Abstract
How oncogenes modulate the self-renewal properties of cancer-initiating cells is incompletely understood. Activating KRAS and NRAS mutations are among the most common oncogenic lesions detected in human cancer, and occur in myeloproliferative disorders (MPDs) and leukemias. We investigated the effects of expressing oncogenic KrasG12D from its endogenous locus on the proliferation and tumor-initiating properties of murine hematopoietic stem and progenitor cells. MPD could be initiated by KrasG12D expression in a highly restricted population enriched for hematopoietic stem cells (HSCs), but not in common myeloid progenitors. KrasG12D HSCs demonstrated a marked in vivo competitive advantage over wild-type cells. KrasG12D expression also increased the fraction of proliferating HSCs and reduced the overall size of this compartment. Transplanted KrasG12D HSCs efficiently initiated acute T-lineage leukemia/lymphoma, which was associated with secondary Notch1 mutations in thymocytes. We conclude that MPD-initiating activity is restricted to the HSC compartment in KrasG12D mice, and that distinct self-renewing populations with cooperating mutations emerge during cancer progression.
Author Summary
Ras proteins act as molecular switches that relay growth signals from outside the cell. This mechanism is often subverted in cancer, and Ras proteins are activated directly by RAS gene mutations in approximately one-third of human malignancies. We have modeled this in mice engineered to have a Ras mutation. These mice develop a disease similar to chronic leukemias in humans called myeloproliferative disorders. It is marked by a fatal accumulation of mature and immature cells in the blood and bone marrow. We investigated whether some or all of these neoplastic cells were immortal. In agreement with the “cancer stem cell” hypothesis, we found that immortal cells were extremely rare in the bone marrow of diseased mice. They were found only in the same cell populations that contain normal bone marrow stem cells. However, these cells had high rates of replication and produced large numbers of daughter cells. Furthermore, many mice went on to develop acute lymphoid leukemia after acquiring additional mutations in maturing lymphoid cells. These studies exemplify the evolution of malignant stem cells during cancer progression. They also highlight the importance of rare, long-lived cells in the genesis and, potentially, therapy of high-risk chronic leukemias caused by abnormal Ras proteins.
TheKrasG12D oncogene causes excessive proliferation of stem cells, promoting their preferential expansion and initiating myeloproliferative disease.KrasG12D does not alter self-renewal, but secondary mutations in lymphoid progenitors allow progression to acute leukemia.
Introduction
Self-renewal is integral to the malignant phenotype [1]. In principle, the ability of cancer cells to self-renew may be intrinsic to the compartment in which the tumor-initiating mutation occurs, or may be acquired as a consequence of mutations in more differentiated cells. The hematopoietic system has proven highly informative for addressing how cancer-associated mutations and cell of origin interact to establish malignant self-renewing populations. Accumulating evidence supports the idea that many hematopoietic malignancies exist in a hierarchy of differentiation with only a minor population capable of propagating and maintaining the disease in vivo [2]. These cells are termed leukemia-initiating cells or leukemia stem cells (LSCs), and manifest some biologic properties of normal hematopoietic stem cells (HSCs). However, the precise relationship between these populations is uncertain and appears to depend, in part, on both the leukemia subtype and on the effects of specific mutations. For example, overexpressing MLL fusion proteins found in human acute myeloid leukemia transforms both murine HSCs and more differentiated progenitors [3,4]. By contrast, inactivation of the JunB transcription factor must occur in the HSC compartment for initiation of myeloid malignancies [5]. These proof-of-concept experiments underscore the importance of understanding how oncogenes and tumor suppressors that are commonly mutated in human cancers perturb self-renewal and growth control. Importantly, the functional characteristics of LSCs that distinguish them from HSCs and how these properties are modulated by oncogenes are poorly understood.
RAS gene mutations are highly prevalent in pancreatic (>80%), colorectal (40%–50%), endometrial (40%), lung (30%), and cervical cancers (20%–30%), as well as in myeloid malignancies (20%–40%) [6]. Of the genes in the canonical RAS family, KRAS accounts for ∼90% of cancer-associated mutations, whereas HRAS mutations are rare. In hematologic cancers, NRAS is mutated 2–3 times more often than KRAS [6]. Cancer-associated RAS mutations, which introduce amino acid substitutions at codons 12, 13, or 61, result in oncogenic Ras proteins that accumulate in the active, GTP-bound conformation because of defective guanine nucleotide hydrolysis [7]. Elevated levels of GTP-bound Ras, in turn, deregulate signaling in cancer cells by altering the activation of effector cascades that include the Raf/MEK/ERK, phosphatidylinositol 3-kinase (PI3K)/Akt, and Ral-GDS pathways [8].
Chronic and juvenile myelomonocytic leukemias (CMML and JMML) are aggressive myeloid malignancies that are classified as myeloproliferative disorders (MPDs) [9]. Both diseases are characterized by leukocytosis with excess monocytes in blood and bone marrow, and by significant infiltration of malignant myeloid cells into the liver, spleen, and other organs. Hyperactive Ras is strongly implicated in the pathogenesis of JMML and CMML. Somatic NRAS and KRAS mutations are found in ∼40% of CMML specimens [10,11], and ∼85% of JMML patients have mutations in KRAS, NRAS, NF1, or PTPN11 (reviewed in [12]). The latter two genes encode proteins that regulate Ras-GTP levels. Importantly, children with germline mutations in NF1 or PTPN11 are at markedly increased risk of developing JMML, which argues strongly that deregulated Ras signaling can initiate this MPD [12]. This hypothesis is further supported by studies of Nf1, Kras, and Ptpn11 mutant mice, all of which develop MPDs that resemble JMML and CMML [13–16]. Mutations that alter other signaling molecules also cause MPD. For example, the BCR-ABL fusion gene is the hallmark of chronic myeloid leukemia, and JAK2 mutations are found in nearly all cases of polycythemia vera (reviewed in [17]). While mutations affecting signaling molecules are found in nearly all types of MPD, additional cooperating mutations are rare. MPDs are therefore genetically straightforward and tractable malignancies for understanding how aberrant signal transduction contributes to cancer by perturbation of stem and progenitor cell fates.
Mice expressing oncogenic KrasG12D in hematopoietic cells develop a fatal MPD with 100% penetrance that is characterized by leukocytosis, splenomegaly, and anemia, with death at approximately 3 mo of age [13,14]. In this system, injecting Mx1-Cre, KrasLSL-G12D mice with polyinosinic-polycytidylic acid (pIpC) induces expression of Cre recombinase, which removes an inhibitory loxP-STOP-loxP (LSL) element and activates the KrasG12D allele (we hereafter refer to Mx1-Cre, KrasLSL-G12D mice that have been treated with pIpC as KrasG12D mice). Mx1-Cre, KrasLSL-G12D mice provide a robust experimental system for investigating how expressing oncogenic Kras from the endogenous promoter affects HSCs and their progeny in vivo. Here, we identify tumor initiating cells in the KrasG12D model of MPD and find that, unlike some myeloid oncogenes, KrasG12D does not confer aberrant self-renewal properties to committed progenitor cells. However, small numbers of primitive KrasG12D cells can initiate MPD. This population shows excessive proliferation and rapidly dominates multilineage hematopoiesis in vivo. These data indicate that hyperactive Ras signaling is sufficient for the competitive advantage demonstrated by mutant HSCs and further implicate the HSC compartment as critical for therapy of JMML and CMML. Transplanted KrasG12D HSCs efficiently initiate T lineage acute lymphoblastic leukemia/lymphoma (T-ALL), which is associated with Notch1 mutations and with acquisition of LSC activity in differentiating thymocytes. These results further demonstrate that distinct self-renewing populations can arise through cooperating oncogenic mutations during cancer progression.
Results
A Small KrasG12D Population Dominates Both Primitive and Differentiated Hematopoietic Compartments in Mx1-Cre, KrasLSL-G12D Mice
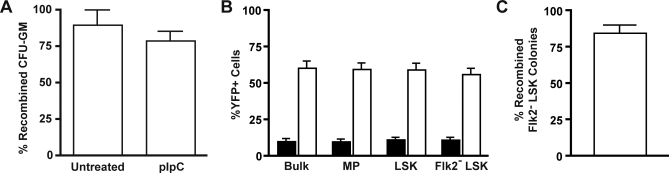
Although basal Mx1-Cre activity is low [18,19], many Mx1-Cre, KrasLSL-G12D mice that are not injected with pIpC ultimately succumb with MPD ([14] and unpublished data). This observation suggests that HSCs and/or progenitor cells that activate KrasG12D expression have a substantial proliferative advantage in vivo. To assess the kinetics of this process, we analyzed recombination in the myeloid progenitors of young Mx1-Cre, KrasLSL-G12D mice that were not treated with pIpC. Bone marrow cells from 3–5-wk-old animals were plated in methylcellulose medium to enumerate granulocyte-macrophage colony forming unit progenitors (CFU-GM), and individual myeloid colonies were genotyped by PCR. Surprisingly, nearly all CFU-GM in untreated animals were recombined as early as 3 wk of age (Figure 1A). We then examined more primitive populations in Mx1-Cre, KrasLSL-G12D mice crossed to a ROSA26 yellow fluorescent protein (YFP) reporter strain (Figure 1B) [20]. In these mice, cells expressing Cre are identified by YFP expression. The frequency of YFP+ cells in wild-type (WT) mice was within the expected range of background Cre expression [18,19]. However, we found a much higher incidence of YFP expression in bone marrow cells of Mx1-Cre, KrasLSL-G12D, ROSA26-YFP mice. This result was consistent among all populations analyzed, including the primitive Flk2− Lin−/lo Sca1+ c-kit+ (Flk2− LSK) compartment, which is highly enriched for HSCs [21]. To confirm that YFP expression correlated with KrasG12D expression, we also directly genotyped colonies formed by single Flk2− LSK cells. This method again revealed a predominance of KrasG12D-expressing cells in naïve Mx1-Cre, KrasLSL-G12D mice (Figure 1C). Together, these data are consistent with an advantage for Cre-expressing cells in untreated Mx1-Cre, KrasLSL-G12D mice.
