Abstract
A tetramer model for HIV-1 integrase (IN) with DNA representing 20 bp of the U3 and U5 long terminal repeats (LTR) termini was assembled using structural and biochemical data and molecular dynamics simulations. It predicted amino acid residues on the enzyme surface that can interact with the LTR termini. A separate structural alignment of HIV-1, simian sarcoma virus (SIV), and avian sarcoma virus (ASV) INs predicted which of these residues were unique. To determine whether these residues were responsible for specific recognition of the LTR termini, the amino acids from ASV IN were substituted into the structurally equivalent positions of HIV-1 IN, and the ability of the chimeras to 3′ process U5 HIV-1 or ASV duplex oligos was determined. This analysis demonstrated that there are multiple amino acid contacts with the LTRs and that substitution of ASV IN amino acids at many of the analogous positions in HIV-1 IN conferred partial ability to cleave ASV substrates with a concomitant loss in the ability to cleave the homologous HIV-1 substrate. HIV-1 IN residues that changed specificity include Val72, Ser153, Lys160–Ile161, Gly163–Val165, and His171–Leu172. Because a chimera that combines several of these substitutions showed a specificity of cleavage of the U5 ASV substrate closer to wild type ASV IN compared with chimeras with individual amino acid substitutions, it appears that the sum of the IN interactions with the LTRs determines the specificity. Finally, residues Ser153 and Val72 in HIV-1 IN are among those that change in enzymes that develop resistance to naphthyridine carboxamide- and diketo acid-related inhibitors in cells. Thus, amino acid residues involved in recognition of the LTRs are among these positions that change in development of drug resistance.
HIV-12 DNA integration is a concerted process that occurs in defined stages. After assembly of a stable complex of the viral integrase (IN) and host cell proteins with specific DNA sequences at the end of the HIV-1 LTR, the terminal dinucleotides are removed from each 3′ end by endo-nucleolytic processing. The viral DNA 3′ ends are then covalently linked to the host cell target DNA in a concerted reaction. Biochemical details concerning the processing and joining steps in the integration process have been reported for several retroviruses including avian sarcoma-leukosis virus (ASV), murine leukemia virus (MuLV), and HIV-1. Specific DNA sequences at the 3′ end of the viral LTR are required for recognition by the assembled viral integrase complex. Typically, the terminal 12–20 nucleotides are sufficient. After 3′ processing of the CAXX sequence from the ends, viral DNA ends are joined by the viral integrase and the gapped intermediates are repaired, and unpaired viral 5′ ends are excised by host nucleases. This results in a 4–6-base pair duplication of host cell DNA, depending upon the virus, flanking the integrated viral DNA. Both the 3′ processing and viral DNA-host DNA joining steps require integrase to be assembled on the specific viral DNA substrate. Integration of viral into host DNA occurs with limited sequence specificity (1). Several cell proteins have been reported to affect the integration process (2–8).
Much of the information available concerning the molecular mechanism of integration comes from the use of reconstituted systems employing duplex oligodeoxyribonucleotides (oligos). The ASV IN, for example, catalyzes specific cleavage at the 3′ end of the strands adjacent to the conserved CA dinucleotide using 15-bp substrates corresponding to the ASV U3 or U5 termini (9, 10). Similar substrates were used to demonstrate the joining reaction in which one oligo integrated into another (11–12). For HIV-1 duplex oligo substrates of comparable size, selected nucleotide substitutions in the U5 and U3 LTR regions were shown to affect one or both of the catalytic functions of HIV-1 integrase. Nucleic acid substitutions in the HIV-1 U5 LTR region, for example, can inhibit 3′ processing (e.g. positions 1–5 and 9–11) or not (e.g. positions 6–8 and 12–14) (13). Other changes at specific nucleic acid positions in the HIV-1 U3 and U5 LTR regions can affect each of the catalytic reactions, although changes in one region can be more pronounced than in the other.
Single LTR end duplex oligos are valuable tools for examining IN catalytic activity. Nevertheless, most do not display the concerted nature of the DNA integration reaction that was first demonstrated by Goff and co-worker (14). In these experiments, deletions were introduced 5′ to the conserved CA dinucleotide in the MuLV U3 LTR region. When cells were infected with the mutant viruses, processing at the ends of both the U3 and U5 LTR regions was affected adversely. This finding implies that the two ends of the viral DNA were brought together at the insertion site such that mutations in one affected the processing of the other. A homodimer of IN supports 3′ processing and one-ended joining, whereas a homotetramer of IN supports these activities in a two-ended concerted DNA integration reaction (15). Thus the biologically important form of IN is likely to be a tetramer. Several assay systems that display concerted DNA integration properties have been described and used to demonstrate that changes in retroviral DNA integration are context-dependent. For example, base pair substitutions in the dominant LTR (HIV-1 U5 and ASV U3) cause significant decreases in the rate of catalysis. In contrast, comparable substitutions in the nondominant LTR, the HIV-1 U3 and ASV U5 LTR regions, are associated with changes in the mechanism from concerted to nonconcerted DNA integration (2, 16–24). Like HIV-1, ASV and MuLV concerted DNA integration is affected by changes in the LTR region, although the critical positions differ among the different retroviruses.
HIV-1 integrase is a 288-amino acid protein composed of three functional domains that are required for each of the catalytic reactions (25). The conserved N-terminal domain (residues 1–50) contains a HHCC zinc-binding site, and binding of Zn2+ to this domain may participate in promoting the formation of IN oligomers (26–29). The C-terminal domain, the least conserved of the three domains in terms of amino acid sequence, is composed of a bundle of three α-helices that form a Src homology-3 fold. This domain has nonspecific DNA binding activity and determinants for multimerization (30). The catalytic core domain contains residues required for catalytic activity, including a conserved DDE motif that binds the required metal cofactor (1). Photo-cross-linking reagents have defined areas of the HIV-1 IN surface in close proximity to the LTR ends (13, 31–34). The viral DNA sequences stimulate high affinity binding of drugs that target the catalytic functions of integrase, suggesting that integrase may assume a distinct enzymatically active conformation following assembly (35, 36). No full-length integrase structure has yet been determined by x-ray crystallography. Therefore, little is known about how the three domains are positioned relative to one another in the active oligomeric state (37, 38) or what specific amino acid residues are involved in recognition of the viral DNA substrate. Some insight into the assembly of a stable complex of integrase with specific DNA sequences can be gleaned from DNA transposable elements. Such elements share a common mechanism of integration with retroviruses, and the catalytic domains of ASV and HIV integrases are highly homologous to those of Mu, Tn5, and Tc3 transposases (39–43). The Tc3 transposase is a 329-amino acid protein with an N-terminal DNA binding domain, a discreet second DNA binding domain flanking the N-terminal domain, and a catalytic core domain with a DDE motif (44). For the following study, we developed a structural model for HIV-1 IN with bound LTR DNA ends. This model was used to predict amino acid interactions between residues on the IN surface and the LTRs. By substituting amino acids from ASV IN into the structurally related positions of HIV-1, we identified a series of residues that participate in the specific recognition of the LTR ends.
EXPERIMENTAL PROCEDURES
Reagents
[γ-33P]ATP (2500 Ci/mmol), HiTrap™ chelating HP resin, and HiTrap™ heparin HP resin were purchased from Amersham Biosciences. T4 polynucleotide kinase was from United States Biochemical (Cleveland, OH). Isopropyl β-D-thiogalactopyranoside was from Roche Applied Science. A Slide-A-Lyzer dialysis cassette was obtained from Pierce. Centriprep centrifugal filter devices with YM-10 MW membranes were from Millipore (Bedford, MA). Acrylamide and bis-acrylamide solutions were from Bio-Rad. Simple Blue Safestain was from Invitrogen. DE81 filters were purchased from Whatman. Unless specified, all restriction enzymes were purchased from New England Biolabs (Beverly, MA). ASV IN was provided by Dr. Ann Skalka (Fox Chase Cancer Center, Philadelphia, PA).
Bacterial Strains and Growth Conditions
The protein expression host cells, BL21 (DE3), were purchased from Novagen (Madison, WI). An expression construct for HIV-1 IN 1–288 residues (p28bIN-3CS-F185H) was obtained from the laboratory of Dr. Ann Skalka and was constructed by Dr. Mark Andrake. The construct contains the wild type NY5 HIV-1 sequence of the IN gene (Parke-Davis clone) from the NdeI to the HindIII site in the pET28b plasmid vector. The IN sequence encodes four substitutions (C56S, C65S, C280S, and F185H) to increase the solubility of HIV-1 IN and a six-amino acid His tag and thrombin cleavage site at the N terminus of the gene. A translation stop codon was added after residue Asp288.
Preparation of Duplex Oligo Substrates
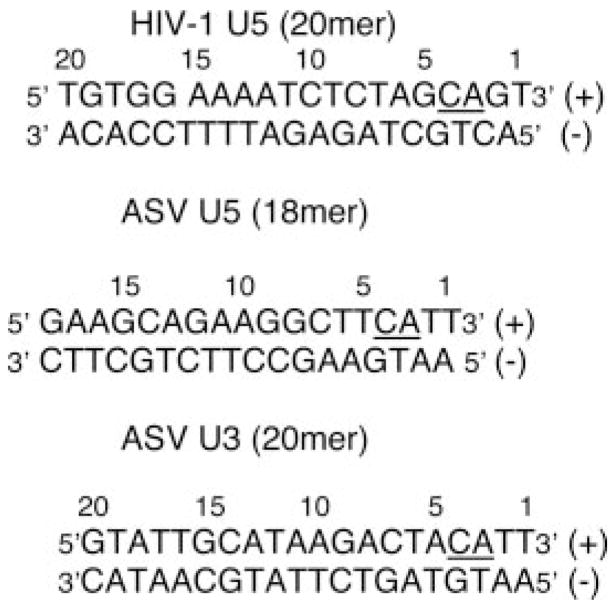
The following oligodeoxyribonucleotides were used in the integrase activity assay shown in Scheme 1.
