Abstract
Radiological images contain a wealth of information, such as anatomy and pathology, which is often not explicit and computationally accessible. Information schemes are being developed to describe the semantic content of images, but such schemes can be unwieldy to operationalize because there are few tools to enable users to capture structured information easily as part of the routine research workflow. We have created iPad, an open source tool enabling researchers and clinicians to create semantic annotations on radiological images. iPad hides the complexity of the underlying image annotation information model from users, permitting them to describe images and image regions using a graphical interface that maps their descriptions to structured ontologies semi-automatically. Image annotations are saved in a variety of formats, enabling interoperability among medical records systems, image archives in hospitals, and the Semantic Web. Tools such as iPad can help reduce the burden of collecting structured information from images, and it could ultimately enable researchers and physicians to exploit images on a very large scale and glean the biological and physiological significance of image content.
Introduction
Images produced by radiology modalities are a critical data type in biomedicine because they provide rich visual information about disease, complementary to non-image data such as genomics, proteomics, histopathology, and clinical laboratory results. While the raw images themselves can be useful in some computer processing applications, much of the critical semantic content in images is not explicit and available to computers, hindering discovery in large-volume image data sets.
The “semantic content” in images is the information extracted by radiologists who view them: the visual appearance of anatomic structures and abnormalities contained in those structures (“image findings” such as a mass in the liver or an enlarged heart). The radiologist-extracted semantic information is rarely linked or embedded within the images, instead being recorded in text reports or in case report forms as part of clinical trials. Thus, much critical radiological image information is not directly accessible to machines, unlike other biomedical data such as genomics, proteomics, and electronic medical records data. If the semantic information in images were recorded in a structured manner, researchers and clinical practitioners could more efficiently search and analyze large image repositories according to particular features of images (e.g., discover characteristic image appearances of particular diseases, compute the average lesion size, and summarize types of abnormalities in different diseases). In addition, it would be possible to combine images with complementary non-image data to enhance biomedical discovery.
Schemas to make the semantic contents of images explicit are being developed,1–3 however, experts who interpret images do not generally record their observations in the highly structured format of such schemas. Images are viewed and interpreted in Picture Archiving and Communication Systems (PACS) or in a variety of image viewers for image manipulation. While these systems provide tools to label images (Figure 1), they do not permit users to record the key semantic image content in a standard machine-interpretable format. A tool permitting researchers to describe the semantic information in images in a manner that fits within the research workflow could help them to collect the necessary structured image data.
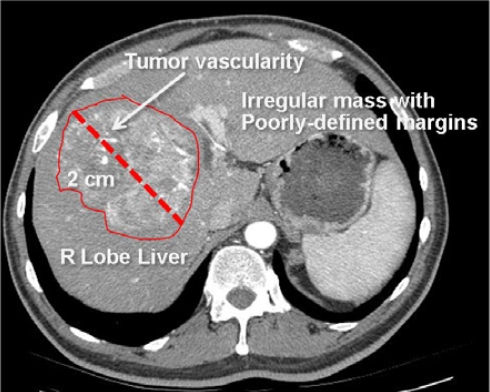
Figure 1.
Radiological image with semantic annotations drawn in an image annotation tool.
This image is annotated to convey the major semantic content, including anatomy (right lobe of liver), pathology (irregular mass, tumor vascularity), and imaging observations (2 cm in size, irregular shape, and poorly-defined margins). These image labels are human-readable, but not machine-accessible.
Several groups have previously developed annotation tools in biomedicine and radiology, focusing on acquisition of the graphical symbols or on focused application needs,4, 5 however, there are several challenges that have not yet been addressed by the prior work on image annotation tools. The first challenge is that current annotation tools lack an information model for semantic image annotations. Such models are important for defining standard data representations, the types of information that image annotations convey, and the types of queries that are possible, similar to the Microarray Gene Expression Object Model (MAGE-OM) in the functional genomics community.6
A second challenge for current annotation tools is that the terminology and syntax for describing the semantic content in images varies across existing systems, with no widely-adopted standards, resulting in limited interoperability. Standard terminologies for describing medical image contents—the imaging observations, the anatomy, and the pathology—are generally not incorporated into current image annotation tools. In terms of syntax, schemes for annotating images have been proposed in non-medical domains;3, 7, 8 however, no comprehensive standard appropriate to radiological imaging has yet been developed. Current standard syntaxes in use include (1) Digital Imaging and Communications in Medicine (DICOM), (2) HL7, and (3) HTML and RDF. DICOM-SR is emerging as a standard for representing non-image data, but it does not yet specifically address semantic annotation content.
