Abstract
Numerous DNA mismatches and lesions activate MutS homologue (MSH) ATPase activity that is essential for mismatch repair (MMR). We have found that a mismatch embedded in a nearest-neighbor sequence context containing symmetric 3′-purines (2 × 3′-purines) enhanced, whereas symmetric 3′-pyrimidines (2 × 3′-pyrimidines) reduced, hMSH2-hMSH6 ATPase activation. The 3′-purine/pyrimidine effect was most evident for G-containing mispairs. A similar trend pervaded mismatch binding (KD) and the melting of unbound oligonucleotides (Tm; ΔG). However, these latter measures did not accurately predict the hierarchy of MSH ATPase activation. NMR studies of imino proton lifetime, solvent accessibility, and NOE connectivity suggest that sequence contexts that provoke improved MSH-activation displayed enhanced localized DNA flexibility: a dynamic DNA signature that may account for the wide range of lesions that activate MSH functions.
Keywords: ATPase, hereditary nonpolyposis colon cancer, human, mismatch repair, MutS homologs
Mismatch repair (MMR) corrects a wide spectrum of DNA mismatches and lesions that may lead to spontaneous mutations (1). Several prototypical MMR genes have been identified in Escherichia coli that include MutS, MutL, MutH, and UvrD(MutU). Only MutS homologs (MSH) and MutL homologs (MLH/PMS) appear to have been conserved throughout evolution. Alteration of the hMSH2 and hMLH1 genes cause the majority of hereditary nonpolyposis colon cancer (HNPCC) and are frequently associated with numerous types of sporadic cancers (ref. 2 and references therein).
Both MSH and MLH/PMS proteins contain consensus ATP-binding and hydrolysis (ATPase) activities required for MMR (3–5). A wide range of DNA lesions activate the MSH ATPase, including the 8 possible DNA mismatches, small insertion/deletion loops, and numerous damaged/modified nucleotides (6–9). This broad spectrum of DNA substrates is remarkable compared with glycosylases that also recognize nucleotide mismatch/lesions but demonstrate a very narrow range of substrate specificity (for review see ref. 10). Structural analysis of MSH proteins bound to several mismatched nucleotides (11) have revealed a number of consistent features including a 45–60° DNA bend at the mismatch and the formation of an incipient clamp in which the MSH protein both grasps and interrogates the mismatch (12–14). Yet all of the MSH structures contact and interrogate the mismatch region similarly regardless of the mismatch and/or MSH-adenosine nucleotide configuration (11). These results underline the lack of any molecular basis for the extensive range of MSH recognition or the transition from smooth to bent DNA.
Both thermal instability and altered DNA flexibility of the mismatched region have been suggested factors that might influence MSH recognition (15, 16). The idea that altered DNA flexibility may be a determinant is based on picosecond motions inferred from NMR 13C relaxation data, although MSH recognition and activation or MMR appears to occur in the millisecond–minute time scale (16). Nucleotide-stacking interactions serve an important role in stabilizing DNA helical structure (17) and have been used to account for the thermal stability of mismatched DNA (16, 18). The energetic components that account for altered DNA thermal stability and flexibility are controversial (19, 20).
The type of mismatched nucleotides, lesions, and nearest-neighbor nucleotides influence MSH recognition and activation (21–23). Here, we have systematically examined the effect of nearest-neighbor sequence context on hMSH2-hMSH6 mispair recognition and ATPase activation as a model for understanding the extensive range of MSH recognition/activation properties.
Results
Activation of the hMSH2-hMSH6 ATPase.
MSH proteins display a mismatch-dependent ATPase activity that is essential for MMR (24). We examined the hMSH2-hMSH6 ATPase activation by 6 different single-nucleotide mismatches containing all 16 possible nearest-neighbor sequence contexts [96 oligonucleotides; Fig. 1A; Fig. S1; Table S1, Table S2, Table S3, Table S4, Table S5 and Table S6). Because the Km (average ≈30 μM) is well below the cellular ATP concentration (1–3 mM), the catalytic rate constant (kcat) is the most accurate reflection of MSH ATPase activation efficiency for MMR (7, 25–28); where kcat/2 appears to indicate the time-dependent loading of MSH sliding clamps. Moreover, kinetic derivations have suggested that the Km contains numerous embedded poorly defined DNA-binding, ATP-binding, sliding, and dissociation rate constants, such that deliberations of a “specificity constant” (kcat/Km) would appear complicated at best (7, 49).
Fig. 1.