SCHEME 1.
The plus-strand substrates (100 pmol, containing the conserved “CA” dinucleotides) were 5′ end-labeled using T4 polynucleotide kinase (30 U) and 2 μl of [γ-33P]ATP as described previously (45). The specific activity of the radiolabeled substrates was diluted to 105 cpm/pmol using an unlabeled processed strand oligo, and the mixture was purified and recovered from a 20% denaturing polyacrylamide gel. Duplex oligos were formed by annealing to a molar excess of unlabeled complementary strand as described (46).
Construction of Chimera HIV-1/ASV INs
Mutagenesis oligodeoxyribonucleotides were obtained from Integrated DNA Technologies Inc. (Coralville, IA) and are listed in the table found in the supplemental data. The mutations were constructed using the QuikChange site-directed mutagenesis kit from Stratagene (La Jolla, CA) according to manufacturer’s directions. Codon preferences for Escherichia coli were used in the design of the oligos. For chimeras S54–57, the mutations were introduced by the PCR overlap extension method (47). The presence of all mutations was confirmed by sequencing the complete individual DNA clones. A gel extraction kit (Qiagen, Valencia, CA) was used to purify all PCR products. The Minipreps DNA purification system (Pro-mega, Madison, WI) was used to prepare DNAs.
Purification of HIV-1 IN and Chimeras
His-tagged HIV-1 IN and chimeras were purified as described previously (48) with some modification. Briefly, expression of proteins was induced in BL21 (DE3) cells at 20 °C by adding isopropyl β-D-thiogalactopyranoside to 0.5 mM after the bacteria had grown to optical density at an A600 of 0.8. Bacteria were lysed in 25 mM bis-Tris, pH 6.1, 1 M KCl, 1 M urea, 1% thiodiglycol, and 5 mM imidazole and then filtered through an 0.2-μm membrane from Nalgene (Rochester, NY). The lysates fraction was applied to a HiTrap™ chelating HP nickel-affinity column (5 ml) (Amersham Biosciences), and IN was eluted with a 5 mM to 1.0 M linear imidazole gradient. Fractions containing partially purified IN, detected by absorbency at 280 nm, were pooled and applied to a HiTrap™ heparin HP column (5 ml), and His-tagged IN was eluted with a 0.25–1.0 M linear KCl gradient. The pooled heparin IN fractions were concentrated using a Centriprep filter with YM-10 MW membrane and then dialyzed against 25 mM bis-Tris, pH 6.1, 0.5 M KCl, 1% thiodiglycol, 1 mM dithiothreitol, 0.1 mM EDTA, 40% glycerol. The purified protein was aliquoted and stored at −80 °C. The protein concentration was determined using a Bio-Rad protein assay as described by the manufacturer. The N-terminal poly His tag was removed from S160–161 and S153 chimeras using a thrombin kit from Novagen as described by the manufacturer. Briefly, the cleavage reaction in 50 μl contained cleavage buffer, 10 μg of IN, and 0.1 unit of thrombin (diluted 1:10) and was incubated at room temperature for 30 min. An additional 0.1 unit of thrombin was then added, and the reaction was incubated for an additional 30 min. Twenty μl of a 50% slurry of nickel-nitrilotriacetic acid resins (Novage) equilibrated with 25 mM bis-Tris, pH 6.1, was added to the reaction mixture and stirred gently for 30 min at 4 °C. The resin was collected by centrifugation at 15,000 × g for 1 min. The supernatant containing the thrombin was discarded. The resin was washed with 25 mM bis-Tris buffer, pH 6.1, and then the processed IN was eluted with 150 mM imidazole buffer.
Integrase 3′ End Processing Assay Using Duplex Oligodeoxyribonucleotide Substrates
The processing reactions for the HIV-1 U5 LTR substrate were carried out as described previously (46). Reactions were in a volume of 10 μl with 25 mM MOPS, pH 7.2, 10 mM dithiothreitol, 15 mM potassium glutamate, 5% polyethylene glycol 8000, 5% Me2SO, 500 ng of HIV-1, or HIV-1 chimeras and 1 pmol of labeled HIV-1 U5 duplex substrate as indicated. Reaction mixtures were assembled from individual components and preincubated on ice overnight. To start the processing reaction, MgCl2 was added to a final concentration of 10 mM, and reaction mixtures were incubated at 37 °C for 90 min. The reactions were stopped by the addition of 2 μl of stop buffer (95% formamide, 20 mM EDTA, 0.1% xylene cyanol, 0.1% bromphenol blue), heated at 95 °C for 5 min, and then placed on ice. Products of the reaction were separated through a 20% polyacrylamide denaturing gel. Labeled reaction products were visualized using Kodak MR film exposed overnight. For reactions containing ASV U5 or U3 LTR substrate, the final reaction mixture contained 20 mM MOPS, pH 7.2, 3 mM dithiothreitol, 100 μg/ml bovine serum albumin, 500 ng of ASV IN, and 1 pmol of labeled duplex substrates as indicated.
Structural Model for HIV-1 IN with Bound LTR DNA
The initial model for the full-length three-domain IN structure was constructed using the separate two-domain crystal structures of the N-terminal domain and catalytic core (PDB code 1K6Y (49)) and the catalytic core and C-terminal domain (PDB code 1EX4 (50)). The two-domain structures were combined to form a three-domain structure by superimposing the catalytic core domains. The 1K6Y tetramer built from crystal lattice contacts was the basis for forming the tetramer of full-length IN. The zinc coordination complex was covalently modeled using 2.25-Å bond length restraints to the coordinating histidines and 2.35 Å to the coordinating cysteines. Residues 271–288 are disordered at the C terminus (50) and were omitted from the model. Residues 47–55 between the N-terminal and catalytic domains and the loop of residues 140–148 are disordered in the N-terminal two-domain structure (49). The missing loops were produced by obtaining initial Cα–Cα distances from a dynamic programming search of overlapping 30-mers generated from the whole PDB protein structure data base (51). The program AMMP (52) was used with the current all-atom sp4 potential set (53, 54). The charge generation parameters were taken from Bagossi et al. (55). The new atoms were built using the Kohonen and analytic model-building features of AMMP (56), minimized with conjugate gradients. The amortized fast multipole algorithm in AMMP was used for the long-range terms in the nonbonded and electrostatic potentials so that no cut-off radius was employed. The model contained 4 potassium and 4 phosphate ions from the crystal structures. The tetramer model was solvated in 16,195 water molecules. This solvated integrase was subjected to molecular dynamics simulation for 1.0 ns on a 1-GHz Linux PC. Frames were saved every 1 ps.
The initial model for viral DNA was taken from the crystal structure of 1K61, which contained 20 base pairs of B-DNA. The DNA was converted to the HIV U5 sequence using the AMMP molecular mechanics and dynamics program (52). AMMP was used to generate new atomic positions by means of an analytic coordinate generator (56) followed by conjugate gradients minimization for each substituted base pair. Then the entire DNA molecule was minimized using conjugate gradients with all nonbonded and geometric terms. The DNA was positioned approximately using the DNA at the catalytic site of Tn5 transposase as a guide to place the processed 3′-OH at the catalytic site of IN. The two processed deoxyribonucleotides GT were removed from the 3′ end. A magnesium ion was placed between the 3′-OH, and the side chains of Asp64 and Asp116 at the catalytic site of each subunit. The DNA was rotated and translated into the groove formed between the catalytic domain of subunit B and the N- and C-terminal domains of subunit D approximately using the program O (57). The fit to IN was optimized using the GDock routine of AMMP with harmonic restraints to atomic positions for critical atoms (tethers) and restraints on the DNA interatomic distances followed by conjugate gradients minimization. The GDock routine uses a genetic algorithm to search rotational and translational space where the rotational components are represented with unit quaternions. The use of unit quaternions instead of orthogonal rotation matrices avoids artificial singularities in the search space and greatly enhances the convergence of the algorithm. The two unpaired nucleotides 1 and 2 were moved out of the B-form using torsion searches to eliminate collisions with the protein. The 3′-OH was restrained to coordinate with the magnesium ion at the catalytic site. Once the first DNA was placed in the groove, the second LTR end was placed by symmetry in the adjacent groove between the catalytic domains of subunit D and the N- and C-terminal domains of subunit B followed by alternating rigid body movements of the LTR and C-terminal domain using GDock to overcome collisions (as the tetramer is not completely symmetric). Finally, the complex was minimized by conjugate gradients to ensure good nonbonded interactions.
