Abstract
Through targeted homologous recombination, we developed a panel of matched colorectal cancer cell lines that differ only with respect to their endogenous TP53 status. We then used these lines to define the genes whose expression was altered after DNA damage induced by ionizing radiation. Transcriptome analyses revealed a consistent up-regulation of polo-like kinase 1 (PLK1) as well as other genes controlling the G2/M transition in the cells whose TP53 genes were inactivated compared with those with WT TP53 genes. This led to the hypothesis that the viability of stressed cells without WT TP53 depended on PLK1. This hypothesis was validated by demonstrating that stressed cancer cells without WT TP53 alleles were highly sensitive to PLK1 inhibitors, both in vivo and in vitro.
In cancers, the TP53 tumor suppressor gene is inactivated more frequently than any of the other ≈21,000 protein-encoding genes in the human genome (1–6). TP53 has been estimated to be altered by point mutation in approximately half of all human cancers (2, 7–9). In cancers without such intragenic mutations, the p53 protein is often functionally inactivated by binding to proteins encoded by viral or cellular oncogenes such as E6 or MDM2, respectively (10–15). In normal cells, the p53 protein is a key node in the network controlling the response to stress, particularly those associated with damage to DNA such as oxidation and irradiation (16–20). One of its major functions is the activation of a transcriptional response to such stresses that determine whether cells arrest and repair the damage or undergo apoptosis. In the absence of TP53, both cell cycle arrest and apoptosis are compromised, presumably allowing these cancer cells to proliferate under situations wherein normal cells would die. Several of the transcriptionally activated genes that mediate the cell cycle arrest and apoptotic functions of TP53 have been identified, including the cyclin-dependent kinase inhibitor p21 and the proapoptotic protein PUMA (21–23).
In light of its extraordinarily high mutation rate across many different tumor types, TP53 provides a uniquely attractive target for drug development. However, like any tumor suppressor gene, TP53 is not itself easily “druggable:” Drugs generally inhibit the function of proteins rather than restore normal function to defective proteins. It has thereby been challenging to develop small molecules that restore transcriptional activation to mutant p53 proteins, although some promising compounds of this type have been developed (24–27).
It therefore may be useful to attempt to target elements in the pathways that TP53 regulates, rather than p53 itself, for therapeutic purposes. Indeed, clever strategies for exploiting the absence of functional p53 in cells have been devised and some are in clinical trials (28). Because loss of G1 arrest in p53 mutants prevents repair of DNA damage, a number of synthetic lethal strategies have been proposed. Notably, some of these strategies involve compounds that disrupt the G2/M checkpoint, the major repair checkpoint that remains at least partially functional in cells without normal p53 function (27, 29–36).
In an effort to better understand the pathways regulated by p53, we constructed a variety of human colorectal cancer cell lines that were isogenic except at the TP53 locus. Human colorectal cancer cells, rather than mouse cells, were chosen for these experiments because the pathways that are regulated by p53 are likely to vary between species and cell types (37–39). We then analyzed the transcriptome of these cells after subjecting them to the stress imposed by DNA damage.
Most similar transcriptome analyses have in the past been performed on cells with overexpressed p53 genes (21–23, 40–42). Overexpression can induce a variety of effects that may play little role under more physiologic conditions (43, 44). Through the use of cells in which the WT or mutant TP53 genes were under the control of their normal regulatory elements, we hoped to uncover pathways that provided leads for therapeutic development. Indeed, we found that a common signature of cells with inactive TP53 genes, regardless of the precise nature of the mutant allele, was a relative up-regulation of genes controlling the G2/M checkpoint. This in turn suggested a specific therapeutic approach for cancers without WT TP53 genes, as described below.
Results
Generation of a Panel of Isogenic Cell Lines Differing in p53 Status.
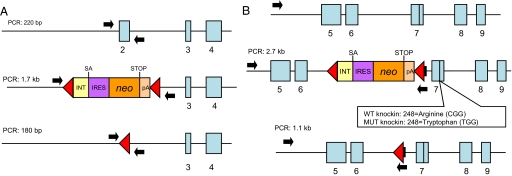
We used 3 recombinant adenoassociated virus (rAAV) vectors to alter the endogenous alleles of 4 commonly used colorectal cancer cell lines. One vector resulted in the deletion of TP53 exon 2 in the targeted lines (Fig. 1A). The 2 others were “knockin” vectors, resulting in the creation of either mutant (tryptophan) or WT (arginine) codons at amino acid 248 in exon 7 (Fig. 1B). The codon 248 mutation is among the most common observed in human cancers. The isogenic cell lines used in this study derived from SW48, DLD-1, RKO, and HCT116 cells, are listed in Table 1. In general, at least 2 independent clones of each knockout or knockin cell line were derived. Independent clones with the same genotype always behaved identically in the assays described below.