A third challenge for medical imaging is that annotations often have particular information requirements: there may be restrictions on annotations in terms of terminology, or a user may need to specify more specific information for particular annotations. For example, if a user makes a statement about the liver, the user should include one or more observations (e.g., the user could state “2cm abnormality in right lobe of liver” but should not simply state “right lobe of liver”). Another example of an annotation information requirement is that if a modifier of an observation is provided, the observation it modifies should also be stated (e.g., the user could state “irregular mass” but not just “irregular”). Image annotation tools should automatically check the image annotations and alert the user about annotation requirements.
A final challenge is that annotations on images convey semantic information that is not generally computationally accessible. Many annotation tools enable users to create graphical symbols on images, but the semantics is not explicit and difficult to process computationally (Figure 1).
In this paper, we describe an image Physician Annotation Device (iPad) that tackles the challenges detailed above.
Methods
The iPad application comprises three components: (1) an information model for semantic image annotations, (2) a user interface for collecting the annotations from users, and (3) a storage back-end to save annotations as XML, and to serialize the data to other standard formats. A screenshot of the iPad application is shown in Figure 2. The user selects an image to be annotated and enters a description using a syntax and grammar that is similar to English (e.g., “an enhancing, irregular mass in the right lobe of the liver”). iPad simultaneously provides user feedback with text and, if desired, voice of the text. Once the content of the annotation is valid, iPad stores the annotation as an XML file, subsequently serialized to a variety of standard formats such as DICOM-SR or HL7-CDA. The image itself remains separate from the annotation (the “image metadata”), though the annotation is linked to the image through the image unique identifier (e.g., DICOM UID).
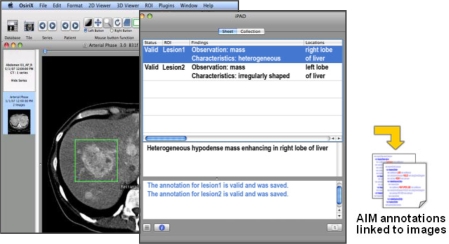
Figure 2.
The iPad application. iPad is a plug-in to OsiriX. An image is loaded in OsiriX (left screen) and geometric regions in the image are drawn (shown as rectangles in the image). The user types their observations about anatomy and abnormality in the iPad window (middle screen) while simultaneously receiving feedback from iPad about their annotation (e.g., controlled terms and spelling errors are highlighted). The annotations are saved as XML files in the AIM format (right).
1. Information Model for Semantic Image Annotation
We adapted our previously-described information model (“Annotation and Image Markup (AIM) schema”)2 to describe the minimal information necessary to record an image annotation (Figure 3), inspired in concept by the MIAME project to describe the universe of information in microarray experiments.9 The AIM schema distinguishes image “annotation” and “markup.” Annotations describe the meaning in images, while markup is the visual presentation of the annotations. In the AIM schema, all annotations are either an Image-Annotation (annotation on an image) or an Annotation-of-Annotation (annotation on an annotation). Image annotations include information about the image as well as their semantic contents (anatomy, imaging observations, etc).2 The AIM schema is an XML schema (XSD file) which enables validation of instances of AIM XML files.
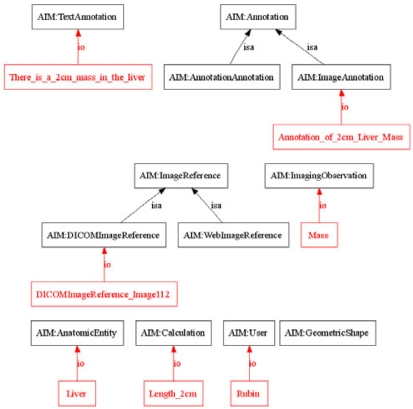
Figure 3.
