Abstract
Pathogenic bacteria deliver effector proteins into host cells through the type III secretion apparatus to modulate the host function. We identify a family of proteins, homologous to the type III effector Cif from enteropathogenic Escherichia coli, in pathogens including Yersinia, Photorhabdus, and Burkholderia that contain functional type III secretion systems. Like Cif, this family of proteins is capable of arresting the host cell cycle at G2/M. Structure of one of the family members, Cif homolog in Burkholderia pseudomallei (CHBP), reveals a papain-like fold and a conserved Cys-His-Gln catalytic triad despite the lack of primary sequence identity. For CHBP and Cif, only the putative catalytic Cys is susceptible to covalent modification by E-64, a specific inhibitor of papain-like cysteine proteases. Unlike papain-like enzymes where the S2 site is the major determinant of cleavage-site specificity, CHBP has a characteristic negatively charged pocket occupying surface areas corresponding to the S1/S1′ site in papain-like proteases. The negative charge is provided by a conserved aspartate, and the pocket best fits an arginine, as revealed by molecular docking analysis. Mutation analysis establishes the essential role of the catalytic triad and the negatively charged pocket in inducing cell cycle arrest in host cells. Our results demonstrate that bacterial pathogens have evolved a unique papain-like hydrolytic activity to block the normal host cell cycle progression.
Keywords: Burkholderia, crystal structure, cyclomodulin, type III secretion, EPEC
Gram-negative bacterial pathogens, such as Shigella, Salmonella, Yesinia, and enteropathogenic Escherichia coli (EPEC), employ a specialized type III secretion system (TTSS) to inject effector proteins into host cells (1). These effectors function to interfere with various host signal transduction pathways, most frequently the innate immune system (2), for the benefits of pathogen survival and systemic infection (3, 4). Elucidating biochemical functions of type III effectors, particularly those conserved among different pathogens, has greatly promoted our understanding of novel yet common mechanisms of bacterial pathogenesis (5–7).
In addition to the innate immune pathway, recent studies suggest that the host cell cycle machinery represents an emerging target of bacterial toxins, including some type III effectors (8–11). The Shigella type III effector IpaB binds directly to Mad2L2 to release its inhibitory effect on the anaphase-promoting complex/cyclosome, thereby causing a delayed mitotic progression or cell cycle arrest at the G2/M phase (9). This serves as an important strategy for Shigella to colonize the gut epithelium efficiently. Another bacterial toxin called cytolethal distending toxin (CDT) uses a DNase I-like activity to trigger the DNA damage checkpoint and block the host cell cycle at the G2/M transition (12, 13). Similarly, a G2/M arrest phenotype is observed upon infection of epithelial cells by certain strains of EPEC because of the activity of a newly identified type III effector called Cif (cycle-inhibiting factor) (14). In contrast to CDT, Cif does not activate the DNA damage checkpoint (15). Interestingly, a recent study shows that post-G2 cells infected with Cif-expressing EPEC are instead arrested in the G1 phase, not the G2/M phase (16). Despite the strikingly abnormal cell cycle distribution induced by Cif, its biochemical activity and the underlying mechanism of Cif function remain completely unknown.
In this work, we identify a family of known and putative type III effectors from Yersinia, Photorhabdus, and Burkholderia showing sequence homology to Cif, and we demonstrate that they all harbor a sufficient and potent G2/M arrest activity. The crystal structure of Cif homolog in Burkholderia pseudomallei (CHBP) reveals a papain-like fold with a well-conserved Cys-His-Gln catalytic triad. The Cys is susceptible to covalent modification by E-64, a papain family-specific inhibitor. A characteristic negatively charged pocket that could best fit an arginine likely serves as a distinct substrate-binding subsite. Mutations disrupting the catalytic triad or the negatively charged pocket abolish cell cycle arrest function of the Cif family. Our results demonstrate that bacterial pathogens have evolved a unique papain-like hydrolytic activity to target and paralyze the eukaryotic cell cycle machinery.
Results
Identification of a Type III Effector Family of Cell Cycle Modulators.
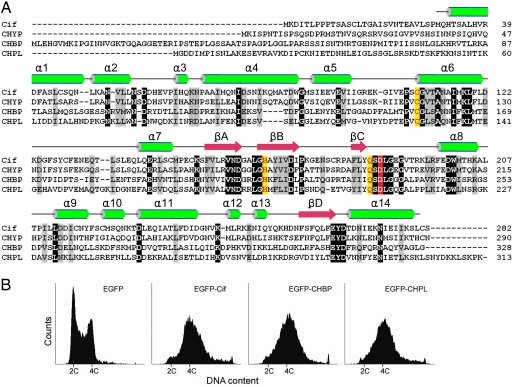
EPEC Cif contains 282 aa with no domains recognized by SMART (http://smart.embl-heidelberg.de/) or Pfam (http://pfam.janelia.org/). Extensive sequence analysis employing PSI-BLAST iterations (17) revealed no homologous proteins with known functions and instead led to identification of three new homologous bacterial proteins. They are named CHYP (Cif homolog in Yersinia pseudotuberculosis), CHPL (Cif homolog in Photorhabdus luminescens), and CHBP. As shown in Fig. 1A, CHYP has the highest degree of sequence homology to Cif (≈54% identity and ≈70% similarity); CHPL and CHBP are distantly related and only exhibit ≈20% sequence identity to Cif. The sequence conservation among the entire Cif family lies mainly in the middle and C-terminal regions, and their more diverse N-terminal sequences agree with the prediction that they are all TTSS effectors (see below).