Fig. 1.
TP53 locus before and after targeting. (A) (Top) TP53 genomic locus including exons 2, 3, and 4 (blue boxes). (Middle) Same locus after insertion of the targeting vector (62), resulting in replacement of exon 2 and surrounding intronic sequences with a cassette containing intronic sequences (INT), splice acceptor site (SA), internal ribosomal entry sequence (IRES), neomycin phosphotransferase gene (neo), stop codon (STOP), and polyadenylation site (pA). Red triangles indicate loxP recombination sites. (Bottom) The same locus after Cre-mediated excision of the targeting construct. (B) (Top) TP53 genomic locus including exons 5 to 9 (blue boxes). (Middle) Same locus after insertion of the rAAV targeting vectors, one containing the WT sequence in exon 7 and the other a mutation (R248W) in exon 7. (Bottom) The same locus after Cre-mediated excision of the knockin constructs. In both A and B, arrows indicate the position of the PCR primers used to screen clones for the desired recombination. The sizes of the PCR products generated with these primers are also indicated.
Table 1.
TP53 genotypes of the panel of isogenic cell lines used in this study
| Tumor of origin | Genotype | Allele 1 | Allele 2 |
|---|---|---|---|
| SW48 | +/+ | WT | WT |
| SW48 | +/− | WT | Inactivated |
| SW48 | −/− | Inactivated | Inactivated |
| RKO | +/+ | WT | WT |
| RKO | +/− | WT | Inactivated |
| RKO | −/− | Inactivated | Inactivated |
| RKO | R248W/+ | R248W | WT |
| HCT116 | +/+ | WT | WT |
| HCT116 | +/− | WT | Inactivated |
| HCT116 | −/− | Inactivated | Inactivated |
| HCT116 | R248W/− | R248W | Inactivated |
| HCT116 | R248W/+ | R248W | WT |
| DLD-1 | S241F/SIL | S241F | Silent |
| DLD-1 | −/SIL | Inactivated | Silent |
| DLD-1 | +/SIL | WT | Silent |
| DLD-1 | S241F/− | S241F | Inactive |
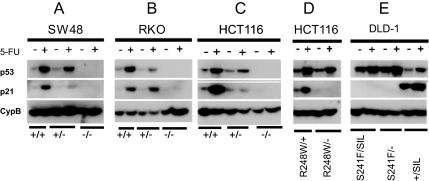
Examples of the PCR and sequencing results that were used to confirm the targeting events are illustrated in Fig. S1 A–D. Western blot analyses were performed before and after treatment with 5-FU, a cancer chemotherapeutic drug known to activate p53 (Fig. 2). SW48 parental cells have 2 WT TP53 alleles. When 1 of them is disrupted by targeted homologous recombination, creating a heterozygote, there was less p53 protein in the cell after 5-FU treatment (Fig. 2A). When both alleles were disrupted, there was of course no p53 protein. Identical results were observed in HCT116 and RKO cells, both of which normally have 2 WT alleles of TP53 (Fig. 2 B and C). The p21 protein (product of the CDKN1A gene) is one of the most well-characterized targets of p53 transcriptional activation (45). This protein was induced in the parental cells after 5-FU activation of p53 but not in the cells with both alleles disrupted (Fig. 2 A–C). In the heterozygotes with 1 WT p53 allele and 1 disrupted allele, p21 induction by 5-FU was detectable but somewhat variable, perhaps reflecting TP53-gene dosage effects that varied among the cell lines.
Fig. 2.
TP53 and p21 protein expression in isogenic cells lines of various TP53 genotypes. Cells were cultured in the absence or presence of 5-FU. The CypB blots were used as loading controls.
When 1 WT allele of HCT116 cells was replaced with a mutant (R248W) allele, there was still induction of p53 by 5-FU and consequent activation of p21 (R248W/+ line in Fig. 2D). However, when 1 WT allele of HCT116 cells was inactivated by homologous recombination, and the other was replaced with a mutant (R248W) allele, induction of p53 by 5-FU was less marked, and induction of p21 was completely eliminated (R248W/− line in Fig. 2D). DLD-1 cells normally have 1 allele that is mutant (S241F) and 1 allele that is not detectably expressed, as assessed by RT-PCR analysis (silent). We detected no mutations of TP53 in this second allele upon sequencing of all of the exons and intron–exon boundaries of the gene, so the silencing may have been epigenetic. In parental DLD-1 cells (genotype S241F/SIL), there was some increase in p53 upon 5-FU treatment but no induction of p21 (Fig. 2E). When the silent allele was disrupted through homologous recombination, there was no change either in p53 or p21 levels, as expected (genotype S241F/−). However, when the mutant S241F allele of DLD-1 was replaced with a WT allele, the level of p53 protein was reduced and p21 was induced upon 5-FU treatment (S241F/+ line in Fig. 2E).
