Abstract
The mucin 1 (MUC1) oncoprotein is aberrantly overexpressed in human breast cancers. Although MUC1 modulates the activity of estrogen receptor α (ER), there is no information regarding the effects of MUC1 on global gene expression patterns and the potential role of MUC1-induced genes in predicting outcome for breast cancer patients. We have developed an experimental model of MUC1-induced transformation that has identified the activation of genes involved in cholesterol and fatty acid metabolism. A 38-gene set of experimentally derived MUC1-induced genes associated with lipid metabolism was applied to the analysis of ER+ breast cancer patients treated with tamoxifen. The results obtained from 2 independent databases demonstrate that patients overexpressing MUC1 and the lipid metabolic pathways are at significantly higher risk for death and recurrence/distant metastasis. By contrast, these genes were not predictive in untreated patients. Furthermore, a positive correlation was found between expression of the 38-gene set and the ER signaling pathway. These findings indicate that (i) MUC1 regulates cholesterol and fatty acid metabolism, and (ii) activation of these pathways in ER+ breast cancers predicts failure to tamoxifen treatment.
Keywords: breast cancer, expression profiling, lipid metabolism, cholesterol biosynthesis, risk factors
The mucin 1 (MUC1) transmembrane glycoprotein is expressed on the apical borders of normal secretory mammary epithelial cells (1). With loss of polarity in association with transformation, MUC1 is expressed at high levels over the entire cell surface, allowing MUC1 to associate with members of the ErbB family of receptor tyrosine kinases (1, 2). The MUC1 C-terminal subunit (MUC1-C), and specifically its 72-aa cytoplasmic domain (MUC1-CD), also interacts with diverse effectors, such as c-Src (3), β-catenin (3, 4), and IKKβ/NF-κB (5), that have been linked to transformation. Other work on human breast cancer cells has demonstrated that MUC1-C accumulates in the cytosol and is transported to the nucleus (6), where it interacts with estrogen receptor α (ER) (7). MUC1-C associates with ER complexes on estrogen-responsive promoters, increases recruitment of p160 coactivators, and antagonizes the inhibitory effects of the antiestrogen tamoxifen (7). Tamoxifen is used as an adjuvant treatment to prevent breast cancer recurrence and as a therapy to extend the lives of patients with metastatic disease (8, 9). Levels of ER and progesterone receptor (PR) protein expression are predictors of tamoxifen response; however, 25% of ER+/PR+ tumors, 66% of ER+/PR− tumors, and 55% of ER−/PR+ tumors fail tamoxifen treatment (10). The mechanisms responsible for these treatment failures remain unclear. Here, we report that overexpression of the cytoplasmic domain of MUC1 (MUC1-CD) leads to altered expression of genes involved in cholesterol and fatty acid metabolism. These pathways are strongly associated with ER-dependent signaling and significantly predict response to tamoxifen treatment, as measured by disease-free and overall survival.
Results
MUC1 Is Associated with Alterations in Genes That Regulate Lipid Metabolism.
By using Ingenuity Pathway Analysis (IPA) to functionally classify the genes differentially expressed in MUC1-CD-transformed 3Y1 cells growing in vitro and in vivo, we identified a set of genes encoding proteins involved in cholesterol and fatty acid metabolism. As determined by Fisher's exact test, the incidence of genes involved in lipid metabolism among the differentially expressed genes was highly significant, with P values of 10−6 and 10−10 for cells grown in vitro and in vivo, respectively. Based on these findings, we obtained a 38-gene set that we designated the MUC1-induced lipid metabolism signature (MLMS; Table 1). These genes form a specific network that is largely represented by enzymes and transporters (Fig. 1). These genes also converge on a central node that contains the gene encoding the sterol regulatory element-binding protein 1 (SREBP1; Fig. 1 and Table 1). SREBP1 is a potent activator of SREBP-responsive genes that control the synthesis of cholesterol, fatty acids, and triglycerides (11). In addition, the gene encoding INSIG1, a transcriptional activator of SREBP1 (12), was identified in the MLMS (Table 1).
Table 1.
