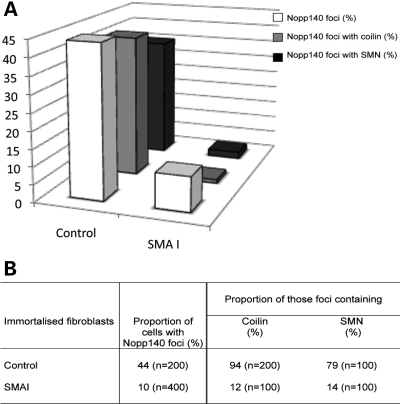
We also examined the distribution of fibrillarin, GAR1, NAP57/dyskerin and Nopp140, and nuclear proteins associated with snoRNPs that are shared between the nucleoli and CBs. Fibrillarin is a core protein associated with the C/D box snoRNPs, such as U3 snoRNPs (29). GAR1 and NAP57/dyskerin are H/ACA box snoRNP core proteins (38). Moreover, SMN has been shown to interact directly with fibrillarin and GAR1 (30,31), but was not found associated with the mature snoRNPs (37). Nopp140 is a snoRNP chaperone that transiently associates with the mature nucleolar H/ACA and C/D box snoRNPs (38). In addition, Nopp140 has been shown to bind directly to the CB marker coilin (39). First, the nucleolar distribution of those proteins appeared similar in SMA and control cells (Fig. 1C–F). Double immunolabelling experiments showed that in 17% of SMA cells fibrillarin accumulated with coilin in CBs when compared with 37% of control cells (Fig. 1C and Table 1). In the remainder of cells, the CBs did not contain fibrillarin. GAR1 localized with coilin in CBs from 6% of SMA and from 23% of control cells, and NAP57/dyskerin in CBs from 5% of SMA and from 36% of control cells (Fig. 1D and E, Table 1). Unexpectedly there were only 10% of SMA cells with Nopp140 foci compared with 44% of control cells (Fig. 2 and Table 1). Double immunolabelling analyses with SMN or coilin (two CB markers) revealed that 94% of those Nopp140 foci contain coilin and 79% of them contain SMN. However, only 12 and 14% of those SMA cells with Nopp140 foci showed the protein in CBs (∼1% of the cell culture, Fig. 2B). The remainder of SMA cells with Nopp140 foci showed structures that lack the two CB marker coilin and SMN, indicating that Nopp140 and CBs form two distinct nuclear structures in those cells. The Nopp140 foci without coilin are reminiscent of residual nuclear bodies enriched in nucleolar proteins that have been reported in fibroblast cells derived from coilin knockout mice (40).
Figure 2.
Analyses of the subnuclear localization of the snoRNP chaperone Nopp140 in CBs from control and SMA cells. The results in the histogram (A) and table (B) demonstrated the marked difference between control and Type I SMA-derived cells. Note the right-hand part of the table in (B) refers to the Nopp140-positive foci (44% in control and 10% in SMAI) as 100%.
In summary, our results revealed that PHAX, CRM1 and fibrillarin are associated with CBs in similar proportion (14–23%) of SMA cells (Table 1). The H/ACA core snoRNP proteins GAR1 and NAP57/dyskerin showed a marked reduction of CB localization in SMA compared with control cells. In addition, Nopp140 was only occasionally associated with CBs and rather accumulated in residual nuclear bodies.
CBs actively recruit Nopp140 in cells overexpressing SMN
Because SMN levels are markedly reduced in SMA cells (41), we tested whether SMN levels might play a role in the localization of Nopp140 to CBs. Therefore, the SMA fibroblast cells were transiently transfected with the expression vector encoding eGFP-SMN and were analysed with antibodies against Nopp140 (Fig. 3). We previously showed that eGFP-SMN concentrates in CBs of transfected cells (36). Cells overexpressing the transgene showed Nopp140 and eGFP-SMN colocalize in CBs from 34% of transfected cells, which is similar to the proportion of control fibroblasts (35%, Table 1). These data confirmed that increased SMN levels contribute to the localization of Nopp140 in CBs.
Figure 3.
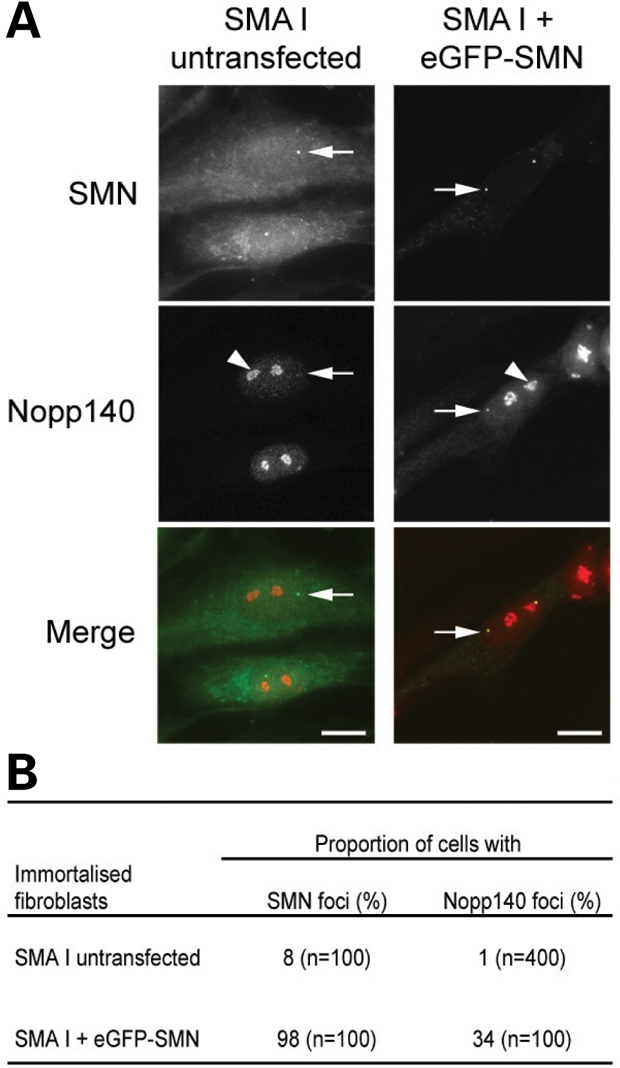
Subnuclear localization of SMN and Nopp140 observed upon transient transfection of immortalized Type I SMA-derived fibroblast cells. The SMA cells were transiently transfected with eGFP-SMN (A). The arrowhead and arrow point to one of the nucleoli and one of the CBs, respectively. The distribution of SMN and Nopp140 foci were compared with untransfected cells (B). Scale bars, 3 µm.