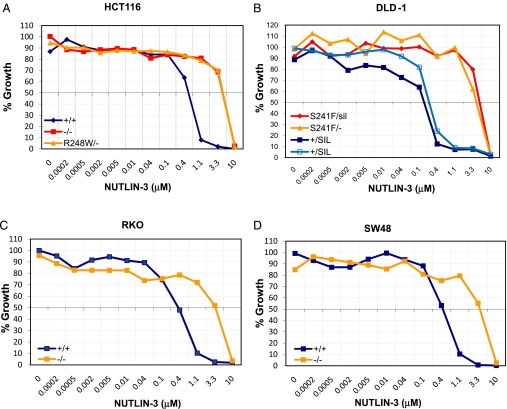
As another test of the functionality of the knockin and knockout clones, we evaluated the effects of Nutlin-3, a small molecule that binds to MDM2 and disrupts the interaction between MDM2 and p53 proteins (46, 47). This drug retards the ability of MDM2 to ubiquinate p53 and mark it for degradation. We found that all clones harboring WT p53 were more growth-inhibited by Nutlin-3 than clones without WT p53 (Fig. 3). Clones with completely inactive TP53 genes had Nutlin-3 sensitivities identical to those with point mutations of p53.
Fig. 3.
TP53 genotype-dependent effect of Nutlin-3 on isogenic cancer cell lines. The indicated lines were exposed to increasing doses of Nutlin-3 for 96 h, and their growth was evaluated by assessing cell number in a SYBR green growth assay. All values were normalized to the number of cells of untreated controls. The 2 lines marked +/SIL were independently generated clones.
Transcriptional Profile of Cells With and Without WT TP53.
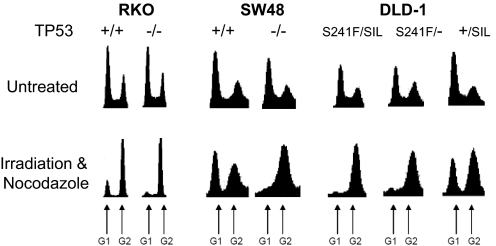
Ionizing radiation such as that produced by γ-irradiation induces a p53-dependent G1 arrest via the induction of DNA double-strand breaks (48–50). Using the TP53 isogenic panel, we first confirmed that the predicted G1 arrest was observed in the lines containing WT TP53 genes (Fig. 4). In the isogenic lines in which both TP53 alleles were inactivated by targeted disruption or point mutation, minimal G1 arrest was observed (Fig. 4).
Fig. 4.
G1 arrest in cell lines containing WT TP53 genes. Flow cytometry profiles before and after exposure of the indicated lines to ionizing radiation are shown. After irradiation, the cells were treated with nocodazole to block them from undergoing mitosis and entering into a subsequent G1. Peaks corresponding to G1 and G2/M are indicated.
We next compared the transcriptional profiles of these cells after γ-irradiation. As expected, a number of known p53 target genes were found to be present at higher levels in the cells containing WT TP53 alleles than in their isogenic counterparts without WT TP53 alleles (Table S2). The induced transcripts included those encoded by CKDN1A, BBC3 (PUMA), MDM2, FDXR, CCNG1, and PPM1D. The differential expression was much more prominent after irradiation than in the absence of irradiation (Table S2). Interestingly, a number of highly up-regulated transcripts that had not been identified in previous studies were identified in these experiments. Among these, one of the strongest up-regulated transcripts was a large noncoding RNA of unknown function (LOC401131 in Table S2).
We were most interested in transcripts that were up-regulated in the cells without WT TP53 genes, because their identification could lead to therapeutic approaches to inhibit the growth of cancer cells with inactive TP53 genes. After irradiation, the expression levels of 35 genes were consistently higher (by at least 2-fold) in all lines without WT TP53 alleles than in those with WT TP53 alleles (Table S2). In all 35 cases, the differential expression was confirmed by quantitative PCR (Table S3). This up-regulation was largely due to a repression of expression in the lines with WT TP53 (Table S4). Surprisingly, the majority of the up-regulated genes were components of the G2/M and/or spindle assembly checkpoints (Table S1). Moreover, many other genes associated with the G2/M and spindle checkpoints were also up-regulated, but less than the 2-fold required for inclusion in Table S1 (Table S4).
Drug-Targeting of Cells with Inactive TP53 Alleles.