The 38-gene MLMS
| Gene symbol | Function |
|---|---|
| ABCA1 | Cholesterol efflux pump |
| ACAT2 | Formation of cholesteryl esters |
| ACLY | Synthesis of acetyl-CoA |
| ACSL1 | Lipid biosynthesis and fatty acid degradation |
| ACSL3 | Lipid biosynthesis and fatty acid degradation |
| APOC1 | Lipid and cholesterol transport |
| APOE | Lipid and cholesterol transport |
| CYP51A1 | Biosynthesis of cholesterol |
| DHCR24 | Biosynthesis of cholesterol |
| DHCR7 | Biosynthesis of cholesterol |
| ECHDC1 | β-Oxidation of fatty acids |
| ELOVL6 | Fatty acid elongation |
| FADS1 | Desaturation of fatty acids |
| FASN | Synthesis of saturated fatty acids |
| FDFT1 | Biosynthesis of cholesterol |
| FDPS | Biosynthesis of cholesterol |
| HMGCS1 | Biosynthesis of cholesterol |
| HSD11B1 | Interconversion of cortisol and cortisone |
| IDH1 | Production of NADPH |
| IDI1 | Biosynthesis of cholesterol |
| INSIG1 | Transcriptional activator of SREBP1 |
| LSS | Biosynthesis of cholesterol |
| NSDHL | Biosynthesis of cholesterol |
| PCYT2 | Synthesis of major membrane phospholipids |
| PLA2G2A | Hydrolysis of phosphoglycerides |
| PMVK | Biosynthesis of cholesterol |
| PSAP | Precursor of sphingolipid activator proteins |
| PSAT1 | Estrogen and progesterone-regulated enzyme |
| PTGS1 | Synthesis of prostaglandins |
| PTGS2 | Synthesis of prostaglandins |
| SC4MOL | Biosynthesis of cholesterol |
| SC5DL | Biosynthesis of cholesterol |
| SCD | Synthesis of unsaturated fatty acids |
| SLC16A7 | Pyruvate/lactate transporter |
| SOAT1 | Biosynthesis of cholesterol |
| SQLE | Biosynthesis of cholesterol |
| SREBP1 | Transcriptional regulator lipid biosynthesis |
| VLDLR | Mediates entry of fatty acids and triglycerides |
Fig. 1.
The MLMS forms a specific network largely represented by enzymes and transporters that are linked to the sterol regulatory element-binding protein 1 (SREBP1/SREBF1). Red color indicates presence in the MLMS; solid line indicates activation; dashed line indicates deactivation.
MLMS Comprises Multiple Genes Involved in Cholesterol and Fatty Acid Metabolism.
The MLMS includes the gene encoding ATP citrate lyase (ACLY), an enzyme that mediates synthesis of acetyl CoA, the common precursor for the cholesterol and fatty acid pathways (Table 1). The differentially expressed genes also represented the biosynthesis of cholesterol and its derivatives, as well as the transport of these molecules; for example, ABCA1, APOC1, APOE, and VLDLR (Table 1 and Fig. S1). Other genes in the MLMS are involved in the synthesis of fatty acids (FASN) and their metabolism (ACSL1, ACLS3, and ECHDC1; Table 1).
MLMS Predicts Response of Human Breast Cancers to Tamoxifen Treatment.