RNAi knockdown experiments confirm a functional link between Nopp140 and SMN
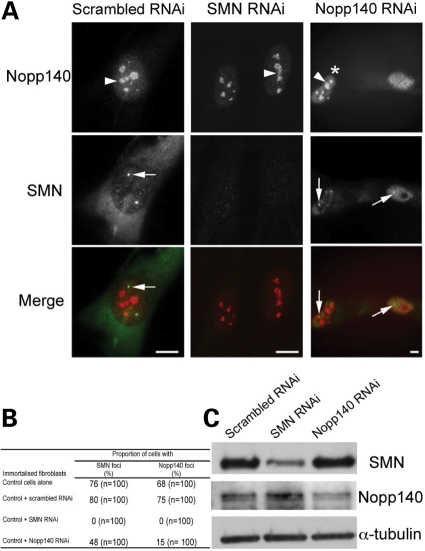
To further investigate the relationship between Nopp140 and SMN, we used RNAi-mediated depletion of SMN or of Nopp140 in control fibroblasts (Fig. 4A–C). Double labelling for coilin and SMN revealed CB localization of SMN in 80% of cells transfected for 48 h with control scrambled or GL2 (firefly luciferase) siRNA. Similar results were obtained when costaining of coilin with Nopp140 was used (75% of cells; Fig. 4B). However, very different observations were made with cells transfected with either SMN or Nopp140 siRNA. Although numerous small coilin foci were detectable in the nucleoplasm of transfected cells, they were never found coincident with SMN or Nopp140 in SMN-depleted cells. Treatment of cells with Nopp140 siRNA slightly reduced the number of cells with SMN or Nopp140 localized in CBs (48 or 15%, respectively). Immunoblotting analyses were used to assay the amount of the two proteins in total extracts from cells transfected with control, SMN or Nopp140 siRNA (Fig. 4C). We observed that the siRNAs reduced the expression levels by 80 and 60% of the target proteins SMN and Nopp140, respectively. The expression levels of Nopp140 or SMN remained constant when not targeted by the siRNA, indicating the specificity of the RNAi-mediated depletion. Taken together our findings further argued that SMN is required for proper Nopp140 accumulation in CBs.
Figure 4.
SMN and Nopp140 RNA interference. The double immunolabelling experiments for SMN and Nopp140 were performed in immortalized control human fibroblasts 48 h after transfected with SMN or Nopp140 siRNA (A). The arrowhead and arrow point to one of the nucleoli and one of the CBs, respectively. The asterisk points to an unsilenced cell. The statistical analyses are presented in table (B). Western blot analyses of protein extracts from cells treated with siRNA to scrambled, SMN or Nopp140 revealed the specificity of knockdown (C). The α-tubulin incubation served as loading control. Scale bars, 3 µm.
SMA mutant proteins fail to enhance the CB localization of Nopp140
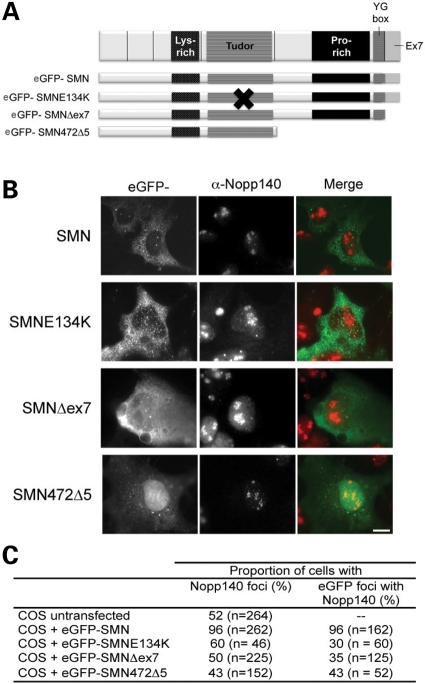
Because our results favoured the view that SMN levels are important for Nopp140 localization to CBs, we tested the effects of eGFP-SMN overexpression in COS cells, which already expressed high amount of endogenous SMN (Fig. 5A–C). We showed that transient overexpression of SMN in COS cells still enhances the localization of Nopp140 to CBs of 96% of transfected compared with 52% of untransfected COS cells. This observation prompted us to test the effects on Nopp140 localization in CBs of COS cells transiently transfected with SMA mutants SMNE134K, SMNΔex7 and SMN472Δ5, respectively (Fig. 5A). Although coilin showed CB colocalization with SMA mutants in transfected cells (36), Nopp140 showed colocalization with the GFP-tagged mutant SMN foci in 30, 35 and 43% of cells expressing SMNE134K, SMNΔex7 and SMN472Δ5, respectively (Fig. 5). This contrasted with the complete colocalization of Nopp140 and wild-type eGFP-SMN in all foci. These data indicated that all three SMA mutants are defective in promoting the recruitment of Nopp140 in CBs.