Figure 1. Spontaneous KrasG12D Activation in CFU-GM and HSCs from Mx1-Cre, KrasLSL-G12D Mice.
(A) Bone marrow was harvested at 24 d or 5 wk of life from mice that either received a single dose of pIpC at 21 d of life or were left untreated, and plated in methylcellulose medium with GM-CSF. Individual colonies were isolated and genotyped by PCR to assess for Cre-mediated excision of the LSL cassette (n = 18 colonies from two uninjected animals, and 34 colonies from two injected animals, compiled from two independent experiments; error bars show SEM).
(B) Mx1-Cre (filled bars) and Mx1-Cre, KrasLSL-G12D (open bars) mice that inherited an LSL-YFP reporter were sacrificed at 5 wk of life, without ever being injected with pIpC. Expression of YFP requires excision of the LSL cassette from the reporter gene by Cre recombinase. The percentage of YFP+ cells in all bone marrow cells (Bulk), Lin− c-kit+ Sca1− myeloid progenitors (MP), LSK cells, and Flk2− LSK cells was assessed by flow cytometry. (n = 4 for WT and for Mx1-Cre, KrasLSL-G12D mice compiled from three independent experiments; error bars show SEM).
(C) Flk2− LSK cells were sorted from bone marrow of 4-wk-old Mx1-Cre, KrasLSL-G12D mice that had not received pIpC, and plated in methylcellulose medium for a culture period of 14 d. Individual colonies were genotyped by PCR (n = 44 colonies from 6 mice in 3 independent experiments; error bar shows SEM).
Increased Cell Cycle Entry of KrasG12D HSCs
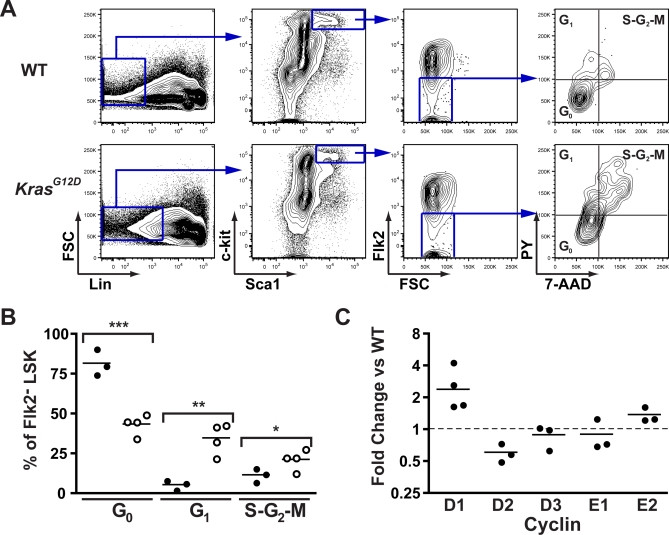
The apparent outgrowth of KrasG12D Flk2− LSK cells in mice that were not injected with pIpC suggested that KrasG12D expression might increase proliferation in this compartment. To test this hypothesis, we stained bone marrow cells collected from KrasG12D mice and WT littermates 2 wk after pIpC treatment with antibodies to cell surface proteins and with dyes that stain DNA and RNA (7-aminoactinomycin D [7-AAD] and pyronin Y, respectively). These studies revealed a significant reduction in the number of quiescent Flk2− LSK cells in KrasG12D animals, which are identified by having a 2n DNA content and low pyronin Y staining (Figure 2A and 2B). Whereas the Flk2− subset of WT LSK contains roughly 80% cells in the G0 phase of the cell cycle, KrasG12D Flk2− LSK are only 50% quiescent. As an initial exploration of mechanisms regulating cell cycle progression, we analyzed D- and E-type cyclin expression by quantitative PCR in sorted Flk2− LSK cells from KrasG12D and WT mice. KrasG12D Flk2− LSK cells had significantly higher expression of cyclin D1 (Figure 2C).
Figure 2. Increased Proliferation of KrasG12D HSC.
Bone marrow from 5-wk-old WT and KrasG12D mice was stained with 7-AAD and pyronin Y (PY) for DNA and RNA quantitation, along with surface markers for Flk2− LSK cells.
(A) Gating is shown for cell cycle analysis of WT and KrasG12D Flk2− LSK cells.
(B) Summary of replicate samples from WT (closed circles) and KrasG12D (open circles) mice (n = 3 or 4 as shown; compiled from two independent experiments). Means and SEM are WT: G0 81.8 ± 4.72, G1 5.66 ± 1.76, S-G2-M 11.7 ± 2.56; and KrasG12D: G0 43.6 ± 3.19, G1 34.9 ± 4.85, S-G2-M 21.4 ± 3.14. p-Values by unpaired t-test are indicated: ***, p < 0.001; **, p < 0.01; *, p < 0.05.
(C) RNA from doubly sorted Flk2− LSK cells was isolated, and quantitative PCR performed on cDNA to test expression levels of selected cyclins (n = 3 or 4 as shown; bar shows geometric mean). Results are expressed as fold change in expression compared to WT Flk2− LSK cells, after normalization to β-actin expression. These results represent three or four independent experiments as shown, each performed with pooled bone marrow from three to five animals. The geometric mean of cyclin D1 expression is 2.4-fold over WT (95% confidence interval 1.2–4.9). Purity of sorted cells is shown in Figure S1.
Self-Renewal Is Restricted to Primitive Hematopoietic Cells in KrasG12D Mice
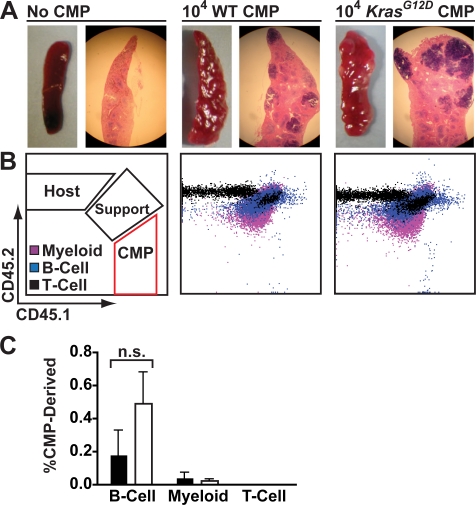
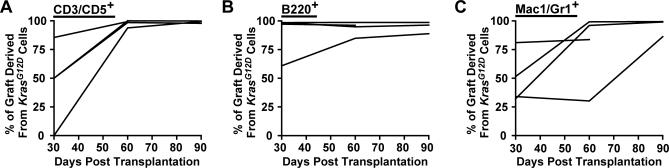
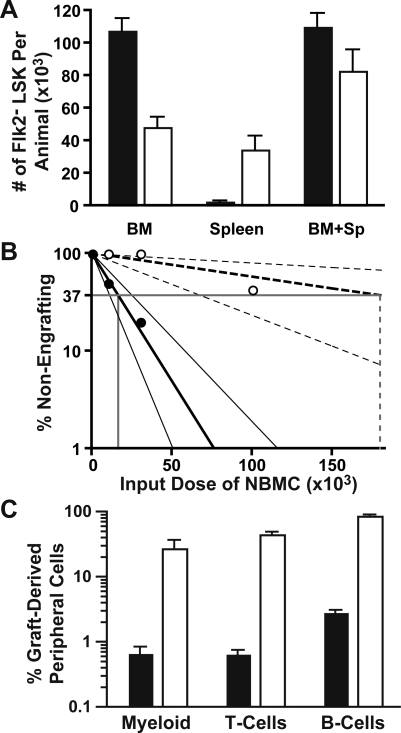
Bone marrow mononuclear cells from KrasG12D mice display a hypersensitive pattern of CFU-GM progenitor growth, which is a cellular hallmark of JMML [12–14]. We found that this abnormal CFU-GM activity resides primarily in the common myeloid progenitor (CMP) compartment, and that these cells demonstrate enhanced proliferation in vivo ([22] and unpublished data). To investigate if this population could initiate and maintain MPD, we collected Lin−/lo Sca1− c-kit+ CD34+ FcγRII/III− CMPs from 5-wk-old KrasG12D mice and WT littermates by FACS, and transferred 10,000 of these cells into lethally irradiated recipients with 106 WT marrow cells for radioprotective support. Transplanted tester cells, recipient cells, and support cells were marked by expression of different isoforms of CD45, allowing them to be distinguished by flow cytometry. Transplanted KrasG12D or WT CMPs demonstrated robust day 8 spleen colony-forming unit (CFU-S8) activity, with colony size somewhat larger for KrasG12D input CMPs (Figure 3A). However, we detected less than 0.1% of circulating myeloid cells derived from transplanted KrasG12D or WT CMPs 1 mo after transplantation (Figure 3B and 3C). As expected from previous studies [23,24], KrasG12D and WT CMPs made minor contributions to the circulating B cell compartment with a statistically insignificant trend towards greater B cell production from KrasG12D cells. Taken together, these data indicate that KrasG12D CMPs do not initiate a hematologic disease. By contrast, transferring 500 Flk2− LSK cells from KrasG12D animals into lethally irradiated recipients rapidly resulted in durable multilineage reconstitution. KrasG12D cells dominated the T cell and B cell compartments sooner and to a higher degree than the progeny of WT Flk2− LSK cells in control animals. The myeloid series demonstrated a more variable time course, but KrasG12D derived cells also eventually out-competed WT cells (Figure 4).