Nearest-neighbor sequence context affects the mismatch-dependent steady-state ATPase activity of hMSH2-hMSH6. (A) Michaelis–Menten analysis of hMSH2-hMSH6 in the presence of 240 nM DNA substrate and increasing concentrations of ATP (see Table S1, Table S2, Table S3, Table S4, Table S5, and Table S6 (47). Average ATP turnover (kcat) of the 2 × 3′ purine sequence context (red bar); average kcat of the 1 × 3′ purine plus 1 × 3′ pyrimidine sequence context (green bar); average kcat of the 2 × 3′ pyrimidine sequence context (blue bar). Error bars indicate the ± standard deviation for kcat. The gray box shows ATPase activation by a homoduplex DNA containing an A/T or G/C base pair at the mismatch site (± standard deviation; see text). The dashed line indicates ATPase activity of biotin-streptavidin blocked-end mismatched or homoduplex DNA (29). (B) Nearest-neighbor sequence context affects dissociation equilibrium binding constants (KD) for hMSH2-hMSH6. IAsys total internal reflectance (TIR) real-time binding studies using 41-mer single 3′-biotinylated mismatched DNA substrates immobilized on the TIR surface via biotin–streptavidin linkage (47). Model 2 × 3′-purine (CxA; red bar) and 2 × 3′-pyrimidine (AxC; blue bar) sequence contexts extremes were examined. Association rate constants (kON) and dissociation rate constants (kOFF) were derived from binding isotherms (Table 1). Error bars (some within the colored bar) represent the standard deviation calculated from multiple experiments. (C) Nearest-neighbor sequence context affects thermal denaturation of mismatched DNA. Melting curves for mismatched 10-mer duplexes embedded in model 2 × 3′-purine (CxA; red bar) and 2 × 3′-pyrimidine (AxC; blue bar) sequence contexts extremes were examined. The G/T* mismatch sequence context was CxG (red bar) and GxC (blue bar). Error bars (some within the colored bar) represent the standard deviation calculated from multiple experiments.
A substantial effect of nearest-neighbor sequence context was observed for DNA substrates containing a G/T, G/G, and G/A mismatch (Fig. 1; Table S1, Table S2, Table S3, Table S4, Table S5, and Table S6). An empirical canon for sequence context effects emerged from the data: 2 × 3′-purines flanking to the mismatch enhanced the hMSH2-hMSH6 ATPase, while 2 × 3′-pyrimidines flanking to the mismatch reduced the hMSH2-hMSH6 ATPase (Fig. 1A, Table S1, Table S2, Table S3, Table S4, Table S5, and Table S6). The exception to this general observation was the relatively poor C/T mismatch MMR substrate in which there was no significant effect of sequence context (Fig. 1A and Table S6). A nearest-neighbor sequence containing 1 × 3′-purine and 1 × 3′-pyrimidine appeared intermediate in its stimulatory effect on the hMSH2-hMSH6 ATPase with the G/G and G/A mismatch (Fig. 1A; Table S3, Table S4). A lack of association with Km is consistent with previous kinetic derivations (7); Fig. S2.
Within several single mismatches, there appeared to be additional order to the hMSH2-hMSH6 ATPase stimulatory effect. For example, the least stimulation by the G/T and G/A mismatches occurred when the mismatched nucleotides were flanked by 2 × 3′-pyrimidines (Fig. 1A; Table S1 and Table S4). However, if the G was flanked by a 3′-pyrimidine but the T was flanked by a 3′-purine, the stimulation of the hMSH2-hMSH6 ATPase approaches the maximum observed with the G/T mismatch flanked by symmetric 3′-purines (kcat·ave ≈ 21.1 min−1 and kcat·ave ≈ 21.7 min−1, respectively; Table S1). In contrast, the least stimulation of the hMSH2-hMSH6 ATPase is observed when the A in the G/A mismatch is flanked by a 3′-pyrimidine regardless of the 3′ context of the G (kcat·ave ≈ 11.5 min−1 and kcat·ave ≈ 11.7 min−1; Table S4). The complement to this observation was when the A in a G/A mismatch was flanked by a 3′-purine, where the stimulation appeared nearly identical regardless of the 3′-nucleotide adjacent to the G (kcat·ave ≈ 17.8 min−1 and kcat·ave ≈ 17.7 min−1; Table S4). The 2 × 3′-pyrimidines produced the least stimulation when surrounding an A/A, whereas all other sequence contexts appeared largely identical (Fig. 1A and Table S5).
Historical comparisons have revealed the hMSH2-hMSH6 kcat in the presence of homoduplex DNA varied 4–7 min−1. The protein preparation used in this study displayed a kcat of 6.4 ± 1.2 min−1 (see Fig. 1A, gray bar). This background hydrolysis in the presence of homoduplex DNA may be further reduced 6-fold with biotin–streptavidin blocked-end DNA substrates to a level equivalent to the hMSH2-hMSH6 in the absence of DNA (≈1 min−1; Fig. 1A, dark bar (29). These and other studies have concluded that DNA ends are a significant contributor to MSH ATPase activation, a condition that has confounded mechanistic misinterpretations and is rare in vivo (30, 31). Analysis of the C/C and T/T mismatched nucleotides were not included because they displayed a kcat ≈ 6–8 min−1, consistent with their significantly reduced recognition and repair (25, 26, 32). Subtraction of the “open-end” background from the mismatch-stimulated ATPase suggests a 2- to 3-fold difference in stimulatory activity within and between mismatches. This translates to an ability to load between 3 and 9 MSH sliding clamps per minute during MMR (29, 30). The poorly repaired C/C and T/T mismatches appear capable of loading ≈1 MSH sliding clamps per minute, which may be largely below the threshold of efficient MMR. These kinetic yields are consistent with titration studies that suggest a minimum of 4–16 MSH proteins per completed MMR in vitro (28).