Figure 5.
Comparison of the localization of Nopp140 in CBs of cells transfected with wild-type SMN and SMA mutants. The SMN protein is depicted with the Lys-rich, Tudor, Pro-rich, oligomerization YG-box and ex7 encoded domains. SMN and SMA mutants were fused to the N-terminal end of eGFP (A, 36). The constructs indicated were transiently transfected into COS cells (B). Overexpression of eGFP-SMN (green) enhances the localization of Nopp140 to CBs (red), as observed in yellow (merge). The arrow points to one of the CBs. While the SMA mutants SMNE134K, Δex7 and 472Δ5 do not. The data are quantified in table (C). Scale bars, 3 µm.
The disease severity correlates with the CB localization of Nopp140
Previous IF studies showed that SMA disease severity correlates with the number of fibroblasts with SMN foci (4,36). A similar correlation was established with the localization of SMN in CBs from motor neurons of spinal cord sections from SMA fetuses (3). These observation along with the reduced accumulation of Nopp140 in CBs from SMA fibroblasts prompted us to examine the potential relationship between Nopp140 and SMA severity. Therefore, we analysed primary cultures of skin fibroblasts from all three types of SMA patients (Fig. 6 and Table 2). Double labelling for Nopp140 and the CB marker coilin revealed that Nopp140 localized in CBs of 63% CB-containing cells from a control infant (Fig. 6C and Table 2). Nopp140 accumulated in CBs of ∼20, 40 and 65% of CB-containing cells obtained from Type I, II or III SMA patients, respectively (Fig. 6C and Table 2). These results revealed a correlation between disease severity and altered composition of CBs in fibroblasts from SMA patients.
Figure 6.
Subnuclear localization of Nopp140 in primary fibroblasts derived from control and Type I, II and III SMA patients. Primary fibroblasts were immunostained for two CB markers SMN (A) or coilin (B) in green and Nopp140 in red. Colocalization in CBs (arrow) is shown in yellow (merge). The arrowhead and arrow point to one of the nucleoli and one of the CBs, respectively. The statistical analyses show a correlation between Nopp140-positive CBs and SMA disease severity and are presented in a histogram (C) and in Table 2. Scale bars, 3 µm.
Table 2.
Analyses of Nopp140 localization in CBs from SMA patient skin fibroblasts
| Individual | Phenotype | Total number of cellsa | Proportion of cells with Nopp-CBsb (% ± SEM) |
|---|---|---|---|
| ctrl | Control | 642 | 63 ± 5 |
| bro | Type I | 850 | 16 ± 3 |
| cou | Type I | 829 | 24 ± 6 |
| sav | Type I | 853 | 22 ± 3 |
| gz | Type II | 635 | 40 ± 6 |
| mi | Type II | 1112 | 43 ± 6 |
| sa | Type III | 935 | 63 ± 8 |
| ga | Type III | 1049 | 67 ± 8 |
The values of Types I and II primary skin fibroblasts were statistically different from control cells (Pχ2≤ 0.001).
aAbout a hundred cells per experiment were counted single-blinded by two people (S.C. and S.L.).
bCBs were visualized using anti-coilin antibodies.
DISCUSSION
We have analysed here the localization of a number of protein constituents of CBs in fibroblast cells derived from infants affected with SMA. We show that the disease severity inversely correlates with the number of cells containing the snoRNP chaperone Nopp140-positive CBs. Moreover, the functional deficiency of the SMA mutant proteins in transiently transfected COS cells is reflected by their failure to stimulate the accumulation of Nopp140 in CBs.
The low levels of SMN in SMA-derived fibroblasts that prevent the accumulation of snRNPs in CBs (36) do not impair the CB localization of two factors that are implicated in the spliceosomal U snRNA export from the nucleus (42), PHAX and CRM1 (Table 1). We observe here an increased localization of CRM1 in CBs of SMA cells. This might reflect a change in its intranuclear trafficking and/or in its nucleocytoplasmic transport. Indeed, the newly synthesized U1, U2, U4 and U5 snRNAs are initially exported to the cytoplasm where they are assembled into pre-mature snRNPs by the SMN complex (26,29). The U snRNA nuclear export requires a monomethyl cap structure on the RNA for association with the nuclear cap-binding complex (CBC). The adaptor PHAX binds both CBC and U snRNA forming the pre-complex that interacts with the nuclear export signal receptor CRM1 for translocation into the cytoplasm. Our results are consistent with the observation that the levels of the most abundant snRNPs are unchanged in SMA fibroblasts (43). However, the depletion of SMN in SMA mouse models affects the nuclear accumulation of snRNPs in motor neurons (44) and the repertoire of snRNAs (43,45) and the quality of splicing in a tissues-specific manner (45).