AIM Schema and Annotation Instance. A portion of the AIM schema (black) and example instance of Image-Annotation (red) are shown. Only is-a and instance-of relations are depicted. The figure shows that the annotation describes an image (Image 112), which visualizes the liver, and is seen to contain a mass in the liver measuring 2cm in size.
To tackle the challenge of providing checks on annotation content, the iPad application is linked to an ontology of radiology knowledge. Text entered into iPad is parsed into tokens (typically, 1–3 word phrases) and matched to entities in the ontology. iPad determines particular annotation requirements by consulting the ontology. Currently, iPad incorporates the RadLex ontology of radiology,10 constraining the image annotation only to terms in the ontology and alerting the user to spelling errors (Figure 2). iPad also checks to ensure that radiological descriptions are complete; specifically, (1) completeness: if an anatomic entity is mentioned, iPad checks to ensure that one or more observations about that entity are recorded (e.g., a mass in the liver); and (2) dangling modifiers: if a modifier of an observation is mentioned, iPad checks to ensure that an observation is also mentioned. These checks are implemented using the RadLex ontology to determine which tokens are anatomic entities, observations, and modifiers.
2. User Interface for Collecting Annotations
We created iPad as a plugin to OsiriX.11 Osirix is an open-source imaging platform for multimodality and multidimensional radiological image display and visualization. The program provides image access and display functionality and enables image viewing workflow similar to commercial PACS workstations. However, since the program is open source and provides a plug-in-architecture, its functionality can be extended through applications such as iPad.
The iPad tool extends the native OsiriX annotation object. When a user draws a region on an image in OsiriX, iPad accesses the geometric information in the drawn region and the pixels it contains, storing that information in the AIM semantic annotation information model (Figure 3). Users interpreting images type their observations into the iPad, and iPad provides feedback to users by highlighting relevant portions of their entries (Figure 4A). In addition, users can access the RadLex ontology in a tree viewer and explore the term hierarchy for appropriate standard descriptive terms (Figure 4A). Users can browse the structured image metadata being collected by iPad, both the pixel data in the image region and the descriptive information the user creates in the annotation (Figure 4B).
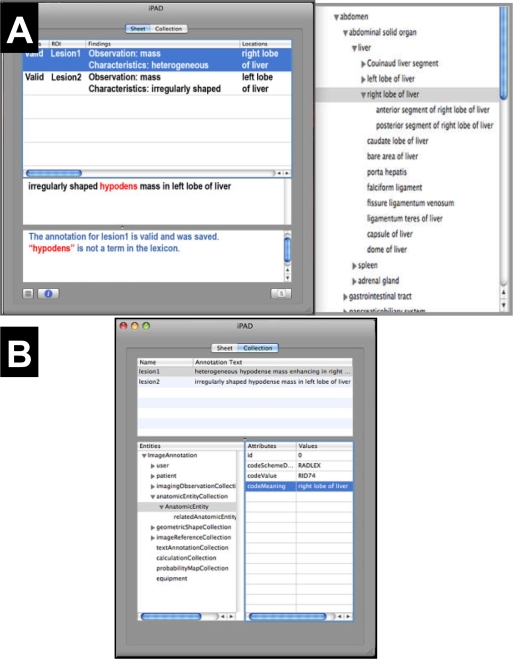
Figure 4.
iPad user interface and structured semantic annotation information. (A) The iPad annotation screen (left) is a simple text box into which the user types their image observations (middle panel). Ontologies such as RadLex linked to iPad provide visual feedback, such as prompting for the appropriate controlled terms for the user entry (right) or possible misspellings (highlighted terms in iPad annotation screen). (B) Users can view the structured semantic image annotation data in an expandable tree, organized according to types of image metadata iPad collects.
