Abstract
Disaccharide modified glycopeptides with hydrophobic sidechains are active against vancomycin-resistant enterococci and vancomycin-resistant S. aureus. The activity depends on the length of the sidechain. The benzyl sidechain of N-(4-fluorobenzyl)vancomycin (FBV) has the minimal length sufficient for enhancement in activity against vancomycin-resistant pathogens. The conformation of FBV bound to the peptidoglycan in whole cells of S. aureus has been determined using rotational-echo double resonance NMR by measuring internuclear distances from the 19F of FBV to 13C and 15N labels incorporated into the cell-wall peptidoglycan. The hydrophobic sidechain and aglycon of FBV form a cleft around the pentaglycyl bridge. FBV binds heterogeneously to the peptidoglycan as a monomer with the 19F positioned near the middle of the pentaglycyl bridge, approximately 7 Å from the bridge-link. This differs from the situation for N-(4-(4-fluorophenyl)benzyl)vancomycin complexed to the peptidoglycan where the 19F is located at the end of pentaglycyl bridge, 7 Å from the cross-link.
Keywords: Dipolar coupling, glycopeptide antibiotic, magic-angle spinning, peptidoglycan, solid-state NMR, transglycosylase
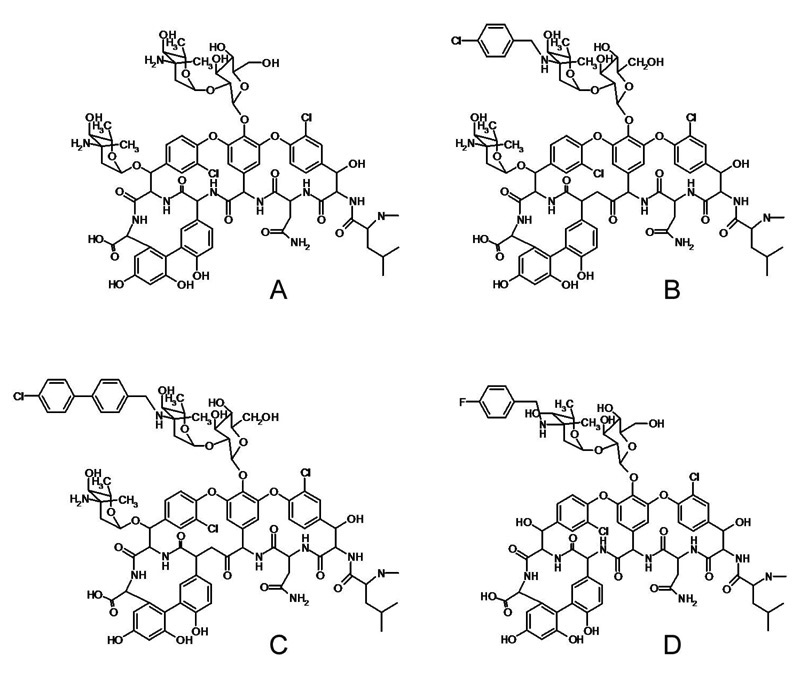
Structure-activity relationship studies of chloroeremomycin (Figure 1A) have revealed that the mono-alkylation of the vancosamine on the disaccharide positioned at the 4th amino acid of the glycopeptide aglycon with a hydrophobic moiety significantly increases drug antimicrobial activity against Gram-positive pathogens, including vancomycin-susceptible enterococci (VSE) and vancomycin-resistant enterococci (VRE) (1,2). Because the site of the hydrophobic sidechain is at the disaccharide, away from the D-Ala-D-Ala-binding cleft of the aglycon structure, the sidechain does not improve drug binding by interacting directly with the D-Ala-D-Ala or D-AlaD-Lac termini of peptidoglycan (PG) stem structures found in VSE or VRE, respectively. The antimicrobial activity of N-(4-chlorobenzyl)chloroeremomycin (LY191145), a chloroeremomycin derivative with p-chlorobenzyl sidechain shown in Figure 1B, has a minimum inhibitory concentration (MIC) of 16 µg/mL (3) against the Van-A type VRE, E. faecium. This drug is approximately 30 times more potent than vancomycin against VRE, yet the dissociation constant (Kd) of LY19145 for the complex with the tripeptide L-Lys-D-Ala-D-Lac (a VRE PG-mimic) is only 0.7 mM, compared with 2.4 mM for vancomycin (3). In general, disaccharide-modified glycopeptides that exhibit improvement in drug efficacies against VRE as high as a hundred fold, show no improvement in binding to PG-stem mimics of VRE (4).
Figure 1.