Interestingly, PHAX was shown to be involved in the complex intranuclear trafficking of snoRNAs that guide post-transcriptional modifications of ribosomal rRNAs and spliceosomal snRNAs (46). Indeed, the accumulation of the precursor U3 box C/D snoRNA in CBs requires the interaction with PHAX. In CBs, the precursor snoRNA assembles with box C/D core proteins Nop56, Nop58 and fibrillarin, and the 3′ end is subjected to modifications prior that the mature snoRNP is targeted to the nucleolus by CRM1 (34). SMN has been shown to interact with fibrillarin (30,31), but was not found associated with functional snoRNPs, suggesting that SMN might be involved in a late assembly step (37). Nevertheless, the accumulation of the U3 snoRNA depends on SMN levels (37). Consistent with the idea that the remaining function of CBs involving PHAX and CRM1 might not be affected in SMA cells, we detected fibrillarin, a protein marker of the fully assembled box C/D snoRNPs, in both nucleoli and CBs of fibroblasts obtained from a patient affected with the most severe form of the disease. Our results indicate that the reduced SMN levels suffice for the assembly and localization of the mature snoRNPs to the nucleolus. This is also in agreement with the observation that SMN is not essential for the assembly of snoRNPs in yeast Schizosaccharomyces cerevisiae, which lack an SMN ortholog (12).
In yeast, Srp40p and Nsr1p regulate the accumulation of box C/D snoRNA in the nucleolar body, which is the functional counterpart of CBs in higher eukaryotic cells (47). Nopp140, the mammalian ortholog of Srp40p, is an abundant phosphoprotein detected predominantly in nucleoli and is among the first proteins that have been identified in CBs (48,49). Nopp140 associates with the functional H/ACA snoRNP particles that are formed of a H/ACA RNA and four core proteins, NAP57/dyskerin, GAR1, NHP2 and NOP10 (50). It has been shown that SMN binds directly to GAR1 (30), which is not essential for the structural intergrity of H/ACA RNPs (38). Among the different types of H/ACA RNAs, there are the small CB-specific RNAs (scaRNA), which form the scaRNPs that guide snRNA modifications in CBs (33). In this regard, it will be interesting to analyse in a future study the RNA composition of CBs in SMA cells.
Nopp140 is not an integral part of snoRNPs and the association occurs through simultaneous interactions of Nopp140 with several components of the assembled snoRNPs (38). Nopp140 also associates with precursor and mature box C/D snoRNPs in the nucleoplasm and not with the functional snoRNPs in the nucleolus, indicating that Nopp140 acts during the final maturation steps before the localization of box C/D snoRNPs in nucleoli (37). Therefore, Nopp140 appears as chaperone of both box H/ACA and box C/D RNPs (38). Our analyses here reveal that Nopp140 localization in nucleoli is unaffected in fibroblasts from all three forms of SMA patients. However, the localization of Nopp140 in CBs is altered in primary SMA cells and shows a correlation with disease severity. This is even more surprising given that Nopp140 has been shown to interact directly with coilin (39) and that the Nopp140-binding domain of coilin plays a role in the dynamic retention of coilin in CBs (51). We were able to restore the accumulation of Nopp140 in CBs of SMA cells by transient expression of SMN, suggesting that the interaction between Nopp140 and coilin can occur in SMA cells. This indicates that Nopp140 localization in CBs is associated with SMN levels and, probably, a CB activity. In agreement with our results, other studies using RNA interference to deplete SMN from HeLa cells reported that SMN plays a role in the formation and composition of CBs (52–55). The role of SMN in CB formation was also demonstrated in motor neurons from a conditional SMN knockout mouse model (56). Indeed, the different behaviour of Nopp140 in SMA cells expressing constitutively low levels of SMN might reflect a change in the stability of the association with CBs when UsnRNP biogenesis is inhibited (36).
An additional functional link between Nopp140 and SMN comes from our findings in COS cells transfected with three SMA mutants SMNE134K, SMNΔex7 and SMN472Δ5. The mutant SMNE134K harbours a glutamic acid (E) to lysine (K) substitution at position 134 in the Tudor domain. The mutants SMNΔex7 and SMN472Δ5 lack the last 16 C-terminal amino acid residues and the C-terminal half of the protein, respectively. We previously showed that all three mutant proteins concentrate in CBs (36). In addition, overexpression of SMA deletion mutants resulted in depletion of snRNPs from CBs, whereas SMNE134K was colocalized with snRNPs in CBs. Here, we show that overexpression of wild-type SMN protein promotes the accumulation of Nopp140 in CBs, while all three SMA mutants are defective in stimulating the CB accumulation of Nopp140. Because CBs are depleted of snRNPs in cells transfected with SMNΔex7 and SMN472Δ5, our results suggest that the CB localization of Nopp140 is not determined by the presence of snRNPs in CBs. Nevertheless, transient overexpression of SMN increases the rate of UsnRNP biogenesis and maturation in CBs (36) and, therefore, the requirement of box H/ACA and C/D scaRNPs for modifications of the spliceosomal snRNAs in CBs. Although, the function of Nopp140 and SMN in CBs is still unknown, our observations suggest that SMN might be important for some aspects of snoRNP biogenesis in CBs that involve Nopp140. The specific reduction of H/ACA versus C/D RNPs in CBs of SMA cells mirrors that of Nopp140 (Table 1). This finding may indicate a tighter association of Nopp140 with H/ACA RNPs whose major core protein NAP57/dyskerin was originally identified as being Nopp140-associated (48). It is also possible that the presence of Nopp140 in CBs is associated with a novel function of CBs that remains to be identified.