3. Annotation Storage and Serialization
When the user completes an annotation, iPad stores it as an XML document, an instance of the AIM schema.2 The image being annotated remains unchanged; the image annotation metadata is stored in an XML file that is linked to the image via the DICOM UID. A code module processes the AIM instances to serialize them to DICOM, HL7 CDA (XML), or OWL to enable semantic integration and to allow agents to access the image annotations across hospital systems and the Web.2
Evaluation
We performed a preliminary evaluation of iPad by asking two radiologists to use it to annotate 10 radiological images acquired as part of a research study. They used iPad to describe the features of abnormalities seen in each image. The radiologists were asked to qualitatively evaluate iPad in terms of usability in the image interpretation workflow and utility of the feedback iPad provides, compared with their experience in unassisted collection of image metadata (the current paradigm in radiology research).
Results
iPad provides a simple interface to adding structured semantic information to images, simplifying the process of associating semantic information with images or image regions. In current practice without iPad, radiologists view images in a workstation such as OsiriX or PACS, they define image regions of interest, and they describe the features of those regions in a text report or in a data collection form. The iPad tool enables a similar workflow, with users accessing images and defining image regions in OsiriX; however, iPad also enables the user to collect descriptive information about the image in the same tool used to view it (Figure 2). While users can browse and choose controlled terms from an ontology viewer (Figure 4), the iPad tokenizing and term matching feature allows users to directly type in their descriptions, reducing potential disruption in the workflow.
iPad provides visual guidance to users as they record their observations, giving feedback as to the required semantic annotation content. As they enter information, iPad highlights terms matching the RadLex ontology and flags possible misspellings (Figure 4). iPad also provides visual cues about the completeness of radiological descriptions (mentions of anatomy also include descriptions of observations, and mentions of modifiers also include an associated image observation). For example, if the radiologist enters “Liver,” iPad flags this as incomplete, while “Mass in the Liver” would not be flagged. Similarly, the user would be alerted that “Enlarged” is an incomplete description, while “Enlarged Liver” is not incomplete.
The radiologists who evaluated iPad reported that the tool enabled them to efficiently annotate the study images in a similar workflow to which they are currently accustomed. While iPad collects detailed structured information, the manner in which it accomplishes this task did not qualitatively hinder the radiologists in describing the images and recording their observations. Based on this encouraging preliminary evaluation, we are enhancing iPad to allow recording of one or many annotations by enabling single-click phrase insertion as well as contextual search of the RadLex ontology during typing. These enhancements will enable much larger, formal qualitative or quantitative evaluations of iPad.
Discussion
While radiological images contain a wealth of information, much of it is not in the raw pixels themselves, but in observations and interpretations radiologists and researchers make when viewing them. Ideally, this additional information should be connected directly to the image (through annotation) to enable analyses of images with non-image data. Most of the image viewing tools used in research and clinical practice incorporate graphical annotation palettes, but the semantics of image annotation is not recorded, and annotation information from images is not easily accessed and analyzed. Tools that support the collection of structured semantic information from information models generally provide data entry screens too detailed and complex to be usable in the imaging research workflow. Thus, there is need for a tool for collecting semantic annotation information in images in a streamlined manner.
iPad enables researchers to create semantic annotations on radiological images. The iPad user interface provides guidance to required annotation content as users enter information, enabling iPad to accommodate structured image annotation while being less disruptive to the research workflow than unassisted annotation methods. In addition, iPad is extensible, and we anticipate the tool will have utility in a broad range of biomedical imaging applications where collecting structured information from images is needed.
A useful feature of iPad is annotation content checks. The spectrum of image metadata that can be collected can be large, and certain combinations of information may be appropriate in particular contexts. iPad accesses knowledge in an ontology to provide feedback to the user on the annotation content. Admittedly, the current content checks are simplistic, focusing on spelling and matching terms in the RadLex ontology. However, richer ontological knowledge could be accessed to support more complex annotation checks in the future. iPad has been architected as an extensible tool, and we are planning to support richer annotation checks by extending the ontologies iPad accesses.