Fig. 1.
Identification of the Cif family of cell cycle modulators. (A) Multiple sequence alignment of the Cif family of effectors. The Cif family was identified by BLAST searches using the sequence of Cif from EPEC. Alignment was generated by using the ClustalW program with manual adjustments based on predicted secondary structures. The protein names are indicated at the beginning of the alignment: CHYP, from Y. pseudotuberculosis YPIII (ORF name, YPK_1971); CHPL, from P. luminescens TTO1 (ORF name, plu2515); CHBP, from B. pseudomallei K96243 (ORF name, BPSS1385). Secondary structure elements determined from the CHBP crystal (see Fig. 2) are indicated on top of the sequence with α-helix in green and β-strand in pink. Identical residues are highlighted in black, and similar residues are in gray. Functionally important residues (see Figs. 3B and 5) are colored in yellow or red. (B) Effects of the Cif family members on eukaryotic cell cycle progression. Shown is the flow cytometry analysis of DNA contents of EGFP-positive 293T cells transfected with indicated EGFP-tagged constructs.
Similarly to Cif-harboring EPEC, Y. pseudotuberculosis causes gastroenteritis in rodents, and CHYP can only be identified in Y. pseudotuberculosis YPIII strain. P. luminescens is a symbiont of nematodes, and the infection kills a broad spectrum of insects. The gene encoding CHPL is present in the genome of P. luminescens TTO1 strain, which contains an unprecedented large number of potential virulence genes (18) and a TTSS cluster essential for colonization in insects (19). B. pseudomallei, classified as a category B agent by the US Centers for Disease Control and Prevention, is a potential bioterrorism agent. Infection with the bacteria causes melioidosis, a severe human disease with a high mortality (20–50%) and endemic to areas of Southeast Asia and Northern Australia (20). Notably, sequenced genomes of >10 different B. pseudomallei clinical isolates all contain the CHBP gene, which is absent from genomes of closely related Burkholderia thailandensis and Burkholderia mallei that usually do not cause human melioidosis. Comparative genomic analysis of B. pseudomallei and other related bacteria reveals numerous horizontally acquired genomic islands and three TTSSs (21). Among them, TTSS3 shares homology with the Inv/Mxi-SpaTTSS cluster of Shigella flexneri and Salmonella typhimurium and is important for infection of cultured cells and virulence in mice (22, 23).
Cif is carried in a prophage-acquired region outside of the type III locus in EPEC (14, 24). The CHYP ORF in Y. pseudotuberculosis YPIII is next to a gene predicted to encode a transposase fragment. The CHBP ORF in B. pseudomallei K96243, located adjacent to TTSS1 (21), is also flanked by transposase-encoding DNA fragments. The CHPL ORF is predicted to reside in a phage-like genomic region in P. luminescens TTO1 (18). Because Cif is a TTSS effector and its gene is acquired through phage-mediated horizontal transfer (14, 24), it is not surprising that Cif homologs are also candidate substrates of their TTSS, and their genes may evolve through lateral transfer during evolution. With the ongoing microbial genome-sequencing efforts, more Cif family members will likely be found in other TTSS-containing bacterial pathogens.
Despite the limited overall sequence identity, all of the Cif family members harbor several conserved patches of residues, including Cys-109 and His-165 (numbered in Cif) and similar predicted secondary structure arrangements (Fig. 1A). We therefore tested whether Cif homologs indeed share Cif activity of inhibiting eukaryotic cell cycle progression. Similar to that observed with EPEC infection, ectopically expressed EGFP-Cif induced marked accumulation of 293T cells with 4C DNA content (Fig. 1B). Cells transfected with EGFP-CHBP or EGFP-CHPL were also arrested at the G2/M transition (Fig. 1B). These results suggest that CHBP from B. pseudomallei and CHPL from P. luminescens are functional homologs of Cif from EPEC, and the entire Cif family effectors likely employ a similar biochemical mechanism in blocking the host cell cycle progression.
Overall Structure of CHBP.
To understand the nature of the Cif family of cell cycle modulators and gain mechanistic insights into their functions, we purified recombinant CHBP-N48 (truncation of the N-terminal 47 residues) and determined the 2.1-Å crystal structure by Se-SAD (single-wavelength anomalous diffraction) (Fig. 2). Details of the crystallographic analysis are listed in supporting information (SI) Table S1. The structure contains a dimer of CHBP in one asymmetric unit (Fig. S1). Dimerization buries ≈10% of the total solvent-accessible area for one monomer. Few specific interactions exist between the two CHBP monomers, and recombinant CHBP, CHPL, or Cif exclusively stays as monomers in solution. These findings suggest that the CHBP dimer observed in the structure likely results from crystal packing and lacks biological relevance. Structures of the two monomers are nearly identical with a Cα backbone rmsd of 0.32 Å. Thus, hereafter, we limit our discussion to one CHBP molecule (Chain A, Fig. S1). The final model contains residues 78–327 of CHBP and misses the C-terminal last amino acid (Gly-328), residues 221–224 in a loop region, and the N-terminal 30 residues because of the lack of electron density. The N-terminal region in the Cif family shows little sequence conservation (Fig. 1A) and is expected to serve as the type III secretion signal. When directly delivered into HeLa cells, recombinant CHBP-N48 was fully active in inducing a G2 arrest (Fig. S2A). Progressive deletion of the N terminus of Cif until residue 37 (Cif-N38), approximately corresponding to Arg-85 in CHBP, had no effects on its G2 arrest activity, but further truncation of the N-terminal 51 residues (Cif-N52) resulted in a nonfunctional Cif (Fig. S2B). These data suggest that the N-terminal region of the Cif family is dispensable for cell cycle arrest and that the obtained structure of CHBP (residues 78–327) contains a fully functional effector entity.