Figure 3. KrasG12D Does Not Lead to CMP Self-Renewal.
Sorted CD45.1 CMPs were transplanted into lethally irradiated CD45.2 recipients.
(A) Spleens were harvested for CFU-S8 colony analysis 8 d after transplantation of 104 KrasG12D or WT CMPs. Gross images and hematoxylin/eosin stained sections are shown.
(B) Peripheral blood of animals analyzed 1 mo after transplantation of 104 KrasG12D or WT CMPs along with 106 CD45.1/CD45.2 heterozygous nucleated bone marrow cells for radioprotection (Support). Costaining with surface markers allowed detection of contributions to myeloid (purple), B (blue), and T (black) lineages.
(C) Percentages of CMP-derived progeny within the indicated lineages found in peripheral blood 1 mo after transplantation of WT (filled bars) or KrasG12D (open bars) CMPs (n = 4 in two independent sorts).
Figure 4. Transplanted KrasG12D Flk2− LSK Cells Outperform WT LSK Cells in Competitive Reconstitution.
Lethally irradiated recipients (n = 4 in a single experiment) were transplanted with 500 each of KrasG12D and WT Flk2− LSK cells, then bled monthly until sacrifice. Graphs show the proportion of graft-derived cells that came from the KrasG12D donor in circulating (A) T cell (CD3/CD5+), (B) B cell (B220+), and (C) myeloid (Mac1/Gr1+) populations. This was calculated by (1) gating for graft-derived cells (CD45.1 and CD45.1/CD45.2) within a lineage, and then (2) determining the fraction of KrasG12D-derived cells (CD45.1) within this subset.
Recipients of KrasG12D Flk2− LSK cells that were euthanized 3 mo after transplantation had mild to moderate MPD, manifested as leukocytosis and splenomegaly with myeloid and erythroid infiltration (Figure 5). As discussed below, we also found that all recipients of KrasG12D Flk2− LSK cells developed T-ALL 2–4 mo after adoptive transfer. Although early mortality from T-ALL precluded analyzing recipient mice beyond 3 mo, KrasG12D Flk2− LSK cells recapitulate the essential features of MPD seen in the original Mx1-Cre, KrasLSL-G12D model [13,14].
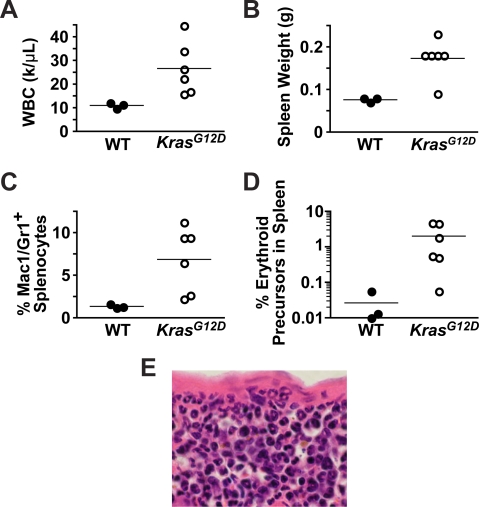
Figure 5. Transplanted KrasG12D Flk2− LSK Cells Induce MPD.
Mice were analyzed 3 mo after transplantation with either KrasG12D or WT Flk2− LSK cells for evidence of MPD (n = 3 WT, n = 6 KrasG12D in a single experiment).
(A) Numbers of circulating leukocytes (WBC).
(B) Spleen weights.
(C) Percentages of myeloid cells (Mac1+ and/or Gr1+) in the spleen.
(D) Percentages of erythroid precursors (nucleated and TER119+) in the spleen.
(E) A representative splenic section demonstrating infiltration by mature myeloid cells (ring-shaped nuclei) and erythroblasts (small dense nuclei).
HSCs Are Less Frequent in KrasG12D Mice but Have Enhanced Repopulating Ability
We next asked how KrasG12D expression affects HSC function. Immunophenotypic analysis revealed a 2-fold reduction in the number of marrow Flk2− LSK cells in KrasG12D mice 2 wk after pIpC injection (Figure 6A). This was mostly offset by an increased number of splenic Flk2− LSK cells. The reduction in marrow Flk2− LSK cells persisted in older animals (Figure S2). We also performed limit dilution studies to assess functional HSC activity. In these experiments, lethally irradiated recipients received decreasing numbers of whole bone marrow cells from either KrasG12D mice or WT littermates that had been injected with pIpC 2 wk earlier. Recipients were bled monthly, and flow cytometry was performed to assess whether CD45.1+ donor cells were able to provide durable (>2 mo) multilineage (myeloid, B cell, and T cell) engraftment. These studies demonstrated a striking 10-fold decrease in the number of long-term repopulating stem cells in KrasG12D animals compared to the WT littermate controls (Figure 6B; Table S1).
Figure 6. HSC Number and Proliferative Capacity in KrasG12D Mice.
HSCs were analyzed in KrasG12D mice or WT littermates that were treated with pIpC at 21 d of age and then sacrificed at 35 d of age.
(A) Total numbers of Flk2− LSK cells in KrasG12D mice (open bars) or WT mice (filled bars) were quantified by flow cytometry in bone marrow (p < 0.001, t-test) and spleen (p < 0.01); total numbers (spleen + marrow) are also shown (p = 0.12). n = 6 mice per genotype, and error bars show SEM; data are pooled from two independent experiments. Frequencies of Flk2− LSK cells among viable nucleated cells were multiplied by nucleated bone marrow cell counts in femurs and tibias and then scaled to estimate total bone marrow numbers using published distributions [75].
(B) Whole bone marrow from KrasG12D (open circles) or WT littermates (filled circles) was tested for repopulating activity in a limit dilution transplantation assay. The calculated values for frequencies of repopulating units were 1 in 16,610 nucleated bone marrow cells (NMBC) for WT marrow and 1 in 180,404 for KrasG12D marrow. Narrow lines designate 95% confidence intervals (1:7,338 to 1:37,598 for WT and 1:68,953 to 1: 471,998 for KrasG12D; frequencies in WT versus KrasG12D marrow are significantly different with p = 0.0003 by two-tailed t-test). Outcomes of individual experiments are described in Table S1.
(C) Mice that engrafted after transplantation with limiting numbers of whole bone marrow cells (1 × 104 for WT, 1 × 105 for KrasG12D) were bled at 2 mo to determine the percentage of circulating myeloid (Mac1/Gr1+), B-lineage (B220+), and T-lineage (CD3/CD5+) cells that were derived from the transplanted marrow. Filled bars represent mice receiving WT marrow (n = 2), open bars represent mice receiving KrasG12D marrow (n = 3; error bars show SEM; note logarithmic scale).
Progression of MPD despite a reduction in the size of the HSC compartment suggests an increased production of mature cells by each KrasG12D HSC. To address this possibility, we examined the patterns of reconstitution from either KrasG12D or WT HSCs in mice that received the limit dilution dose (1 × 105 KrasG12D and 1 × 104 WT bone marrow cells). As ∼50% of these recipients were engrafted with donor cells, the Poisson distribution predicts that approximately 70% of the engrafting mice received a single HSC and ∼24% received two HSC. By 2 mo after transplantation, donor KrasG12D marrow cells made a markedly greater contribution to recipient hematopoiesis than WT cells (Figure 6C).
We were able to compare repopulation of the stem cell compartment by WT and KrasG12D HSCs in a few lethally irradiated recipient mice before the onset of T-ALL. In these experiments, lethally irradiated recipients (CD45.2) were reconstituted with equal numbers of KrasG12D (CD45.1) and WT (CD45.1/CD45.2) Flk2− LSK cells, as well as 106 CD45.2 whole bone marrow cells (CD45.2) for radioprotection. Whereas only one of six recipients euthanized 2 mo after transplantation showed a clear bias towards KrasG12D-derived Flk2− LSK cells, mice that survived for 3 mo without evidence of diffuse T-ALL demonstrated an overwhelming bias towards KrasG12D-derived Flk2− LSK cells, myeloid progenitors, and mature myeloid cells (Figure S3).
T-ALL Arises from Transplanted KrasG12D HSCs in Thymocytes That Acquire Somatic Notch1 Mutations
Recipients that were injected with KrasG12D Flk2− LSK cells, either alone or in a 1:1 ratio with WT Flk2− LSK cells, uniformly became moribund 8–14 wk after transplantation. Examination of euthanized mice revealed massive thymic enlargement with an arrest in T cell development at the CD4/CD8 double positive stage, and variable infiltration of blast cells within the liver, spleen, and bone marrow (Figure 7A). Importantly, two of three animals from the bone marrow limit dilution transplantation assay that were engrafted with a single KrasG12D repopulating unit developed T-ALL that was identical to that seen in animals repopulated with 500 KrasG12D Flk2− LSK cells (unpublished data).