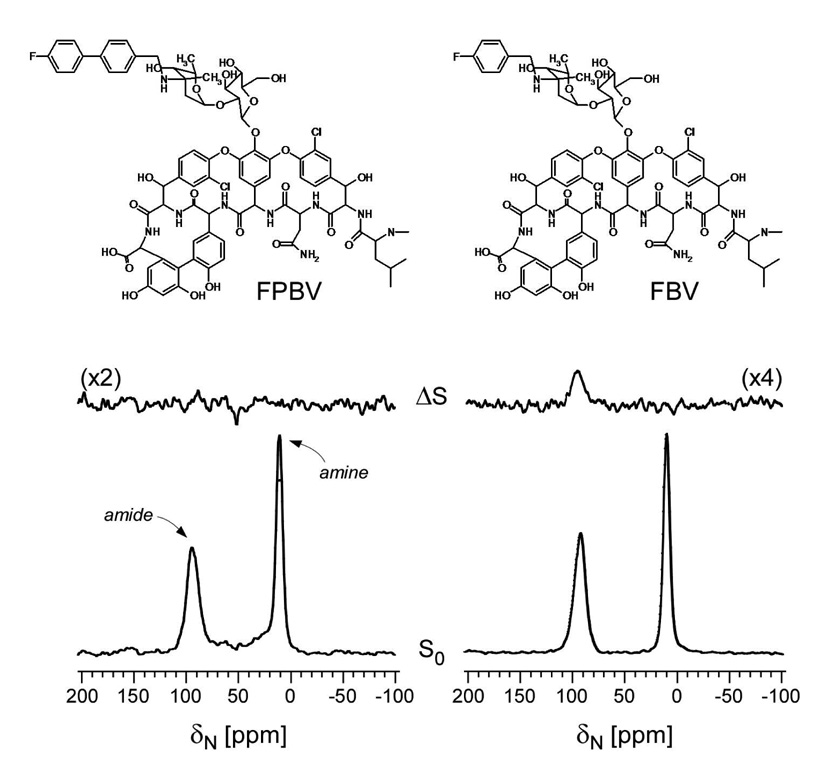
Chemical structures of (A) chloroeremomycin, (B) N-(4-chlorobenzyl)chloroeremomycin (LY191145), (C) N-(4-(4-chlorophenyl)benzyl)chloroeremomycin (oritavancin), and (D) N-(4-fluorobenzyl)vancomycin (FBV); the latter differs from chlorobenzyl-chloroeremomycin by the absence of 4-epi-vancosamine at the 6th amino acid position of aglycon structure and substitution of F for Cl in the hydrophobic sidechain. Minimal inhibitory concentrations against vancomycin-resistant E. faecium are (A) 32 µg/mL, (B) 16 µg/mL, and (C) 0.5 µg/mL.
Chloroeremomycin, LY191145, and N-(4-(4-chlorophenyl)benzyl)chloroeremomycin (oritavancin) (Figure 1C) have sidechain lengths of 0-, 5-, and 9 equivalent carbons (5), respectively, and MICs against vancomycin-resistant E. faecium (Van-A type) of 32, 16, and 0.5 µg/mL, respectively (3). In general, chloroeremomycin derivatives with sidechain lengths equivalent to 9–14 carbons exhibit maximum activity against enterococcal bacteria. We have previously characterized the homogeneous binding of disaccharide-modified chloroeremomycins with hydrophobic sidechain lengths equivalent to 7 and 9 carbons complexed to whole cells of S. aureus using solid-state NMR (6,7). In this report we characterize the in situ binding of fluorobenzyl-vancomycin FBV (Figure 1D) with a hydrophobic sidechain length equivalent to 5 carbons using rotational-echo double resonance (REDOR) NMR (8). The shorter length of the hydrophobic sidechain of FBV results in conformational heterogeneity in its cell-wall complex which is directly related to its reduced antimicrobial activity.
Material and Methods
Synthesis of N-(4-fluorobenzyl)vancomycin (FBV)
FBV was synthesized by reaction of the vancosamine sugar amine with 4-fluorbenzaldehyde, as described by Nagarajan et al (2). Briefly, 8.13 µL of 4-fluorobenzaldehyde (79 µmol) was added to 100 mg of vancomycin (66 µmol) dissolved in 200 mL of methanol. The reaction mixture was stirred under N2 gas at 65 C. After an hour, 5.4 mg of NaBH3CN was added. The reaction ran for two days with progress monitored by HPLC, using a C18 column, Zorbax SB-Aq (Agilent Technologies, Palo Alto, CA) 4.6 mm ID × 50 mm (100 Å particle size), with a flow rate of 1 mL/min. Triethylammonium phosphate buffer (pH 3.2) was the basis for two mobile phases. The retention time for vancomycin was 0.9 min, and for FBV, 2.6 min. The FBV was purified from the reaction mixture using a semipreparative reverse phase C18 column, Microsorb C18 (Varian, Palo Alto, CA) 21.4 ID × 250 mm (100 Å particle size), at a flow rate of 10 mL/min. The fraction containing FBV was concentrated using a rotary evaporator and then lyophilized. The MS-MALDI calculated mass for FBV, C73H80Cl2FN9O24, [M + H+] was 1557.4 and the found mass was 1557.0.
Complexes of FBV to Whole Cells of S. aureus
FBV was complexed to whole cells of S. aureus that had been harvested at the end of exponential growth or during stationary growth; that is, the drug was added after not during bacterial growth. The detailed protocols for the defined medium (SASM) for growing S. aureus (ATCC 6538P) and preparing glycopeptides complexed to whole cells of S. aureus are described elsewhere (9). In one sample, 6.0 mg of FBV was added to whole cells grown in SASM containing natural-abundance L-form amino acids (0.1 g/L), with the addition of D-[1-13C]alanine (0.1 g/L) and alanine racemase inhibitor alaphosphin (5 µg/mL). The estimated (6) glycopeptide binding occupancy in the cell wall was 68%. Three samples of FBV-whole-cell complexes were prepared from S. aureus grown in SASM in which the natural abundance glycine and L-lysine were replaced by [1-13C]glycine and L-[ε-15N]lysine. FBV was complexed to whole cells harvested after 7, 8 and 33 hours of growth, with FBV binding-site occupancies of 70, 50 and 62%, respectively. The cells harvested at 7 hours of growth were at the end of the exponential-growth phase, whereas cells harvested after 8 and 33 hours were in stationary-growth phase. The FBV-whole cell complexes were formed by addition of FBV to the harvested whole cells suspended in 15 mL of 40 mM triethanolamine buffer (pH 7.0). The FBV whole-cell complexes were incubated on ice for 5 min, followed by flash freezing and lyophilization. The triethanolamine buffer served as a cryoprotectant and lyoprotectant during the lyophilization. The lyophilized FBV-whole cell complexes were compressed and packed into zirconia rotors for solid-state NMR experiments.