RESULTS
Structural Model of HIV-1 IN with LTR DNA
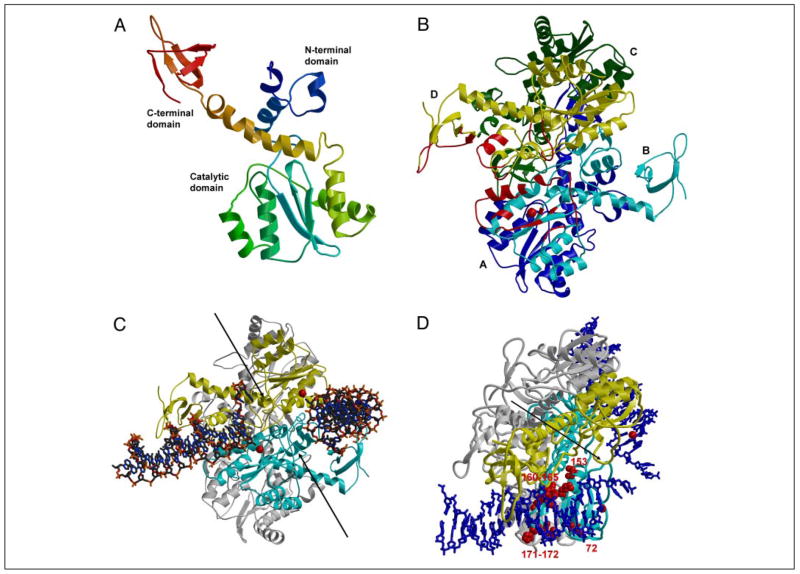
Although a crystal structure for full-length HIV-1 integrase is not yet available, we developed a model for an IN homotetramer bound to its LTR DNA substrate using knowledge of existing integrase two-domain structures, the retrotransposon transposase (Tc3 and Tn5), and the deduced amino acid sequence alignment of known retroviral integrases. Fig. 1 shows our structural model for HIV-1 integrase. We assembled the three functional domains of integrase into an integrated structure by superimposing the separate two-domain crystal structures for integrase: the N-terminal domain and catalytic core (PDB code 1K6Y (44, 49, 59)) and the catalytic core and C-terminal domain (1EX4 (50)) (Fig. 1A). A tetramer model of IN was assembled using crystal lattice contacts (Fig. 1B). In this model the N- and C-terminal domains were arranged on the same side of the subunit and lay relatively close to each other. Molecular dynamics simulations of the solvated IN tetramer model were used to explore the conformational variation of the three domains in the tetramer. During the simulation there was a significant conformational change of the C-terminal domains that were rotated toward the catalytic domains. This conformational change formed a narrower groove that appeared to be more favorable for enclosing viral DNA between the C-terminal and catalytic domains of different subunits.
FIGURE 1. HIV-1 IN homotetramer model with bound HIV-1 LTR substrates.
Target DNA is not placed in this model. Bobscript (58) was used to make the figure. A, monomer assembled from N-terminal/core and core/C-terminal structures. The ribbon backbone is colored from blue for the N-terminal domain to red for the C-terminal domain. B, IN tetramer model with subunits A–D colored blue, cyan, green, and yellow, respectively. Only the cyan and yellow subunits are predicted to interact with the two LTRs. The regions of IN predicted to interact with the LTR binding on the left are indicated in red. The red sphere indicates Mg2+at the catalytic site. C, IN tetramer with two LTRs. The view is similar to that in B. The cyan and yellow subunits interact with LTR; the other two subunits are shown in gray. The DNA is in a stick representation, and the red spheres for Mg2+ indicate the two catalytic sites. The two arrows indicate the pair of grooves on the enzyme surface that may be the site of target DNA binding. D, amino acids in IN tetramer involved in determining specificity for the LTR ends. The view of the tetramer is rotated downward relative to panel C to highlight one of the LTR DNA ends. The DNAs are in a blue stick representation. Amino acids involved in determining the specific recognition of one of the two LTR ends are presented in a space-filling model (red). All other notations are as described in C.
We based the initial placement of the viral DNA substrate in the integrase catalytic site on the crystal structure of the complex between Tn5 transposase and its DNA substrate (PDB code 1F3I (39)). We rotated the DNA to fit the groove that was formed between the N-terminal, catalytic core, and C-terminal domains (see Fig. 1C). Information from cross-linking studies (13, 31–34) helped to orient the viral DNA in the correct groove. A second DNA substrate was positioned symmetrically in the adjacent groove to model the U3 substrate. This model complex consists of an IN tetramer with subunits A–D and two viral DNA substrates. The pairs of dimers are arranged asymmetrically. The IN-LTR model represents an intermediate stage after the processing of the two nucleotides from the 3′ end of the viral DNA. Another pair of grooves (indicated by the arrows in Fig. 1C) is visible in the IN-LTR model, where we would predict that target DNA can bind.
Predictions of Amino Acids in HIV-1 IN Subsites Near the LTR Termini
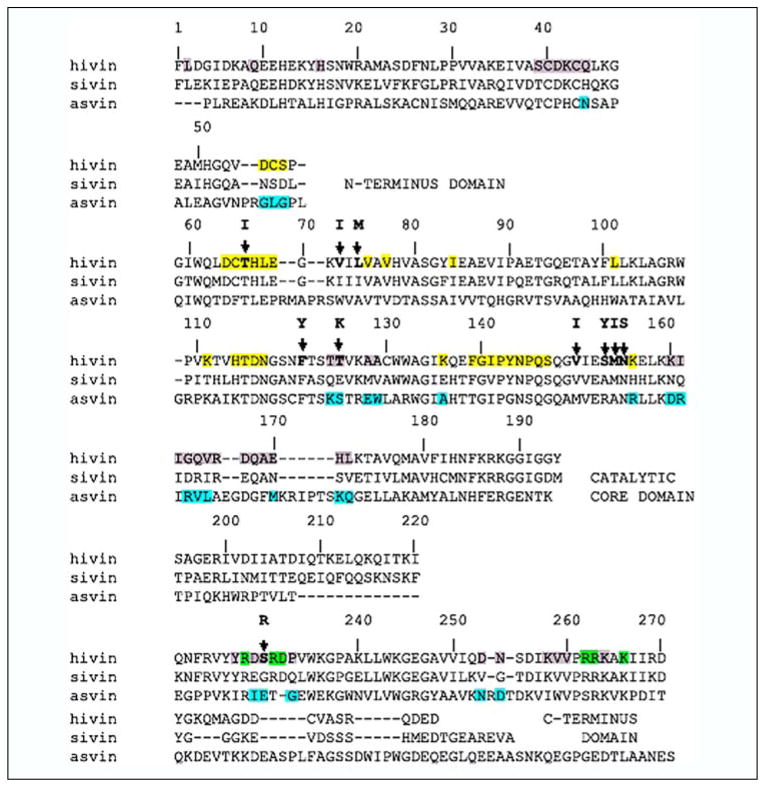
Our structural model of an active HIV-1 integrase-DNA complex shows multiple contacts between IN and the donor LTR DNA substrate. Of note, the processed end of the LTR lies in the catalytic site of subunit B, and this end of the LTR interacts with IN residues from the catalytic domains of the B and D subunits, whereas the more distal regions of the LTR interact with residues of the N- and C-terminal domains of subunit D. There are numerous amino acid residues in subunits B and D of the tetramer structure, which lie within 10 Å of the LTR ends and could therefore interact with the LTR DNA ends (illustrated by the red color on the ribbon backbone in Fig. 1B for one of the two LTR ends). The amino acid residues predicted to be on the IN tetramer surface and near the CA dinucleotide portion of the LTR ends include residues 64–69, 75, 77, 84, 114–117, 139–145, 148–152, and 156 (subunit B) and 55–57 and 140–148 (subunit D) (Fig. 2, yellow highlight). Those predicted to lie near LTR positions 5–15 are residues 66–69, 158–172, and 175–176 (subunit B) and 13–16, 34–44, 227–233, 236, 253–254, and 258–264 (subunit D) (Fig. 2, gray shading), and those near the 16–20 bp regions of the LTRs are 228, 231–232, 235, 262–263, 266 (subunit D) (Fig. 2, green highlight). Because the model is unlikely to predict the exact conformation of the LTRs or IN tetramer, more distal potential interacting residues were noted; these included residues 2 (subunit C), 9 (subunit D), and 124–125, 128–129 (subunit A).
FIGURE 2. Structural alignment of the primary amino acid sequence of HIV-1, SIV, and ASV INs.
Residues shaded in yellow, gray, and green in HIV-1 IN are proposed from the structural model presented in Fig. 1 to be in close proximity to base pairs 1– 4, 5–15, and 16 –20, respectively, of the LTR DNA ends. The HIV-1 LTR end sequence is: 5′-TGTGGAAAATCTCTAGCAGT-3′. Blue-shaded residues in ASV IN mark those unique to all three INs. A dash indicates no amino acid in the corresponding structures. Bold letters above the HIV-1 sequence indicate residues that change in drug-resistant IN mutants (60 – 64). The alignment was compiled by Snasel et al. (73).
The consensus alignment of HIV-1, SIV, and ASV IN amino acid sequences was then used to identify which predicted DNA contact residues are unique for each retrovirus (Fig. 2, blue highlight on the ASV sequence). We prepared a series of HIV-1 IN derivatives (compiled in Table 1), which substituted amino acids found in ASV IN into the structurally equivalent position of HIV-1 IN, to determine whether these substitutions change the specificity for recognition of the LTR ends. We refer to each derivative by the HIV-1 IN position from the N terminus (i.e. S171–172 substitutes amino acids at positions 171 and 172). The base pair regions in the LTRs that are predicted to interact with these amino acids are listed in the second column of Table 1. These predictions represent our best structural estimate of the closest positional contacts. Where amino acids are adjacent to one another in the linear sequence, they have been changed as a contiguous block to reduce the number of derivatives to be analyzed (i.e. see S160–161). Two types of control substitutions were tested. First, S124–125 and S128–130 test changes in the distal region that may provide additional LTR contacts. Second, S193, S197, and S200–201 test changes in the region that is predicted to be occupied by target DNA. We have introduced ASV amino acids into the HIV-1 IN because the specific activity of the RSV enzyme is greater than that of HIV-1. Also ASV IN is more soluble than HIV-1 IN. In some cases, there are structural insertions or deletions of amino acids observed between the enzymes. For one example, the region around residues 54–57 has an insertion in ASV compared with HIV-1 IN. In these cases, we have analyzed the effect of the amino acid exchanges on either side of the insertion independent of one another.