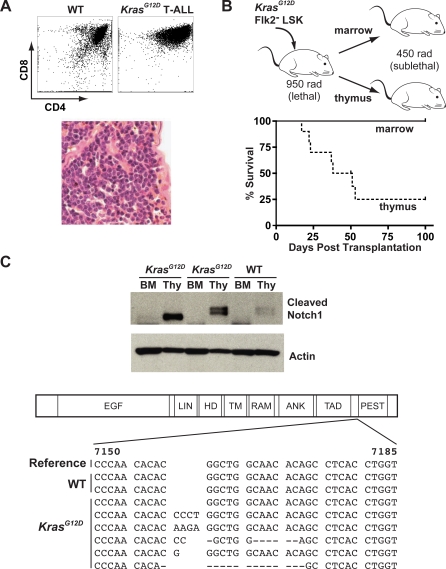
Figure 7. Transplanted KrasG12D Flk2− LSK Cells Initiate T-ALLs That Contain Notch1 Mutations.
(A) Flow cytometry of thymocytes harvested from moribund recipients of KrasG12D Flk2− LSK cells shows an abnormal accumulation of CD4/CD8 double positive and immature CD8 single positive cells. Spleen histology demonstrates infiltration by monomorphic cells with open chromatin (hematoxylin/eosin).
(B) Primary recipients (n = 5) received 500 KrasG12D Flk2− LSK cells with or without an equal number of WT Flk2− LSK cells after lethal irradiation (950 rad). These primary recipients were euthanized 2–3 mo later, and 106 bone marrow cells or thymocytes were transferred into sublethally irradiated (450 rad) secondary recipients (two per primary mouse). Sublethal irradiation selectively permits transfer of acute leukemia but not KrasG12D HSCs or MPD [13]. Thymocytes, but not bone marrow cells, transferred T-ALL.
(C) Cell lysates from thymocytes and bone marrow of animals euthanized 3 mo after transplantation with KrasG12D or WT HSCs were blotted with an antibody specific for cleaved Notch1. Three independent primary recipients are shown. Sequence analysis demonstrates frameshift mutations near the PEST domain of Notch1 in thymocytes from five of six mice that received KrasG12D HSC, but not in recipients receiving WT HSCs alone (reference sequence from GenBank [http://www.ncbi.nlm.nih.gov/Genbank] accession number NM_008714).
Next, we asked if T-ALL arose within the bone marrow or the thymus of mice transplanted with KrasG12D HSCs. To do this, we took advantage of our prior observation that recipients conditioned with sublethal irradiation fail to engraft with KrasG12D bone marrow [13]. Therefore, sublethally irradiated recipients can exclusively select for hematologic malignancies with more aggressive biologic behavior. We isolated bone marrow cells and thymocytes from primary recipients of KrasG12D Flk2− LSK cells that developed T-ALL, and injected each population into sublethally irradiated secondary recipients (Figure 7B). As expected, no animals demonstrated multilineage engraftment or MPD. Animals transplanted with thymocytes quickly succumbed with an identical T-ALL as primary recipients; however, none of the mice transplanted with bone marrow developed leukemia. Thus, whereas bone marrow-derived KrasG12D HSCs efficiently give rise to T-ALL, the T-ALL LSC population is initially restricted to the thymus in primary recipient mice.
These results suggested that one or more secondary mutations might have developed in a novel T-lymphoid clone. Somatic NOTCH1 mutations are common in human and murine T-ALL [25–28]. To determine if a similar mechanism might contribute to the evolution of KrasG12D HSCs to T-ALL LSCs, we performed Western blot analysis to detect cleaved (activated) Notch1 protein. Cleaved Notch1 was observed in thymocytes from diseased primary recipients, but not in bone marrow cells (Figure 7C), a finding that is consistent with the secondary transplant data. Direct sequencing around the PEST domain uncovered frameshift mutations in exon 34 of Notch1 in thymocytes from five of six animals transplanted with KrasG12D Flk2− LSK cells that developed T-ALL, but no mutations in thymocytes from control mice that received WT HSCs alone and remained well. The presence of a somatically acquired Notch1 mutation in a large fraction of the tumor provides a molecular indication of clonality.
Thymic Abnormalities in KrasG12D Mice
The propensity of KrasG12D HSCs to generate T-ALL led us to investigate the effects of KrasG12D expression in early T-lineage cells. Flow cytometry of the bone marrow revealed no expansion of Lin− Flk2+ IL-7Rα+ c-kitint Sca1int common lymphoid progenitors (Figure 8A) [29]. However, KrasG12D mice demonstrated consistent thymic enlargement compared to age-matched littermate controls, even prior to the onset of T-ALL (Figure 8B). Immunophenotyping of primary thymocytes demonstrated an essentially normal distribution of CD4+ and CD8+ expression, with a slight trend toward an increased number of CD4/CD8 double negative (DN) cells (after excluding Mac1+ and Gr1+ infiltrating myeloid cells) (Figure 8B). By contrast, further examination of the DN compartment using the cell surface markers CD44 and CD25 uncovered skewed development in Kras mutant mice (Figure 8C). Together, these data demonstrate that oncogenic KrasG12D perturbs thymic homeostasis, particularly in early stages of thymocyte maturation.
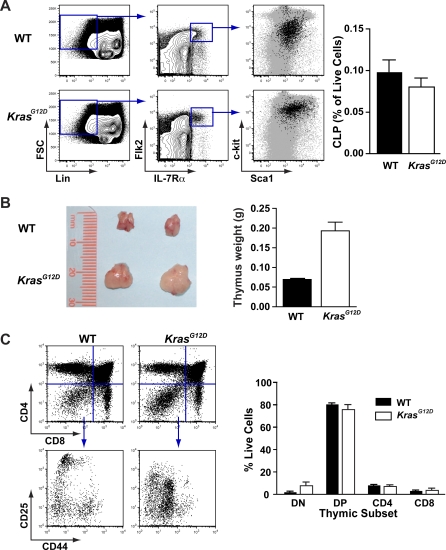
Figure 8. Abnormal T-lymphoid Development in KrasG12D Mice.
(A) Enumeration of Lin− Flk2+ IL-7Rα+ c-kitint Sca1int common lymphoid progenitors (CLPs) in bone marrow of WT and KrasG12D mice. A typical comparison and gating strategy is shown, with the far right panel showing expression of c-kit and Sca1 in CLPs (black dots) compared to the larger population of Lin− cells (gray dots). CLP frequencies are graphed (n = 5 WT and 6 KrasG12D; error bars show SEM and difference is not statistically significant by unpaired t-test).
(B) Enlarged thymi in 7-wk-old KrasG12D mice compared with WT littermates; photograph shows a typical example, and graph shows data from a representative cohort (n = 3 mice and error bars show standard deviation; p < 0.05).
(C) Flow cytometry of primary thymocytes for expression of CD4 and CD8, and also CD25 and CD44 expression within DN (CD4− CD8−) cells; figure shows a representative example. Myeloid cells were excluded using Mac1 and Gr1 staining. Graph represents the frequency of thymocytes within in the live gate in DN, double positive (CD4+D8+, DP), and CD4 or CD8 single positive cells (n = 3 mice per genotype and error bars show SEM; p > 0.05 for all populations. Data are representative of three independent experiments).
Discussion
We find that oncogenic KrasG12D expression in HSCs confers a strong in vivo growth advantage, increases proliferation, and results in MPD and T-ALL. In MPD, as in normal marrow, stem cell activity is restricted to the Flk2− LSK population, which represents less than 0.1% of nucleated marrow cells. While pathologic behaviors of more mature cells may contribute to tissue infiltration, anemia, and organomegaly, self-renewal is confined to this very primitive population. Therefore, hyperactive Ras signaling promotes excess proliferation in multiple hematopoietic compartments without immortalizing non–self-renewing cells. Similar data have been described in murine models of MPD based on BCR-ABL overexpression or loss of JunB, both of which also deregulate cytoplasmic signaling networks [5,30]. By contrast, recent experiments have provided direct evidence that some oncogenic transcription factors allow committed myeloid progenitors to acquire self-renewal ability [3,4]. Taken together, studies of myeloid oncogenes performed to date support the general idea that mutations that predominately alter cytoplasmic signaling networks and those that affect transcription factors controlling cell fate decisions comprise discrete complementation groups for the fully transformed phenotype [31,32].
Our limit dilution transplantation data demonstrate that oncogenic Kras confers a dramatic growth advantage in the HSC compartment. Under stringent conditions in which the contribution of a single WT HSC can barely be detected, the progeny of one (or at most three) KrasG12D HSC comprise a substantial fraction of the hematopoietic compartment. These studies provide direct experimental evidence that the outgrowth of malignant cells in MPD can be attributed to hyperactive Ras signaling in HSC. Our data suggest a pathogenic model in which JMML or CMML is initiated by a somatic mutation that deregulates Ras signaling in a single HSC. This idea is consistent with limited data from human patients and xenograft studies that implicate the HSC as the cell of origin for JMML [33–38].