Mismatch Binding Affinity of hMSH2-hMSH6.
To examine the effects of nearest-neighbor sequence on hMSH2-hMSH6 mismatch binding affinity, we performed IAsys total internal reflectance (TIR) real-time binding studies (Fig. 1B and Table 1). To reduce the complexity of this analysis, we focused on the CxA (2 × 3′ purine) and AxC (2 × 3′ pyrimidine) sequence contexts because they represented activation extremes of the hMSH2-hMSH6 ATPase (where x is the mismatch; Fig. 1 B; Table S1, Table S2, Table S3, Table S4, Table S5, and Table S6).
Table 1.
Effect of nearest-neighbor sequence context on hMSH2-hMSH6 and mismatch binding kinetics and oligonucleotide melting
| Mismatch | Sequence context | kon (M−1s−1 × 105) | koff (s−1 × 10−3) | KD (M × 10−9) |
Tm,°C |
ΔH, kcal/mol | ΔG, kcal/mol | |
|---|---|---|---|---|---|---|---|---|
| Predicted | Observed | |||||||
| G/T | 2 × 3′ purines | 7.5 (+/−1.6) | 1.4 (+/−0.1) | 1.9 (+/−0.4) | 35.1 | 31.3 (+/−0.5) | 59.6 | 8.9 |
| 2 × 3′ pyrimidines | 9.4 (+/−1.9) | 4.1 (+/−1.2) | 4.6 (+/−2.2) | 35.4 | 31.7 (+/−0.6) | 56.5 | 8.9 | |
| G/T* | 2 × 3′ purines | 37.4 | 34.0 (+/−0.3) | 53.7 | 9.2 | |||
| 2 × 3′ pyrimidines | 45.6 | 42.6 (+/−0.1) | 69.7 | 11.5 | ||||
| C/A | 2 × 3′ purines | 8.7 (+/−1.9) | 5.5 (+/−1.7) | 6.7 (+/−3.5) | 11.9 | 11.7 (+/−2.8) | 34.4 | 5.4 |
| 2 × 3′ pyrimidines | 6.8 (+/−0.1) | 4.6 (+/−0.3) | 6.8 (+/−0.4) | 29.3 | 24.4 (+/−0.5) | 50.0 | 7.7 | |
| G/G | 2 × 3′ purines | 5.1 (+/−0.2) | 4.9 (+/−1.5) | 9.5 (+/−3.2) | 29.4 | 24.9 (+/−0.3) | 55.1 | 7.8 |
| 2 × 3′ pyrimidines | 5.3 (+/−0.6) | 23.1 (+/−0.7) | 43.0 (+/−8.0) | 40.8 | 36.2 (+/−0.4) | 64.1 | 10.2 | |
| G/A | 2 × 3′ purines | 10.7 (+/−1.1) | 7.5 (+/−2.6) | 7.1 (+/−3.2) | 31.5 | 27.5 (+/−0.5) | 57.1 | 8.3 |
| 2 ×3′ pyrimidines | 11.3 (+/−0.1) | 23.6 (+/−0.1) | 20.9 (+/−3.1) | 39.6 | 35.8 (+/−0.2) | 68.4 | 10.3 | |
| A/A | 2 × 3′ purines | 8.6 (+/−0.4) | 12.8 (+/−0.2) | 14.9 (+/−3.6) | 26.5 | 21.0 (+/−0.8) | 45.7 | 7.2 |
| 2 × 3′ pyrimidines | 4.4 (+/−0.5) | 10.1 (+/−0.2) | 23.5 (+/−6.6) | 31.5 | 25.6 (+/−0.2) | 43.6 | 7.9 | |
| C/T | 2 × 3′ purines | 3.9 (+/−0.6) | 28.5 (+/−0.9) | 72.3 (+/−13.0) | 23.3 | 18.6 (+/−1.5) | 42.3 | 6.9 |
| 2 × 3′ pyrimidines | 4.5 (+/−0.2) | 29.1 (+/−0.9) | 65.6 (+/−22) | 24.5 | 19.5 (+/−0.1) | 38.4 | 6.9 | |
| T/A | 2 × 3′ purines | 41.5 | 38.3 (+/−0.5) | 65.8 | 10.5 | |||
| 2 × 3′ pyrimidines | 45.7 | 42.7 (+/−0.4) | 73.7 | 11.7 | ||||
| G/C | 2 × 3′ purines | 4.1 (+/−0.5) | 63.0 (+/−5.8) | 146.6 (+/−6.8) | 50.3 | 47.4 (+/−0.2) | 72.1 | 12.6 |
| 2 × 3′ pyrimidines | 49.1 | 46.4 (+/−0.1) | 78.1 | 12.9 | ||||
*The 2 × 3′ purine context is CxG, and the 2 × 3′ pyrimidine context is GxC.