Dipolar Recoupling
Rotational-echo double resonance (REDOR), a solid-state NMR method that recouples dipolar interactions under magic-angle spinning, (8) was used to determine heteronuclear dipolar couplings and hence inter-nuclear distances. REDOR is a difference experiment in which two spectra are collected, one in the absence of heteronuclear dipolar coupling (full echo, S 0 spectrum), and the other in the presence of the coupling (dephased echo, S spectrum). In the S 0 spectrum, dipolar dephasing is refocused over a single rotor period due to the spatial averaging by the motion of the rotor under magic-angle spinning. In the S spectrum, the spin part of the dipolar interaction was manipulated by the application of rotor-synchronized dephasing π-pulses to prevent full refocusing. Dipolar evolution over the rotor period in the S spectrum results in a reduced peak intensity for spin pairs that are dipolar coupled. The difference in signal intensity (REDOR difference, ΔS = S 0 - S) for the observed spin (13C or 15N) in the two parts of the REDOR experiment is directly related to the heteronuclear dipolar coupling from which the corresponding distance to the dephasing spin (19F) is determined.
Solid-state NMR Spectrometer
REDOR was performed using a transmission-line probe (10) with a 17-mm long, 8.6-mm inside-diameter analytical coil and a Chemagnetics/Varian ceramic stator (Fort Collins, CO/Palo Alto, CA). Four radiofrequencies, proton (200 MHz), fluorine (188 MHz), carbon (50.3 MHz), and nitrogen (20.3 MHz) were mutually isolated by the positions of their tuning circuits along the transmission line, and simultaneously tuned to the single solonoidal coil of the probe. Lyophilized whole-cell samples were contained in Chemagnetics/Varian 7.5-mm outside-diameter zirconia rotors. The rotors were spun at 5000 Hz with the speed under active control to within ±2 Hz. A Tecmag (Houston, TX) pulse programmer controlled the spectrometer. Power amplifiers for each radio frequency were under active control to eliminate long-term drifts in the performance of the spectrometer due to component aging and minor changes in the temperature of the room, spinning gas, and the amplifiers (11). The S and S 0 alternate-scan strategy compensated for short-term drifts. The π-pulse lengths were 10 µs for 19F, 13C, and 15N. Standard XY-8 phase cycling (12) was used for all refocusing and dephasing pulses. An 89-mm bore Oxford (Cambridge, England) superconducting solenoid provided a 4.7-T static magnetic field. Proton-carbon matched cross-polarization transfers were made in 2 ms at 50 kHz. The single-frequency proton dipolar decoupling was 98 kHz throughout dipolar evolution and data acquisition.
Calculated REDOR Dephasing
REDOR dephasing was calculated using the modified Bessel function expressions given by Mueller et al. (13) and de la Caillerie and Fretigny (14) for a spin-½ pair. For FBV 19F coupling to D-[1-13C]Ala, the parameters (mean and width) describing the 19F-13C distance distribution were allowed to vary to minimize the root-mean-square deviation (RMSD) between the experimental and calculated dephasing (6,15). For all possible 19F positions of FBV relative to a model pentaglycyl bridge, the RMSD values were calculated using an error function (χ2[x,y,z,j]) for coordinate space at 0.2 Å resolution.
where Rexp(j) is the experimental 13C{19F}REDOR dephasing (ΔS/S0) for the tjth dephasing time, and Rcalc(x,y,z,j) is the calculated dephasing for 19F positioned at (x, y, z) and the tjth dephasing time. The Rcalc(x,y,z,j) function was normalized to account for the five carbonyl carbons in the pentaglycyl bridge that are dipolar coupled to the fluorine,
where ωD(i) is the dipolar coupling for the 19F of FBV to the ith carbonyl carbon of the pentaglycyl bridging segment with carbonyl-carbon coordinates, (xi, yi, zi). The pentaglycyl α-helix was assembled and energy minimized using Insight II (MSI, San Diego, CA) (6).
Molecular Modeling
For the dephasing calculation of the bridge labeled by [1-13C]glycine, a five-glycyl α-helix was assembled. The grid position of a single 19F label relative to the pentaglycyl helix was varied, and the RMSD was minimized between experimental and calculated dephasing.
Results
FBV Complexed to D-[1-13C]Alanine-Labeled Whole Cells
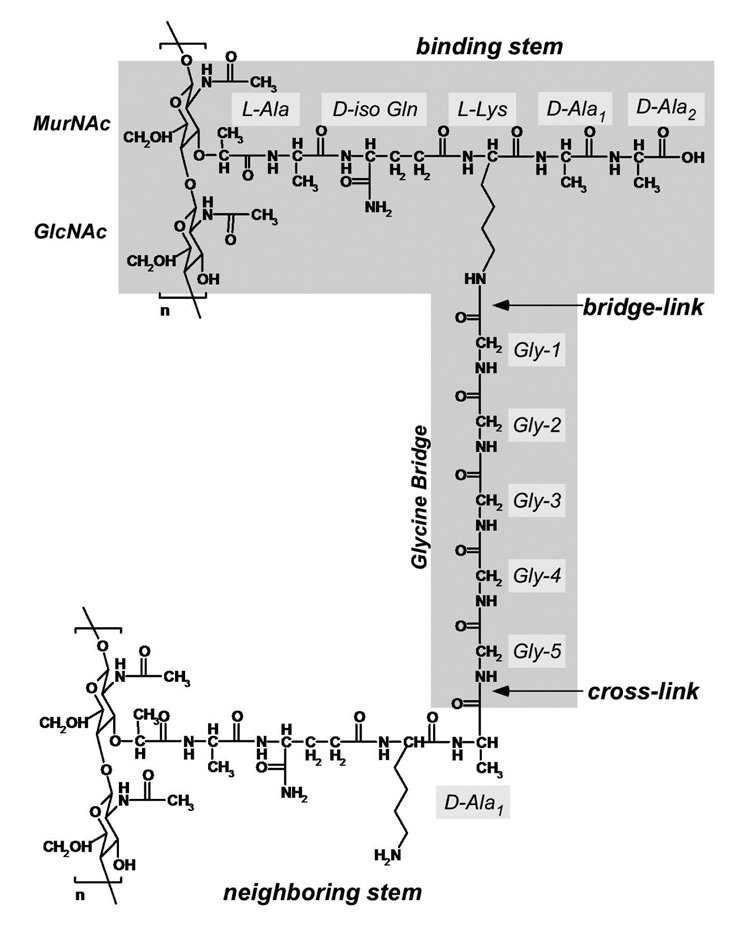
The 13C{19F} REDOR spectra of FBV complexed to whole cells of S. aureus grown in medium containing D-[1-13C]alanine and alanine racemase inhibitor have a REDOR difference at 175 ppm (not shown). Because FBV does not cross the membrane (16), the ΔS at 175 ppm is exclusively from the carbonyl carbons of D-Ala-1 at cross-links (Figure 2) that are dipolar coupled to the 19F of FBV bound in the cell wall. The 175-ppm full-echo peak consists of contributions from the D-[1-13C]alanine incorporated into Park’s Nucleotide (a cytoplasmic PG precursor), teichoic acids, and the PG itself. (The racemase inhibitor prevents incorporation of label in cytoplasmic proteins.) These three contributions are not resolved and are referred to together as the 175-ppm REDOR full echo (S0). They can be separated experimentally by deconvolution using REDOR methods (17) but that was not attempted here.