TABLE 1.
Summary of activity in HIV-1/ASV chimeras
| Name of derivative | Predicted LTR base pair interaction | Substitutionsa | Outcomea |
|---|---|---|---|
| S54 | 1–4 | V54N | Decrease in 3′ processing |
| S54–57 | 1–4 | V54N, D55G, S56L, S57G | Decrease in 3′ processing |
| S72 | 1–4 | V72W | Change in LTR specificity |
| S125 | 5–15 (distal) | T125S | Increase in 3′ processing of HIV-1 substrate |
| S124–125 | 5–15 (distal) | T124K, T125S | Decrease in 3′ processing |
| S128–130 | 5–15 (distal) | A128E, A129W, C130L | Decrease in 3′ processing |
| S153 | 5 | S153R | Change in LTR specificity |
| S156–157 | 4–5 | K156R, E157L | Decrease in 3′ processing |
| S160–161 | 5–6 | K160D, I161R | Change in LTR specificity |
| S163–165 | 6–8 | G163R, Q164V, V165L | Change in LTR specificity |
| S171–172 | 7–10 | H171K, L172Q | Change in LTR specificity |
| S193 | None (host DNA) | G193K | Decrease in 3′ processing |
| S197 | None (host DNA) | G197I | Decrease in 3′ processing |
| S200–201 | None (host DNA) | I200H, V201W | Decrease in 3′ processing |
| S255 | 7–11 | S255D | No change in activity |
| S4C | 1–10 | V72W, S153R, G163R, Q164V, V165L, H171K, L172Q | Cumulative change in LTR specificity |
Italics indicate that the amino acid was derived from the structurally related position of ASV IN. Source of amino acids is as described in the legend for Fig. 2. Bold characters indicate amino acid positions in HIV-1 IN where drug resistance changes have been detected (60–64).
Change in specificity means cleavage of the ASV U5 duplex oligo substrate. A decrease in processing is observed with both HIV-1 and ASV U5 substrates.
Purification of Chimeric HIV-1 INs
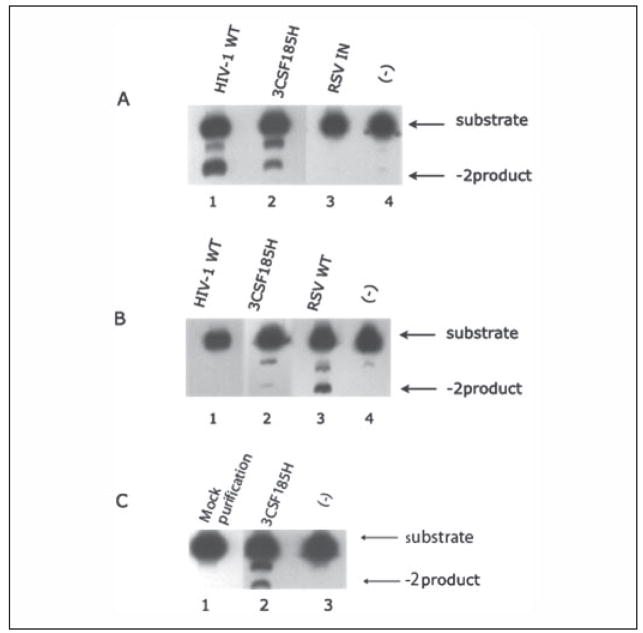
To test the contribution of specific amino acids in HIV-1 integrase to its function, we constructed and purified a series of cognate recombinant derivatives, which substitute amino acids from the ASV IN into the structurally related position of HIV-1 (see Table 1). The purification procedure involves expressing HIV-1 IN with an N-terminal linked poly-His tail, purifying IN successively on a Ni2+ column followed by a heparin column. The poly-His tail was removed from S160–161 and S153 by thrombin cleavage, and the IN was further purified by binding and eluting from the Ni2+ resin. Each of the purified chimeras was analyzed by SDS-PAGE, 10 of which are shown in Fig. 1S in the supplemental data. A single protein band of protein was detected after Coomassie Brilliant Blue staining, indicating that in each case the protein preparations were more than 95% homogenous. This purification procedure is identical to that used for the wild type HIV-1 IN. The wild type HIV-1 IN cleaves its homologous HIV-1 duplex substrate but does not cleave the comparable ASV substrate (Fig. 3, A and B). This indicates that the chimeras are free of nonspecific nucleases. As a further control for contamination, we carried out the same purification procedure but did not induce the expression of a chimera (mock purification). The final fraction does not 3′-process the HIV-1 U5 LTR duplex (Fig. 3C).
FIGURE 3. The 3′ processing activity of HIV-1 and ASV U5 LTR end substrates by wild type HIV-1, ASV, and 3CSF185H HIV-1 INs.
The processing of 5′ 33P-labeled HIV-1 (A and C) or ASV U5 (B) duplex oligos was as described under “Experimental Procedures.” Products were separated by polyacrylamide gels under denaturing conditions to separate the starting substrates from the 3′-processed products. In A and B: HIV-1 IN, 500 ng (lane 1); HIV-1 IN 3CSF185H, 500 ng (lane 2); ASV IN, 500 ng (lane 3); no enzyme (−) (lane 4). In C: mock purification, 500 ng (lane 1); 3CSF185H, 500 ng (lane 2); no enzyme (lane 3).
In constructing the HIV-1/ASV IN chimeras we used an HIV-1 IN derivative, p28bIN-3CS-F185H. This enzyme contains Ser substituted for Cys56, Cys65, and Cys280, and His substituted for Phe185, all of which improve the solubility properties of the enzyme. Positions 56 and 280 have different amino acids among the structurally aligned enzymes (see Fig. 2). Therefore, it seems possible that these substitutions could alter the specificity for recognition of the LTR substrates. To test this possibility, we compared the 3′ processing activity catalyzed by 3CSF185H to wild type IN using duplex oligo substrates representing both the HIV-1 U5 LTR and the ASV U5 LTR ends (Fig. 3). Both enzymes processed the HIV-1 U5 LTR to yield the −2 product (Fig. 3A). The main difference detected was an ~5-fold lower specific activity for the 3CSF185H enzyme compared with wild type IN. ASV IN does not cleave the HIV-1 substrate. With the ASV substrate, no activity was detected with the wild type HIV-1 IN, whereas the 3CSF185H had barely detectable activity (Fig. 3B). As expected, the wild type ASV IN cleaved the ASV duple substrate. Taken together, these results indicate that the substitutions introduced into the HIV-1 IN to enhance its solubility had little effect on the specificity for processing of the LTR substrates. We therefore used the plasmid encoding 3CSF185H HIV-1 IN as the base construct for introducing codons for ASV amino acids in structurally related positions.
The Effect of Specific Amino Acid Changes on Integrase Function
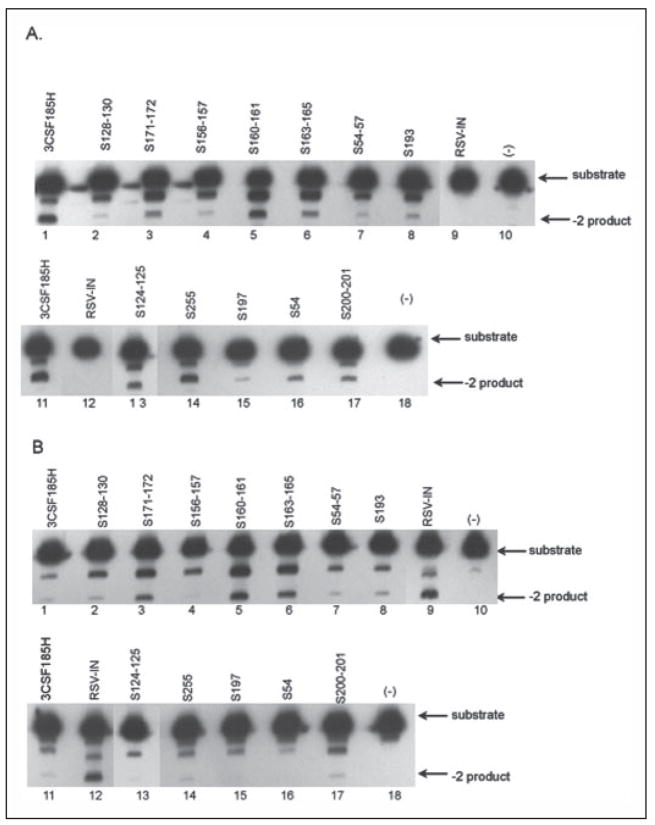
We screened the purified chimeras for changes in their 3′ processing activity with the U5 HIV-1 (20-mer) and the U5 ASV (18-mer) LTR duplex oligo substrates (Fig. 4). Fig. 4A shows that HIV-1 IN processes HIV-1 U5 duplex oligo to produce the −2 product (lanes 1 and 11), whereas the ASV IN does not (lanes 9 and 12). Of the 12 chimeric HIV-1/ASV enzymes analyzed, 10 showed some loss in specific 3′ processing to different extents than the HIV-1U5 LTR substrates compared with the parental protein, whereas two derivatives (S160–161, S255) showed little or no change. The activity of these same enzymes for the duplex U5 ASV LTR substrate is shown in Fig. 4B. In this case, the ASV IN cleaves the homologous substrate (Fig. 4B, lanes 9 and 12), whereas the HIV-1 IN has very limited activity (lanes 1 and 11). Nine of the chimeras had no specific cleavage activity with the ASV U5 duplex substrates. In contrast, the S171–172, S160–161, and S163–165 derivatives appear to 3′ process the ASV substrate (Fig. 4B, lanes 3, 5, and 6, respectively) to form the −2 product, which is not seen with the wild type HIV-1 IN (lane 1). These results were repeated but using the ASV U3 20-mer duplex oligo substrate, and the results were the same as shown in Fig. 4B using the ASV U5 duplex substrate (data not shown).