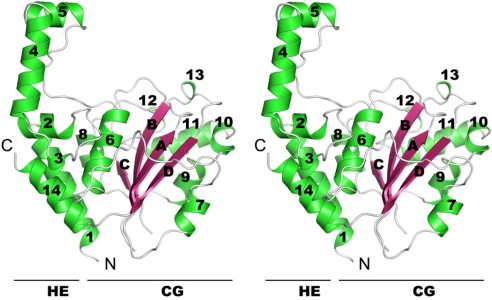
Fig. 2.
Stereoview of the overall structure of CHBP. Secondary structure elements are numbered from the N to the C terminus. α-Helices are labeled 1–14 and drawn as green coils, β-strands are labeled A–D and shown as flat arrows in pink, and other structural elements are drawn as thick gray lines. The structure can be divided into two lobes, the N-terminal HE lobe and the C-terminal CG lobe, marked underneath the corresponding region of the structure.
The CHBP structure with an overall dimension of 56 Å × 37 Å × 46 Å belongs to the α + β class according to the SCOP nomenclature (http://scop.mrc-lmb.cam.ac.uk/scop/). The structure is composed of 14 α-helices (α1–α14) and four β-strands (βA–βD) that form an antiparallel β-sheet and can be divided into two lobes. The β-sheet, together with several surrounding long linking loops and α6–α13, form a compact core globular lobe named the CG lobe. Extensive hydrophobic interactions play important roles in stabilizing the CG lobe structure. α1–α5, together with the extreme C-terminal α14, compose the other lobe named helical extension lobe (HE lobe). In the HE lobe, α1 and α14 form a helix pair and pack against the CG lobe at its bottom left side. The short helices α2 and α3 further stack on top of the α1/α14 helix pair and connect it with α4 and α5. The long α4 projects upward and away from the CG lobe into the solvent. α5, perpendicular to α4, is connected to α6 in the CG lobe through an extended long loop (residues 144–155), which generates a tunnel between the HE and CG lobes. Three interactions could be found between the CG and HE lobes. Tyr-110 in the loop linking α2 and α3 uses its carbonyl oxygen and phenol oxygen to make two hydrogen bond contacts with the Nε of Lys-165 and the side-chain amide nitrogen of Asn-161 from α6, respectively. The side-chain amide nitrogen of Asn-320 from the middle of α14 is also hydrogen-bonded to the carbonyl oxygen of Leu-166 in α6. Certain EPEC strains harboring missense Cif mutants truncated from Asn-273 (equivalent to Asn-320 in CHBP) to the C terminus are deficient in inducing cell cycle arrest (14). This could be explained by the possible role of Asn-320 and α14 in stabilizing the CG lobe (the catalytic domain, see below) or possibly the whole CHBP molecule. Indeed, forced high-level expression of the natural missense mutant could still induce cell cycle arrest in 293T cells (Fig. S2B). Interestingly, deletions of α4 and α5 in the HE lobe of CHBP, which is unlikely to affect the structural integrity of the CG lobe, abolished the cell cycle arrest function (Fig. S2A), implicating a possible role of the protruding α4 and α5 and the HE lobe in mediating substrate recognition.
Papain-Like Fold and Potential Catalytic Triad.
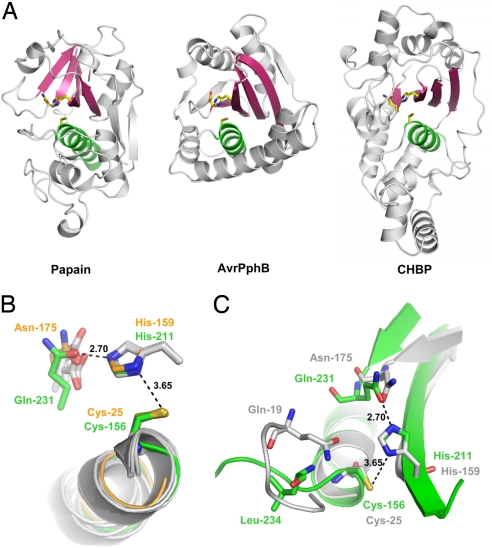
We further subjected the structure of the CG lobe to DALI searches (25). Remarkably, hydrolytic enzymes with a similar papain-like fold dominate the list of hits returned from the DALI server with the top five unique ones (Z-score >4.0) being Pseudomonas avirulence protein AvrPphB protease [Protein Data Base (PDB) ID code 1UKF] (26), Ufm1-specific protease 1 (PDB ID code 2Z84) (27), the N-terminal papain-like catalytic domain of Phytochelatin synthase (ALR0975 from cyanobacterium Nostoc; PDB ID code 2BU3) (28), herpesvirus-encoded ubiquitin-specific protease M48 (PDB ID code 2J7Q) (29), and the papain-like domain of a Vibrio toxin secretion protein (PDB ID code 3B79). The first four have an intact Cys-His-Asp catalytic triad that exhibit cysteine protease activities in vitro. Generally, the N-terminal region of papain-like proteases is largely helical whereas the C-terminal part is enriched in β-strands (7). These characteristics are also present in the CG lobe of CHBP (Fig. 3A). Furthermore, the papain-like fold features a common structural core comprising one helix (green in Fig. 3A) and one four-strand antiparallel β-sheet (pink in Fig. 3A), which provides a framework to support and position the catalytic triad residues precisely within a V-shape active-site cleft. The CHBP structure evidently bears a similar structural core (α6 and βA–D, Fig. 3A), and the corresponding residues are within the most conserved region (Fig. 1A). Despite these similarities, structural organization of CHBP outside of the core helix and four-strand sheet does not resemble that of any other papain-like enzyme.