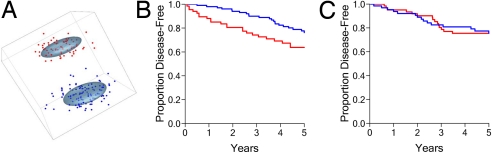
The findings that MUC1 (i) interacts with ER, (ii) blocks the effects of tamoxifen in vitro, and (iii) regulates lipid metabolism suggested that MUC1 expression could have an effect on the response of ER+ breast cancers to tamoxifen treatment. In this context, we examined a database from 176 patients with ER+ breast tumors who were treated with tamoxifen in the adjuvant setting (13). Using k-means clustering to distinguish patients with differential MLMS expression and principal component analysis to visualize the clusters, we found segregation into MLMS+ and MLMS− groups, in which MLMS+ significantly overexpressed MUC1 (2-tailed t test, P < 0.0001) and the entire 38-gene set [mean expression score ± SEM (see Materials and Methods): MLMS+ score, 1.22 ± 0.018; MLMS− score, −0.33 ± 0.015; 2-tailed t test, P < 0.0001; Fig. 2A]. Group 1 includes 67 patients with MLMS+ tumors, and group 2 represents 109 patients with MLMS− tumors (Fig. 2A). Recurrence/distant metastasis-free (disease-free) survival was significantly lower (P = 0.031) in the MLMS+ compared with the MLMS− group (Fig. 2B). This association between MLMS status and response to tamoxifen treatment was also confirmed by a univariate Cox proportional hazard analysis of recurrence/distant metastasis-free survival (hazard ratio, 1.88; 95% C.I., 1.04–3.39; P = 0.036). This database also includes 125 patients with ER+ (n = 85) and ER− tumors who were not treated with tamoxifen. For these patients, recurrence/distant metastasis-free (disease-free) survival was identical (P = 0.98) in the MLMS+ and MLMS− groups (Fig. 2C). Overall survival is not available for this database. These findings thus indicate that MLMS+ status is highly predictive of the response to tamoxifen with respect to disease-free survival.
Fig. 2.
Overexpression of the MLMS is predictive of response to tamoxifen treatment with respect to disease-free survival. (A) The k-means clustering of 176 ER+ tamoxifen-treated breast tumors based on differential MLMS expression and visualization of clusters by using principal components analysis demonstrates distinct partitioning of tumors into MLMS+ (overexpressors; red) and MLMS− (blue) groups. (B) Kaplan–Meier survival curves for recurrence/distant metastasis-free (disease-free) survival indicate significantly (log-rank P = 0.031) decreased survival in the tamoxifen-treated MLMS+ group compared with the MLMS− group. (C) Kaplan–Meier survival analysis for recurrence/distant-metastasis-free (disease-free) survival indicates no survival difference (log-rank P = 0.98) between the MLMS+ and MLMS− groups within the 125 untreated breast cancer patients.
Confirmation of the Predictive Capability of the MLMS.
A second database derived from 147 patients with ER+ breast tumors who were treated with adjuvant tamoxifen also demonstrated that MLMS status identifies 2 distinct groups. Group 1 includes 25 patients with MLMS+ tumors, and group 2 represents 122 patients with MLMS− tumors [mean expression score ± SEM (see Materials and Methods): MLMS+ score, 2.24 ± 0.059; MLMS− score, −0.076 ± 0.015; 2-tailed t test, P < 0.0001; Fig. 3A]. MUC1 was overexpressed in tumors with MLMS+ status (2-tailed t test, P < 0.0001; Fig. 3B). As found in the first database, the MLMS+ group had a highly significant (P = 2.2E-7) decrease in recurrence/distant metastasis-free (disease-free) survival compared with that for the MLMS− group (Fig. 3C). Moreover, and notably, overall survival was significantly (P = 4.0E-7) decreased for patients with MLMS+ tumors (Fig. 3D). Univariate Cox proportional hazard analyses confirmed that MLMS+ status is associated with statistically increased risks for recurrence/distant metastasis [hazard ratio (HR), 4.24; 95% C.I., 2.18–8.00; P = 5.8E-5] and death (HR, 5.19; 95% C.I. 2.25–11.84; P = 2.1E-4). Furthermore, a multivariate analysis demonstrated that MLMS+ status has the greatest HR for both disease-free and overall survival (Table 2).
Fig. 3.
The MLMS is coexpressed with MUC1 and predicts poor response to tamoxifen treatment with respect to disease-free and overall survival. (A) Hierarchical clustering of 147 ER+ tamoxifen-treated breast tumors based on differential MLMS expression demonstrates distinct partitioning of tumors into MLMS+ (overexpressors) and MLMS− groups. Expression values are indicated in log2 scale. The genes having significant overexpression across both databases are shown. (B) MUC1 is overexpressed in the MLMS+ group. + indicates overexpression. (C and D) Kaplan–Meier survival curves for recurrence/distant metastasis-free (disease-free) (C) and overall (D) survival indicate significantly decreased survival in the MLMS+ compared with the MLMS− group (disease-free survival log-rank P = 2.2E-7; overall survival log-rank P = 4.0E-7).