We do not necessarily advocate iPad for routine clinical radiological practice; iPad is targeted primarily for enabling imaging research, where collection of rich semantic information is needed for radiological discovery and to advance imaging science. In clinical practice, few cases require detailed image annotation, though it could be useful for radiologists to selectively annotate clinical images to enable discovery in large clinical image archives. Enhancements to the user interface would be needed to make iPad practical to use in high-volume image interpretation paradigms, such as incorporating voice recognition (obviating the need for the user to type in the findings).
While the focus of iPad is on annotating radiological images, we believe this tool could be applicable and useful to all of biomedical imaging. If the application domain could benefit from describing structured semantic information in the image or image regions, then iPad may be valuable, provided extensions to the iPad ontology are made to support the terminology and annotation needs of that domain.
Conclusion
We have developed iPad, a semantic image annotation tool for radiology. This tool extends the functionality of a currently-available image viewing platform, enabling users to add semantic content to images via a simple user interface and that can be incorporated into the imaging research workflow.
The tool has the capability to check the validity of the annotations and it could enable imaging researchers to access and analyze the semantic content of images in large imaging archives in the future.
Acknowledgments
This work is supported by a grant from the National Cancer Institute (NCI) through the cancer Biomedical Informatics Grid (caBIG) Imaging Workspace, subcontract from Booz-Allen & Hamilton, Inc. 0970 370 X277 1390 and the National Institutes of Health, CA72023.
References
- 1.Lober WB, Trigg LJ, Bliss D, Brinkley JM. IML: An image markup language. J Am Med Inform Assn. 2001:403–7. [PMC free article] [PubMed] [Google Scholar]
- 2.Rubin DL, Mongkolwat P, Kleper V, Supekar K, Channin DS, Medical Imaging on the Semantic Web: Annotation and Image Markup 2008AAAI Spring Symposium Series, Semantic Scientific Knowledge Integration 2008(in press)Stanford University; http://stanford.edu/~rubin/pubs/Rubin-AAAI-AIM-2008.pdf; 2008(in press) [Google Scholar]
- 3.Troncy R, van Ossenbruggen J, Pan JZ, Stamou G.Image Annotation on the Semantic Web W3C Incubator Group Report14 August 2007.2007[updated 2007; cited March 1, 2008]; Available from: http://www.w3.org/2005/Incubator/mmsem/XGR-image-annotation/
- 4.Armato SG., 3rd Image annotation for conveying automated lung nodule detection results to radiologists. Academic radiology. 2003 Sep;10(9):1000–7. doi: 10.1016/s1076-6332(03)00116-8. [DOI] [PubMed] [Google Scholar]
- 5.Zheng Y, Wu M, Cole E, Pisano ED. Online annotation tool for digital mammography. Academic radiology. 2004 May;11(5):566–72. doi: 10.1016/S1076-6332(03)00726-8. [DOI] [PubMed] [Google Scholar]
- 6.Whetzel PL, Parkinson H, Causton HC, Fan L, Fostel J, Fragoso G, et al. The MGED Ontology: a resource for semantics-based description of microarray experiments. Bioinformatics. 2006 Apr 1;22(7):866–73. doi: 10.1093/bioinformatics/btl005. [DOI] [PubMed] [Google Scholar]
- 7.Halaschek-Wiener C, Golbeck J, Schain A, Grove M, Parsia B, Hendler J. Annotation and provenance tracking in semantic web photo libraries. Provenance and Annotation of Data. 2006;4145:82–9. [Google Scholar]
- 8.Khan L. Standards for image annotation using Semantic Web. Computer Standards & Interfaces. 2007 Feb;29(2):196–204. [Google Scholar]
- 9.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001 Dec;29(4):365–71. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 10.Langlotz CP. RadLex: a new method for indexing online educational materials. Radiographics. 2006 Nov-Dec;26(6):1595–7. doi: 10.1148/rg.266065168. [DOI] [PubMed] [Google Scholar]
- 11.Rosset A, Spadola L, Ratib O. OsiriX: an open-source software for navigating in multidimensional DICOM images. J Digit Imaging. 2004 Sep;17(3):205–16. doi: 10.1007/s10278-004-1014-6. [DOI] [PMC free article] [PubMed] [Google Scholar]