FIGURE 4. The processing of HIV-1 and ASV U5 duplex oligo substrates by wild type and HIV-1/ASV IN chimeras.
The processing of U5 duplex oligo substrates by IN was as described in the legend to Fig. 3. The starting substrates with a 5′33P label introduced on the CA dinucleotide strand of the duplex and the specific cleavage product with two bases removed from the end are marked by arrows on the right. A, HIV-1 U5 LTR (20-mer) duplex oligo. B, ASV U5 LTR (18-mer) duplex oligo. HIV-1 IN (3CSF185H), lanes 1 and 11; ASV IN, lanes 9 and 12; no enzyme (−), lanes 10 and 18. HIV-1/ASV chimeras (see Table 1 for specific amino acid substitutions): S128 –130, lane 2; S171–172; lane 3; S156 –157, lane 4; S160 –161, lane 5; S163–165, lane 6; S54 –57, lane 7; S193, lane 8; S124 –125, lane 13; S255, lane 14; S197, lane 15; S54, lane 16; S200 –201, lane 17.
We performed enzyme titration curves and time courses of reaction for formation of the −2 product with the ASV U5 substrate for several of the chimeras. The titration for the S160–161 derivative shows an increase in −2 products with the addition of increasing amounts of the chimera to the reaction (Fig. 2S–A in the supplemental data). A similar increase in accumulation of the −2 product was observed with a fixed enzyme concentration but increased time of incubation (data not shown). In contrast, the S156–157 chimera shows no concentration dependence for formation of the very small amount of −2 product observed (Fig. 2S–B in the supplemental data). This result indicates that the inability to detect the specific 3′ processing product is not related to the amount of enzyme added but is due to an amino acid substitution that inactivates the catalytic processing of the enzyme.
Inhibitors of HIV-1 Integrase Can Define the Enzymatically Active Structure
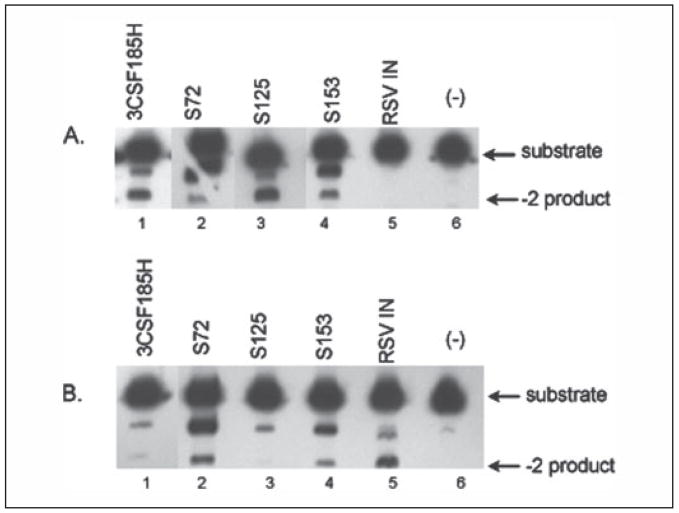
Naphthyridine carboxamide, L870,810, inhibits the strand transfer activity of HIV-1 integrase (60). It was reported that when L870,810 was added to cells infected with different strains of HIV-1 in vivo, mutations encoding F121Y, T125K, V150I, and V72I substitutions arose in the integrase coding region. In animals challenged with SHIV 89.6p and then treated with L870,812, another napthyridine carboxamide derivative, there was the rapid appearance of a mutation encoding a N155H substitution in the integrase coding region that induced resistance to the drug (61). No genetic changes were found in the U5 or U3 LTR regions. Integrase inhibitors L731,988 and L708,906, which inhibit IN with a mechanism similar to naphthyridine carboxamide, also selected for viruses with resistance to the drugs. The mutant viruses encode integrase substitutions T66I, L74M, S153Y, M154I, N155S, and S230R (62–64). The 10 IN positions associated with drug resistance are listed in Fig. 2 (bold letters above the HIV-1 sequence). Our sequence comparisons show that amino acids at positions 72, 74, 125, 153, and 230 are unique among the aligned HIV-1, SIV, and ASV protein sequences. We therefore prepared three chimeric HIV-1/ASV enzymes, S72, S153, and S125, and analyzed their activity with HIV-1 and ASV U5 substrates (Fig. 5). The activity for the S125 chimera was greater than the 3CSF185H HIV-1 IN for the HIV-1 substrate. In contrast, the S72 and the S153 enzymes showed decreased activity with the HIV-1 substrate to different extents with a concomitant detectable activity for the ASV substrate. These results indicate that two of the three sites tested in which amino acid positions are associated with drug resistance also affect specific recognition of the LTR termini.
FIGURE 5. The processing of HIV-1 and ASV U5 duplex oligo substrates by HIV-1/RSV chimeras with amino acid exchanges at drug-resistant sites.
The processing of U5 duplex oligo substrates by IN was as described in the legend to Fig. 3. A, HIV-1 U5 LTR duplex oligo. B, ASV U5 LTR duplex oligo. HIV-1 IN (3CSF185H), lane 1; ASV IN, lane 5; no enzyme (−), lane 6. HIV-1/ASV chimeras (see Table 1 for specific amino acid substitutions): S72, lane 2; S125; lane 3; S153, lane 4. Notations used are as in Fig. 3.
Specificity of the 3′ Processing Reaction
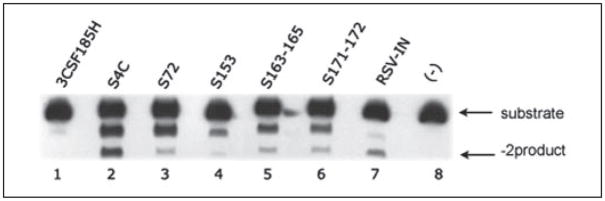
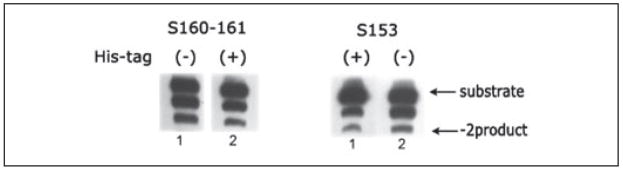
In most of the chimeras analyzed to date (except S54, S125, S197, S200–201, and S255), we observed a significant increase in the nonspecific −1 product with both the ASV and the HIV-1 substrates compared with 3CSF185H (Figs. 4 and 5). This indicates a loss in specificity compared with parental protein even though the correct −2 products are detected for some of the enzymes. As noted above, this nonspecific cleavage activity is not due to a nuclease contaminant in the IN preparations. Rather, we believe that it is related to the fact that there are multiple contacts between the amino acids on the HIV-1 IN surface and the LTR substrates and that the chimeras change only a few of these contacts in any given enzyme tested. This could cause a loosening of the specificity. If this hypothesis is correct, as we combine multiple amino acid substitutions that are involved in specific recognition of the LTR ends into a single construct, we would predict that there will be an increase in the specific cleavage of the ASV LTR substrate. To test this hypothesis, we combined the S171–172, S153, S72, and S163–165 substitutions in an enzyme referred to as S4C. This enzyme is catalytically active with the ASV U5 LTR substrate (Fig. 6). Furthermore, the amount of the −2 product has increased significantly relative to the nonspecific −1 cleavage product that was observed with the individual enzymes (Fig. 6). As a further control for nonspecific nuclease, we combined the S197 derivative, which inactivates HIV-1 IN, with the S4C enzyme in 3CSF185H backbone and found that this mutant lost its ability to specifically 3′ process the ASV and HIV-1 duplex substrates to form the −2 product (data not shown). Finally, Fig. 7 shows the activity of the S160–161 and the S153 enzymes with the ASV U3 LTR duplex substrate is the same before and after removal of the poly-His sequence from the enzyme. Therefore, the change in specificity observed with the above chimeras is not related to the presence of the poly-His sequence on the N terminus.
FIGURE 6. 3′ processing activity of ASV U5 LTR substrates by the S4C HIV-1/ASV IN chimera compared with that of individual chimeras.
The S72, S153, S171–172, and S163–165 substitutions were combined into a single chimera, S4C, and the 3′ processing activity with ASV U5 duplex oligo substrates was compared with the activity of the individual chimeras. HIV-1 IN (3CSF185H), lane 1; ASV IN, lane 7; no enzyme (−), lane 8. Combined HIV-1/ASV chimera S4C, lane 2. HIV-1/ASV chimeras: S72, lane 3; S153, lane 4; S171–172, lane 5; S163–165, lane 6. Notations used are same as in Fig. 3.
FIGURE 7. Comparison of activities with ASV U3 LTR substrates of HIV-1/ASV IN chimeras with and without poly-His tails.