To begin to address the mechanism by which mutant HSCs outgrow their WT counterparts, we analyzed the cell cycle in KrasG12D Flk2− LSK cells and found they are preferentially in cycle. The overexpression of cyclin D1 in KrasG12D cells we observed is consistent with many prior studies in cultured cell lines engineered to overexpress oncogenic Ras [39,40]. Intriguingly, HSCs in mice lacking D-type cyclins demonstrate severe proliferative defects and accumulate in the G0 and/or G1 phases of the cell cycle [41]. If increased cyclin D1 levels conversely result in excessive proliferation of HSC, then MPD in KrasG12D mice may be mediated in part by cyclin D1, similar to the requirement for cyclin D1 in a model of Ras-mediated breast cancer [42].
We demonstrate a substantial early growth advantage of HSCs that express KrasG12D; however, it is also possible that oncogenic Ras expression has a negative long-term impact on HSC function. Increased proliferation or oncogenic stress may ultimately detract from self-renewal capacity. Reduced HSC fitness was observed in Pten−/− mice, in which phosphatidylinositol 3-kinase signaling is hyperactive [43,44]. Similar effects of KrasG12D are suggested by the reduced numbers of HSCs in KrasG12D mice, although this finding could also reflect changes in the composition of the Flk2− LSK population or cell-extrinsic effects related to alteration of the marrow microenvironment. The rapid demise of primary KrasG12D mice from MPD, and of transplant recipients from T-ALL, precluded serial transplantation experiments to test the long term fitness of KrasG12D HSC. In addition, the 5-fold discrepancy between HSC numbers that were measured by flow cytometry versus limit dilution transplantation suggests a defect in engraftment of KrasG12D HSC. This idea is consistent with a prior report in which retroviral transduction of mutant NRAS appeared to reduce engraftment potential [45], and with extensive data showing that proliferating HSCs fail to engraft efficiently [46–50].
Despite the reduced number of HSC in the bone marrow of KrasG12D mice, our data are not entirely consistent with the idea that KrasG12D is a cell-intrinsic negative regulator of HSC self-renewal. If it were, we would expect specific loss of KrasG12D cells and outgrowth of WT or KrasLSL-G12D cells, because conditional models using Mx1-Cre typically retain a small pool of cells with the unrearranged locus [51,52]. However, we observed preferential retention of KrasG12D cells, even within the diminishing Flk2− LSK compartment. This observation suggests that residual WT HSCs are unable to compensate for the reduced HSC number. Therefore, we favor the hypothesis that the reduction in HSC number in KrasG12D mice reflects a disordered bone marrow microenvironment with reduced supportive capacity rather than a purely cell-intrinsic effect of KrasG12D in HSC.
The natural history of hematologic disease was different in primary KrasG12D mice than in recipients of transplanted KrasG12D HSC. A subtle but important finding is that MPD is established earlier in primary KrasG12D mice than in transplanted recipients. (Figure 5 and [13]; also see [16,53]). There are several possible explanations for this observation. The hematopoietic microenvironment in young mice may be more permissive for MPD than the irradiated bone marrow of an adult. Additionally, pIpC administration in Mx1-Cre mice may quickly create a field of KrasG12D myeloid progenitors that contributes to the rapid evolution of MPD in primary mice through cytokine-mediated autocrine and/or paracrine mechanisms [54]. It is also possible that nonhematopoietic stromal cells in Mx1-Cre, KrasLSL-G12D mice express K-RasG12D and contribute to the rapid onset of MPD in primary KrasG12D mice.
The kinetics of MPD development relate directly to the high frequency of T-ALL we observed in transplanted recipients as compared to primary KrasG12D mice. The apparent incidence of T-ALL is highly subject to selection bias, because animals that die from MPD cannot be evaluated for subsequent emergence of T-ALL. For example, we have observed ∼10%–15% of KrasG12D mice develop T-ALL on an inbred C57BL/6 strain background (unpublished data), but median time to death from MPD is shorter than the typical latency of T-ALL, and lymphoid tumors exclusively appear in mice with a relatively late onset of MPD. Similarly, we have not observed spontaneous T-ALL in F1 (C57BL/6 × 129Sv/Jae) mice, which die from MPD at a younger age than the C57BL/6 strain described here ([13] and unpublished data). In transplant recipients, aggressive T-ALL arose in mice that also had evidence of underlying MPD that was not yet severe enough to kill the animal. Together, these observations suggest that the attenuation of MPD in transplant recipients was central to the apparent increase in the incidence of T-ALL in the transplant setting.
Cooperation of hyperactive Ras and deregulated Notch signaling in T-ALL has recently been shown [55–57]. Our data extend these studies by demonstrating the remarkable efficiency with which KrasG12D HSCs can initiate T-ALL, and delineating how multiple cell types may participate in the stepwise acquisition of oncogenic mutations in hematologic cancers. In the Mx1-Cre, KrasLSL-G12D model, T-ALL is initiated by oncogenic Kras expression in HSC, but full transformation occurs when cooperating Notch1 mutations arise in a T-lineage cell. In this sense, both KrasG12D HSCs and the fully transformed thymocytes can be considered different types of malignant stem cells with distinct leukemogenic potentials.
One potential implication of these results is that the initiating Kras mutation creates conditions favorable for acquisition of cooperating mutations by increasing the size of susceptible lymphoid progenitor pools and/or conferring resistance to apoptotic signals during thymic selection. KrasG12D appears to most greatly affect the DN population that is characteristically undergoing TCR rearrangement, selection, and proliferation [58,59]. Interestingly, K-RasG12D protein expression may substitute for the pre-T cell receptor rearrangement at this critical checkpoint, thereby allowing propagation of thymocytes that would normally be edited [60]. Consistent with this idea, a patient with impaired lymphoid homeostasis and multiple lymphoid malignancies was recently reported to have a germline NRASG13D mutation, and oncogenic NRAS suppressed apoptosis of lymphocytes after cytokine withdrawal [61]. By contrast, Kindler et al. recently reported reduced thymic cellularity in Mx1-Cre, KrasG12D mice [56]. We speculate that the proliferative effects of K-RasG12D in the T cell compartment were obscured in those studies by the short interval between pIpC injection, which induces systemic interferon production, and histologic analysis.
The idea that patients may harbor a variety of genetically distinct LSC is consistent with studies of patients with chronic myeloid leukemia in blast transformation [62], and has important therapeutic implications. The need to eliminate partially transformed but self-renewing cells, like KrasG12D HSCs, will depend on their propensity to initiate a life-threatening disease. Targeted therapies that are directed against onco-proteins such as K-RasG12D will effectively eliminate premalignant clones only if the targeted lesions are initiating rather than secondary mutations. For example, inhibition of Notch signaling is an attractive therapeutic strategy for T-ALL that is being investigated in the clinic. However, if these cancers arise from aberrant HSCs that do not contain a NOTCH1 mutation and are not eradicated by treatment, relapse could occur through the acquisition of distinct cooperating mutations in a self-renewing preleukemic population. Consistent with this idea, studies of human T-ALL suggest that NOTCH1 mutation occurs as a secondary mutation in at least some cases, with some patients developing recurrent disease having distinct NOTCH1 alleles [63].
Finally, our data have implications for understanding the nature of cancer stem cell populations in nonhematopoietic malignancies. KRAS is the most frequent target of dominant oncogenic mutations in human cancer, and it is particularly important in carcinomas of the lung, pancreas, and colon. Analogous cancers arise in strains of mice expressing conditional oncogenic Kras alleles in these tissues [64–68]. Importantly, whereas oncogenic Kras expression efficiently initiates tumorigenesis in murine lung and pancreas, colon cancer is observed only when the tumor suppressor Apc is inactivated as well [68]. These data are consistent with studies of human patients, which imply that KRAS mutation occurs early in pancreatic cancer but typically after APC mutation in colon carcinoma [69–71]. Lung cancer in KrasG12D mice appears to be initiated in a distinct bronchio-alveolar stem cell population [72]. On the basis of these observations and our data, we speculate that, like HSCs, cells initiating pancreatic and lung cancer will possess inherent self-renewal potential, and that KRAS mutations only contribute to colon tumorigenesis in cells that have already acquired a mutation that enhances self-renewal. Uncovering specific proteins and pathways that are essential for the self-renewal and survival of Kras mutant cancer stem cell populations may reveal novel targets for therapeutic intervention in a variety of human cancers.
Materials and Methods
Ethics statement.
All animals were handled in strict accordance with good animal practice as defined by the relevant national and local animal welfare bodies, and all animal work was approved by the institutional animal research committee (University of California, San Francisco IACUC).
Mouse strains.
Animals were housed in a barrier facility at the University of California, San Francisco. All mice were of the C57BL/6 strain. As described [13], 21-d-old Mx1-Cre, KrasLSL-G12D were injected IP with 250 μg pIpC (Sigma). ROSA26 LSL-YFP reporter mice were a gift from C. Lowell. Ptprc (CD45) congenic mice were from Jackson Labs.
Colony assays.
To assay CFU-GM, nucleated bone marrow cells (105) or splenocytes (2 × 105) were suspended in 1 ml methylcellulose medium (M3231, StemCell Technologies) with 10 ng/ml murine GM-CSF (PeproTech). Colonies were counted after 8 d. To assay Flk2− LSK cells, 100 sorted cells were plated in 1 ml methylcellulose medium (M3434, StemCell Technologies), which is provided containing IL-3, IL-6, stem cell factor, and erythropoietin, and incubated at 37 °C for 14 d. For Kras genotyping, individual colonies were picked with a 10 μl pipette and frozen overnight in 10 μl ddH2O prior to PCR analysis [13].
Flow cytometry.