Among the G2/M checkpoint genes up-regulated in the cells without WT TP53 genes, 2 were particularly intriguing: PLK1 and AURKB. These genes have well-defined, sequential roles in the G2/M checkpoint, are highly specific in their function, and have enzymatic activities that can be inhibited by previously described small molecules. The expression data (Table S1) coupled with the responses to DNA damaging agents among the isogenic cell line panel (Fig. 4) suggested a therapeutic approach that would exploit the differences in PLK1 and AURKB expression observed in cancer cells without WT TP53 alleles. In particular, we hypothesized that the up-regulation of PLK1 and AURKB was required for the continued viability of cells without WT TP53 after stress. Moreover, we expected that a portion of normal (WT TP53 gene-containing) cells would be specifically arrested in G1 by the stress and would be partially spared the toxicity of these drugs, which presumably act largely on cells in the G2 phase of the cell cycle.
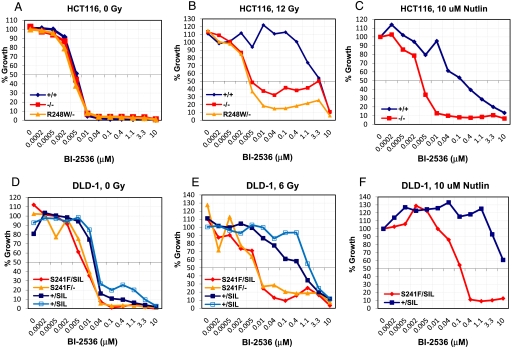
To test this hypothesis, we synthesized 3 AURKB inhibitors (VX-680, AZD1152, and MLN8054) and 2 PLK1 inhibitors (BI-2536 and ON01910) and evaluated them in isogenic lines derived from HCT116 and DLD-1 cells. In each case, we compared a cell line with 2 WT TP53 alleles to its isogenic twin containing 2 inactivated TP53 alleles. In the presence of any of the 4 tested drugs alone, only minor differences in the sensitivities of the isogenic cell lines were observed (examples in Fig. 5 A and D). However, when the cells were first irradiated to induce a G1 arrest, the cells without WT TP53 genes proved much more sensitive to the 2 PLK1 inhibitors, particularly BI-2536 (Fig. 5 B and E and Fig. S2). The 3 AURKB inhibitors did not appreciably alter the growth of cells with WT TP53 alleles compared with those with inactive TP53 alleles (Fig. S3).
Fig. 5.
Evaluation of BI-2536 on the growth of isogenic cell lines with or without WT TP53 genes. Growth of the indicated lines to increasing doses of the PLK1 inhibitor BI-2536 alone, in the presence of ionizing radiation, or in the presence of Nutlin-3. The growth was assessed by a SYBR green-based growth assay and was normalized to the growth of untreated controls (A and D), in the presence of irradiation (B and E) or the presence of Nutlin-3 (C and F). The 2 lines marked +/SIL were independently generated clones.
To explore the generality of WT TP53-mediated protection from PLK inhibitor toxicity, we used Nutlin-3 instead of γ-irradiation to induce a stress-like state, i.e., to block a portion of cells with WT TP53 genes in G1. Nutlin-3 in combination with BI-2536 was indeed able to selectively inhibit the growth of TP53 mutant cell lines in a fashion comparable with that observed with irradiation plus BI-2536 (Fig. 5 C and F). This selective toxicity was observed in clones derived from all 4 parental colorectal cancer cell lines (SW48, RKO, HCT116, and DLD-1) and was independent of the nature of the mutation that inactivated TP53 [missense mutation vs. targeted disruption (Fig. 5 and Fig. S4)].
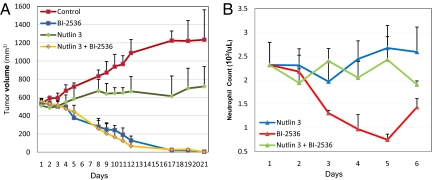
Finally, we attempted to determine whether this therapeutic approach would be efficacious in an experimental animal model. Treatment with BI-2536 at 100 mg/kg twice a week resulted in dramatic tumor regression in nude mice harboring relatively large xenografts of HCT116 cells in which the WT TP53 genes were disrupted (Fig. 6A). Notably, oral administration of Nutlin-3 (200 mg/kg) did not decrease the efficacy of BI-2536. This was expected from our in vitro experiments on the same cell line, wherein we found that Nutlin-3 could not rescue cells without WT TP53 genes from the effects of BI-2536 (Fig. 5C). However, unlike the situation in vitro, we could not protect WT TP53 HCT-116 tumor cells from BI-2536-mediated cell death with Nutlin-3 in vivo. Whether this was because of inadequate concentrations of Nutlin-3 in vivo for prolonged periods is not known. On the other hand, the major toxicity of BI-2536 in Phase I trials has been hematopoietic, with dangerous levels of neutropenia observed after treatment with this agent in the majority of patients (51). To determine whether Nutlin-3 could rescue this toxicity, we administered the combination of oral Nutlin-3 (200 mg/kg) and BI-2536 (100 mg/kg) to BALB/c mice. BALB/c rather than nude mice were used in these experiments because BI-2536 caused bone marrow toxicity in BALB/c mice at doses that did not cause such toxicity in nude mice. BALB/c mice treated with BI-2536 (100 mg/kg) developed neutropenia within 48 h after treatment with BI-2536. Oral administration of Nutlin-3 (200 mg/kg) efficiently protected the mice from this neutropenia (Fig. 6B).