The localization of maturation factors in CBs is important for RNP biogenesis (28). Recent reports show that during their biogenesis, RNPs undergo remodelling that requires a complex interplay of transient interactions with processing machinery (57). The role of the Hsp90 chaperone in the biogenesis of U3 box C/D snoRNP, U4 snRNA and the telomerase box H/ACA scaRNP exemplifies this (58). It remains to be established whether this remodelling machinery requires SMN and Nopp140 to function.
Collectively, our analyses of the composition of CBs in SMA-derived cells identify subnuclear localization as an important biomarker that, at the cellular level, will allow screening of pharmacological agents for therapeutic approaches in SMA.
MATERIALS AND METHODS
Cell culture and transfections
Human fibroblasts and COS cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, penicillin (100 U/ml) and streptomycin (100 mg/ml). Cells were plated in either 60 mm or eight-chamber culture-slides (Becton Dickson Laboratory) and transiently transfected with purified DNA plasmids using either FuGENE (Roche diagnostics) or electroporation (Amaxa) for 48 h and with siRNAs (25 nm) using HiPerFect transfection reagent (Qiagen) for 48 h according to the manufacturer's instructions. The siRNA duplexes have been predesigned from Qiagen. The accession number is indicated. The sequences were as follows: SMN1 (NM_000344), 5′-CTG GCATAGAGCAGCACTAAA-3′ (SI00727181) and Nopp140 (NM_004741), 5′-CAGGTCAATTCTATTAAGTTT-3′ (SI00660226). The construction of eGFP-SMN and eGFP-SMN mutants has been previously described (36).
Cell fixation and IF microscopy
Cells were fixed for 20 min in freshly made PBS/4% paraformaldehyde and permeabilized for 10 min with 1% Triton X-100 in PBS at room temperature. After three washes, the samples were blocked with 10% horse serum (HS) in PBS for 1 h and then incubated with primary antibodies diluted in 1% HS as described in Table 3. The fluorescently labelled secondary anti-rabbit and anti-mouse Cy3 (Jackson Laboratories) and Alexa Fluor 488 (Molecular Probes) antibodies were used at 1:200 dilution. The samples were counterstained with 4,6-diamidino-2-phenylindole (0.1 µg/ml) and mounted in either AF1 (Cityfluor) or Mowiol (Hoechst) and observed by non-confocal (Leica DMR, objective 63×/1.32) microscopy. The images were acquired with a cooled CCD camera (Micromax, Princetown Instruments, Inc.) using MetaView Imaging System and prepared with Adobe Photoshop.
Table 3.
Antibodies used in the IF studies
| Antibody | Host | Fixation | IF dilution | Source |
|---|---|---|---|---|
| Anti-SMN | Mouse | Formaldehyde (FAH) | 1:500 | Transduction Laboratories |
| Anti-coilin | Mouse | FAH | 1:1000 | Transduction Laboratories or Sigma-Aldrich |
| Rabbit | FAH | 1:1000 | Santa Cruz | |
| Anti-Nopp140, RS8 | Rabbit | FAH | 1:5000 | (59) |
| Anti-NAP57 | Rabbit | FAH | 1: 250 | (50) |
| Anti-GAR1 | Rabbit | FAH | 1: 250 | From W. Filipowicz |
| Anti-fibrillarin | Rabbit | FAH and MetOH | 1:1000 | Abcam |
| Anti-PHAX | Mouse | FAH and MetOH | 1:1000 | From I. Mattaj |
| Anti-CRM1 | Rabbit | FAH and MetOH | 1:2000 | Abcam |
Cell lysate preparation and immunoblotting
The cells were detached from culture flasks in ice-cold PBS using cell scraper, washed once in the same buffer and centrifuged at 500g for 5 min in a refrigerated bench centrifuge (Eppendorf 5417R) as previously described (34). The cellular pellets were either kept frozen at −80°C or immediately proceeded for immunoblotting analyses. The pellets were resuspended in ice-cold Tris–HCl buffer (25 mm, pH 7.4) in the presence of EDTA-free protein inhibitors (Roche). Following a Bradford assay (Bio-Rad Laboratories) using BSA as standard, the protein extracts were diluted in Laemmli sample buffer, separated by SDS–PAGE using Prosieve polyacrylamide (FMC BioProducts) and electrotransferred onto Immobilon-P membranes (PVDF, Millipore). The membranes were incubated with primary antibodies diluted in 5% non-fat milk in PBS for overnight at 4°C and proceed with horseradish peroxidase-conjugated secondary antibody (1:10 000) and detected using chemiluminescent reagents (General Electric). The dilution of the antibodies used was as follows: anti-SMN (1:1000 from Transduction Laboratories), anti-Nopp140 (RS8, 1:5000, 59) and anti-α-tubulin (1:10 000 from Sigma-Aldrich). The X-ray films were scanned as a grayscale image at medium resolution (300–400 dpi) and the relative quantitation of knockdown proteins was determined using the gel analysis method from ImageJ.
FUNDING
This study was supported by INSERM, CNRS and AFM. B.R. was a recipient of an undergraduate fellowship from Ministère de l'Education Nationale, de la Recherche et de la Technologie and from AFM. U.T.M. is supported by a grant from National Institutes of Health (HL079566).