Flow cytometry was performed as described [21,24]. Staining was carried out in FACS Staining Buffer (FSB; HBSS with 2% heat inactivated FCS) at 0 °C unless otherwise stated. HSCs were defined as Lin−/lo Sca1+ c-kit+ Flk2−, and CMP as Lin−/lo Sca1− c-kit+ CD34+ FcγR−. Antibodies were from eBioscience except as specified. To identify HSCs, cells were first stained for 1 h with unconjugated lineage antibodies: CD3 (17A2, BioLegend); CD4 (RM4–5); CD5 (53–7.3); CD8 (53–6.7); B220 (RA3-6B2); Ter119 (TER-119); Mac1 (M1/70); Gr1 (RB6-8C5). Cells were washed then incubated for 30 min with TriColor goat-anti-rat F(ab′)2 (Invitrogen) and murine IgG (Sigma). Cells were washed again, stained for 30 min with Pacific Blue anti-Sca1 (D7, BioLegend); APC anti-c-kit (2B8, BioLegend); PE anti-Flk2 (A2F10, BioLegend); and 7-AAD (Sigma) 5 μg/ml for dead cell exclusion. IL-7Rα was detected using a PE conjugate (eBioscience). In this case, or when analyzing YFP+ cells, Flk2 was detected with biotinylated anti-Flk2 and APC-Alexa Fluor 750 streptavidin (Invitrogen). We found no difference in Flk2− LSK staining when Mac1+ cells were identified an on independent channel but not excluded. For myeloid progenitor analysis, PE anti-Flk2 was replaced with PE anti-FcγRII/III (93) and FITC anti-CD34 (RAM34), and cells were stained for an additional 30 min prior to analysis. Mature cells were assigned lineage using Pacific Blue anti-Mac1 and anti-Gr1 (myeloid), FITC anti-CD3 and anti-CD5 (T), and PE anti-B220 (B). T cell subsets in mice with T-ALL were distinguished with Pacific Orange anti-CD4 (Invitrogen), APC anti-CD8, FITC anti-CD3, and PE anti-CD5. To discern origin of cells in chimeric mice, antibodies to CD45.1 (A20; PE-Cy7) and CD45.2 (104; Alexa Fluor 700) were added. Primary thymocytes were analyzed with PE-Cy7 anti-CD4, Alexa 647 anti-CD8, PE anti-CD25, Pacific Blue anti-CD44, and FITC anti-Mac1 and anti-Gr1.
Prior to sorting, c-kit+ cells were enriched with anti-CD117 microbeads and an AutoMACS (Miltenyi). Cell sorting was performed on a FACSAria and flow cytometry on an LSRII, both using FACSDiva software (BD). Data were analyzed using FlowJo software (TreeStar).
Transplantation.
Recipients received a single fraction of 950 rads for lethal irradiation, or 450 rads for sublethal irradiation, from a cesium source. Donor cells were prepared in 100 to 200 μl of FSB, and injected retro-orbitally into anesthetized mice. Recipients received water with neomycin and polymyxin for 2 wk. Blood counts were monitored monthly using a Hemavet 950FS (Drew Scientific).
Limit dilution was performed as described [73]. Lethally irradiated recipients were transplanted with 1 × 104, 3 × 104, or 1 × 105 nucleated bone marrow cells along with 106 unfractionated WT bone marrow cells for radioprotection. Peripheral blood was analyzed monthly by flow cytometry; mice with detectable engraftment in myeloid, B and T lineages 2 mo after transplantation were scored as positive and L-Calc software (StemCell Technologies) was used for statistical analysis.
For the CFU-S8 assay, lethally irradiated mice received 105 CMPs without support cells. Spleen colonies were observed 8 d later by gross examination at harvest and after 48 h fixation in 10% formalin. Sections were stained with hematoxylin and eosin.
Notch1 analysis.
Cells were lysed in 1% NP-40 with 30 mM NaF, 30 mM β-glycerophosphate, 20 mM Na4P2O7, 1 mM Na3VO4, and Complete (Roche) and analyzed after SDS-PAGE using cleaved Notch1 and β-actin antibodies (Cell Signaling). Genomic DNA was PCR-amplified sequenced bidirectionally using 5′-ATAGCATGATGGGGCCACTA-3′ and 5′-GCCTCTGGAATGTGGGTGAT-3′.
Cell cycle analysis.
Staining with 7-AAD and pyronin Y was performed as described [74] in nucleic acid staining solution (NASS; 0.1M phosphate-citrate buffer [pH 6.0] [Sigma], 5mM EDTA, 0.15M NaCl, 0.5% BSA), with 0.02% saponin (Sigma). Nucleated bone marrow cells were stained for 30 min with FITC lineage antibodies (CD3, CD4, CD5, CD8, B220, Ter119, and Gr1), Pacific Blue anti-Sca1, unconjugated anti-CD16/32 (2.4G2, UCSF hybridoma core), and biotinylated anti-Flk2. Cells were washed and then stained with APC-Alexa Fluor 750 streptavidin. Cells were washed again and resuspended in 500 μl of NASS with 1 μg/ml 7-AAD (Sigma), and incubated at room temperature for 30 min, then on ice for 5 min. Pyronin Y (Sigma) was then added to 1 μg/ml, and cells were incubated for an additional 10 min before being washed and resuspended in 200 μL of FSB. Cells were finally stained with APC anti-c-kit and FITC anti-Mac1. Differences in the quiescent fraction of HSCs were analyzed using an unpaired t-test.
Quantitative real-time PCR.
The assay was performed as described [50]. Flk2− KLS cells from animals pooled by genotype were double-sorted directly into RNA binding/lysis buffer from the RNEasy kit (Qiagen), and total RNA was extracted per instructions. First strand cDNA synthesis was performed using a SuperScript III kit (Invitrogen) per manufacturer's instructions. Reactions were performed in an ABI-7900 sequence detection system using SYBR green according to manufacturer's instructions (Applied Biosystems). Each amplification was performed in 10 μl with a template cDNA equivalent of 100 sorted HSCs. Each sample was tested in triplicate with each primer pair, and normalized to β-actin expression. Due to limiting numbers of doubly sorted cells, final cell purity was not analyzed; however the staining characteristics of c-kit-enriched and singly sorted cells are presented in Figure S1.
Poisson statistics.
To analyze repopulation by a single HSC, we identified a cohort of mice receiving a cell dose yielding engraftment in only 50% of recipients. The Poisson distribution indicates that the probability of a mouse receiving k HSCs is given by
where n is the average HSC number per mouse. The average that yields k = 0 HSC at a rate of 0.5 is given by 0.5 = e −n, which is solved to give n = 0.693. Using this value for the average number of HSC per mouse, the Poisson distribution can be used to estimate the likelihood of any mouse receiving a given number of HSC: f(0) = e −ln 2 = 0.5; f(1) = (ln 2)e −ln 2 = 0.346; f(2) = 1/2 (ln 2)2 e −ln 2 = 0.120; f(3) = 1/6 (ln 2)3 e −ln 2 = 0.028. The proportion of mice expected to receive four or more HSCs is the remainder, 1 − (0.5 + 0.346 + 0.12 + 0.028) = 0.006, or 0.6%.
Supporting Information
Cells expressing c-kit were enriched by magnetic bead isolation, then doubly sorted by FACS. FACS data for the first and second sorts for typical examples of WT and KrasG12D mice are shown.
(740 KB TIF)
Flow cytometry was performed to enumerate Flk2− LSK cells in bone marrow (n = 4; p < 0.05) and spleens (n = 6; p < 0.05) harvested from 8-wk-old KrasG12D or WT mice. Data were analyzed as in Figure 2. Combined numbers in marrow and spleen were not significantly different between genotypes (p = 0.27). Data are pooled from two independent experiments; error bars show standard error of the mean (SEM). Statistical significance was evaluated with the unpaired t-test.
(80 KB TIF)
Mice transplanted with 500 CD45.1 KrasG12D and 500 CD45.1/CD45.2 WT HSCs were sacrificed (A) 2 mo (n = 6) or (B) 3 mo (n = 2) after transplantation, and their chimerism was assessed by flow cytometry. The percentage of graft cells derived from the KrasG12D HSCs is shown for Flk2− LSK cells (black bars), CMPs (dark grey bars), granulocyte-macrophage progenitors (GMP, light grey bars), and mature (Mac1/Gr1+) circulating myeloid cells (open bars). GMPs were defined as Lin−/lo Sca1− c-kit+ FcγRII/III+. Animals lacking any graft-derived contribution to the HSC compartment have been omitted.
(168 KB TIF)
The indicated numbers of mice were transplanted with 10,000, 30,000, or 100,000 nucleated bone marrow cells, and the proportions with detectable multilineage engraftment were determined 2 mo later. The number of repopulating units was calculated using the totals from these four experiments with L-Calc software (StemCell Technologies).
(47 KB PDF)
Acknowledgments
We thank David A. Tuveson and Tyler Jacks for generously providing KrasLSL-G12D mice.