Fig. 6.
In vivo effects of BI-2536 and Nutlin-3. (A) BI-2536 treatment of nude mice bearing HCT116 TP53−/− xenografts results in regression of the tumors. The mice were treated for 3 weeks as follows: twice a week with 100 mg/kg BI-2536 and twice a day 2 times a week with 200 mg/kg Nutlin-3 before BI-2536 treatment. The volume of the tumor was measured on the days indicated. (B) BALB/c mice were given 200 mg/kg Nutlin-3 4 and 24 h before administering a single i.v. dose of 100 mg/kg BI-2536. Neutrophils were counted before initial treatment and every 24 h thereafter just before BI-2536 administration. Means and SD of the counts from 5 mice in each treatment arm are shown.
Discussion
Our observations on the isogenic cell lines generated for this study are consistent with previous work (16–19, 46) showing that (i) mutant p53 proteins are more stable than the normal p53 protein (Fig. 2E); (ii) p21 is induced by 5-FU only when WT p53 is present in the cell (Fig. 2); and (iii) Nutlin-3 activates WT TP53 and results in growth arrest (Fig. 3). The data also indicate that this large and varied isogenic panel provides a valuable model for determining chemical sensitivities based on TP53 status. One advantage of the approach taken in this work, involving homologous recombination to delete or insert specific sequences in endogenous genes, is that the only major difference between cell line pairs is in the sequence of TP53. The fact that several independently derived lines with the same TP53 genotype behaved identically in the assays supports this conclusion. Although we cannot exclude differential expression of other p53 isoforms, they are therefore not likely to play a role in the phenotypes observed in this study.
One of the most interesting observations made in this study was that the majority of the genes whose expression was higher in cells lacking WT TP53 than in those with WT TP53 genes were involved in the G2/M transition. Although we do not know the basis for the up-regulation of these genes in cells with inactivated p53 genes, there are at least 3 possibilities. First, these genes could be directly repressed by WT p53 binding to their promoters. There is indeed evidence that p53 can directly repress genes rather than activate them (52). Second, it is possible that the relatively higher expression of these genes simply reflects a larger fraction of cells arrested in G2/M after DNA damage when p53 is inactivated. However, the data in Fig. 4 show there is only a modest increase in the fraction of cells in G2/M in cells with inactive TP53 genes compared with those with WT TP53; the major difference in the cell cycle profiles is in the presence or absence of a G1 block that generally affects only a minority of the cell population. The third possibility, and the one we favor, is that the up-regulation represents an indirect downstream effect of the altered regulatory pathways resulting from the absence of WT TP53. Regardless of the explanation, the differential expression of specific G2/M checkpoint genes such as PLK1 and AURKB provides a rationale for developing new therapeutic approaches.
The approach described in Figs. 5 and 6, employing an inhibitor of PLK1 together with an agent that protects cells with WT TP53 genes, builds on previous work in basic and applied research. In particular, previous studies have shown that TP53 is required for the G1 arrest after DNA damage and that the CDK2NA gene is essential for this arrest (53). The potential to exploit the defective checkpoint status of cells with inactive TP53 genes has also been widely recognized and in part stimulated the discovery of drugs that can inhibit PLK1, AURKB, and other proteins that regulate the G2/M checkpoint (27, 29–36, 51, 54–60). Our results expand on these seminal observations in several ways. First, we confirm the requirement for WT TP53 in isogenic pairs derived from 4 different human cancer cell lines. The biochemical and functional distinctions between these lines, which differ in some cases by only a single base pair, are remarkable. Second, we show that the relatively high expression of G2/M checkpoint genes is a consistent feature of the cancer cell lines without WT TP53. Although many previous studies have uncovered genes up-regulated in cells with WT TP53 genes after stress, the discovery of a large class of functionally related genes that is specifically up-regulated in cells without WT TP53 genes is unique. Third, our studies suggest that coupling BI-2536, a powerful therapeutic, with an agent that protects a portion of normal cells from entering G2 is a promising strategy for reducing the toxicity associated with PLK1 inhibitors. Although this strategy might appear to be applicable to any protein controlling the G2/M checkpoint, our data suggest otherwise. In particular, inhibitors of AURKB showed no differential effects on lines with WT vs. without WT TP53 genes (Fig. S4) even though AURKB functions in the same pathway as PLK1 (61). The reason for this difference between the cytotoxic effects of AURKB and PLK1 inhibitors is unknown. Regardless of the reason, our data suggest that coupling BI-2536 or related drugs with agents that normally induce G1 arrest is worthy of further investigation.