Fig. 3.
CHBP adopts a papain-like fold and shares a catalytic triad. (A) Structure comparison of CHBP with papain-like enzymes. (Left) Papain (PDB ID code 1PPN). (Center) AvrPphB (PDB ID code 1UKF). (Right) CHBP. Relative orientation of the structures is based on least-squares superimposition of the catalytic Cys and His residues. The catalytic triad residues are shown as sticks. The conserved core helix and β-sheets are colored green and pink, respectively. (B) Comparison of the active sites of CHBP and papain-like enzymes. The superimposition was based on Cys-156 and His-211 in CHBP and the corresponding residues in papain (Cys-25 and His-159) and four other papain family members (PDB ID codes 1UKF, 1XD3, 1EVU, and 2BSZ). Active-site residues of CHBP and papain are shown as stick models in green and orange, respectively; active-site residues in the other four papain family members are shown as gray stick models. (C) Potential oxyanion hole in CHBP. The catalytic triad of CHBP shown in green is superimposed onto that of papain (gray). The associated secondary structures are colored accordingly. Upon superimposition, the main-chain amide of Leu-234 in CHBP is closest to the side-chain amide of Gln-19 in papain that forms an oxyanion hole with the backbone amide of Cys-25.
For papain-like hydrolytic enzymes, a reactive Cys acts as a catalytic nucleophile, and a nearby His functions as a general base to deprotonate the Cys sulfhydryl group and increase its nucleophilicity. The His imidazolium ring is further hydrogen-bonded to a third catalytic residue, usually an Asp or Asn. The Cys, His, and Asp/Asn, collectively referred to as the catalytic triad, are positioned by the common structural core; the Cys is usually located at the N terminus of the helix, and the His and the Asp/Asn are provided by the β-sheet and linking loops (Fig. 3A). CHBP also contains conserved Cys and His at corresponding positions, i.e., Cys-156 at the N terminus of α6 and His-211 in βB. Superimposition of Cys-156 and His-211 of CHBP onto Cys-25 and His-159 of papain or equivalent catalytic residues of other papain-like enzymes showed a remarkable fit, brought the core α-helices to a nearly identical position (Fig. 3B), highlighting the structural similarity between CHBP and papain-like enzymes. Moreover, the Cys and His in CHBP and papain-like enzymes adopt a similar geometry, indicating that CHBP likely uses the two residues to carry out a hydrolytic reaction. In accordance, Cys-156 is solvent-exposed, and the sulfur atom is surrounded by extra electron density that results from oxidation of the sulfhydryl group into sulfonic acid (Fig. S3). Similar oxidation has been observed in the structure of AvrPphB (26) and other papain-like enzymes (30). This phenomenon supports the expected lowered pKa and high reactivity of Cys-156 as a catalytic nucleophile.
Upon superimposition, Gln-231 in CHBP was found to be located at a position similar to that occupied by the third catalytic residue in papain-like enzymes (Fig. 3B). Although Cα of Gln-231 is 3 Å apart from that of Asn-175 in papain, their side chains point toward the same direction, and the amide oxygen atoms are proximal to each other. These structural features should allow for Gln-231 to polarize His-211 during catalysis, analogous to the situation in papain-like enzymes. Taken together, Cys-156, His-211, and Gln-231 of CHBP form a potential catalytic triad that resembles that of papain-like hydrolytic enzymes. Notably, all of the three residues are conserved among the entire Cif family (Fig. 1A), suggesting that other family members likely share a similar catalytic triad and the associated hydrolytic activity.
During catalysis by papain-like enzymes, the Cys-mediated nucleophilic attack on a substrate carbonyl carbon turns the carbonyl oxygen into an oxyanion. In papain, the oxyanion is held by the backbone amide of Cys-25 and the side-chain amide of Gln-19 that form the oxyanion hole. In CHBP, the main-chain amide of Leu-234 is the structural counterpart of the side-chain amide of Gln-19 in papain (Fig. 3C), suggesting its potential role in oxyanion hole formation. Gln-19 in papain and equivalent residues in other papain family enzymes are usually located on the loop preceding the core helix harboring the catalytic Cys. The loop turns almost 180° to bring Gln-19 and Cys-25 into proximity to allow for oxyanion hole formation (Fig. 3C). However, the corresponding loop in CHBP apparently lacks such a turn, and Leu-234 is instead provided by another loop linking βC and α8.
Evidence for Cysteine Protease Activity and Distinct Substrate-Binding Sites.