Abbreviations
- 7-AAD
7-aminoactinomycin D
- CFU-GM
granulocyte-macrophage colony forming unit
- CFU-S8
day 8 spleen colony-forming unit
- CMML
chronic myelomonocytic leukemia
- CMP
common myeloid progenitor
- DN
double negative
- HSC
hematopoietic stem cell
- JMML
juvenile myelomonocytic leukemia
- LSC
leukemia stem cell
- LSK
Lin−/lo Sca1+ c-kit+
- LSL
loxP-STOP-loxP
- MPD
myeloproliferative disorder
- pIpC
polyinosinic-polycytidylic acid
- SEM
standard error of the mean
- T-ALL
T lineage acute lymphoblastic leukemia/lymphoma
- WT
wild type
- YFP
yellow fluorescent protein
Footnotes
Author contributions. AJS, LSC, MD, HCK, MS, MLH, EP, KS, and BSB conceived and designed the experiments. AJS, LSC, MD, HCK, MLH, and BSB performed the experiments. AJS, LSC, MD, HCK, MS, MLH, KS, and BSB analyzed the data. MD, HCK, MS, MLH, EP, KS, and BSB contributed reagents/materials/analysis tools. AJS, KS, and BSB wrote the paper.
Funding. This work was supported by National Institutes of Health grants R37 CA72614, U01 CA84221, and K08 CA103868; by a Specialized Center of Research award from the Leukemia and Lymphoma Society (LLS 7019–04); by a translational grant from the V Foundation for Cancer Research; by Concern Foundation; by the Connor Foundation; and by the Frank A. Campini Foundation. MS is supported by a fellowship from Fonds de la recherche en santé du Québec. MLH is supported by a career development award from the St. Baldrick's Foundation. BSB and EP are Scholars of the American Society of Hematology. AJS is a Medical Fellow of the Howard Hughes Medical Institute. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Competing interests. The authors have declared that no competing interests exist.
References
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Dick JE. Acute myeloid leukemia stem cells. Ann N Y Acad Sci. 2005;1044:1–5. doi: 10.1196/annals.1349.001. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- Cozzio A, Passegue E, Ayton PM, Karsunky H, Cleary ML, et al. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev. 2003;17:3029–3035. doi: 10.1101/gad.1143403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passegue E, Wagner EF, Weissman IL. JunB deficiency leads to a myeloproliferative disorder arising from hematopoietic stem cells. Cell. 2004;119:431–443. doi: 10.1016/j.cell.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- Van Etten RA, Shannon KM. Focus on myeloproliferative diseases and myelodysplastic syndromes. Cancer Cell. 2004;6:547–552. doi: 10.1016/j.ccr.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Hirsch-Ginsberg C, LeMaistre AC, Kantarjian H, Talpaz M, Cork A, et al. RAS mutations are rare events in Philadelphia chromosome-negative/bcr gene rearrangement-negative chronic myelogenous leukemia, but are prevalent in chronic myelomonocytic leukemia. Blood. 1990;76:1214–1219. [PubMed] [Google Scholar]
- Onida F, Kantarjian HM, Smith TL, Ball G, Keating MJ, et al. Prognostic factors and scoring systems in chronic myelomonocytic leukemia: a retrospective analysis of 213 patients. Blood. 2002;99:840–849. doi: 10.1182/blood.v99.3.840. [DOI] [PubMed] [Google Scholar]
- Lauchle JO, Braun BS, Loh ML, Shannon K. Inherited predispositions and hyperactive Ras in myeloid leukemogenesis. Pediatr Blood Cancer. 2006;46:579–585. doi: 10.1002/pbc.20644. [DOI] [PubMed] [Google Scholar]
- Braun BS, Tuveson DA, Kong N, Le DT, Kogan SC, et al. Somatic activation of oncogenic Kras in hematopoietic cells initiates a rapidly fatal myeloproliferative disorder. Proc Natl Acad Sci U S A. 2004;101:597–602. doi: 10.1073/pnas.0307203101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan IT, Kutok JL, Williams IR, Cohen S, Kelly L, et al. Conditional expression of oncogenic K-ras from its endogenous promoter induces a myeloproliferative disease. J Clin Invest. 2004;113:528–538. doi: 10.1172/JCI20476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki T, Mohi MG, Ismat FA, Bronson RT, Williams IR, et al. Mouse model of Noonan syndrome reveals cell type- and gene dosage-dependent effects of Ptpn11 mutation. Nat Med. 2004;10:849–857. doi: 10.1038/nm1084. [DOI] [PubMed] [Google Scholar]
- Le DT, Kong N, Zhu Y, Lauchle JO, Aiyigari A, et al. Somatic inactivation of Nf1 in hematopoietic cells results in a progressive myeloproliferative disorder. Blood. 2004;103:4243–4250. doi: 10.1182/blood-2003-08-2650. [DOI] [PubMed] [Google Scholar]
- Tefferi A, Gilliland DG. Oncogenes in myeloproliferative disorders. Cell Cycle. 2007;6:550–566. doi: 10.4161/cc.6.5.3919. [DOI] [PubMed] [Google Scholar]
- Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- Kemp R, Ireland H, Clayton E, Houghton C, Howard L, et al. Elimination of background recombination: somatic induction of Cre by combined transcriptional regulation and hormone binding affinity. Nucleic Acids Res. 2004;32:e92. doi: 10.1093/nar/gnh090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen JL, Weissman IL. Flk-2 is a marker in hematopoietic stem cell differentiation: a simple method to isolate long-term stem cells. Proc Natl Acad Sci U S A. 2001;98:14541–14546. doi: 10.1073/pnas.261562798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Meter ME, Diaz-Flores E, Archard JA, Passegue E, Irish JM, et al. K-RasG12D expression induces hyperproliferation and aberrant signaling in primary hematopoietic stem/progenitor cells. Blood. 2007;109:3945–3952. doi: 10.1182/blood-2006-09-047530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traver D, Miyamoto T, Christensen J, Iwasaki-Arai J, Akashi K, et al. Fetal liver myelopoiesis occurs through distinct, prospectively isolatable progenitor subsets. Blood. 2001;98:627–635. doi: 10.1182/blood.v98.3.627. [DOI] [PubMed] [Google Scholar]
- Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- Weng AP, Ferrando AA, Lee W, Morris JP, 4th, Silverman LB, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- Lin YW, Nichols RA, Letterio JJ, Aplan PD. Notch1 mutations are important for leukemic transformation in murine models of precursor-T leukemia/lymphoma. Blood. 2006;107:2540–2543. doi: 10.1182/blood-2005-07-3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neil J, Calvo J, McKenna K, Krishnamoorthy V, Aster JC, et al. Activating Notch1 mutations in mouse models of T-ALL. Blood. 2006;107:781–785. doi: 10.1182/blood-2005-06-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji H, Ishii-Ohba H, Ukai H, Katsube T, Ogiu T. Radiation-induced deletions in the 5′ end region of Notch1 lead to the formation of truncated proteins and are involved in the development of mouse thymic lymphomas. Carcinogenesis. 2003;24:1257–1268. doi: 10.1093/carcin/bgg071. [DOI] [PubMed] [Google Scholar]
- Karsunky H, Inlay M, Serwold T, Bhattacharya D, Weissman I. Flk2+ common lymphoid progenitors possess equivalent differentiation potential for the B and T lineages. Blood. 2008;111:5562–5570. doi: 10.1182/blood-2007-11-126219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntly BJ, Shigematsu H, Deguchi K, Lee BH, Mizuno S, et al. MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer Cell. 2004;6:587–596. doi: 10.1016/j.ccr.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Land H, Parada LF, Weinberg RA. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- Gilliland DG, Griffin JD. The roles of FLT3 in hematopoiesis and leukemia. Blood. 2002;100:1532–1542. doi: 10.1182/blood-2002-02-0492. [DOI] [PubMed] [Google Scholar]
- Miles DK, Freedman MH, Stephens K, Pallavicini M, Sievers E, et al. Patterns of hematopoietic lineage involvement in children with neurofibromatosis, type 1, and malignant myeloid disorders. Blood. 1996;88:4314–4320. [PubMed] [Google Scholar]
- Cooper LJ, Shannon KM, Loken MR, Weaver M, Stephens K, et al. Evidence that juvenile myelomonocytic leukemia can arise from a pluripotential stem cell. Blood. 2000;96:2310–2313. [PubMed] [Google Scholar]
- Matsuzaki S, Matsuda K, Miki J, Nakazawa Y, Sakashita K, et al. Development of two cytogenetically abnormal clones from multipotential hematopoietic stem cells in a patient with juvenile myelomonocytic leukemia. Leuk Res. 2005;29:1069–1072. doi: 10.1016/j.leukres.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Ito M, Yamamoto T, Yan XY, Yagasaki H, et al. Engraftment of NOD/SCID/gammac(null) mice with multilineage neoplastic cells from patients with juvenile myelomonocytic leukaemia. Br J Haematol. 2005;130:51–57. doi: 10.1111/j.1365-2141.2005.05578.x. [DOI] [PubMed] [Google Scholar]
- Freedman MH, Hitzler JK, Bunin N, Grunberger T, Squire J. Juvenile chronic myelogenous leukemia multilineage CD34+ cells: aberrant growth and differentiation properties. Stem Cells. 1996;14:690–701. doi: 10.1002/stem.140690. [DOI] [PubMed] [Google Scholar]
- Lapidot T, Grunberger T, Vormoor J, Estrov Z, Kollet O, et al. Identification of human juvenile chronic myelogenous leukemia stem cells capable of initiating the disease in primary and secondary SCID mice. Blood. 1996;88:2655–2664. [PubMed] [Google Scholar]
- Marshall C. How do small GTPase signal transduction pathways regulate cell cycle entry. Curr Opin Cell Biol. 1999;11:732–736. doi: 10.1016/s0955-0674(99)00044-7. [DOI] [PubMed] [Google Scholar]