A significant effect of nearest-neighbor sequence context was observed on the hMSH2-hMSH6 mismatch binding affinity to G/A and G/G mismatched duplexes (Fig. 1B and Table 1). When the G/A mismatch contained 2 × 3′-purines the observed equilibrium binding affinity was substantially greater (KD·CxA = 7.15 ± 3.20 nM) than the same DNA substrate containing 2 × 3′-pyrimidines (KD·AxC = 20.94 ± 3.13 nM). Similarly, when the G/G mismatch contained 2 × 3′-purines, the binding affinity was significantly greater (KD·CxA = 9.53 ± 3.23 nM) compared with the same DNA substrate containing 2 × 3′-pyrimidines (KD·AxC = 42.98 ± 8.04 nM). A more modest effect of nearest-neighbor sequence context was observed for G/T (KD·CxA = 1.90 ± 0.35 nM; KD·AxC = 4.64 ± 2.22 nM) and A/A (KD·CxA = 14.90 ± 3.57 nM; KD·AxC = 23.52 ± 6.60 nM) mismatched DNAs. No significant nearest-neighbor effect on hMSH2-hMSH6 binding affinity was observed for substrates containing either a C/A or C/T mismatch.
Examination of the component rate constants revealed that the kOFF was considerably slower when the G/T, G/G, and G/A mismatches were flanked by 2 × 3′-purines (Table 1). The kON was not significantly affected by sequence context with one exception: the kON of hMSH2-hMSH6 with the A/A mismatch appeared to increase when flanked by 2 × 3′-purines (Table 1). These results may indicate the intrinsic error in the system or a difference in the conformation of the mismatched DNA that depends on sequence context. Taken as a whole, these results are consistent with the hypothesis that the sequence context affects the ability of hMSH2-hMSH6 to maintain a stable binding, which is largely reflected by a reduced kOFF. However, the binding data suggests that the C/A mismatch should activate the hMSH2-hMSH6 greater than the best G/A or G/G sequence context, which is not the case for ATPase activity or MMR (7, 25–28).
Melting Temperature of Mismatched DNA.
Because nucleotides flanking a mismatch affect the melting profile (18, 33–35), we examined the melting temperature (Tm) of the 6 model mismatched nucleotides in the CxA (2 × 3′-purines) and AxC (2 × 3′-pyrimidines) sequence context extremes and compared these results with predicted Tm values (see www.bioinfo.rpi.edu/applications/mfold/). The Tm of the G/T, C/A, G/G, A/A, and G/A mismatched duplexes were significantly affected by sequence context and were in general agreement with the predicted values (Table 1). The homoduplex DNA containing a central T/A produced a reproducible 4 °C difference in Tm between the CxA and AxC sequence context compared with similar oligonucleotides containing a central G/C (Table 1). In all cases, the lowest Tm as well as calculated ΔG were found for mismatches flanked by symmetric 3′-purines (red bar).
The most significant effect on thermal stability was found with the C/A and A/A mismatched oligonucleotides where a nearly 30 °C difference in the Tm between the corresponding homoduplex was observed. (Table 1). This observation is in contrast to the apparent lack of a C/A or A/A mismatch sequence context effect on hMSH2-hMSH6 ATPase activation or binding affinity; suggesting that the thermal instability of a mismatched DNA alone is not predictive of hMSH2-hMSH6 mismatch recognition, ATPase activation, or MMR.
Nearest-Neighbor Nucleotides Alter the Chemical Environment Surrounding the Mismatch.
We performed a preliminary examination of the CxA or AxC nearest-neighbor sequence context extremes on the G/A and G/T mismatch by NMR. We picked these mismatches based on their large sequence context effect and their frequent generation by replication misincorporation in vivo. All of the DNA duplex substrates displayed the expected number of aromatic (H8, H2, H5/H6), H1′, H2′, H2″, and H3′ and imino-1H resonances (selected assignments are shown in Figs. 2A and 3A). We did not observe additional resonances that would indicate slowly exchanging conformers. Both the G/T and G/A mismatched duplexes exhibit greater chemical shift dispersion in the AxC sequence context, a general indicator of a more structured environment (36).
Fig. 2.