Table 2.
Multivariate Cox proportional hazard analysis of recurrence/distant metastasis (disease) and death for 147 ER+, tamoxifen-treated breast cancer patients
| Effect | Disease-free survival |
Overall survival |
||||
|---|---|---|---|---|---|---|
| HR | 95% C.I. | P | HR | 95% C.I. | P | |
| Age (per year) | 1.01 | 0.98–1.05 | 0.48 | 1.02 | 0.97–1.07 | 0.48 |
| Grade (3 vs. 1, 2) | 2.12 | 1.00–4.36 | 0.048 | 3.91 | 1.45–10.84 | 0.0074 |
| Tumor size (per mm) | 1.02 | 0.98–1.06 | 0.31 | 0.98 | 0.91–1.04 | 0.46 |
| Lymph node+ | 1.55 | 0.66–3.70 | 0.32 | 1.76 | 0.49–7.18 | 0.39 |
| PR+ | 0.34 | 0.16–0.72 | 0.0054 | 0.39 | 0.14–1.06 | 0.064 |
| MLMS+ | 3.90 | 1.68–8.88 | 0.0020 | 4.57 | 1.61–13.02 | 0.0047 |
The indicated model effects were used in the analysis. Age and tumor size were considered continuous variables. Grade, positive lymph node status, progesterone receptor expression, and MLMS status were considered binary variables.
We expanded this multivariate analysis by stratifying grade 3 and lymph node-positive patients by using the MLMS. For lymph node-positive patients, those expressing the MLMS had a 2.66-fold (95% C.I., 1.21–5.78; P = 0.016) greater risk for disease and a 2.86-fold (95% C.I., 1.09–7.61; P = 0.033) greater risk for death compared with nonexpressors. For patients with grade 3 tumors, those expressing the MLMS had a 5.40-fold (95% C.I., 1.13–20.88; P = 0.037) greater risk for disease and a 5.47-fold (95% C.I., 1.16–20.11; P = 0.034) greater risk for death compared with nonexpressors. Taken together with the previous data, these results confirm that MLMS status enhances the identification of patients at greater risks for disease and death.
MLMS Is Coexpressed with ER-Dependent Genes.
ER protein expression is not necessarily reflective of active estrogen signaling (14). To determine whether MLMS status is associated with ER-mediated transcription, we examined both databases for activation of ER-dependent genes (15, 16). The results demonstrate that MLMS+ status correlates significantly (r > 0.97; P < 0.0001) with ER-dependent gene expression (Fig. 4).
Fig. 4.
Overexpression of the MLMS is associated with ER-dependent gene expression. Hierarchical clustering of breast tumors based on differential ER-dependent gene expression for 147 (A) and 176 (C) cases demonstrates distinct partitioning into 2 groups (+, MLMS+; −, MLMS−). Genes having significantly different expression between the 2 groups are shown. (B and D) ER-dependent gene expression (ER score) and MLMS expression (MLMS score) are significantly correlated in both databases (B, r = 0.97, P < 0.0001; D, r = 0.98, P < 0.0001).
Discussion
The present findings provide a previously uncharacterized, biologically derived, MUC1-induced gene signature that is highly predictive of response to tamoxifen. The MUC1 oncoprotein is aberrantly overexpressed in about 90% of human breast tumors (1, 17). MUC1 overexpression is conferred, at least in part, by up-regulation of MUC1 mRNA levels at the transcriptional level (18–21). MUC1 interacts with ER and certain other transcription factors, and thereby contributes to the regulation of gene expression (7, 22). However, little is otherwise known about the effects of MUC1 on global gene expression patterns. Our analysis of MUC1-transformed cells demonstrated induction of the SREBP1 gene, which encodes a transcription factor involved in the activation of genes in the cholesterol and fatty acid synthetic pathways (11). In this context, MUC1-induced transformation is associated with activation of the ACLY gene that confers the synthesis of acetyl CoA needed for both the cholesterol and fatty acid pathways. MUC1 also activated multiple genes involved in cholesterol and fatty acid synthesis. Importantly, one of the genes activated by MUC1 encodes fatty acid synthase (FASN), an enzyme that is up-regulated in diverse human malignancies and has been linked to the transformed phenotype (23). Based on these findings, we identified a 38-gene, MUC1-induced signature associated with cholesterol and fatty acid synthesis. The results demonstrate that this 38-gene MLMS is highly predictive for decreased recurrence/metastasis-free survival and overall survival in patients treated with tamoxifen.