HIV-1/ASV chimeras S160 –161 and S153 (as indicated) were purified as described under “Experimental Procedures,” and the poly-His tail was removed by thrombin cleavage. The 3′ processing activity of the enzyme with (+) or without (−) the poly-His tail on ASV U3 LTR ends was carried out as described under “Experimental Procedures.”
DISCUSSION
A model of an HIV-1 IN tetramer with two bound LTR termini (20 base pairs) was assembled using published structural and molecular dynamics simulations. This model predicts that LTR DNA binds between the catalytic domain of one subunit and the C-terminal domain of another subunit. We modeled 20 base pairs of the LTR termini into the structural model because Li and Craigie have isolated MuLV preintegration complexes and demonstrated by nuclease sensitivity studies that at least 16 base pairs of the LTR ends are protected from digestion,3 and in vitro reconstituted IN reactions suggested that at least 16 bp of the LTR ends were required to support a concerted DNA integration reaction (16–17). The positioning of the 3′ ends of the LTR DNA places them adjacent to the active site Asp residues. This positioning is in agreement with the reported cross-linking of viral DNA to peptides 49–69, 139–152, 247–270, 271–288, and 153–167 (13, 31–32, 34). For example, the 5′ A1 end of the LTR is predicted to lie close to Tyr143 and Gln148, and C2 is near residues in peptides 51–64, as shown by photo-cross-linking results of Esposito and Craigie (13). Gao et al. (66) showed by disulfide cross-linking that Glu246 is close to A7; however, in our model this residue would be closer to the bp 10–15 region. This discrepancy may be due to bending and other conformational changes in the DNA that have not been modeled, as the exact change is difficult to predict. Furthermore, there is good agreement of predicted interactions of DNA with Lys156, Lys159, and Lys160 and mutational and cross-linking analysis with nucleotide analogs that identified these residues (33). The predicted IN interactions with viral DNA in the structural model are also in agreement with the results of many mutagenesis studies. Sayasith et al. (67) used site-directed mutagenesis to show the importance of residues Glu152, Pro145, and Lys156 for 3′ processing, integration, and disintegration, and these residues are all close to the LTR in our model. These investigators also showed that Pro90 mutation affected mainly integration and disintegration. In our model Pro90 is in a turn near Glu93 and His67 at the catalytic site. Lu et al. (68) showed defective viral replication for mutations of Gln62, His67, Asn120, Asn144, Gln148, Glu152, and Asn155. These residues are located in or near the catalytic site (amino acid residues 62, 67,120, 152, and 155) or in the predicted DNA-binding loop of 140–148 (amino acids 144 and 148) in our model. The predicted interactions of the C-terminal domain with viral DNA are consistent with loss of DNA binding for mutations of Arg262 and Leu234 (69).
Several other groups have developed predictions and models for the IN complex(es) with DNA that differ from our model in the orientation of the DNA or the number of subunits forming the binding site. Predictions for the location of DNA binding sites were made in conjunction with the description of the two different two-domain IN crystal structures (49–50). These predictions suggested that viral DNA bound more similarly to the DNA in complex with Tn5 transposase (39). However, in our model the LTRs are oriented at a different angle and interact with a different surface of the C-terminal domain. Theoretical models also have been produced, and several were compared recently (70). Some models were based on the crystal structure of the catalytic and C-terminal domains of SIV IN (71), in which two C-terminal domains are adjacent in the dimer, and both were predicted to interact with DNA. Our model predicts that each LTR binds to one C-terminal domain. Other models, such as that of Podtelezhnikov et al. (72), predict that the viral DNA binds to one subunit, whereas our model predicts that LTRs bind two subunits in the IN tetramer. Furthermore, our model is the only one that has been evaluated by mutational analysis to change the LTR specificity.
We tested our IN/LTR DNA model by substituting ASV for HIV-1 IN amino acid residues predicted to be near the LTR ends and that are in structurally related positions. We identified five chimeras in which substitutions left a catalytically active integrase with a changed specificity preference for the ASV IN substrate. The fact that such a high percentage of the total chimeras tested resulted in a change in specificity for the LTR substrates argues that the placement of the LTRs in our structural model is reasonably accurate. A fourth IN sequence, MMPV, has now been added to the structural alignment (73). Those residues that change specificity are also unique to the MMPV sequence. All of the amino acid residues identified so far that determine specificity reside in the central core domain of IN, which contains the catalytic DDE triad. Although more than 35 amino acids were identified in the initial analysis as potential candidates for interaction with the LTR termini, we would expect only some of them to be involved in site-specific recognition. When changes are introduced at key amino acids that interact with the base pairs in substrate DNA, we expect to observe changes in the enzyme that affect specificity without catalytically inactivating the enzyme. In contrast, substitutions at sites near but not directly interacting with the substrate may result in spatial alterations in the structure of IN that distort the active site or prevent key residues from interacting with substrate. In this case, we would expect to lose catalytic activity with both the HIV-1 and the ASV DNA substrates. With two exceptions, this appears to be the case with the chimeras that do not cleave the ASV substrate.
In the structural model, the two LTRs are placed in different grooves on the surface of the IN tetramer, and the two processed ends face one another (Fig. 1C). This positioning would facilitate the nucleophilic attack of the 3′-hydroxyl ends of the CA strand on respective strands of the target DNA. In examining the model, we noticed that there is a second trench on the enzyme surface that is almost perpendicular to the grooves accommodating the LTRs (indicated by the black arrow in Fig. 1C). We speculate that the target DNA would fit into this second groove, which would bring it adjacent to the catalytic site residues of IN and both 3′-hydroxyl ends of the LTR ends. The flexible loop of residues 140–150 may move to provide a gateway for correct binding of the target DNA. Consistent with this hypothesis, we find that there are a series of charged residues along the length of the trenches predicted to bind target DNA, which could be involved in nonspecific binding of the DNA to the enzyme surface. Many of these residues are structurally conserved between the different HIV-1, SIV, and ASV INs, which would be consistent with the fact that integrase uses many sites for integration into the target DNA. Moreover, cross-linking experiments by Heuer and Brown (31, 32) using model disintegration substrates indicate that the target DNA portion of the model substrate cross-link to IN peptides that contain many of the basic residues described above.
Although the LTR ends face one another, the 3′ processed hydroxyl ends are ~33 Å apart. Because the double helix of B-DNA is about 20 Å in diameter, the separation of the LTR termini would position them for insertion into the two strands of target DNA in a staggered manner, thus providing a mechanistic explanation for the 5-bp duplication of the cell DNA flanking the HIV-1 DNA after integration. We would predict that in ASV IN the spacing of the ends of the LTRs would be wide enough to produce the 6-bp duplication of the target DNA. Further studies are required to test the effects of the chimeric substitutions on the spacing associated with the joining reaction.
Of the five chimera HIV-1/ASV INs that change sequence specificity, the amino acids at positions 160–161, 163–165, and 171–172 are predicted to be near base pairs 5–11 of the LTR (see Fig. 1D). The amino acid at positions 72 and 153 are adjacent to the active site residues near the CA dinucleotide end region of the LTRs. This indicates that there are multiple contact points along the LTRs with various amino acids on the HIV-1 IN surface that provide the specificity for the binding and processing reactions. The 5–11-bp region of the LTRs was previously shown in several studies to be important for 3′ processing, strand transfer, and concerted DNA integration (13, 16, 20). As a control, when amino acid substitutions were made in the HIV-1 IN at positions not predicted to be near the LTRs, such as S124–125, S128–130, S193, S197, and S200–201, there was a significant decrease in 3′ processing without any detectable change in specificity for the LTR substrates.
The substitution of amino acids that result in a change in specificity for the LTR DNA termini generally, but not exclusively, involves a change in a charged or polar amino acid side chain that could alter specific ionic or hydrogen bond interactions with the DNA. For example, S171–172 contains a Lys for His and a Gln for Leu change, S163–165 contains an Arg for Gly change and a Val for Gln, S160–161 contains a Asp for Arg and an Arg for Ile change, and S153 contains an Arg for Ser change. In contrast, S72 contains a Trp for Val change, which could interact with the aromatic base on the DNA. Note that in cases where multiple substitutions of contiguous residues were made such as at the positions of residues 163–165, we do not know whether the specificity change is attributed to just one or to more than one residue that has been changed. Also, when amino acid substitutions are made in HIV-1 IN, they are present in all four subunits of the tetramer. Therefore, we cannot distinguish which subunits of the tetramer are contributing the amino acids that affect the specificity of LTR recognition by the enzyme. The structural model also predicts contacts between amino acid residues at the C terminus of IN and the 14–16-base pair region of the LTRs (Fig. 1C). These residues, such as Arg231, Asn232, and Arg263, are conserved among integrase proteins and therefore may not be important as a determinant of specificity for the LTRs. However, Asp229 and Ser230 are unique. Therefore, these residues may be involved in specific recognition of the LTR ends, and this is being tested.
None of the five chimeras that alter the LTR recognition has the strict specificity for 3′ processing associated with the wild type enzymes in an Mg2+-dependent reaction. This is evident by the large amount of the −1 product that can be seen among the cleavage products, which is far in excess of what is observed with wild type or parental IN protein. There are also other smaller cleavage products detected on the gels (data not shown). The formation of these nonspecific cleavage products is most likely related to the fact that there are multiple contacts between the amino acids in the HIV-1 IN and the LTR ends and the chimeras change only some of these contacts. Evidence in support of this hypothesis is our observation that an enzyme that combines the substitutions of four chimeras shows a significant increase in the −2 product relative to the −1 product compared with the individual chimeras (Fig. 6), and the smaller products are no longer observed. We would therefore anticipate that once all of the amino acid residues that are in contact with the LTR termini are identified and the substitutions are combined into a single construct, the specificity of this enzyme would more closely mimic that of wild type RSV IN.