Materials and Methods
For detail of targeting vector construction, cell lines and targeting, compounds and γ-irradiation, Western blot analysis, microarray and qPCR procedures, and mouse studies, see SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Wayne Yu for assisting with the microarray experiments, Evangaline Watson for expert technical assistance with the animal experiments, Leslie Meltzer for assistance with flow cytometry, Nadine Forbes for assisting with the CBC analyses on Hemavet 950, and Jihye Yun, Christine Gan, Christoph Lengauer, Devin Dressman, and Ian Cheong for helpful discussions. This work was supported by The Virginia and D. K. Ludwig Fund for Cancer Research and National Institutes of Health Grants CA 43460, CA 09243, and CA 62924.
Footnotes
Conflict of interest statement: Under separate licensing agreements between Genzyme Corporation and The Johns Hopkins University, the authors are entitled to a share of royalties received by the University on sales of products described in this article. The terms of this arrangement are being managed by the Johns Hopkins University in accordance with its conflict of interest policies.
Data deposition: Microarray expression data have been deposited in National Center for Biotechnology Information Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE13886).
This article contains supporting information online at www.pnas.org/cgi/content/full/0813333106/DCSupplemental.
References
- 1.Olivier M, Hussain SP, Caron de Fromentel C, Hainaut P, Harris CC. TP53 mutation spectra and load: A tool for generating hypotheses on the etiology of cancer. IARC Sci Publ. 2004;157:247–270. [PubMed] [Google Scholar]
- 2.Soussi T, Wiman KG. Shaping genetic alterations in human cancer: The p53 mutation paradigm. Cancer Cell. 2007;12(4):303–312. doi: 10.1016/j.ccr.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Sjoblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314(5797):268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 4.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318(5853):1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 5.Jones S, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321(5897):1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soussi T, Beroud C. Assessing TP53 status in human tumours to evaluate clinical outcome. Nat Rev Cancer. 2001;1(3):233–240. doi: 10.1038/35106009. [DOI] [PubMed] [Google Scholar]
- 8.Olivier M, et al. The IARC TP53 database: New online mutation analysis and recommendations to users. Hum Mutat. 2002;19(6):607–614. doi: 10.1002/humu.10081. [DOI] [PubMed] [Google Scholar]
- 9.Hainaut P, Hollstein M. p53 and human cancer: The first ten thousand mutations. Adv Cancer Res. 2000;77:81–137. doi: 10.1016/s0065-230x(08)60785-x. [DOI] [PubMed] [Google Scholar]
- 10.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63(6):1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 11.Momand J, Wu HH, Dasgupta G. MDM2–master regulator of the p53 tumor suppressor protein. Gene. 2000;242(1–2):15–29. doi: 10.1016/s0378-1119(99)00487-4. [DOI] [PubMed] [Google Scholar]
- 12.Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75(3):495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 13.Momand J, Jung D, Wilczynski S, Niland J. The MDM2 gene amplification database. Nucleic Acids Res. 1998;26(15):3453–3459. doi: 10.1093/nar/26.15.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 15.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69(7):1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 16.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408(6810):307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 17.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8(4):275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 18.Prives C, Hall PA. The p53 pathway. J Pathol. 1999;187(1):112–126. doi: 10.1002/(SICI)1096-9896(199901)187:1<112::AID-PATH250>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 19.Oren M. Decision making by p53: Life, death and cancer. Cell Death Differ. 2003;10(4):431–442. doi: 10.1038/sj.cdd.4401183. [DOI] [PubMed] [Google Scholar]