Figure 2.
Chemical structure of S. aureus peptidoglycan (PG). The uncross-linked PG-stem structure consists of five amino acids, L-Ala, D-iso-Gln, L-Lys, D-Ala, and D-Ala. The PG-bridge structure consists of five glycine residues in a compact helical conformation. The pentaglycyl bridging segment is attached to the ε-nitrogen of L-Lys of the PG stem to form the bridge-link. The cross-link is a peptide bond between the C-terminus of the D-Ala of the 4th amino acid on one stem to the N-terminus of the pentaglycyl bridging segment of the adjacent PG stem. The terminal D-Ala, the 5th amino acid of the PG-stem, is cleaved upon formation of the cross-link.
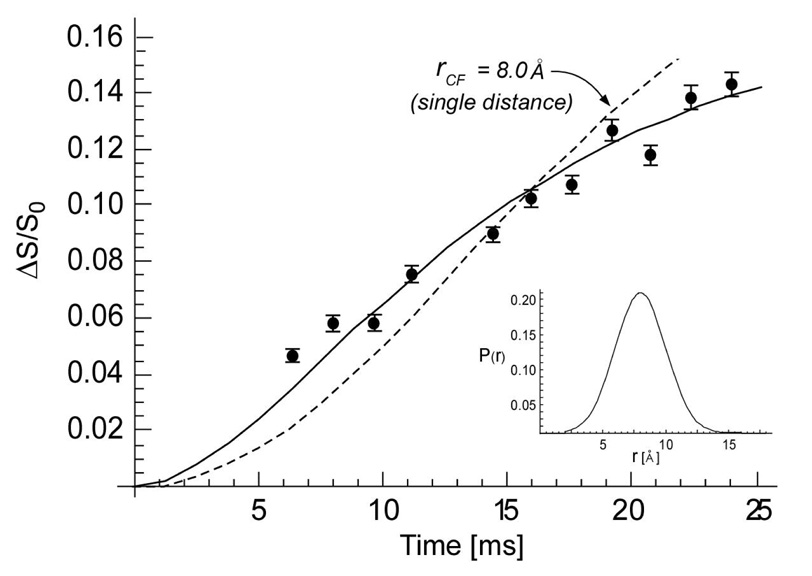
The combined 175-ppm13C{19F} REDOR dephasing (Δ S/S0) for dipolar evolution times between 5 and 25 msec is represented by solid circles in Figure 3. The solid line is the calculated 13C{19F} dephasing fit to the data assuming a distribution of 13C-19F distances (inset) and an asymptotic dephasing limit. The calculated dephasing for a single distance was not a good match to experiment (dotted line). The average 13C-19F distance of the distribution is 8.0 Å. This is the distance from the 19F of FBV to the cross-link of an adjacent stem. Intramolecular 13C-19F distances from the D-Ala-D-Ala bound to the FBV aglycon exceed 12 Å (7) and result in negligible contributions to the dephasing.
Figure 3.
13C{19F} REDOR dephasing (ΔS/S0) of the D-Ala carbonyl-carbon peak at 175 ppm for a complex of FBV and whole cells of S. aureus grown on media containing D-[1-13C]alanine in the presence of an alanine racemase inhibitor. Each closed circle resulted from the accumulation of 40,000 scans. The solid line is the calculated dephasing assuming the distribution of 13C-19F distances shown in the inset and a total dephasing of 17%. The average 13C-19F distance is 8.0 Å.
The asymptotic limit for the dephasing from the fit is 17%. Estimation of an asymptotic dephasing limit in whole cells of S. aureus is calculated as follows. The expected ΔS is equal to the drug D-Ala-D-Ala primary binding-site occupancy (68%) times the relative number (46%) of PG-stems having an uncross-linked D-Ala-D-Ala terminus (18). As described above, the full echo (S0) has contributions from D-alanine of cross-linked stems (54%), uncross-linked stems (46%), and teichoic acids. The ratio of D-alanine in teichoic acid to D-alanine in cross-linked stems is sensitive to the growth conditions and can vary from 1.0 to 2.0 (18). Assuming an average ratio of D-alanine in teichoic acid to D-alanine in cross-linked stems of 1.5, the estimated ΔS/S0 = (0.68)(0.46)/[(0.54)(1) + (0.46)(2) + (0.54)(1.5)] = 17%, in agreement with the observed dephasing limit.