The error-prone reverse transcription process of retroviruses caused by the lack of the 3′-to-5′ exonuclease associated with reverse transcriptase leads to a population of viruses containing a large number of base substitutions in the genome sequence. When suboptimal concentrations of drugs targeting viral enzymes are administered to virus-infected cells or occur in patients who do not follow drug protocols, there is a rapid selection for drug-resistant enzymes. Like reverse transcriptase and protease (PR), we would predict that drug-resistant enzymes would appear for drugs targeting IN. The amino acids that change in developing the drug resistance are predicted to be among those that are involved in specific recognition of the substrates. For PR, for example, the amino acids involved in specific recognition of the polyprotein cleavage sites were found to be among those that change when drug resistance was selected (65). Inhibitors of HIV-1 IN that function in the nanomolar range to inhibit the joining of the cellular and viral DNAs have been described (62–64). Macrophage- and T cell line-tropic strains of HIV-1 propagated in cells in the presence of L731,988 and L708,906, two diketo acid inhibitors of strand transfer, acquire specific mutations in the integrase coding region (M154I, S153Y, N155S, L74M, S230R, and T66I) that confer anti-retroviral resistance. L870,810 (60), a naphthyridine carboxamide inhibitor of HIV-1 integrase, selected the F121Y, T125K, V150I, and V72I mutations in the integrase coding region. The occurrence of specific drug-resistant mutations can be interpreted in relation to our structural model. Mutations F121Y, T125K, V150I, V72I, T66I, S153Y, M154I, and N155S have been observed when drug resistance arises to IN inhibitors in infected cells (60, 62–64). The majority of these mutations are predicted to lie near the IN catalytic site, which is expected to be very sensitive to structural distortions produced by mutation. Met154, Ser153, and Thr66 are conserved amino acids located proximal to the Asp64 and Glu152 active site residues. Also, residues Val150, and Asn155 are located in the catalytic site or next to residues 64 and 152 in the catalytic site. The side chains of residues Val72, Leu74, and Phe121 lie in an internal hydrophobic region at the back of the catalytic site where even small structural distortions due to conservative changes in amino acid side chain can have significant effects on activity. Other mutations such as S230R alter residues in the C-terminal domain that are predicted to interact with the LTR, whereas Thr125 lies about 10 Å from the LTR in the 10–15-bp region. Interestingly, both of these residues were Lys in the resistant mutant, and the basic lysine side chain can enhance the binding of the negatively charged DNA.
Of the 10 positions identified in drug-resistant INs, five contain unique amino acids in the structurally related positions of HIV-1, SIV, and ASV IN. This finding suggested that some of these positions might be involved in specific recognition of the LTR DNA. To test this hypothesis, we constructed three chimeras that substituted RSV amino acids (not the drug-resistant amino acid) into HIV-1 IN positions 72, 125, and 153. Positions 72 and 153 are near, whereas position 125 is distal to, the LTRs in the structural model. The S72 and S153 chimeras, but not the S125 chimera, demonstrated a change in specificity for recognition of the LTR ends. Therefore, the mechanism of inhibition by these drugs is not strictly related to the strand transfer reaction but can affect the early steps in the integration mechanism in recognition of the LTR termini.
In the case of a position 153 chimera, it gains the ability to 3′ process the RSV LTR duplex with only a small decrease in its ability to 3′ process the U5 HIV-1 duplex substrate (Fig. 5). As such, we would predict that this mutation by itself would have a small effect on replication of HIV-1 in cells. Lee and Robinson (63) have analyzed the HIV-1 IN mutant S153Y and demonstrated that this mutation causes only a small delay in virus replication. The S153R, studied in this report, has been analyzed recently in the context of the virus system in cells and found to have little or no detectable affect on viral replication (68). In contrast to position 153, the substitution at position 72 (V72W) changed to a larger extent the specificity for 3′ processing from the HIV-1 to the RSV LTR (Fig. 5); we noted a 10-fold decrease in processing of the U5 HIV-1 LTR duplex substrate by this chimera. On this basis, we would predict that the V72I drug-resistant mutation would be found only in the presence of other substitutions. This appears to be the case because the V72I substitution is reported only in combination with the F121Y and T125K substitutions (60). This finding argues that the presence of these second site mutations compensate for the affects of the V72I substitution on LTR recognition. As shown in Fig. 5, we have already demonstrated that the T125S substitution increases the 3′ processing of U5 HIV-1 LTR duplex so that it could compensate for the low activity caused by the V72I mutation
Supplementary Material
Acknowledgments
We thank Alla Gustchina and Alex Wlodawer for sharing the structural alignment of integrases before publication. We thank Yuan Fang Wang and Yunfeng Tie for help with the figures of the IN model. We also thank members of the Skalka laboratory, G. Merkel and M. D. Andrake, for the ASV IN protein, the HIV-1 IN construct, and advice in its expression and purification. We are also grateful for helpful comments on the manuscript offered by Drs. Skalka and Andrake.
Footnotes
This work was supported in part by United States Public Health Service Grants CA52047 (to J. L.), GM62920, and GM065762, the Georgia Cancer Coalition, and the Georgia Research Alliance (to I. T. W. and R. W. H.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental material.
The abbreviations used are: HIV, human immunodeficiency virus; IN, integrase; LTR, long terminal repeat; MuLV, murine leukemia virus; ASV, avian sarcoma virus; RSV, Rous sarcoma virus; SIV, simian sarcoma virus; oligo, oligodeoxyribonucleotide; PDB, Protein Data Bank; bis-Tris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol; MOPS, 4-morpholinepropanesulfonic acid.
M. Li and R. Craigie, personal communication.
References
- 1.Kulkosky J, Skalka AM. Pharmacol Ther. 1994;61:185–203. doi: 10.1016/0163-7258(94)90062-0. [DOI] [PubMed] [Google Scholar]
- 2.Aiyar A, Hindmarsh P, Skalka AM, Leis J. J Virol. 1996;70:3571–3580. doi: 10.1128/jvi.70.6.3571-3580.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farnet CM, Bushman FD. Cell. 1997;88:483–492. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- 4.Hindmarsh P, Ridky T, Reeves R, Andrake M, Skalka AM, Leis J. J Virol. 1999;73:2994–3003. doi: 10.1128/jvi.73.4.2994-3003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen H, Engelman A. Proc Natl Acad Sci U S A. 1998;95:15270–15274. doi: 10.1073/pnas.95.26.15270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris D, Engelman A. J Biol Chem. 2000;275:39671–39677. doi: 10.1074/jbc.M002626200. [DOI] [PubMed] [Google Scholar]
- 7.Kalpana G, Reicin A, Cheng G, Sorin M, Paik S, Goff S. Virology. 1999;259:274–285. doi: 10.1006/viro.1999.9767. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki Y, Craigie R. J Virol. 2002;76:12376–12380. doi: 10.1128/JVI.76.23.12376-12380.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katzman M, Katz RA, Skalka AM, Leis J. J Virol. 1989;63:5319–5327. doi: 10.1128/jvi.63.12.5319-5327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kukolj G, Skalka AM. Genes Dev. 1995;20:2556–2567. doi: 10.1101/gad.9.20.2556. [DOI] [PubMed] [Google Scholar]
- 11.Craigie R, Fujiwara T, Bushman F. Cell. 1990;62:829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- 12.Katz RA, Merkel G, Kulkosky J, Leis J, Skalka AM. Cell. 1990;63:87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- 13.Esposito D, Craigie R. EMBO J. 1998;17:5832–5843. doi: 10.1093/emboj/17.19.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murphy JE, Goff SP. J Virol. 1992;66:5092–5095. doi: 10.1128/jvi.66.8.5092-5095.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faure A, Calmels C, Desjobert C, Castroviejo M, Caumont-Sarcos A, Tarrago-Litvak L, Litvak S, Parissi V. Nucleic Acids Res. 2005;33:977–986. doi: 10.1093/nar/gki241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brin E, Leis J. J Biol Chem. 2002;277:10938–10948. doi: 10.1074/jbc.M108116200. [DOI] [PubMed] [Google Scholar]