Nearest-neighbor sequence context alters the NMR of a G/T mismatch. Model 2 × 3′-purine (CxA) and 2 × 3′-pyrimidine (AxC) sequence contexts extremes were examined (shown in middle). (A) 1H NMR spectra of the aromatic region; CxA (Upper) and AxC (Lower) sequence context recorded at 298 K in D2O solutions. Resonances at the mismatch (G5; T16, maroon), flanking sequence (red), and additional residues (gray) are labeled. (B) 31P spectra (proton decoupled) of CxA (Upper) and AxC (Lower) duplexes recorded at 298 K in D2O solutions. Phosphodiester resonances were assigned by using 2D 31P-1H correlation spectra. (C) Imino proton spectra of CxA (Upper) and AxC (Lower) duplexes recorded at 288 K in H2O solutions. Imino protons at the mismatch (G5; T16, maroon), flanking sequences (red), and additional residues (black) are labeled. (D) Individual base pair lifetime τop of AxC and CxA duplexes at 276 K and 288 K. Note that some of the imino protons are nearly insensitive to the catalyst at this temperature, notably G7 and G17 for CxA and G7 and G15 for AxC. In case of G7 in the AxC duplex, this behavior persists at 288 K.
Fig. 3.
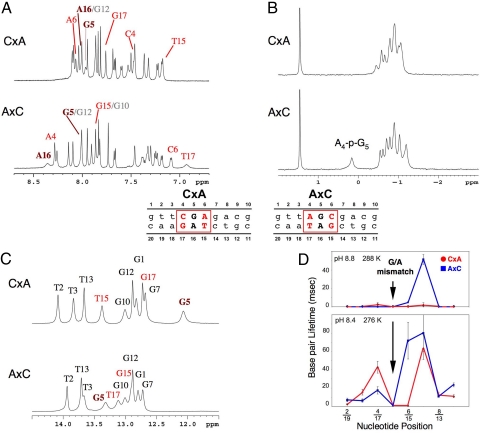
Nearest-neighbor sequence context alters the NMR of a G/A mismatch. Model 2 × 3′-purine (CxA) and 2 × 3′-pyrimidine (AxC) sequence contexts extremes were examined (shown in middle). (A) 1H NMR spectra of the aromatic region; CxA (Upper) and AxC (Lower) sequence context recorded at 298 K in D2O solutions. Resonances at the mismatch (G5; A16, maroon), flanking sequence (red), and additional residues (gray) are labeled. (B) 31P spectra (proton decoupled) of CxA (Upper) and AxC (Lower) duplexes recorded at 298 K in D2O solutions. Phosphodiester resonances were assigned by using 2D 31P-1H correlation spectra. (C) Imino proton spectra of CxA (Upper) and AxC (Lower) duplexes recorded at 288 K in H2O solutions. Imino protons at the mismatch (G5, maroon), flanking sequences (red), and additional residues (black) are labeled. (D) Individual base pair lifetime τop of AxC and CxA duplexes at 276 K and 288 K. Note that G7 of AxC is nearly insensitive to the catalyst at this temperature.
The phosphodiester backbone could be assigned by using 31P-1H 2D NMR analysis (Figs. 2B and 3B). The majority of the phosphodiesters chemical shifts appeared normal, with a narrow shift window of ≈0.8 ppm. These results indicate an overall B-type helix with a BI phosphodiester backbone conformation (37). However, the phosphodiester between T16 and G17 (T16-p-G17) of the G/T mismatched in the CxA sequence context and the phosphodiester between A4 and G5 (A4-p-G5) of the G/A mismatch in the AxC sequence displayed unusual chemical shifts (Figs. 2B and 3B). Similar downfield-shifted resonances have been reported for other DNA mismatch/lesion oligonucleotides and have been attributed to a significant proportion of BII backbone conformation (37, 38). Although nonstandard phosphodiester conformations could conceptually serve as markers for mismatch recognition, they are unlikely to drive MSH recognition/activation since we observe the BII conformation in both well and poorly recognized sequence contexts (Figs. 2B and 3B).
Studies of the imino protons suggested that all base pairs along the length of the oligonucleotide are formed including the G/T wobble pair (Fig. 2C). We observed no extrahelical nucleotides as has been reported by Fazakerley et al. (39). The chemical shifts were nearly identical at the ends of the G/T oligonucleotides (G1, T2, G10, G12, T13) regardless of sequence context (Fig. 2C); suggesting similar and largely unperturbed structures. Several imino protons 3′ of the G5 in the G/A oligonucleotides (G10, G12, T13) also appeared unperturbed (Fig. 3C). However, the T2 imino protons 5′ of mismatched G5 appear shifted when sequence contexts are compared, and the imino proton of mismatched G5 resonates at 13.4 ppm in the AxC sequence context and 12 ppm in the CxA sequence context. These observations appear identical to previous reports (34, 40).
Imino Proton Exchange.