FBV Complexed to [1-13C]Glycine-Labeled Whole Cells
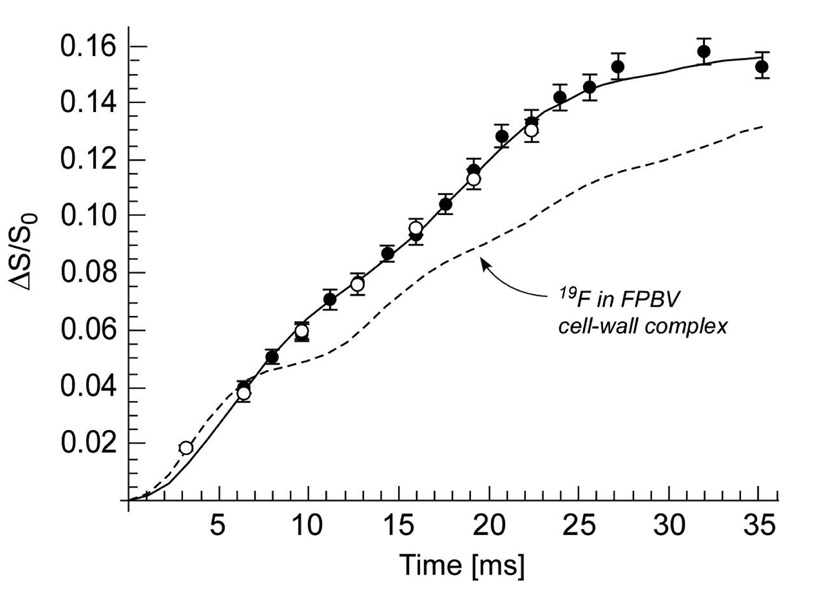
The 13C{19F} REDOR spectra of FBV complexed to whole cells of S. aureus grown in medium containing [1-13C]glycine have a REDOR difference for the carbonyl glycyl-carbon resonance at 172 ppm (spectra not shown). The full-echo REDOR signal is due to equal contributions from glycine labels in cell-wall pentaglycyl bridging segments and in cytoplasmic proteins (6), but the REDOR difference arises from just the cell-wall component. The open circles (Figure 4) represent the 13C{19F} dephasing for FBV complexed to whole cells harvested after 7 hours of growth with a binding-site occupancy of 63%, while the closed circles show the dephasing (scaled by 0.9) for FBV complexed to whole cells harvested after 33 hours of growth with a drug binding-site occupancy of 70%. The 13C{19F} dephasing for the two samples is superimposable.
Figure 4.
13C{19F} REDOR dephasing (ΔS/S0) of the Gly carbonyl-carbon peak at 172 ppm for complexes of FBV to whole cells of S. aureus grown on media containing [1-13C]glycine. The open circles represent the dephasing from whole cells harvested after 7 hours of growth and complexed to FBV with a primary binding-site occupancy of 63%. The closed circles represent the dephasing for whole cells harvested after 33 hours of growth and complexed to FBV with a primary binding-site occupancy of 70%. The latter dephasing has been scaled by a factor of 0.9 to account for the difference in binding-site occupancy. The solid line is the calculated dephasing assuming the five 13C-19F distances shown in Figure 5, with the 19F of bound FBV near the middle of pentaglycyl helix. The dotted line is the calculated dephasing that best matches experiment (reference 7) for the 19F in a FPBV-PG cell-wall complex.
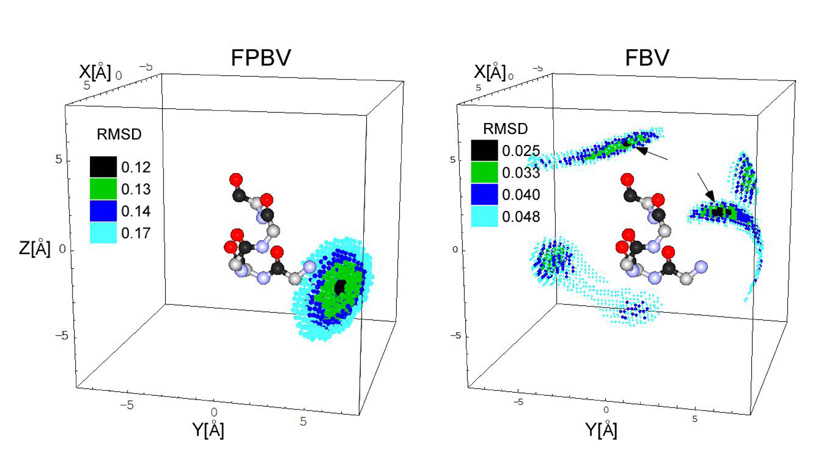
The calculated 13C{19F} dephasing (Figure 4, solid line) is in agreement with experiment, assuming the best-fit 19F positions of FBV relative to the five carbonyl carbons of the pentaglycyl bridging segment in a compact helical conformation (Figure 5, black spheres). Based on the center of the loose grouping of black dots near the middle of the helix (arrow), the distances from the 19F to the carbonyl carbons of Gly-1, Gly-2, Gly-3, Gly-4, and Gly-5 are 5.6, 5.7, 9.4, 8.0, and 8.4 Å respectively. The 19F of FBV is mostly but not exclusively near the middle of the pentaglycyl bridge structure (Figure 5, right, upper arrow). By contrast, the 19F position of N-(4-(4-fluorophenyl)benzyl)vancomycin (FPBV) (with a hydrophobic sidechain length equivalent to 9 carbons compared to 4 carbons for FBV) bound to S. aureus PG is positioned along the helical axis of the pentaglycyl bridge structure near the cross-link (Figure 5, left, tight grouping of black dots).
Figure 5.