- 17.Brin E, Leis J. J Biol Chem. 2002;277:18357–18364. doi: 10.1074/jbc.M201354200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fitzgerald ML, Vora AC, Zeh WG, Grandgenett DP. J Virol. 1992;66:6257–6263. doi: 10.1128/jvi.66.11.6257-6263.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hindmarsh P, Johnson M, Reeves R, Leis J. J Virol. 2001;75:1132–1141. doi: 10.1128/JVI.75.3.1132-1141.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masuda T, Kuroda M, Harada S. J Virol. 1998;72:8396–8402. doi: 10.1128/jvi.72.10.8396-8402.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCord M, Chiu R, Vora A, Grandgenett D. Virology. 1999;259:392–401. doi: 10.1006/viro.1999.9782. [DOI] [PubMed] [Google Scholar]
- 22.Vora AC, Chiu R, McCord M, Goodarzi G, Stahl SJ, Mueser TC, Hyde CC, Grandgenett DP. J Biol Chem. 1997;272:23938–23945. doi: 10.1074/jbc.272.38.23938. [DOI] [PubMed] [Google Scholar]
- 23.Vora AC, McCord M, Fitzgerald ML, Inman RB, Grandgenett DP. Nucleic Acids Res. 1994;22:4454–4461. doi: 10.1093/nar/22.21.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Z, Kang SM, LeBlanc A, Hajduk SL, Morrow CD. Virology. 1996;226:306–317. doi: 10.1006/viro.1996.0658. [DOI] [PubMed] [Google Scholar]
- 25.Engelman A, Englund G, Orenstein J, Martin M, Craigie R. J Virol. 1995;69:2729–2736. doi: 10.1128/jvi.69.5.2729-2736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee SP, Han MK. Biochemistry. 1996;35:3837–3844. doi: 10.1021/bi952056p. [DOI] [PubMed] [Google Scholar]
- 27.Lee SP, Xiao J, Knutson JR, Lewis MS, Han MK. Biochemistry. 1997;36:173–180. doi: 10.1021/bi961849o. [DOI] [PubMed] [Google Scholar]
- 28.Zheng R, Jenkins TM, Craigie R. Proc Natl Acad Sci U S A. 1996;93:13659–13664. doi: 10.1073/pnas.93.24.13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ellison V, Gerton J, Vincent KA, Brown PO. J Biol Chem. 1995;270:3320–3326. doi: 10.1074/jbc.270.7.3320. [DOI] [PubMed] [Google Scholar]
- 30.Andrake M, Skalka AM. J Biol Chem. 1996;271:19633–19636. doi: 10.1074/jbc.271.33.19633. [DOI] [PubMed] [Google Scholar]
- 31.Heuer TS, Brown PO. Biochemistry. 1997;36:10655–10665. doi: 10.1021/bi970782h. [DOI] [PubMed] [Google Scholar]
- 32.Heuer TS, Brown PO. Biochemistry. 1998;37:6667–6678. doi: 10.1021/bi972949c. [DOI] [PubMed] [Google Scholar]
- 33.Jenkins TM, Esposito D, Engelman A, Craigie R. EMBO J. 1997;16:6849–6859. doi: 10.1093/emboj/16.22.6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drake RR, Meamati N, Hong H, Pilon AA, Sunthankar P, Hume SD, Milne GWA, Pommier Y. Proc Natl Acad Sci U S A. 1998;95:4170–4175. doi: 10.1073/pnas.95.8.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pommier Y, Johnson AA, Marchand C. Nature. 2005;4:236–248. doi: 10.1038/nrd1660. [DOI] [PubMed] [Google Scholar]
- 36.Asante-Appiah E, Skalka AM. Adv Virus Res. 1999;52:351–369. doi: 10.1016/s0065-3527(08)60306-1. [DOI] [PubMed] [Google Scholar]
- 37.Craigie R. J Biol Chem. 2001;276:23213–23216. doi: 10.1074/jbc.R100027200. [DOI] [PubMed] [Google Scholar]
- 38.Petit C, Schwartz O, Mammano F. J Virol. 1999;73:5079–5088. doi: 10.1128/jvi.73.6.5079-5088.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davies D, Goryshin I, Reznikoff W, Rayment I. Science. 2000;289:77–85. doi: 10.1126/science.289.5476.77. [DOI] [PubMed] [Google Scholar]
- 40.Mizuuchi K. Annu Rev Biochem. 1992;61:1011–1051. doi: 10.1146/annurev.bi.61.070192.005051. [DOI] [PubMed] [Google Scholar]
- 41.Mizuuchi K. Genes Cells. 1997;2:1–12. doi: 10.1046/j.1365-2443.1997.970297.x. [DOI] [PubMed] [Google Scholar]
- 42.Graig NL. Science. 1995;270:253–254. doi: 10.1126/science.270.5234.253. [DOI] [PubMed] [Google Scholar]
- 43.Douglas RD, Lisa MB, William SR, Ivan R. J Biol Chem. 1999;274:11904–11913. [Google Scholar]
- 44.van Pouderoyen G, Ketting RF, Perrakis A, Plasterk RH, Sixma TK. EMBO. 1997;16:6044–6054. doi: 10.1093/emboj/16.19.6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson M, Morris S, Chen A, Stavnezer E, Leis J. BMC Biol. 2004;2/8 doi: 10.1186/1741-7007-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jenkins TM, Engelman A, Ghirlando R, Craigie R. J Biol Chem. 1996;271:7712–7718. doi: 10.1074/jbc.271.13.7712. [DOI] [PubMed] [Google Scholar]
- 47.Aiyar A, Leis J. BioFeedback. 1993;14:366–367. [Google Scholar]
- 48.Yi J, Asante-Appiah E, Skalka AM. Biochemistry. 1999;38:8458–8468. doi: 10.1021/bi982870n. [DOI] [PubMed] [Google Scholar]
- 49.Wang JY, Ling H, Yang W, Craigie R. EMBO J. 2001;20:7333–7343. doi: 10.1093/emboj/20.24.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen JC, Krucinski J, Miercke LJ, Finer-Moore JS, Tang AH, Leavitt AD, Stroud RM. Proc Natl Acad Sci, U S A. 2000;97:8233–8238. doi: 10.1073/pnas.150220297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harrison RW. J Comp Chem. 1993;14:1112–1122. [Google Scholar]
- 53.Weber IT, Harrison RW. Protein Eng. 1996;9:679–690. doi: 10.1093/protein/9.8.679. [DOI] [PubMed] [Google Scholar]
- 54.Weber IT, Harrison RW. Protein Sci. 1997;11:2365–2374. doi: 10.1002/pro.5560061110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bagossi P, Zahuczky G, Tozser J, Weber IT, Harrison RW. J Mol Model. 1999;5:143–152. [Google Scholar]
- 56.Harrison RW. J Math Chem. 1999;26:125–137. [Google Scholar]
- 57.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Crystallogr Sect A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 58.Esnouf RM. Acta Crystallogr Sect D. 1999;55:938–940. doi: 10.1107/s0907444998017363. [DOI] [PubMed] [Google Scholar]
- 59.Yang Z, Mauser T, Bushman F, Hyde C. J Mol Biol. 2000;296:535–548. doi: 10.1006/jmbi.1999.3463. [DOI] [PubMed] [Google Scholar]
- 60.Hazuda D, Anthony N, Gomez R, Jolly S, Wai J, Zhuang L, Fisher T, Embrey M, Guare J, Egbertson M, Vacca J, Huff J, Felock P, Witmer M, Stillmock K, Danovich R, Grobler J, Miller M, Espeseth A, Jin L, Chen IW, Lin J, Kassahun Kelem, Ellis J, Wong B, Xu W, Pearson P, Schleif W, Cortese R, Emini E, Summa V, Holloway K, Young S. Proc Natl Acad Sci, U S A. 2004;101:11233–11238. doi: 10.1073/pnas.0402357101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hazuda D, Young S, Guare J, Anthony N, Gomez R, Wal J, Vacca J, Handt L, Motzel S, Klein H, Keith A, Wilson A, Tussey L, Schleif W, Gabryelski L, Jin L, Miller M, Casimiro D, Emini E, Shiver J. Science. 2004;305:528–532. doi: 10.1126/science.1098632. [DOI] [PubMed] [Google Scholar]
- 62.Hazuda DJ, Flock P, Witmer M, Wolfe A, Stillmock K, Grobler JA, Espeseth A, Gabryelski L, Schleif W, Blau C, Miller MD. Science. 2000;287:646–650. doi: 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]
- 63.Lee DJ, Robinson WE., Jr J Virol. 2004;78:5835–5847. doi: 10.1128/JVI.78.11.5835-5847.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fikkert V, Maele BV, Vercammen J, Hantson A, Remoortel BV, Michiels M, Gurnari C, Pannecouque C, Maeyer MD, Engelborghs Y, Clercq ED, Debyser Z, Witvrouw M. J Virol. 2003;77:11459–11470. doi: 10.1128/JVI.77.21.11459-11470.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ridky TW, Kikonyogo A, Leis J. Biochemistry. 1998;37:13835–13845. doi: 10.1021/bi980612k. [DOI] [PubMed] [Google Scholar]
- 66.Gao K, Butler SL, Bushman F. EMBO J. 2001;20:3565–3576. doi: 10.1093/emboj/20.13.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sayasith K, Sauve G, Yelle J. Mol Cells. 2000;10:525–532. doi: 10.1007/s10059-000-0525-3. [DOI] [PubMed] [Google Scholar]
- 68.Lu R, Limon A, Ghory HZ, Englman A. J Virol. 2005;79:2493–2505. doi: 10.1128/JVI.79.4.2493-2505.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lutzke RA, Plasterk RH. J Virol. 1998;72:4841–4848. doi: 10.1128/jvi.72.6.4841-4848.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Karki RG, Tang Y, Burke TRJ, Nicklaus MC. J Comput Aided Mol Des. 2004;18:739–760. doi: 10.1007/s10822-005-0365-5. [DOI] [PubMed] [Google Scholar]
- 71.Chen Z, Yan Y, Munshi S, Li Y, Zugay-Murphy J, Xu B, Witmer M, Felock P, Wolfe A, Sardana V, Emini EA, Hazuda D, Kuo LC. J Mol Biol. 2000;296:521–533. doi: 10.1006/jmbi.1999.3451. [DOI] [PubMed] [Google Scholar]
- 72.Podtelezhnikov AA, Gao K, Bushman FD, McCammon JA. Biopolymer. 2003;68:110–120. doi: 10.1002/bip.10217. [DOI] [PubMed] [Google Scholar]
- 73.Snasel J, Krejcik Z, Jencova V, Rosenberg I, Ruml T, Alexandratos J, Gustchina A, Pichova I. FEBS J. 2005;272:203–216. doi: 10.1111/j.1432-1033.2004.04386.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.