We examined imino proton exchange at infinite ammonia exchange catalyst (Figs. 2D and 3D) (41). The terminal base pairs were particularly susceptible to the exchange catalyst and display lifetimes <1 msec (41). Short base-pair lifetimes were also observed for the imino protons within the G/T or G/A mismatch as have been previously reported (42). However, a consistent pattern of enhanced base-pair lifetime stability was observed for imino protons 3′ of the G/T and G/A mismatch in the AxC sequence context (Figs. 2D and 3D). Moreover, this enhanced imino proton stability 3′ of the G/T mismatch appears to be preserved at elevated temperatures (288 K). An important consequence of mismatch recognition by MSH proteins is the insertion of a highly conserved phenylalanine residue 3′ of the mismatch (12–14). An increased stability of 3′ base pairs appears to suggest that on at least 1 strand surrounding a mismatch, this interrogation may be significantly more difficult in the AxC sequence context.
We recorded NOESY spectra to assess the connectivity of base–base and base–H1′. An NOE connectivity network is a hallmark of normal intrastrand base stacking and base–sugar conformation. The NOE intensity was reduced on both DNA strands 5′ of G5 and 3′ of T16 of the G/T mismatch within the CxA sequence context (Figs. S3 and S4). In contrast, a modest reduction in base–base NOE intensity was observed in the G/T mismatch within the AxC sequence context that was confined to nucleotides distant from the mismatch site. The G/A mismatch duplexes exhibited a more marked alteration of NOE connectivity. In the CxA sequence context, breaks or weak base–base NOE are observed on both DNA strands surrounding the G/A mismatch (Figs. S5 and S6). However, the NOE appeared unperturbed on the top strand of the AxC sequence context (Fig. S6). Taken as a whole, these results are consistent with the conclusion that the top-strand of a G/A mismatch in the AxC sequence context is significantly less mobile compared with the CxA sequence context. Combined with 1H-NMR and imino base-pair lifetime data, these results are consistent with the conclusion that a well-recognized sequence context (2 × 3′-purine) increases the localized dynamic DNA flexibility at the mismatch.
Discussion
We describe the effect of nearest-neighbor sequence context on mismatch-dependent activation of hMSH2-hMSH6. Examination of the intrinsic sequences that occur around symmetric mismatched nucleotides suggests little if any effect of non-nearest-neighbor base pairs on hMSH2-hMSH6 mismatch recognition and ATPase activation (20), although longer-range effects have been reported (22). Although a sequence context effect is not novel in MMR (21), the underlying mechanism is unknown. Our studies have suggested that when a significant nearest-neighbor sequence context effect is manifest, 2 × 3′-purines enhanced, and 2 × 3′-pyrimidines reduced hMSH2-hMSH6 ATPase activation (kcat). A similar trend is observed for mismatch binding (KD), whereas an inverse effect was observed for the Tm of unbound mismatched oligonucleotides. Importantly, the KD and Tm do not accurately account for hMSH2-hMSH6 ATPase activation. Interestingly, the effect of sequence context on KD appears associated with alteration of kOFF. This observation appears to suggest that important conformational transition(s) must occur that results in stable MSH mismatch binding and, ultimately, ATPase activation. In the absence of these conformational transition(s), the MSH protein appears to readily dissociate from the DNA, which occurs in the absence of ATP binding/hydrolysis contrary to one proposed model for MMR (ref. 43 and references therein).
Sequence context influenced solvent exposure and imino proton lifetimes of mismatch DNA. MSH mismatch recognition appears to involve ordering of peptide domains, the establishment of a 45–60° bend in the DNA, and the insertion of a conserved phenylalanine residues into an underwound minor groove 3′ to one of the mismatched nucleotides (12–14). Our results suggest that a well-recognized sequence context (2 × 3′-purine) enhances localized DNA flexibility as well as 3′ base pair accessibility. Time-averaged NMR has detailed picosecond–nanosecond dynamics of G/T mismatched DNA (16). Here, the base-pair lifetime measures of localized DNA flexibility occur on the millisecond–second time scale, which appears more comparable with MSH ATPase activation kinetics.
It has been suggested that mismatches and lesions might reduced base stacking in DNA providing a basis for protein–DNA recognition during DNA repair (43). Comparison of the hMSH2-hMSH6 ATPase activation and Tm data suggests otherwise (Fig. 1). Importantly, we find that the predicted and observed Tm correlated well for the mismatched sequence contexts studied here (Table 1). The algorithms used to predict Tm are largely founded on the aromatic, electrostatic, and dispersive free-energy changes associated with nearest-neighbor stacking during DNA helix formation (44). Based on these nearest-neighbor stacking models, one would predict the hierarchy of MSH recognition/activation to be C/A > C/T ≈ A/A > G/G ≈ G/A > G/T. Yet the exact opposite is observed here and in MMR reactions: G/T > G/G ≈ G/A > A/A ≈ C/A > C/T (7, 25, 26, 28, 32). These results suggest that traditional base stacking may account for only a minor portion of the energetic contribution(s) associated with MSH mismatch recognition.