ACKNOWLEDGEMENTS
We thank R. Bordonné (IGM-CNRS, Montpellier, France) for stimulating discussions, J. Cartaud and D. Paulin for the constant support and colleagues for critical reading of manuscript. We are grateful to U. Kutay, I. Mattaj and W. Filipowicz for generously providing antibodies and S. Vasseur and E. Kazmierczak from MyoBank-Association Française contre les Myopathies (AFM) for providing some of the fibroblast cell cultures.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Pearn J. Incidence, prevalence, and gene frequency studies of chronic childhood spinal muscular atrophy. J. Med. Genet. 1978;15:409–413. doi: 10.1136/jmg.15.6.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lefebvre S., Bürglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 3.Lefebvre S., Burlet P., Liu Q., Bertrandy S., Clermont O., Munnich A., Dreyfuss G., Melki J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet. 1997;16:265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- 4.Coovert D.D., Le T.T., McAndrew P.E., Strasswimmer J., Crawford T.O., Mendell J.R., Coulson S.E., Androphy E.J., Prior T.W., Burghes A.H. The survival motor neuron protein in spinal muscular atrophy. Hum. Mol. Genet. 1997;6:1205–1214. doi: 10.1093/hmg/6.8.1205. [DOI] [PubMed] [Google Scholar]
- 5.Monani U.R. Spinal muscular atrophy: a deficiency in a ubiquitous protein; a motor neuron-specific disease (Review) Neuron. 2005;48:885–896. doi: 10.1016/j.neuron.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 6.Lorson C.L., Androphy E.J. An exonic enhancer is required for inclusion of an essential exon in the SMA-determining gene SMN. Hum. Mol. Genet. 2000;9:259–265. doi: 10.1093/hmg/9.2.259. [DOI] [PubMed] [Google Scholar]
- 7.Hofmann Y., Lorson C.L., Stamm S., Androphy E.J., Wirth B. Htra2-beta 1 stimulates an exonic splicing enhancer and can restore full-length SMN expression to survival motor neuron 2 (SMN2) Proc. Natl Acad. Sci. USA. 2000;97:9618–9623. doi: 10.1073/pnas.160181697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cartegni L., Krainer A.R. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat. Genet. 2002;30:377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- 9.Kashima T., Manley J.L. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat. Genet. 2003;34:460–463. doi: 10.1038/ng1207. [DOI] [PubMed] [Google Scholar]
- 10.Vitte J., Fassier C., Tiziano F.D., Dalard C., Soave S., Roblot N., Brahe C., Saugier-Veber P., Bonnefont J.P., Melki J. Refined characterization of the expression and stability of the SMN gene products. Am. J. Pathol. 2007;171:1269–1280. doi: 10.2353/ajpath.2007.070399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Briese M., Esmaeili B., Sattelle D.B. Is spinal muscular atrophy the result of defects in motor neuron processes? Bioessays. 2005;9:946–957. doi: 10.1002/bies.20283. [DOI] [PubMed] [Google Scholar]
- 12.Schmid A., DiDonato C.J. Animal models of spinal muscular atrophy (Review) J. Child Neurol. 2007;22:1004–1012. doi: 10.1177/0883073807305667. [DOI] [PubMed] [Google Scholar]
- 13.Le T.T., Pham L.T., Butchbach M.E.R., Zhang H.L., Monani U.R., Coovert D.D., Gavrilina T.O., Xing L., Bassell G.J., Burghes A.H.M. SMNΔ7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum. Mol. Genet. 2005;14:845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- 14.Gavrilina T.O., McGovern V.L., Workman E., Crawford T.O., Gogliotti R.G., DiDonato C.J., Monani U.R., Morris G.E., Burghes A.H. Neuronal SMN expression corrects spinal muscular atrophy in severe SMA mice while muscle-specific SMN expression has no phenotypic effect. Hum. Mol. Genet. 2008;17:1063–1075. doi: 10.1093/hmg/ddm379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Talbot K., Davies K.E. Is good housekeeping the key to motor neuron survival? Cell. 2008;133:572–574. doi: 10.1016/j.cell.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Eggert C., Chari A., Laggerbauer B., Fischer U. Spinal muscular atrophy: the RNP connection. Trends Mol. Med. 2006;12:113–121. doi: 10.1016/j.molmed.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 17.Murray L.M., Comley L.H., Thomson D., Parkinson N., Talbot K., Gillingwater T.H. Selective vulnerability of motor neurons and dissociation of pre- and post-synaptic pathology at the neuromuscular junction in mouse models of spinal muscular atrophy. Hum. Mol. Genet. 2008;17:949–962. doi: 10.1093/hmg/ddm367. [DOI] [PubMed] [Google Scholar]
- 18.Kariya S., Park G.H., Maeno-Hikichi Y., Leykekhman O., Lutz C., Arkovitz M.S., Landmesser L.T., Monani U.R. Reduced SMN protein impairs maturation of the neuromuscular junctions in mouse models of spinal muscular atrophy. Hum. Mol. Genet. 2008;17:2552–2569. doi: 10.1093/hmg/ddn156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pellizzoni L., Charroux B., Dreyfuss G. SMN mutants of spinal muscular atrophy patients are defective in binding to snRNP proteins. Proc. Natl Acad. Sci. USA. 1999;96:11167–11172. doi: 10.1073/pnas.96.20.11167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bühler D., Raker V., Lührmann R., Fischer U. Essential role for the tudor domain of SMN in spliceosomal U snRNP assembly: implications for spinal muscular atrophy. Hum. Mol. Genet. 1999;8:2351–2357. doi: 10.1093/hmg/8.13.2351. [DOI] [PubMed] [Google Scholar]