- 20.Sherr CJ. Principles of tumor suppression. Cell. 2004;116:235–246. doi: 10.1016/s0092-8674(03)01075-4. [DOI] [PubMed] [Google Scholar]
- 21.Laptenko O, Prives C. Transcriptional regulation by p53: One protein, many possibilities. Cell Death Differ. 2006;13(6):951–961. doi: 10.1038/sj.cdd.4401916. [DOI] [PubMed] [Google Scholar]
- 22.El-Deiry WS. Regulation of p53 downstream genes. Semin Cancer Biol. 1998;8(5):345–357. doi: 10.1006/scbi.1998.0097. [DOI] [PubMed] [Google Scholar]
- 23.Nakamura Y. Isolation of p53-target genes and their functional analysis. Cancer Sci. 2004;95(1):7–11. doi: 10.1111/j.1349-7006.2004.tb03163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bykov VJ, et al. Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat Med. 2002;8(3):282–288. doi: 10.1038/nm0302-282. [DOI] [PubMed] [Google Scholar]
- 25.Bykov VJ, et al. Reactivation of mutant p53 and induction of apoptosis in human tumor cells by maleimide analogs. J Biol Chem. 2005;280(34):30384–30391. doi: 10.1074/jbc.M501664200. [DOI] [PubMed] [Google Scholar]
- 26.Foster BA, Coffey HA, Morin MJ, Rastinejad F. Pharmacological rescue of mutant p53 conformation and function. Science. 1999;286(5449):2507–2510. doi: 10.1126/science.286.5449.2507. [DOI] [PubMed] [Google Scholar]
- 27.Wiman KG. Strategies for therapeutic targeting of the p53 pathway in cancer. Cell Death Differ. 2006;13(6):921–926. doi: 10.1038/sj.cdd.4401921. [DOI] [PubMed] [Google Scholar]
- 28.Bischoff JR, et al. An adenovirus mutant that replicates selectively in p53-deficient human tumor cells. Science. 1996;274(5286):373–376. doi: 10.1126/science.274.5286.373. [DOI] [PubMed] [Google Scholar]
- 29.Nghiem P, Park PK, Kim Y, Vaziri C, Schreiber SL. ATR inhibition selectively sensitizes G1 checkpoint-deficient cells to lethal premature chromatin condensation. Proc Natl Acad Sci USA. 2001;98(16):9092–9097. doi: 10.1073/pnas.161281798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roberge M, et al. High-throughput assay for G2 checkpoint inhibitors and identification of the structurally novel compound isogranulatimide. Cancer Res. 1998;58(24):5701–5706. [PubMed] [Google Scholar]
- 31.Wang Q, et al. UCN-01: A potent abrogator of G2 checkpoint function in cancer cells with disrupted p53. J Natl Cancer Inst. 1996;88(14):956–965. doi: 10.1093/jnci/88.14.956. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y, et al. Radiosensitization of p53 mutant cells by PD0166285, a novel G(2) checkpoint abrogator. Cancer Res. 2001;61(22):8211–8217. [PubMed] [Google Scholar]
- 33.Kawabe T. G2 checkpoint abrogators as anticancer drugs. Mol Cancer Ther. 2004;3(4):513–519. [PubMed] [Google Scholar]
- 34.Dash BC, El-Deiry WS. Cell cycle checkpoint control mechanisms that can be disrupted in cancer. Methods Mol Biol. 2004;280:99–161. doi: 10.1385/1-59259-788-2:099. [DOI] [PubMed] [Google Scholar]
- 35.Wang S, El-Deiry WS. The p53 pathway: Targets for the development of novel cancer therapeutics. Cancer Treat Res. 2004;119:175–187. doi: 10.1007/1-4020-7847-1_9. [DOI] [PubMed] [Google Scholar]
- 36.Dar AA, Belkhiri A, Ecsedy J, Zaika A, El-Rifai W. Aurora kinase A inhibition leads to p73-dependent apoptosis in p53-deficient cancer cells. Cancer Res. 2008;68(21):8998–9004. doi: 10.1158/0008-5472.CAN-08-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grueneberg DA, et al. Kinase requirements in human cells: I. Comparing kinase requirements across various cell types. Proc Natl Acad Sci USA. 2008;105(43):16472–16477. doi: 10.1073/pnas.0808019105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phang BH, Sabapathy K. The codon 72 polymorphism-specific effects of human p53 are absent in mouse cells: Implications on generation of mouse models. Oncogene. 2007;26(21):2964–2974. doi: 10.1038/sj.onc.1210112. [DOI] [PubMed] [Google Scholar]
- 39.Tuveson DA, Jacks T. Modeling human lung cancer in mice: Similarities and shortcomings. Oncogene. 1999;18(38):5318–5324. doi: 10.1038/sj.onc.1203107. [DOI] [PubMed] [Google Scholar]
- 40.O'Farrell TJ, Ghosh P, Dobashi N, Sasaki CY, Longo DL. Comparison of the effect of mutant and wild-type p53 on global gene expression. Cancer Res. 2004;64(22):8199–8207. doi: 10.1158/0008-5472.CAN-03-3639. [DOI] [PubMed] [Google Scholar]