Possible positions of fluorine relative to the bridging pentaglycyl helix in peptidoglycan complexes of FPBV and FBV. The pentaglycyl bridge is shown in an alpha-helical conformation with carbonyl carbons in black, alpha carbons in gray, nitrogens in blue, and oxygens in red. The positions that are consistent with the 13C{19F} REDOR dephasing are indicated by small spheres whose colors indicate the root-mean-square deviation between calculated and experimental dephasing. The position of the fluorine of FPBV bound to the peptidoglycan is restricted to the end of the helix near the cross-link (black dots, left panel). However, the calculated 19F positions for the FBV-PG complex (Figure 5, right) show multiple positions that satisfy the 13C{19F} REDOR dephasing of Figure 4. Two of these positions with the lowest RMSD values are indicated by the arrows in Figure 5 (right); one is near the bridge-link and the other is closer to the middle of bridge. Both positions are proximate to the lysyl amide of L-[ε-15N]lysine at the bridge-link, and so are consistent with the observed 15N{19F} REDOR dephasing of Figure 6 (right).
The observed asymptoptic dephasing of 16% is in agreement with the calculated maximum dephasing of 14%, an estimate based on the product of three factors: (i) the binding-site occupancy (63%), (ii) the fraction of PG-stem structures that terminate in D-Ala-D-Ala (46%), and (iii) the fraction of the cell-wall contribution to the 172-ppm peak in the S0 spectrum (50%). The agreement between observed and calculated asymptotic dephasing limits accounts for all of the complexed 19F and proves that FBV binds to the cell-wall PG as a monomer (6,7).
FBV and FPBV Complexed to [ε-15N]Lysine-Labeled Whole Cells
The 15N{19F} REDOR spectra after 19.2 msec of dipolar evolution (96 Tr) of FBV and FPBV complexed to whole cells of S. aureus grown in defined medium containing L-[ε-15N]lysine are shown in Figure 6. The [ε-15N]lysyl amide peak at 95 ppm in the full-echo spectra is exclusively from the Gly-5-Lys bridge in PG (Figure 2), whereas the [ε-15N]lysyl amine peak at 10 ppm is from the [ε-15N]lysyl amine in Park’s Nucleotide, non-bridge-linked PG, and cytoplasmic proteins (6,9,19). The FBV-PG complex shows 15N{19F} REDOR dephasing of the amide peak (Figure 6, right). Based on an estimated dephasing maximum of 24%, which is the product of binding-site occupancy (62%), the fraction of PG-stems with pentaglycyl bridges attached (85%), and the fraction of PG stems that end in D-Ala-D-Ala (46%), the observed dephasing of 7% means that the fluorine of FBV is about 7.0 Å from the [ε-15N]lysine of the bridge link. This result is consistent with a position near the middle of the pentaglycyl bridge (Figure 5, right panel). For the FPBV-PG complex, no 15N{19F} dephasing is observed (Figure 6, left), which means that the distance from the 19F of the drugs to the [ε-15N]lysyl amide nitrogen of the bridge-link exceeds 9 Å.
Figure 6.
15N{19F} REDOR spectra after 19.2 msec of dipolar evolution (96 Tr) of FBV (right) and FPBV (left) complexed to whole cells of S. aureus grown in defined medium containing L-[ε-15N]lysine. The full-echo spectra are at the bottom of the figure and the corresponding difference spectra at the top. The spectra are each the result of the accumulation of 80,000 scans.
Discussion
Conformation of the FBV-PG Complex
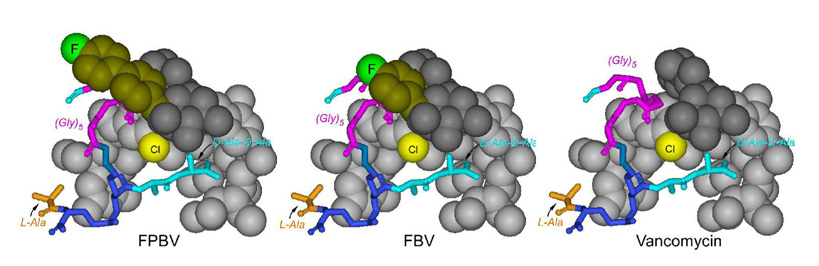
The conformation of the FBV-PG complex is inferred from the proximity of the 19F of FBV to the pentaglycyl-bridge glycines, the bridge-link lysine, and to the PG-stem alanine. This combination positions the 19F near the middle of the pentaglycyl-bridge (Figure 5, right panel), although with a substantial uncertainty. A mid-bridge 19F position for the FBV-PG complex differs from the 19F positions determined for N-(4-(4-fluorophenyl)benzyl)chloroeremomycin ([19F]oritavancin) and FPBV complexed to the PG of S. aureus. For those complexes, the 19F was found along the α-helical axis of the pentaglycyl-bridge (6,7) near the cross-link and distant from the bridge-link (for example, see Figure 5, left panel). Thus, the sidechain of FBV when bound to PG makes less contact with the cross-linked pentaglycyl bridge structure than do the sidechains of [19F]oritavancin and FPBV (Figure 7). This results in FBV-PG conformational heterogeneity and a broad distribution of 19F to D-[1-13C]alanyl distances (Figure 3). By contrast, for the [19F]oritavancin-PG complex, the 13C-19F dephasing was fit to a single distance of 7.6 Å (6). We conclude that the steric match between the hydrophobic sidechain of the drug and the pentaglycyl bridge of the PG is key to homogeneous tight binding and high antimicrobial activity.
Figure 7.
Molecular models of FPBV-PG (left), FBV-PG (middle), and vancomycin-PG (right) cell-wall complexes consistent with the REDOR results of Figure 3–Figure 5.