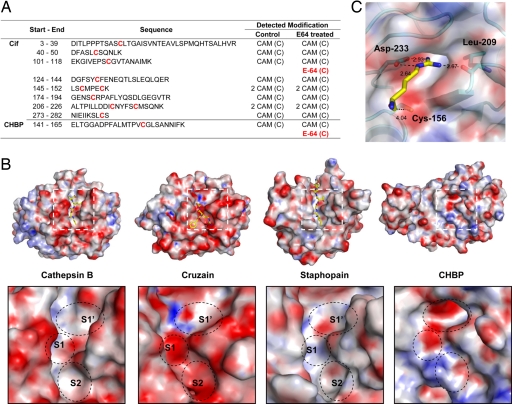
E-64, trans-epoxysuccinyl-l-leucylamido (4-guanidino)butane, potently inhibits the papain-like cysteine protease activity by specifically and covalently modifying the catalytic cysteine sulfhydryl group (7, 31, 32). After incubation of recombinant Cif or CHBP with E-64, a mass increase of 357 Da was observed on the putative catalytic cysteine in both proteins when analyzed by mass spectrometry (Fig. 4A and Fig. S4). This mass increase exactly correlates with E-64 modification. Remarkably, all of the nine deduced cysteine-containing peptides resulting from complete trypsin digestion of Cif and CHBP were identified by mass spectrometry, but only the catalytic cysteines were found to be conjugated by E-64 (Fig. 4A and Fig. S4). Thus, the conserved cysteine in the Cif family harbors chemical properties similar to those of the catalytic cysteine in papain-like proteolytic enzymes, further supporting the idea that the Cif family members are structural homologs of papain-like enzymes.
Fig. 4.
Evidence for catalytic activity and distinct substrate-binding sites. (A) Mass spectrometry analysis of CHBP and Cif upon E-64 treatment. Sequences of all of the deduced cysteine-containing peptides from complete trypsin digestion of CHBP-N48 and Cif are shown in the third column with their start and end residue numbers listed in the second column. All of the cysteines are colored red. Modifications on the cysteine detected by mass spectrometry are listed in the columns to the right of the corresponding peptide sequences. E-64 (C) in red denotes the mass of the cysteine corresponds to that of the E-64 modified form, whereas CAM(C) refers to the (carbamidomethyl)-cysteine generated during sample preparation for MS analysis. The detailed MS/MS spectra are shown in Fig. S4. (B) Comparison of substrate-binding sites in papain-like cysteine proteases and the corresponding regions of CHBP. (Upper Left three). Molecular surface of three papain-like cysteine proteases, cathepsin B (PDB code ID 1QDQ), cruzain (PDB ID code 2AIM), and staphopain (PDB ID code 1PXV) with inhibitors bound to substrate binding sites and shown as sticks. (Lower Left three) Magnified views of their S1, S2, and S1′ sites and the corresponding surface in CHBP (Right). The surface is colored by relative electrostatic potential (obtained by using APBS plug-in in PyMOL), with red and blue denoting negatively and positively charged surfaces, respectively. (C) Molecular docking of an arginine into the negatively charged pocket occupying the S1/S1′ site. A library of 400 dipeptides was searched by in silico molecular docking, and arginine was found to be the only residue that fits into the pocket. Shown as sticks is one of the two possible binding poses of the arginine, in which its guanidinium group makes an ionic and a hydrogen bond contact with Asp-233 and Leu-209, respectively. The pocket is shown in surface presentation.
For papain-like cysteine proteases, certain areas around the catalytic triad cleft are responsible for substrate binding and the cleavage site specificity. Comparison of the surface of CHBP around the catalytic cleft with those of papain-like proteases cathepsin B, cruzain, and staphopain bound with inhibitors occupying the S/S′ sites reveals potential substrate-binding sites and their structural features (Fig. 4B). Usually, papain family proteases harbor a characteristic S2 site, and the P2 residue in the substrate largely contributes to the cleavage site selectivity (26). The corresponding S2 site in CHBP appears to be shallow and lacks distinctive structure. Instead, a deep and highly negatively charged pocket, generated in part by a lid-like loop between βC and α8, occupies the S1/S1′ site. The negative charge is mainly provided by a conserved Asp-233 (Fig. 1A). Neutralization of the negative charge by a D187N mutation in Cif resulted in a largely inactive effector in arresting the host cell cycle (see below). Asp-233 has no interactions with the catalytic triad residues and is likely not directly involved in catalysis. Based on these analyses, we propose that Asp-233 and the negatively charged pocket play a critical role in binding and specificity of the cleavage site in the substrate.
The pocket appears to accommodate a positively charged arginine side chain nicely. To test this prediction in an unbiased way, we searched for dipeptides from all of the 400 possible dipeptides by in silico molecular docking. A two-step protocol using a molecular docking program followed by refining and rescoring with a more computationally intensive molecular mechanics-based energy function (33) was applied to predict the binding poses of those protein–ligand complexes. Regardless of the nature of the other amino acid, arginine is the only residue that fits into the negatively charged pocket (Table S2). Two favorable binding poses of the arginine were identified from the docking simulation, both of which place Cα of the arginine at a close distance of ≈4 Å to the sulfur of Cys-156. In one pose illustrated in Fig. 4C, the arginine residue forms a strong bidentate interaction with Asp-233, donates a hydrogen bond to a backbone carbonyl oxygen of Leu-209, and forms an extra stacking interaction with His-211. In view of results of the docking analysis (Table S2), we generated a recombinant GST protein fused to four tandem repeats of Asn-Gly-Arg-Gly or five copies of Asn-Arg-Gly followed by a C-terminal FLAG tag and assayed whether either of the inserted arginine-rich linkers could serve as an artificial proteolytic substrate for the Cif family of effectors. Unfortunately, no significant proteolytic activities of CHBP or Cif toward either of the purified GST fusion proteins were detected. This finding indicates that additional residues are likely required for efficient substrate recognition assuming that the Cif family indeed comprises proteolytic enzymes rather than another type of hydrolytic enzymes.
Putative Catalytic and Substrate-Binding Sites Are Essential for Cell Cycle Arrest.