Predictive analysis of gene expression has been used to identify signatures that are predictive of recurrence in patients with primary breast tumors treated with tamoxifen (10, 24–26). The findings of a 2-gene signature by Ma et al. (10) have been questioned (27–29). The study by Paik et al. (24) analyzed expression of 21 selected genes in a classifier that was predictive of recurrence and overall survival. This classifier included genes involved in proliferation and ER signaling (24). Using a genome-wide microarray analysis, Chanrion et al. (25) further identified a 36-gene classifier that was predictive of disease-free survival. Eleven genes in the 36-gene signature belong to a proliferation cluster. Moreover, 18 genes were related to ER targets or regulators of ER function (25). This relationship between ER-responsive genes and response to tamoxifen treatment was extended in the study by Musgrove et al. (26). Importantly, the MLMS identified in the present work is distinct from those reported previously (24–26). In this regard, the MLMS represents genes involved in lipid metabolism. Nonetheless, the finding that MLMS+ status is associated with activation of ER-dependent genes indicates that the MLMS genes may function as upstream regulators of the ER response.
Our findings also differ from many of the empirically derived gene signatures in that the present stratification is based on biologically identified lipid pathways associated with MUC1-induced transformation. Using the same approaches, we found that activation of a STAT1-dependent pathway mediates resistance to radiation treatment and predicts response to adjuvant chemotherapy and radiation in breast cancer patients (30–32). Thus, these approaches have allowed the identification of biologically relevant and differentially expressed pathways in experimental models. This “bottom-up” approach has been defined in the literature, but to date has resulted in the identification of a limited number of signatures (30, 33–36). The ability to stratify databases based on known pathways can provide new insights into the development of tumors and the identification of potential therapeutic targets. In this regard, the present findings indicate that targeting of the MUC1 protein could be an effective strategy for attenuating activation of the cholesterol and fatty acid synthetic pathways and for blocking resistance to tamoxifen treatment of breast cancers.
Materials and Methods
Cell Culture and Tumor Models.
Rat 3Y1 embryonic fibroblasts were stably transfected with an empty vector (3Y1/Vector) or one expressing the cytoplasmic domain of MUC1 (3Y1/MUC1-CD) and maintained as described previously (4). Cells were injected s.c. into the hind limbs of female athymic mice (FCRI-Taconic) in increasing concentrations from 102 to 107 cells in 100 μL of PBS per mouse. Ten mice were injected at each cell concentration. Tumor volume was determined by direct measurement with calipers and calculated by using the formula (length × width × depth/2). When the tumor volume was greater than or equal to 2,000 mm3, mice were euthanized by using CO2 followed by cervical dislocation. Tumors were excised, snap-frozen in liquid nitrogen, and stored at −80 °C until RNA extraction. All animal experiments were conducted in accordance with institutional guidelines at The University of Chicago.
Statistical Analysis of DNA Microarrays.
RNA was purified and hybridized with GeneChip Rat Genome 230 2.0 Arrays (Affymetrix) as described previously (37). The selection and analysis of genes differentially expressed in 3Y1/Vector and 3Y1/MUC1-CD cells in vitro and 3Y1/MUC1-CD xenografts were based on previously detailed approaches (30, 37–39). Briefly, each array was hybridized with a pooled sample normalized to total RNA and consisting of RNA obtained from 3 independent xenografts or cell lines. After data retrieval and scaling by using MAS 5.0 suit (Affymetrix), data were rescaled by using “global median normalization” across the entire dataset (37) and filtrated by using a multistep filtration method that involves the application of receiver-operating characteristic analysis (ROC analysis) for the estimation of cutoff signal intensity values (39). Subsequent analysis was based on pairwise comparisons (3Y1/Vector in vitro vs. 3Y1/MUC1-CD in vitro and 3Y1/MUC1-CD in vitro vs. 3Y1/MUC1-CD in vivo) of duplicated arrays by using significance analysis of microarrays (SAM; ref. 40) version 3.0. Differentially expressed probe set IDs were selected by using a 2.0-fold induction cutoff level with a false-discovery ratio of 0. Selected probe set IDs were gene annotated and functionally designated by using IPA (Ingenuity Systems Inc.). Fisher's exact test was used to estimate the significance of the incidence of different functional groups. This method estimates the probability that the association between an experimental and reference gene set is due to random chance. A P value ≤0.05 is considered significant and indicates a nonrandom enrichment of an experimental dataset by members of a specific functional group. Microarray data have been deposited in GEO (accession number GSE14337).