What might account for DNA flexibility-enhanced MSH recognition/activation? A number of investigators have described altered rise, twist, tilt, and roll between adjacent nucleotides that together result in a sequence-dependent slide and shift of nucleotides within DNA (16, 19). Both experiment and modeling suggest that alterations of nucleotide slide and shift produce altered available motions (degrees of freedom) of glycosidic and phosphate bonds (19, 45). The contributions to slide and shift between adjacent nucleotides appear to be primarily electrostatic (45). As might be predicted by an electrostatic-dependent DNA flexibility hypothesis, salt concentrations above (200 mM) or below (50 mM) the physiological range result in reduced MSH ATPase and a lack of discrimination between homoduplex and mismatched DNA (7, 30). Initial tests of MSH recognition/activation using mismatch duplexes containing nucleotides with altered electrostatic properties have been reported (15), although the biochemical conditions make comparisons difficult.
There are at least 2 possible models that might incorporate localized DNA flexibility and the relevant conformational transition(s) required for MSH protein recognition/activation. An induced-bend model would suggest that sequence context influences the “bend-ability” of the mismatched DNA that is exploited by hMSH2-hMSH6 after binding (46). In this model, the MSH protein plays an active role in forcing the mismatched DNA into the 45–60° bent intermediate. It has even been suggested that such an induced bend may provoke mismatched nucleotide extrusion similar to the interrogation mechanism observed for glycosylases (46) although there is no evidence for such an MSH intermediate.
Our data appear to support a second model, capture-bend, where the MSH protein recognizes a dynamically flexible mismatched region provoking further protein conformational transitions that ultimately result in a 45–60° DNA bend. The attraction of the capture-bend model is that the localized DNA flexibility surrounding a mismatch may be considered a fundamental recognition signal for MSH proteins; providing a simple explanation for the wide range of mismatches and lesions that activate MSH proteins. However, because it is doubtful that such localized DNA flexibility results in a spontaneous 45–60° bend, further protein-induced DNA conformational transitions are probable. Alternatively, the 45–60° bend observed in nondynamic structures might reflect an energy minimum that need not be entirely transited for MSH ATPase activation.
Materials and Methods
Protein Purification, DNA Substrate Preparation, ATPase, and IAsys Surface Plasmon Resonance (SPR) Assays.
hMSH2-hMSH6 was overexpressed and purified as previously described (47). All oligonucleotides were synthesized, reverse-phase HPLC purified (Midland Certified Reagent Company), annealed, and purified as previously described (47). DNA substrates were identical except for the bases immediately flanking the respective mismatch (Fig. S1A). ATPase and SPR analysis was performed as previously described (47), except that binding reactions were performed at 35 °C.
UV Thermal Denaturation.
Thermal denaturation experiments were performed with 10-mer duplexes (Fig. S1B) at λ = 260 nm on a CARY 3E spectrophotometer (Varian) using a 0.4 °C/min temperature ramp. Melting temperatures for each of the mismatched duplexes were measured at 10 μM total strand concentration in a buffer comprising 25 mM Hepes (pH 7.8), 100 mM NaCl, and 2 mM MgCl2. The melting constants (Tm, ΔG, ΔH) were obtained from 3 or more independent melting curves by using a 6-parameter fitting routine and assuming a 2-state, helix–coil transition (48).
NMR.
Ten-mer oligonucleotides (Fig. S1B) containing G/T and G/A mismatches in the AxC (2 × 3′-pyrimidines) and CxA (2 × 3′-purines) sequence contexts were prepared in D2O [1.4 μM mismatched oligonucleotide, 10 mM sodium phosphate (pH* 6.6), 100 mM NaCl, 5 mM MgCl2] and H2O solutions [0.8 mM mismatched oligonucleotide, 10 mM sodium phosphate (pH 6.40), 100 mM NaCl, 5 mM MgCl2]. For the exchange measurements in H2O solutions, the buffer was 10 mM sodium phosphate (pH 8.4), 100 mM NaCl; ammonia was added from a stock solution (1–5 M).
NMR experiments were performed at 298 K, 288 K, and 276 K on a Bruker Avance 600 NMR spectrometer at resonance frequencies of 600.1 (1H) and 242.9 (31P) MHz, by using 5-mm QXI probe heads (Bruker). The acquisition and processing parameters for the experiments for resonance assignment are analogous to those described in our earlier studies (48). 1H and 31P spectra were referenced to internal DSS and external 85% H3PO4 (capillary in D2O). Imino proton spectra were obtained by using a jump and return as well as Watergate experiment at lower temperatures. Assignment of 2D spectra and NOESY cross-peak integration were performed by using SPARKY 3.33 (University of California, San Francisco). Exchange rates were determined from imino proton recovery time recorded as a function of the exchange catalyst concentration (NH3) and obtained from semi selective T1 experiments at 276 K and 288 K, using 1.0-msec I- and E-burp pulses centered in the imino proton region (41). Base-pair lifetimes were obtained from a plot of the exchange time vs. 1/catalyst concentration (41).