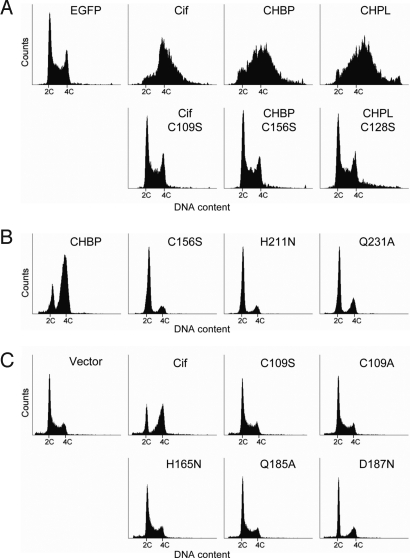
Finally, we investigated the role of the presumed papain-like catalytic activity of the Cif family of type III effectors in inducing eukaryotic cell cycle arrest. Ectopic expression of the catalytic cysteine mutants (Cif C109S, CHBP C156S, or CHPL C128S) in 293T cells failed to induce a G2/M arrest in contrast to their wild-type counterparts (Fig. 5A). Next, when equal amounts of recombinant wild-type CHBP or the catalytic triad mutant (C156S, H211N, or Q231A) were delivered directly into HeLa cells synchronized at the G1/S boundary, all of the catalytic triad mutants appeared to lose the ability to inhibit the cell cycle progression (Fig. 5B). Finally, human EPEC strain E2348/69 harboring wild-type Cif induced an evident G2/M arrest. In contrast, mutations of the catalytic triad of Cif (C109S, C109A, H165N, and Q185A) abolished its cell cycle arrest activity during EPEC infection (Fig. 5C). In the infection assay, we also included the D187N mutant of Cif, and we found that HeLa cells infected with the EPEC harboring the mutant progressed normally from G2 phase to mitosis (Fig. 5C). Taken together, our analysis demonstrates that the papain-like catalytic triad residues and the characteristic negatively charged substrate binding sites are essential for the Cif family of type III effectors to block the host cell cycle progression from G2 to mitosis.
Fig. 5.
The catalytic triad and negatively charged substrate-binding aspartate are essential for cell cycle arrest function of the Cif family. Shown is the cell cycle distribution measured by flow cytometry analysis of DNA contents. (A) Effects of Cys → Ser mutations in the catalytic triad of Cif family effectors on cell cycle progression. Indicated EGFP-tagged constructs were transfected into 293T cells, and EGFP-positive cells were gated out and analyzed. (B) Effects of mutations in the catalytic triad of CHBP. Indicated purified recombinant CHBP proteins were delivered directly into HeLa cells synchronized at the G1/S boundary. (C) Effects of mutations in the catalytic triad and the negatively charged substrate-binding pocket of Cif during EPEC infection.
Discussion
The large majority of papain-like enzymes are cysteine proteases in nature. Nonprotease variants sharing the papain-like fold and catalytic triad have been shown to harbor hydrolytic activities such as arylamine N-acetyltransferase and transglutaminase. Indeed, reactions catalyzed by these nonprotease papain-like enzymes are also initiated by nucleophilic attack of a carbonyl group by the catalytic Cys and involve formation of an acylenzyme intermediate; and they differ from proteases by where the carbonyl group is in the substrate and/or which group further attacks the acylenzyme intermediate and releases the enzyme. For instance, the Cys in transglutaminases attacks the side-chain carbonyl group of a glutamine residue, resulting in release of the ammonium cation; the acylenzyme intermediate is further attacked by the amine of a lysine, which generates an amide linkage between the side chains of the glutamine and lysine. In the case of acetyltransferase, the Cys attacks the carbonyl group in acetyl-CoA. E-64 with a peptide-like structure is remarkably selective for papain-like proteases and has minimal cross-reactivity with other types of papain-like enzymes (31). The fact that the catalytic cysteine in CHBP and Cif is specifically modified by E-64 and the presence of the arginine-fit negatively charged pocket in the equivalent S1/S1′ site favor the idea that the Cif family of effectors are likely cysteine proteases. However, it is certainly a possibility that the Cif family may catalyze a nonproteolytic reaction by using its papain-like catalytic triad. This question can only be answered definitively when the physiological targets/substrates of the Cif family of effectors are identified.
An emerging concept is that pathogenic bacteria of both the animal and plant kingdoms have evolved a large repertoire of effector proteins endowed with proteolytic/hydrolytic activities to interfere with host functions. A growing list of type III effectors or effector families with such activities includes, but is not limited to, the YopT/AvrPphB family (6, 7), the YopJ/AvrA/AvrBsT family (34), AvrRpt2 (35), XopD (36), SseL (37), and the AvrPphE family (38). Interestingly, structure of Pasteurella multocida toxin, a highly potent mitogen and critical virulence factor in the pathogenesis of atrophic rhinitis, reveals a C-terminal AvrPphB-like protease domain required for the cellular function of the toxin (39). Identification of the proteolytic activity and host targets of these effectors has significantly advanced our understanding of bacterial pathogenesis. Here, we add a papain-like hydrolytic activity to the arsenal of pathogenic effectors for thwarting normal functions of the host cell cycle machinery. Determining the nature of the eukaryotic substrate(s) of the Cif family of cell cycle modulators will likely shed new light on our understanding of eukaryotic cell cycle.
Finally, while this manuscript was being submitted for publication, Hsu et al. (40) reported the structure of the C-terminal fragment of Cif. Consistent with our observation, the analysis of Hsu et al. also suggests that Cif harbors a papain-like fold and catalytic triad. Structural comparison indicates a high degree of similarity between the Cif structure and the CG lobe of CHBP, further confirming that the entire Cif or CHBP family of type III effectors likely functions as papain-like enzymes in blocking the host cell cycle progression.