- 21.Selenko P., Sprangers R., Stier G., Bühler D., Fischer U., Sattler M. SMN tudor domain structure and its interaction with the Sm proteins. Nat. Struct. Biol. 2001;8:27–31. doi: 10.1038/83014. [DOI] [PubMed] [Google Scholar]
- 22.Meister G., Bühler D., Pillai R., Lottspeich F., Fischer U. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat. Cell Biol. 2001;3:945–949. doi: 10.1038/ncb1101-945. [DOI] [PubMed] [Google Scholar]
- 23.Pellizzoni L., Yong J., Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science. 2002;298:1775–1779. doi: 10.1126/science.1074962. [DOI] [PubMed] [Google Scholar]
- 24.Narayanan U., Achsel T., Lührmann R., Matera A.G. Coupled in vitro import of UsnRNPs and SMN, the spinal muscular atrophy protein. Mol. Cells. 2004;16:223–234. doi: 10.1016/j.molcel.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 25.Carvalho T., Almeida F., Calapez A., Lafarga M., Berciano M.T., Carmo-Fonseca M. The spinal muscular atrophy disease gene product, SMN: a link between snRNP biogenesis and the Cajal (coiled) body. J. Cell Biol. 1999;147:715–728. doi: 10.1083/jcb.147.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neuenkirchen N., Chari A., Fischer U. Deciphering the assembly pathway of Sm-class U snRNPs (Review) FEBS Lett. 2008;582:1997–2003. doi: 10.1016/j.febslet.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 27.Raska I., Andrade L.E., Ochs R.L., Chan E.K., Chang C.M., Roos G., Tan E.M. Immunological and ultrastructural studies of the nuclear coiled body with autoimmune antibodies. Exp. Cell Res. 1991;195:27–37. doi: 10.1016/0014-4827(91)90496-h. [DOI] [PubMed] [Google Scholar]
- 28.Cioce M., Lamond A.I. Cajal bodies: a long history of discovery. Annu. Rev. Cell Dev. Biol. 2005;21:105–131. doi: 10.1146/annurev.cellbio.20.010403.103738. [DOI] [PubMed] [Google Scholar]
- 29.Matera A.G., Terns R.M., Terns M.P. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 30.Pellizzoni L., Baccon J., Charroux B., Dreyfuss G. The survival of motor neurons (SMN) protein interacts with the snoRNP proteins fibrillarin and GAR1. Curr. Biol. 2001;11:1079–1088. doi: 10.1016/s0960-9822(01)00316-5. [DOI] [PubMed] [Google Scholar]
- 31.Jones K.W., Gorzynski K., Hales C.M., Fischer U., Badbanchi F., Terns R.M., Terns M.P. Direct interaction of the spinal muscular atrophy disease protein SMN with the small nucleolar RNA-associated protein fibrillarin. J. Biol. Chem. 2001;276:38645–38651. doi: 10.1074/jbc.M106161200. [DOI] [PubMed] [Google Scholar]
- 32.Verheggen C., Lafontaine D.L., Samarsky D., Mouaikel J., Blanchard J.M., Bordonné R., Bertrand E. Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO J. 2002;21:2736–2745. doi: 10.1093/emboj/21.11.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jàdy B.E., Darzacq X., Tucker K.E., Matera A.G., Bertrand E., Kiss T. Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm. EMBO J. 2003;22:1878–1888. doi: 10.1093/emboj/cdg187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boulon S., Verheggen C., Jady B.E., Girard C., Pescia C., Paul C., Ospina J.K., Kiss T., Matera A.G., Bordonné R., Bertrand E. PHAX and CRM1 are required sequentially to transport U3 snoRNA to nucleoli. Mol. Cells. 2004;16:777–787. doi: 10.1016/j.molcel.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 35.Sleeman J. A regulatory role for CRM1 in the multi-directional trafficking of splicing snRNPs in the mammalian nucleus. J. Cell Sci. 2007;120:1540–1550. doi: 10.1242/jcs.001529. [DOI] [PubMed] [Google Scholar]
- 36.Renvoisé B., Khoobarry K., Gendron M.C., Cibert C., Viollet L., Lefebvre S. Distinct domains of the spinal muscular atrophy protein SMN are required for targeting to Cajal bodies in mammalian cells. J. Cell Sci. 2006;119:680–692. doi: 10.1242/jcs.02782. [DOI] [PubMed] [Google Scholar]
- 37.Watkins N.J., Lemm I., Ingelfinger D., Schneider C., Hobbach M., Urlaub H., Lührmann R. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol. Cells. 2004;16:789–798. doi: 10.1016/j.molcel.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 38.Meier U.T. The many facets of H/ACA ribonucleoproteins. Chromosoma. 2005;114:1–14. doi: 10.1007/s00412-005-0333-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Isaac C., Yang Y., Meier U.T. Nopp140 functions as molecular link between the nucleolus and the coiled bodies. J. Cell Biol. 1998;142:319–329. doi: 10.1083/jcb.142.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tucker K.E., Berciano M.T., Jacobs E.Y., LePage D.F., Shpargel K.B., Rossire J.J., Chan E.K., Lafarga M., Conlon R.A., Matera A.G. Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J. Cell Biol. 2001;154:293–307. doi: 10.1083/jcb.200104083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lefebvre S., Burlet P., Viollet L., Bertrandy S., Huber C., Belser C., Munnich A. A novel association of SMN with two major non-ribosomal nucleolar proteins and its implication in spinal muscular atrophy. Hum. Mol. Genet. 2002;11:1017–1027. doi: 10.1093/hmg/11.9.1017. [DOI] [PubMed] [Google Scholar]