The Secondary Binding Site in S. aureus Peptidoglycan
We have proposed before that disaccharide-modified lipoglycopeptides like FPBV and oritavancin target a secondary binding site in the cell wall of S. aureus (17,20). The cleft formed between the hydrophobic sidechain and the aglycon structure of the lipoglycopeptides (see Figure 8, bottom left) binds the pentaglycyl bridge of an uncross-linked stem in the cell wall of S. aureus (7). This is a new function for the hydrophobic sidechains of lipoglycopeptides, previously thought to mediate the formation of drug dimers or membrane anchors at the bacterial membrane (21). However, in situ solid-state NMR characterization of binding in S. aureus has shown that lipoglycopeptides with sidechain lengths equivalent to 5- to 9 carbons do not form dimers or membrane anchors (6,7). Even FBV, with a sidechain length equivalent to only 4 carbons, binds to the cell-wall PG as a monomer (a conclusion based on the dephasing asymptotic limits of Figure 3 and Figure 4).
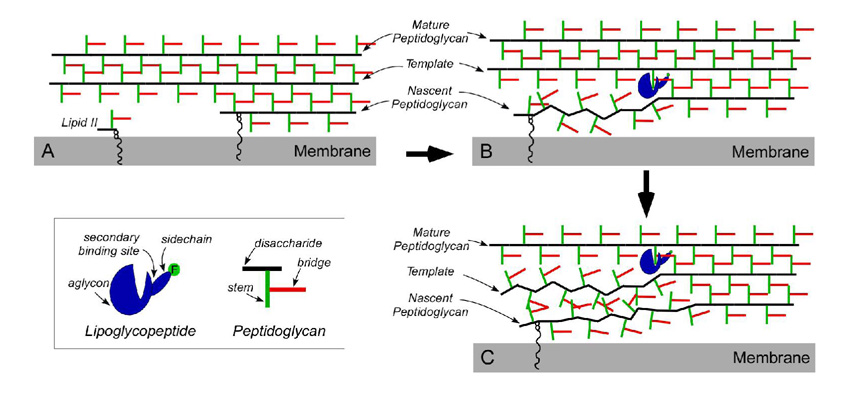
Figure 8.
Schematic representation of the effect on cell-wall biosynthesis (A) of lipoglycopeptide binding to the mature peptidoglycan template (nearest strand to the membrane surface not connected to lipid II) (B), resulting in reductions in cross-linking from propagation of defective templates (C).
Recently we have confirmed the targeting of secondary binding sites in des-N-methylleucyl-FPBV (20) and des-N-methylleucyl-[19F]oritavancin when complexed to whole cells of S. aureus (17). These drugs are Edman degradation products of FPBV and [19F]oritavancin, and each has a damaged D-Ala-D-Ala binding pocket. Despite the loss of the primary binding site, both hexapeptides are highly active antimicrobials and were found to bind to an uncross-linked stem of the PG with a pentaglycyl bridging segment inserted between the hydrophobic sidechain and aglycon wall of the drug (17,20). We believe that FBV has the minimum sidechain length required for such a secondary binding site, although the conformational freedom of the sidechain about the cross-linked bridge indicates a poor steric match, relatively weak binding, and only modest antimicrobial activity.
The Mode of Action of Disaccharide-Modified Glycopeptides
Like vancomycin, disaccharide modified glycopeptides, including FBV, FPBV and oritavancin, target lipid II thereby inhibiting transglycosylase activity (17,19). However, unlike the situation for vancomycin, the secondary binding site in the disaccharide-modified glycopeptides enables these drugs to bind tightly to nascent and mature peptidoglycan. This binding interferes with both transglycosylase and transpeptidase activity (17). The peptidoglycan biosynthesis in S. aureus is fast with the entire cell wall replicated during a cell doubling time of 50 min. This means tight coordination between transglycosylase and transpeptidase activity. The transpeptidases carry out maturation of nascent PG (membrane-associated PG anchored by the lipid transporter C55, Figure 8A) by cross-linking to the surrounding mature PG, in synchronization with PG chain extension by the transglycosylases. To incorporate nascent PG quickly and efficiently into mature PG, we believe that existing mature cell wall serves as a template (17,20). A drug targeting a PG template interferes with recognition (Figure 8B) and therefore decouples transglycosylase and transpeptidase functions. The result is inaccurate future PG templates as the nascent PG is unable to mature correctly. The under-cross-linked PG is then propagated as the new template for subsequent growing layers of nascent PG (Figure 8C), with resultant reductions in cross-linking even in the absence of drug.
Abbreviations
- C55
pyrophosphoryl-undecaprenol
- 13C{19F}
carbon channel observation with fluorine dephasing
- ECU
equivalent carbon units
- [19F]oritavancin
N-(4-(4-fluorophenyl)benzyl)chloroeremomycin
- FBV
N-(4-fluorobenzyl)vancomycin
- FPBV
N-(4-(4-fluorophenyl)benzyl)vancomycin
- lipid II
N-acetylglucosamine-N-acetyl-muramyl-pentapeptidepyrophosphoryl-undecaprenol
- LY191145
N-(4-chlorobenzyl)chloroeremomycin
- MIC
minimum inhibitory concentration
- 15N{19F}
nitrogen channel observation with fluorine dephasing
- oritavancin
N-(4-(4-chlorophenyl)benzyl)chloroeremomycin
- PG
peptidoglycan
- REDOR
rotational-echo double resonance
- SASM
S. aureus standard media
- VRE
vancomycin-resistant enterococci
- VSE
vancomycin-susceptible enterococci
Footnotes
This paper is based on work supported by the National Institutes of Health grant number EB002058.