Materials and Methods
Genes encoding Cif, CHBP, and CHPL were amplified from bacterial genomic DNA. Recombinant proteins for crystallization or functional analysis were expressed and purified from E. coli. The crystal structure of CHBP was determined by Se-SAD. The rest of the information about reagents, detailed crystallization and structural determination, cell culture and cell cycle analysis, bacterial infection, E-64 labeling, and mass spectrometry analysis is presented in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Dr. Yusuke Yamada and his staff at the KEK Synchrotron Facility (Tsukuba, Japan) for assistance with data collection. We are grateful to Dr. Seema Mattoo for reading and editing the manuscript. We thank Dr. Biao Kan (Centers for Disease Control, China) for providing the EPEC strain harboring Cif, Dr. Brendan Kenny (University of Newcastle) for the EPEC E2348/69 strain, and Dr. Todd A. Ciche (Michigan State University) and Dr. Donald E. Woods (University of Calgary) for providing genomic DNA from P. luminescens subsp. laumondii TTO1 and B. pseudomallei K96243, respectively. We also thank members of the Shao laboratory for helpful discussions and technical assistance. This work was supported by Chinese Ministry of Science and Technology 863 Grant 2005AA210950 and 973 National Basic Research Plan of China Grant 2006CB806502 (to F.S.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors for CHBP have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3EIR and 3EIT).
This article contains supporting information online at www.pnas.org/cgi/content/full/0900212106/DCSupplemental.
References
- 1.Galan JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006;444:567–573. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- 2.Roy CR, Mocarski ES. Pathogen subversion of cell-intrinsic innate immunity. Nat Immunol. 2007;8:1179–1187. doi: 10.1038/ni1528. [DOI] [PubMed] [Google Scholar]
- 3.Bhavsar AP, Guttman JA, Finlay BB. Manipulation of host-cell pathways by bacterial pathogens. Nature. 2007;449:827–834. doi: 10.1038/nature06247. [DOI] [PubMed] [Google Scholar]
- 4.Mattoo S, Lee YM, Dixon JE. Interactions of bacterial effector proteins with host proteins. Curr Opin Immunol. 2007;19:392–401. doi: 10.1016/j.coi.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 5.Li H, et al. The phosphothreonine lyase activity of a bacterial type III effector family. Science. 2007;315:1000–1003. doi: 10.1126/science.1138960. [DOI] [PubMed] [Google Scholar]
- 6.Shao F, et al. Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science. 2003;301:1230–1233. doi: 10.1126/science.1085671. [DOI] [PubMed] [Google Scholar]
- 7.Shao F, Merritt PM, Bao Z, Innes RW, Dixon JE. A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell. 2002;109:575–588. doi: 10.1016/s0092-8674(02)00766-3. [DOI] [PubMed] [Google Scholar]
- 8.Huang J, Lesser CF, Lory S. The essential role of the CopN protein in Chlamydia pneumoniae intracellular growth. Nature. 2008;456:112–115. doi: 10.1038/nature07355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iwai H, et al. A bacterial effector targets Mad2L2, an APC inhibitor, to modulate host cell cycling. Cell. 2007;130:611–623. doi: 10.1016/j.cell.2007.06.043. [DOI] [PubMed] [Google Scholar]
- 10.Nougayrede JP, et al. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science. 2006;313:848–851. doi: 10.1126/science.1127059. [DOI] [PubMed] [Google Scholar]
- 11.Oswald E, Nougayrede JP, Taieb F, Sugai M. Bacterial toxins that modulate host cell-cycle progression. Curr Opin Microbiol. 2005;8:83–91. doi: 10.1016/j.mib.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 12.Elwell CA, Dreyfus LA. DNase I homologous residues in CdtB are critical for cytolethal distending toxin-mediated cell cycle arrest. Mol Microbiol. 2000;37:952–963. doi: 10.1046/j.1365-2958.2000.02070.x. [DOI] [PubMed] [Google Scholar]
- 13.Lara-Tejero M, Galan JE. A bacterial toxin that controls cell cycle progression as a deoxyribonuclease I-like protein. Science. 2000;290:354–357. doi: 10.1126/science.290.5490.354. [DOI] [PubMed] [Google Scholar]
- 14.Marches O, et al. Enteropathogenic and enterohaemorrhagic Escherichia coli deliver a novel effector called Cif, which blocks cell cycle G2/M transition. Mol Microbiol. 2003;50:1553–1567. doi: 10.1046/j.1365-2958.2003.03821.x. [DOI] [PubMed] [Google Scholar]
- 15.Taieb F, Nougayrede JP, Watrin C, Samba-Louaka A, Oswald E. Escherichia coli cyclomodulin Cif induces G2 arrest of the host cell cycle without activation of the DNA-damage checkpoint-signalling pathway. Cell Microbiol. 2006;8:1910–1921. doi: 10.1111/j.1462-5822.2006.00757.x. [DOI] [PubMed] [Google Scholar]
- 16.Samba-Louaka A, et al. Bacterial cyclomodulin Cif blocks host cell cycle by stabilizing cyclin-dependent kinase inhibitors p21(waf1) and p27(kip1) Cell Microbiol. 2008;10:2496–2508. doi: 10.1111/j.1462-5822.2008.01224.x. [DOI] [PubMed] [Google Scholar]