- 42.Ohno M., Segref A., Bachi A., Wilm M., Mattaj I.W. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell. 2000;101:187–198. doi: 10.1016/S0092-8674(00)80829-6. [DOI] [PubMed] [Google Scholar]
- 43.Gabanella F., Butchbach M.A.E., Saieva L., Carissimi C., Burghes A.H.M., Pellizzoni L. Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PLOS One. 2007;2:e921. doi: 10.1371/journal.pone.0000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jablonka S., Holtmann B., Meister G., Bandilla M., Rossoll W., Fischer U., Sendtner M. Gene targeting of Gemin2 in mice reveals a correlation between defects in the biogenesis of U snRNPs and motoneuron cell death. Proc. Natl Acad. Sci. USA. 2002;99:10126–10131. doi: 10.1073/pnas.152318699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Z., Lotti F., Dittmar K., Younis I., Wan L., Kasim M., Dreyfuss G. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kiss T., Fayet E., Jàdy B.E., Richard P., Weber M. Biogenesis and intranuclear trafficking of human box C/D/ and H/ACA RNPs. Cold Spring Harb. Symp. Quant. Biol. 2006;71:407–417. doi: 10.1101/sqb.2006.71.025. [DOI] [PubMed] [Google Scholar]
- 47.Verheggen C., Lafontaine D.L., Samarsky D., Mouaikel J., Blanchard J.M., Bordonné R., Bertrand E. Box C/D small nucleolar RNA trafficking involves small nucleolar RNP proteins, nuclear factors and a novel nuclear domain. EMBO J. 2002;20:5480–5490. doi: 10.1093/emboj/20.19.5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meier U.T., Blobel G. NAP57, a mammalian nucleolar protein with a putative homolog in yeast and bacteria. J. Cell Biol. 1994;127:1505–1514. doi: 10.1083/jcb.127.6.1505. (correction appeared in 140:447) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meier U.T. Comparison of the rat nucleolar protein Nopp140 and its yeast homolog SRP40: differential phosphorylation in vertebrates and yeast. J. Biol. Chem. 1996;271:19376–19384. [PubMed] [Google Scholar]
- 50.Darzacq X., Kittur N., Roy S., Shav-Tal Y., Singer R.H., Meier U.T. Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. J. Cell Biol. 2006;173:207–218. doi: 10.1083/jcb.200601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dundr M., Hebert M.D., Karpova T.S., Stanek D., Xu H., Shpargel K.B., Meier U.T., Neugebaueur K.M., Matera A.G., Misteli T. In vivo kinetics of Cajal body components. J. Cell Biol. 2004;164:831–842. doi: 10.1083/jcb.200311121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feng W., Gubitz A.K., Wan L., Battle D.J., Dostie J., Golembe T.J., Dreyfuss G. Gemins modulate the expression and activity of the SMN complex. Hum. Mol. Genet. 2005;14:1605–1611. doi: 10.1093/hmg/ddi168. [DOI] [PubMed] [Google Scholar]
- 53.Shpargel K.B., Matera A.G. Gemin proteins are required for efficient assembly of Sm-class ribonucleoproteins. Proc. Natl Acad. Sci. USA. 2005;102:17372–17377. doi: 10.1073/pnas.0508947102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Girard C., Neel H., Bertrand E., Bordonné R. Depletion of SMN by RNA interference in HeLa cells induces defects in Cajal body formation. Nucleic Acids Res. 2006;34:2925–2932. doi: 10.1093/nar/gkl374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lemm I., Girard C., Kuhn A.N., Watkins N.J., Schneider M., Bordonné R., Lührmann R. Ongoing U snRNP biogenesis is required for the integrity of Cajal bodies. Mol. Biol. Cell. 2006;17:3221–3231. doi: 10.1091/mbc.E06-03-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Frugier T., Tiziano F.D., Cifuentes-Diaz C., Miniou P., Roblot N., Dierich A., Le Meur M., Melki J. Nuclear targeting defect of SMN lacking the C-terminus in a mouse model of spinal muscular atrophy. Hum. Mol. Genet. 2000;9:849–858. doi: 10.1093/hmg/9.5.849. [DOI] [PubMed] [Google Scholar]
- 57.McKeegan K.S., Debieux C.M., Boulon S., Bertrand E., Watkins N.J. A dynamic scaffold of pre-snoRNP factors facilitates human box C/D snoRNP assembly. Mol. Cell Biol. 2007;27:6782–6793. doi: 10.1128/MCB.01097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boulon S., Marmier-Gourrier N., Pradet-Balade B., Wurth L., Verheggen C., Jády B.E., Rothé B., Pescia C., Robert M.C., Kiss T., et al. The Hsp90 chaperone controls the biogenesis of L7Ae RNPs through conserved machinery. J. Cell Biol. 2008;180:579–595. doi: 10.1083/jcb.200708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kittur N., Zapantis G., Aubuchon M., Santoro N., Bazett-Jones D.P., Meier U.T. The nucleolar channel system of human endometrium is related to endoplasmic reticulum and R-rings. Mol. Biol. Cell. 2007;18:2296–2304. doi: 10.1091/mbc.E07-02-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]