- 41.Scian MJ, et al. Modulation of gene expression by tumor-derived p53 mutants. Cancer Res. 2004;64(20):7447–7454. doi: 10.1158/0008-5472.CAN-04-1568. [DOI] [PubMed] [Google Scholar]
- 42.Weisz L, et al. Transactivation of the EGR1 gene contributes to mutant p53 gain of function. Cancer Res. 2004;64(22):8318–8327. doi: 10.1158/0008-5472.CAN-04-1145. [DOI] [PubMed] [Google Scholar]
- 43.Weinmann AS, Farnham PJ. Identification of unknown target genes of human transcription factors using chromatin immunoprecipitation. Methods. 2002;26(1):37–47. doi: 10.1016/S1046-2023(02)00006-3. [DOI] [PubMed] [Google Scholar]
- 44.Wright WE, Shay JW. Cellular senescence as a tumor-protection mechanism: The essential role of counting. Curr Opin Genet Dev. 2001;11(1):98–103. doi: 10.1016/s0959-437x(00)00163-5. [DOI] [PubMed] [Google Scholar]
- 45.El-Deiry WS, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75(4):817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 46.Vassilev LT, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303(5659):844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 47.Secchiero P, di Iasio MG, Gonelli A, Zauli G. The MDM2 inhibitor Nutlins as an innovative therapeutic tool for the treatment of haematological malignancies. Curr Pharm Des. 2008;14(21):2100–2110. doi: 10.2174/138161208785294663. [DOI] [PubMed] [Google Scholar]
- 48.Bunz F, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282(5393):1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 49.Bernhard EJ, Maity A, Muschel RJ, McKenna WG. Effects of ionizing radiation on cell cycle progression. A review. Radiat Environ Biophys. 1995;34(2):79–83. doi: 10.1007/BF01275210. [DOI] [PubMed] [Google Scholar]
- 50.Chang D, Chen F, Zhang F, McKay BC, Ljungman M. Dose-dependent effects of DNA-damaging agents on p53-mediated cell cycle arrest. Cell Growth Differ. 1999;10(3):155–162. [PubMed] [Google Scholar]
- 51.Mross K, et al. Phase I dose escalation and pharmacokinetic study of BI 2536, a novel polo-like kinase 1 inhibitor, in patients with advanced solid tumors. J Clin Oncol. 2008;26(34):5511–5517. doi: 10.1200/JCO.2008.16.1547. [DOI] [PubMed] [Google Scholar]
- 52.Ginsberg D, Mechta F, Yaniv M, Oren M. Wild-type p53 can down-modulate the activity of various promoters. Proc Natl Acad Sci USA. 1991;88(22):9979–9983. doi: 10.1073/pnas.88.22.9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.El-Deiry WS, et al. WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res. 1994;54(5):1169–1174. [PubMed] [Google Scholar]
- 54.Tenzer A, Pruschy M. Potentiation of DNA-damage-induced cytotoxicity by G2 checkpoint abrogators. Curr Med Chem Anticancer Agents. 2003;3(1):35–46. doi: 10.2174/1568011033353533. [DOI] [PubMed] [Google Scholar]
- 55.Zhou BB, Sausville EA. Drug discovery targeting Chk1 and Chk2 kinases. Prog Cell Cycle Res. 2003;5:413–421. [PubMed] [Google Scholar]
- 56.Lenart P, et al. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr Biol. 2007;17(4):304–315. doi: 10.1016/j.cub.2006.12.046. [DOI] [PubMed] [Google Scholar]
- 57.Plyte S, Musacchio A. PLK1 inhibitors: Setting the mitotic death trap. Curr Biol. 2007;17(8):R280–R283. doi: 10.1016/j.cub.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 58.Steegmaier M, et al. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr Biol. 2007;17(4):316–322. doi: 10.1016/j.cub.2006.12.037. [DOI] [PubMed] [Google Scholar]
- 59.Ashwell S, Zabludoff S. DNA damage detection and repair pathways—Recent advances with inhibitors of checkpoint kinases in cancer therapy. Clin Cancer Res. 2008;14(13):4032–4037. doi: 10.1158/1078-0432.CCR-07-5138. [DOI] [PubMed] [Google Scholar]
- 60.Mross KA, et al. Phase I dose escalation and pharmacokinetic study of BI 2536, a novel polo-like kinase 1 inhibitor, in patients with advanced solid tumors. J Clin Oncol. 2008;26:5511–5517. doi: 10.1200/JCO.2008.16.1547. [DOI] [PubMed] [Google Scholar]
- 61.Taylor S, Peters JM. Polo and Aurora kinases: Lessons derived from chemical biology. Curr Opin Cell Biol. 2008;20(1):77–84. doi: 10.1016/j.ceb.2007.11.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.