Statistical Analysis of Breast Cancer Databases.
We analyzed 2 publicly available databases containing expressional data from breast cancer (13, 25, 41) to determine whether the MUC1-induced gene set is predictive in determining response to tamoxifen treatment. Statistical analyses were performed by using samples for which survival data were available, which included 301 cases (13, 41) and 147 cases (25). All statistical analyses were performed by using JMP 7.1 (SAS Institute Inc.). The raw signal intensity for each probe set ID of interest for each patient was normalized to the median value of the probe set ID across the entire database and subsequently log2-transformed. Multiple probe set IDs for a given gene were averaged for each patient sample to obtain a representative expression value for each gene. Expression data were clustered by using hierarchical clustering via Ward's method. The k-means clustering was performed to partition the patient samples into 2 clusters, and principal component analysis was used to visualize these clusters. To identify the genes that were differentially expressed between the 2 patient clusters, we first used F tests to test the null hypothesis of equal variance for each gene between the 2 patient clusters. We entered the result of the F test (equal or unequal variance) into an unpaired 2-tailed Student's t test to test the null hypothesis of equal magnitude of gene expression of each gene between the 2 patient clusters. The α level for each t test was 0.05; however, we corrected for multiple comparisons by using a Bonferroni correction that adjusted the α level to 0.05 divided by the number of comparisons of interest. Gene expression scores for the MUC1-induced lipid metabolism signature (denoted MLMS score) and ER-dependent signature (denoted ER score) were determined for each patient sample by calculating the average log2 level of expression across all differentially expressed genes in the respective gene set. A cutoff score of 0.58 (1.5-fold relative expression) was used to indicate overexpression of the respective gene set. To determine whether clustering based on the differential expression of the MUC1-induced genes could identify patients with decreased survival, we performed Kaplan–Meier survival statistics. Survival analysis was performed on clusters defined by k-means clustering, and log-rank tests were used to test the null hypothesis of no difference in survival functions between the 2 patient clusters.
Supplementary Material
Acknowledgments.
We thank Dr. Samuel Hellman for helpful discussion of the manuscript. These studies were supported by grants from the Lung Cancer Foundation and by National Cancer Institute Grants CA71933, CA78766, and CA97098.
Footnotes
Conflict of interest statement: D.W.K. holds equity in Genus Oncology and is a consultant for the company.
This article is a PNAS Direct Submission.
Data deposition: The microarray data have been deposited in the Gene Expression Omnibus (GEO) Database, www.ncbi.nlm.nih.gov/geo (accession no. GSE14337).
This article contains supporting information online at www.pnas.org/cgi/content/full/0812029106/DCSupplemental.