- Yu Q, Ciemerych M, Sicinski P. Ras and Myc can drive oncogenic cell proliferation through individual D-cyclins. Oncogene. 2005;24:7114–7119. doi: 10.1038/sj.onc.1208853. [DOI] [PubMed] [Google Scholar]
- Kozar K, Ciemerych M, Rebel V, Shigematsu H, Zagozdzon A, et al. Mouse development and cell proliferation in the absence of D-cyclins. Cell. 2004;118:477–491. doi: 10.1016/j.cell.2004.07.025. [DOI] [PubMed] [Google Scholar]
- Yu Q, Geng Y, Sicinski P. Specific protection against breast cancers by cyclin D1 ablation. Nature. 2001;411:1017–1021. doi: 10.1038/35082500. [DOI] [PubMed] [Google Scholar]
- Yilmaz OH, Valdez R, Theisen BK, Guo W, Ferguson DO, et al. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441:475–482. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- Zhang J, Grindley JC, Yin T, Jayasinghe S, He XC, et al. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature. 2006;441:518–522. doi: 10.1038/nature04747. [DOI] [PubMed] [Google Scholar]
- MacKenzie KL, Dolnikov A, Millington M, Shounan Y, Symonds G. Mutant N-ras induces myeloproliferative disorders and apoptosis in bone marrow repopulated mice. Blood. 1999;93:2043–2056. [PubMed] [Google Scholar]
- Habibian HK, Peters SO, Hsieh CC, Wuu J, Vergilis K, et al. The fluctuating phenotype of the lymphohematopoietic stem cell with cell cycle transit. J Exp Med. 1998;188:393–398. doi: 10.1084/jem.188.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glimm H, Oh IH, Eaves CJ. Human hematopoietic stem cells stimulated to proliferate in vitro lose engraftment potential during their S/G(2)/M transit and do not reenter G(0) Blood. 2000;96:4185–4193. [PubMed] [Google Scholar]
- Orschell-Traycoff CM, Hiatt K, Dagher RN, Rice S, Yoder MC, et al. Homing and engraftment potential of Sca-1(+)lin(-) cells fractionated on the basis of adhesion molecule expression and position in cell cycle. Blood. 2000;96:1380–1387. [PubMed] [Google Scholar]
- Fleming WH, Alpern EJ, Uchida N, Ikuta K, Spangrude GJ, et al. Functional heterogeneity is associated with the cell cycle status of murine hematopoietic stem cells. J Cell Biol. 1993;122:897–902. doi: 10.1083/jcb.122.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passegue E, Wagers AJ, Giuriato S, Anderson WC, Weissman IL. Global analysis of proliferation and cell cycle gene expression in the regulation of hematopoietic stem and progenitor cell fates. J Exp Med. 2005;202:1599–1611. doi: 10.1084/jem.20050967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jude C, Climer L, Xu D, Artinger E, Fisher J, et al. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell. 2007;1:324–337. doi: 10.1016/j.stem.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock H, Meade E, Medeiros S, Schindler J, Valk P, et al. Tel/Etv6 is an essential and selective regulator of adult hematopoietic stem cell survival. Genes Dev. 2004;18:2336–2341. doi: 10.1101/gad.1239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Taylor BR, Shannon K, Clapp DW. Quantitative effects of Nf1 inactivation on in vivo hematopoiesis. J Clin Invest. 2001;108:709–715. doi: 10.1172/JCI12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan S, See W, Bonifas J, Stokoe D, Shannon KM. Hyperactivation of protein kinase B and ERK have discrete effects on survival, proliferation, and cytokine expression in Nf1-deficient myeloid cells. Cancer Cell. 2002;2:507–514. doi: 10.1016/s1535-6108(02)00214-3. [DOI] [PubMed] [Google Scholar]
- Chiang M, Xu L, Shestova O, Histen G, L'heureux S, et al. Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras-initiated leukemia. J Clin Invest. 2008;118:3181–3194. doi: 10.1172/JCI35090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler T, Cornejo M, Scholl C, Liu J, Leeman D, et al. K-RasG12D-induced T-cell lymphoblastic lymphoma/leukemias harbor Notch1 mutations and are sensitive to gamma-secretase inhibitors. Blood. 2008;112:3373–3382. doi: 10.1182/blood-2008-03-147587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–226. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- Penit C, Vasseur F, Papiernik M. In vivo dynamics of CD4–8- thymocytes. Proliferation, renewal and differentiation of different cell subsets studied by DNA biosynthetic labeling and surface antigen detection. Eur J Immunol. 1988;18:1343–1350. doi: 10.1002/eji.1830180907. [DOI] [PubMed] [Google Scholar]
- Tourigny M, Mazel S, Burtrum D, Petrie H. T cell receptor (TCR)-beta gene recombination: dissociation from cell cycle regulation and developmental progression during T cell ontogeny. J Exp Med. 1997;185:1549–1556. doi: 10.1084/jem.185.9.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swat W, Shinkai Y, Cheng H, Davidson L, Alt F. Activated Ras signals differentiation and expansion of CD4+8+ thymocytes. Proc Natl Acad Sci U S A. 1996;93:4683–4687. doi: 10.1073/pnas.93.10.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira JB, Bidere N, Niemela JE, Zheng L, Sakai K, et al. NRAS mutation causes a human autoimmune lymphoproliferative syndrome. Proc Natl Acad Sci U S A. 2007;104:8953–8958. doi: 10.1073/pnas.0702975104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson CH, Ailles LE, Dylla SJ, Muijtjens M, Jones C, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med. 2004;351:657–667. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- Mansour M, Duke V, Foroni L, Patel B, Allen C, et al. Notch-1 mutations are secondary events in some patients with T-cell acute lymphoblastic leukemia. Clin Cancer Res. 2007;13:6964–6969. doi: 10.1158/1078-0432.CCR-07-1474. [DOI] [PubMed] [Google Scholar]
- Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L, Mercer K, Breenbaum D, Bronson R, Crowley D, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- Guerra C, Mijimolle N, Dhawahir A, Dubus P, Barradas M, et al. Tumor induction by an endogenous K-ras oncogene is highly dependent on cellular context. Cancer Cell. 2003;4:111–120. doi: 10.1016/s1535-6108(03)00191-0. [DOI] [PubMed] [Google Scholar]
- Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- Sansom OJ, Meniel V, Wilkins JA, Cole AM, Oien KA, et al. Loss of Apc allows phenotypic manifestation of the transforming properties of an endogenous K-ras oncogene in vivo. Proc Natl Acad Sci U S A. 2006;103:14122–14127. doi: 10.1073/pnas.0604130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuveson DA, Hingorani SR. Ductal pancreatic cancer in humans and mice. Cold Spring Harb Symp Quant Biol. 2005;70:65–72. doi: 10.1101/sqb.2005.70.040. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- Kim CF, Jackson EL, Woolfenden AE, Lawrence S, Babar I, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Szilvassy SJ, Humphries RK, Lansdorp PM, Eaves AC, Eaves CJ. Quantitative assay for totipotent reconstituting hematopoietic stem cells by a competitive repopulation strategy. Proc Natl Acad Sci U S A. 1990;87:8736–8740. doi: 10.1073/pnas.87.22.8736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid I, Cole SW, Korin YD, Zack JA, Giorgi JV. Detection of cell cycle subcompartments by flow cytometric estimation of DNA-RNA content in combination with dual-color immunofluorescence. Cytometry. 2000;39:108–116. doi: 10.1002/(sici)1097-0320(20000201)39:2<108::aid-cyto3>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Boggs DR. The total marrow mass of the mouse: a simplified method of measurement. Am J Hematol. 1984;16:277–286. doi: 10.1002/ajh.2830160309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cells expressing c-kit were enriched by magnetic bead isolation, then doubly sorted by FACS. FACS data for the first and second sorts for typical examples of WT and KrasG12D mice are shown.
(740 KB TIF)
Flow cytometry was performed to enumerate Flk2− LSK cells in bone marrow (n = 4; p < 0.05) and spleens (n = 6; p < 0.05) harvested from 8-wk-old KrasG12D or WT mice. Data were analyzed as in Figure 2. Combined numbers in marrow and spleen were not significantly different between genotypes (p = 0.27). Data are pooled from two independent experiments; error bars show standard error of the mean (SEM). Statistical significance was evaluated with the unpaired t-test.
(80 KB TIF)
Mice transplanted with 500 CD45.1 KrasG12D and 500 CD45.1/CD45.2 WT HSCs were sacrificed (A) 2 mo (n = 6) or (B) 3 mo (n = 2) after transplantation, and their chimerism was assessed by flow cytometry. The percentage of graft cells derived from the KrasG12D HSCs is shown for Flk2− LSK cells (black bars), CMPs (dark grey bars), granulocyte-macrophage progenitors (GMP, light grey bars), and mature (Mac1/Gr1+) circulating myeloid cells (open bars). GMPs were defined as Lin−/lo Sca1− c-kit+ FcγRII/III+. Animals lacking any graft-derived contribution to the HSC compartment have been omitted.
(168 KB TIF)
The indicated numbers of mice were transplanted with 10,000, 30,000, or 100,000 nucleated bone marrow cells, and the proportions with detectable multilineage engraftment were determined 2 mo later. The number of repopulating units was calculated using the totals from these four experiments with L-Calc software (StemCell Technologies).
(47 KB PDF)