- 17.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duchaud E, et al. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat Biotechnol. 2003;21:1307–1313. doi: 10.1038/nbt886. [DOI] [PubMed] [Google Scholar]
- 19.Brugirard-Ricaud K, et al. Site-specific antiphagocytic function of the Photorhabdus luminescens type III secretion system during insect colonization. Cell Microbiol. 2005;7:363–371. doi: 10.1111/j.1462-5822.2004.00466.x. [DOI] [PubMed] [Google Scholar]
- 20.Wiersinga WJ, van der Poll T, White NJ, Day NP, Peacock SJ. Melioidosis: Insights into the pathogenicity of Burkholderia pseudomallei. Nat Rev Microbiol. 2006;4:272–282. doi: 10.1038/nrmicro1385. [DOI] [PubMed] [Google Scholar]
- 21.Holden MT, et al. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc Natl Acad Sci USA. 2004;101:14240–14245. doi: 10.1073/pnas.0403302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pilatz S, et al. Identification of Burkholderia pseudomallei genes required for the intracellular life cycle and in vivo virulence. Infect Immun. 2006;74:3576–3586. doi: 10.1128/IAI.01262-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stevens MP, et al. Attenuated virulence and protective efficacy of a Burkholderia pseudomallei bsa type III secretion mutant in murine models of melioidosis. Microbiology. 2004;150:2669–2676. doi: 10.1099/mic.0.27146-0. [DOI] [PubMed] [Google Scholar]
- 24.Loukiadis E, et al. Distribution, functional expression, and genetic organization of Cif, a phage-encoded type III-secreted effector from enteropathogenic and enterohemorrhagic Escherichia coli. J Bacteriol. 2008;190:275–285. doi: 10.1128/JB.00844-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 26.Zhu M, Shao F, Innes RW, Dixon JE, Xu Z. The crystal structure of Pseudomonas avirulence protein AvrPphB: A papain-like fold with a distinct substrate-binding site. Proc Natl Acad Sci USA. 2004;101:302–307. doi: 10.1073/pnas.2036536100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ha BH, et al. Structural basis for Ufm1 processing by UfSP1. J Biol Chem. 2008;283:14893–14900. doi: 10.1074/jbc.M708756200. [DOI] [PubMed] [Google Scholar]
- 28.Vivares D, Arnoux P, Pignol D. A papain-like enzyme at work: Native and acyl-enzyme intermediate structures in phytochelatin synthesis. Proc Natl Acad Sci USA. 2005;102:18848–18853. doi: 10.1073/pnas.0505833102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schlieker C, et al. Structure of a herpesvirus-encoded cysteine protease reveals a unique class of deubiquitinating enzymes. Mol Cell. 2007;25:677–687. doi: 10.1016/j.molcel.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jia Z, et al. Crystal structures of recombinant rat cathepsin B and a cathepsin B–inhibitor complex: Implications for structure-based inhibitor design. J Biol Chem. 1995;270:5527–5533. doi: 10.1074/jbc.270.10.5527. [DOI] [PubMed] [Google Scholar]
- 31.Evans MJ, Cravatt BF. Mechanism-based profiling of enzyme families. Chem Rev. 2006;106:3279–3301. doi: 10.1021/cr050288g. [DOI] [PubMed] [Google Scholar]
- 32.Varughese KI, et al. Crystal structure of a papain–E-64 complex. Biochemistry. 1989;28:1330–1332. doi: 10.1021/bi00429a058. [DOI] [PubMed] [Google Scholar]
- 33.Huang N, Kalyanaraman C, Irwin JJ, Jacobson MP. Physics-based scoring of protein–ligand complexes: Enrichment of known inhibitors in large-scale virtual screening. J Chem Inf Model. 2006;46:243–253. doi: 10.1021/ci0502855. [DOI] [PubMed] [Google Scholar]
- 34.Mukherjee S, et al. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science. 2006;312:1211–1214. doi: 10.1126/science.1126867. [DOI] [PubMed] [Google Scholar]
- 35.Axtell MJ, Chisholm ST, Dahlbeck D, Staskawicz BJ. Genetic and molecular evidence that the Pseudomonas syringae type III effector protein AvrRpt2 is a cysteine protease. Mol Microbiol. 2003;49:1537–1546. doi: 10.1046/j.1365-2958.2003.03666.x. [DOI] [PubMed] [Google Scholar]
- 36.Hotson A, Chosed R, Shu H, Orth K, Mudgett MB. Xanthomonas type III effector XopD targets SUMO-conjugated proteins in planta. Mol Microbiol. 2003;50:377–389. doi: 10.1046/j.1365-2958.2003.03730.x. [DOI] [PubMed] [Google Scholar]
- 37.Rytkonen A, et al. SseL, a Salmonella deubiquitinase required for macrophage killing and virulence. Proc Natl Acad Sci USA. 2007;104:3502–3507. doi: 10.1073/pnas.0610095104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nimchuk ZL, Fisher EJ, Desveaux D, Chang JH, Dangl JL. The HopX (AvrPphE) family of Pseudomonas syringae type III effectors require a catalytic triad and a novel N-terminal domain for function. Mol Plant Microbe Interact. 2007;20:346–357. doi: 10.1094/MPMI-20-4-0346. [DOI] [PubMed] [Google Scholar]
- 39.Kitadokoro K, et al. Crystal structures reveal a thiol protease-like catalytic triad in the C-terminal region of Pasteurella multocida toxin. Proc Natl Acad Sci USA. 2007;104:5139–5144. doi: 10.1073/pnas.0608197104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsu Y, et al. Structure of the cyclomodulin Cif from pathogenic Escherichia coli. J Mol Biol. 2008;384:465–477. doi: 10.1016/j.jmb.2008.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.