References
- 1.Nagarajan R. Structure-activity relationships of vancomycin-type glycopeptide antibiotics. J. Antibiot. 1993;46:1181–1195. doi: 10.7164/antibiotics.46.1181. [DOI] [PubMed] [Google Scholar]
- 2.Nagarajan R, Schabel AA, Occolowitz JL, Counter FT, Ott JL, Felty-Duckworth AM. Synthesis and antibacterial evaluation of N-alkyl vancomycins. J. Antibiot. 1989;42:63–72. doi: 10.7164/antibiotics.42.63. [DOI] [PubMed] [Google Scholar]
- 3.Allen NE, LeTourneau DL, Hobbs JN., Jr. Molecular interactions of a semisynthetic glycopeptide antibiotic with D-alanyl-D-alanine and D-alanyl-D-lactate residues. Antimicrob. Agents Chemother. 1997;41:66–71. doi: 10.1128/aac.41.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen NE, LeTourneau DL, Hobbs JN., Jr. The role of hydrophobic side chains as determinants of antibacterial activity of semisynthetic glycopeptide antibiotics. J. Antibiot. 1997;50:677–684. doi: 10.7164/antibiotics.50.677. [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez MJ, Snyder NJ, Zweifel MJ, Wilkie SC, Stack DR, Cooper RD, Nicas TI, Mullen DL, Butler TF, Thompson RC. Novel glycopeptide antibiotics: N-alkylated derivatives active against vancomycin-resistant enterococci. J. Antibiot. 1998;51:560–569. doi: 10.7164/antibiotics.51.560. [DOI] [PubMed] [Google Scholar]
- 6.Kim SJ, Cegelski L, Studelska DR, O'Connor RD, Mehta AK, Schaefer J. Rotational-echo double resonance characterization of vancomycin binding sites in Staphylococcus aureus. Biochemistry. 2002;41:6967–6977. doi: 10.1021/bi0121407. [DOI] [PubMed] [Google Scholar]
- 7.Kim SJ, Cegelski L, Preobrazhenskaya M, Schaefer J. Structures of Staphylococcus aureus cell-wall complexes with vancomycin, eremomycin, and chloroeremomycin derivatives by 13C{19F} and 15N{19F} rotational-echo double resonance. Biochemistry. 2006;45:5235–5250. doi: 10.1021/bi052660s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gullion T, Schaefer J. Detection of weak heteronuclear dipolar coupling by rotational-echo double-resonance nuclear magnetic resonance. Adv. Magn. Reson. 1989;13:57–83. [Google Scholar]
- 9.Tong G, Pan Y, Dong H, Pryor R, Wilson GE, Schaefer J. Structure and dynamics of pentaglycyl bridges in the cell walls of Staphylococcus aureus by 13C-15N REDOR NMR. Biochemistry. 1997;36:9859–9866. doi: 10.1021/bi970495d. [DOI] [PubMed] [Google Scholar]
- 10.Schaefer J, McKay RA. Multi-tuned single coil transmission line probe for nuclear magnetic resonance spectrometer. U.S. Patent. 1999;5:861, 748.
- 11.Stueber D, Mehta AK, Chen Z, Wooley KL, Schaefer J. Local order in polycarbonate glasses by 13C{19F} Rotational-Echo Double-Resonance NMR. J. Polym. Sci., Part B: Polym. Phys. 2006;44:2760–2775. [Google Scholar]
- 12.Gullion T, Baker DB, Conradi MS. New, compensated Carr-Purcell sequences. J. Magn. Reson. 1990;89:479–484. [Google Scholar]
- 13.Mueller KT, Jarvie TP, Aurentz DJ, Roberts BW. The REDOR transform: direct calculation of internuclear couplings from dipolar-dephasing NMR data. Chem. Phys. Lett. 1995;242:535–542. [Google Scholar]
- 14.de la Caillerie J-BdE, Fretigny C. Analysis of the REDOR signal and inversion. J. Magn. Reson. 1998;133:273–280. doi: 10.1006/jmre.1998.1462. [DOI] [PubMed] [Google Scholar]
- 15.O'Connor RD, Schaefer J. Relative CSA-dipolar orientation from REDOR sidebands. J. Magn. Reson. 2002;154:46–52. doi: 10.1006/jmre.2001.2461. [DOI] [PubMed] [Google Scholar]
- 16.Perkins HR, Nieto M. The preparation of iodinated vancomycin and its distribution in bacteria treated with the antibiotic. Biochem. J. 1970;116:83–92. doi: 10.1042/bj1160083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim SJ, Cegelski L, Stueber D, Singh M, Dietrich E, Tanaka KS, Parr TR, Far AR, Schaefer J. Oritavancin exhibits dual mode of action to inhibit cell-wall biosynthesis in Staphylococcus aureus. J. Mol. Biol. 2008;377:281–293. doi: 10.1016/j.jmb.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cegelski L, Steuber D, Mehta AK, Kulp DW, Axelsen PH, Schaefer J. Conformational and quantitative characterization of oritavancin-peptidoglycan complexes in whole cells of Staphylococcus aureus by in vivo 13C and 15N labeling. J. Mol. Biol. 2006;357:1253–1262. doi: 10.1016/j.jmb.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 19.Cegelski L, Kim SJ, Hing AW, Studelska DR, O'Connor RD, Mehta AK, Schaefer J. Rotational-echo double resonance characterization of the effects of vancomycin on cell wall synthesis in Staphylococcus aureus. Biochemistry. 2002;41:13053–13058. doi: 10.1021/bi0202326. [DOI] [PubMed] [Google Scholar]
- 20.Kim SJ, Matsuoka S, Patti GJ, Schaefer J. Vancomycin derivative with damaged D-Ala-D-Ala binding cleft binds to cross-linked peptidoglycan in the cell wall of Staphylococcus aureus. Biochemistry. 2008;47:3822–3831. doi: 10.1021/bi702232a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beauregard DA, Williams DH, Gwynn MN, Knowles DJ. Dimerization and membrane anchors in extracellular targeting of vancomycin group antibiotics. Antimicrob. Agents Chemother. 1995;39:781–785. doi: 10.1128/AAC.39.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]