References
- 1.Kufe D, et al. Differential reactivity of a novel monoclonal antibody (DF3) with human malignant versus benign breast tumors. Hybridoma. 1984;3:223–232. doi: 10.1089/hyb.1984.3.223. [DOI] [PubMed] [Google Scholar]
- 2.Li Y, et al. Heregulin targets gamma-catenin to the nucleolus by a mechanism dependent on the DF3/MUC1 oncoprotein. Mol Cancer Res. 2003;1:765–775. [PubMed] [Google Scholar]
- 3.Li Y, Kuwahara H, Ren J, Wen G, Kufe D. The c-Src tyrosine kinase regulates signaling of the human DF3/MUC1 carcinoma-associated antigen with Gsk3 beta and beta-catenin. J Biol Chem. 2001;276:6061–6064. doi: 10.1074/jbc.C000754200. [DOI] [PubMed] [Google Scholar]
- 4.Huang L, et al. MUC1 oncoprotein blocks glycogen synthase kinase 3beta-mediated phosphorylation and degradation of beta-catenin. Cancer Res. 2005;65:10413–10422. doi: 10.1158/0008-5472.CAN-05-2474. [DOI] [PubMed] [Google Scholar]
- 5.Ahmad R, et al. MUC1 oncoprotein activates the Ikappab kinase beta complex and constitutive NF-kappab signalling. Nat Cell Biol. 2007;9:1419–1427. doi: 10.1038/ncb1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leng Y, et al. Nuclear import of the MUC1-C oncoprotein is mediated by nucleoporin Nup62. J Biol Chem. 2007;282:19321–19330. doi: 10.1074/jbc.M703222200. [DOI] [PubMed] [Google Scholar]
- 7.Wei X, Xu H, Kufe D. MUC1 oncoprotein stabilizes and activates estrogen receptor alpha. Mol Cell. 2006;21:295–305. doi: 10.1016/j.molcel.2005.11.030. [DOI] [PubMed] [Google Scholar]
- 8.Jordan VC. Chemoprevention of breast cancer with selective oestrogen-receptor modulators. Nat Rev Cancer. 2007;7:46–53. doi: 10.1038/nrc2048. [DOI] [PubMed] [Google Scholar]
- 9.Jensen EV, Jordan VC. The estrogen receptor: A model for molecular medicine. Clin Cancer Res. 2003;9:1980–1989. [PubMed] [Google Scholar]
- 10.Ma XJ, et al. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell. 2004;5:607–616. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 11.Horton JD, Goldstein JL, Brown MS. SREBPs: Activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heemers HV, Verhoeven G, Swinnen JV. Androgen activation of the sterol regulatory element-binding protein pathway: Current insights. Mol Endocrinol. 2006;20:2265–2277. doi: 10.1210/me.2005-0479. [DOI] [PubMed] [Google Scholar]
- 13.Loi S, et al. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol. 2007;25:1239–1246. doi: 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]
- 14.Tonini G, Schiavon G, Fratto ME, Vincenzi B, Santini D. Hormono-biological therapy in metastatic breast cancer: Preclinical evidence, clinical studies and future directions. Expert Opin Biol Ther. 2008;8:221–234. doi: 10.1517/14712598.8.2.221. [DOI] [PubMed] [Google Scholar]
- 15.Kristensen VN, et al. Gene expression profiling of breast cancer in relation to estrogen receptor status and estrogen-metabolizing enzymes: Clinical implications. Clin Cancer Res. 2005;11:878s–883s. [PubMed] [Google Scholar]
- 16.Weisz A, et al. Molecular identification of ERalpha-positive breast cancer cells by the expression profile of an intrinsic set of estrogen regulated genes. J Cell Physiol. 2004;200:440–450. doi: 10.1002/jcp.20039. [DOI] [PubMed] [Google Scholar]
- 17.Hareuveni M, et al. A transcribed gene, containing a variable number of tandem repeats, codes for a human epithelial tumor antigen. cDNA cloning, expression of the transfected gene and over-expression in breast cancer tissue. Eur J Biochem. 1990;189:475–486. doi: 10.1111/j.1432-1033.1990.tb15512.x. [DOI] [PubMed] [Google Scholar]
- 18.Abe M, Kufe D. Transcriptional regulation of DF3 gene expression in human MCF-7 breast carcinoma cells. J Cell Physiol. 1990;143:226–231. doi: 10.1002/jcp.1041430205. [DOI] [PubMed] [Google Scholar]
- 19.Abe M, Kufe D. Characterization of cis-acting elements regulating transcription of the human DF3 breast carcinoma-associated antigen (MUC1) gene. Proc Natl Acad Sci USA. 1993;90:282–286. doi: 10.1073/pnas.90.1.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kovarik A, Peat N, Wilson D, Gendler SJ, Taylor-Papadimitriou J. Analysis of the tissue-specific promoter of the MUC1 gene. J Biol Chem. 1993;268:9917–9926. [PubMed] [Google Scholar]
- 21.Gaemers IC, Vos HL, Volders HH, van der Valk SW, Hilkens J. A STAT-responsive element in the promoter of the episialin/MUC1 gene is involved in its overexpression in carcinoma cells. J Biol Chem. 2001;276:6191–6199. doi: 10.1074/jbc.M009449200. [DOI] [PubMed] [Google Scholar]
- 22.Wei X, Xu H, Kufe D. Human MUC1 oncoprotein regulates p53-responsive gene transcription in the genotoxic stress response. Cancer Cell. 2005;7:167–178. doi: 10.1016/j.ccr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 23.Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 24.Paik S, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 25.Chanrion M, et al. A gene expression signature that can predict the recurrence of tamoxifen-treated primary breast cancer. Clin Cancer Res. 2008;14:1744–1752. doi: 10.1158/1078-0432.CCR-07-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Musgrove EA, et al. Identification of functional networks of estrogen- and c-Myc-responsive genes and their relationship to response to tamoxifen therapy in breast cancer. PLoS ONE. 2008;3:e2987. doi: 10.1371/journal.pone.0002987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan C, et al. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med. 2006;355:560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 28.Goetz MP, et al. A two-gene expression ratio of homeobox 13 and interleukin-17b receptor for prediction of recurrence and survival in women receiving adjuvant tamoxifen. Clin Cancer Res. 2006;12:2080–2087. doi: 10.1158/1078-0432.CCR-05-1263. [DOI] [PubMed] [Google Scholar]
- 29.Reid JF, et al. Limits of predictive models using microarray data for breast cancer clinical treatment outcome. J Natl Cancer Inst. 2005;97:927–930. doi: 10.1093/jnci/dji153. [DOI] [PubMed] [Google Scholar]
- 30.Khodarev NN, et al. STAT1 is overexpressed in tumors selected for radioresistance and confers protection from radiation in transduced sensitive cells. Proc Natl Acad Sci USA. 2004;101:1714–1719. doi: 10.1073/pnas.0308102100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khodarev NN, et al. Signal transducer and activator of transcription 1 regulates both cytotoxic and prosurvival functions in tumor cells. Cancer Res. 2007;67:9214–9220. doi: 10.1158/0008-5472.CAN-07-1019. [DOI] [PubMed] [Google Scholar]
- 32.Weichselbaum RR, et al. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc Natl Acad Sci USA. 2008;105:18490–18495. doi: 10.1073/pnas.0809242105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang HY, et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc Natl Acad Sci USA. 2005;102:3738–3743. doi: 10.1073/pnas.0409462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang HY, et al. Gene expression signature of fibroblast serum response predicts human cancer progression: Similarities between tumors and wounds. PLoS Biol. 2004;2:E7. doi: 10.1371/journal.pbio.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chi JT, et al. Gene expression programs in response to hypoxia: Cell type specificity and prognostic significance in human cancers. PLoS Med. 2006;3:e47. doi: 10.1371/journal.pmed.0030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest. 2005;115:1503–1521. doi: 10.1172/JCI23412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimchi ET, et al. Progression of Barrett's metaplasia to adenocarcinoma is associated with the suppression of the transcriptional programs of epidermal differentiation. Cancer Res. 2005;65:3146–3154. doi: 10.1158/0008-5472.CAN-04-2490. [DOI] [PubMed] [Google Scholar]
- 38.Khodarev NN, Kataoka Y, Murley JS, Weichselbaum RR, Grdina DJ. Interaction of amifostine and ionizing radiation on transcriptional patterns of apoptotic genes expressed in human microvascular endothelial cells (HMEC) Int J Radiat Oncol Biol Phys. 2004;60:553–563. doi: 10.1016/j.ijrobp.2004.04.060. [DOI] [PubMed] [Google Scholar]
- 39.Khodarev NN, et al. Receiver operating characteristic analysis: A general tool for DNA array data filtration and performance estimation. Genomics. 2003;81:202–209. doi: 10.1016/s0888-7543(02)00042-3. [DOI] [PubMed] [Google Scholar]
- 40.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loi S, et al. Predicting prognosis using molecular profiling in estrogen receptor-positive breast cancer treated with tamoxifen. BMC Genomics. 2008;9:239. doi: 10.1186/1471-2164-9-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.