Supplementary Material
Acknowledgments.
We thank Samir Acharya, Pete Spielmann, Mark Foster, Myron Goodman, John Petruska, and Christopher Hunter for insightful discussions. This work was supported by National Institutes of Health (NIH) Training Grant CA09662 (to A.M.), NIH Grants CA67007 (to R.F.) and AI/GM47459 (to M.W.G.), and the Georgia Cancer Coalition (M.W.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808572106/DCSupplemental.
References
- 1.Kolodner R. Biochemistry and genetics of eukaryotic mismatch repair. Genes Dev. 1996;10:1433–1442. doi: 10.1101/gad.10.12.1433. [DOI] [PubMed] [Google Scholar]
- 2.Boland CR, Fishel R. Lynch syndrome: Form, function, proteins, and basketball. Gastroenterology. 2005;129:751–755. doi: 10.1016/j.gastro.2005.05.067. [DOI] [PubMed] [Google Scholar]
- 3.Ban C, Yang W. Crystal structure and ATPase activity of MutL: Implications for DNA repair and mutagenesis. Cell. 1998;95:541–552. doi: 10.1016/s0092-8674(00)81621-9. [DOI] [PubMed] [Google Scholar]
- 4.Haber LT, Walker GC. Altering the conserved nucleotide binding motif in the Salmonella typhimurium MutS mismatch repair protein affects both its ATPase and mismatch binding activities. EMBO J. 1991;10:2707–2715. doi: 10.1002/j.1460-2075.1991.tb07815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gradia S, Acharya S, Fishel R. The human mismatch recognition complex hMSH2-hMSH6 functions as a novel molecular switch. Cell. 1997;91:995–1005. doi: 10.1016/s0092-8674(00)80490-0. [DOI] [PubMed] [Google Scholar]
- 6.Su S-S, Modrich P. Escherichia coli mutS-encoded protein binds to mismatched DNA base pairs. Proc Natl Acad Sci USA. 1986;83:5057–5061. doi: 10.1073/pnas.83.14.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gradia S, Acharya S, Fishel R. The role of mismatched nucleotides in activating the hMSH2-hMSH6 molecular switch. J Biol Chem. 2000;275:3922–3930. doi: 10.1074/jbc.275.6.3922. [DOI] [PubMed] [Google Scholar]
- 8.Karran P, Marinus MG. Mismatch correction at O6-methylguanine residues in E. coli DNA. Nature. 1982;296:868–869. doi: 10.1038/296868a0. [DOI] [PubMed] [Google Scholar]
- 9.Ni TT, Marsischky GT, Kolodner RD. MSH2 and MSH6 are required for removal of adenine misincorporated opposite 8-oxo-guanine in S. cerevisiae. Mol Cell. 1999;4:439–444. doi: 10.1016/s1097-2765(00)80346-9. [DOI] [PubMed] [Google Scholar]
- 10.Lindahl T. Keynote: Past, present, and future aspects of base excision repair. Prog Nucleic Acid Res Mol Biol. 2001;68:xvii–xxx. doi: 10.1016/s0079-6603(01)68084-x. [DOI] [PubMed] [Google Scholar]
- 11.Natrajan G, et al. Structures of Escherichia coli DNA mismatch repair enzyme MutS in complex with different mismatches: A common recognition mode for diverse substrates. Nucleic Acids Res. 2003;31:4814–4821. doi: 10.1093/nar/gkg677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamers MH, et al. The crystal structure of DNA mismatch repair protein MutS binding to a G x T mismatch. Nature. 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- 13.Obmolova G, Ban C, Hsieh P, Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- 14.Warren JJ, et al. Structure of the human MutSalpha DNA lesion recognition complex. Mol Cell. 2007;26:579–592. doi: 10.1016/j.molcel.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 15.Schofield MJ, et al. The Phe-X-Glu DNA binding motif of MutS. The role of hydrogen bonding in mismatch recognition. J Biol Chem. 2001;276:45505–45508. doi: 10.1074/jbc.C100449200. [DOI] [PubMed] [Google Scholar]
- 16.Isaacs RJ, Rayens WS, Spielmann HP. Structural differences in the NOE-derived structure of G-T mismatched DNA relative to normal DNA are correlated with differences in (13)C relaxation-based internal dynamics. J Mol Biol. 2002;319:191–207. doi: 10.1016/S0022-2836(02)00265-6. [DOI] [PubMed] [Google Scholar]
- 17.Kool ET. Hydrogen bonding, base stacking, and steric effects in DNA replication. Annu Rev Biophys Biomol Struct. 2001;30:1–22. doi: 10.1146/annurev.biophys.30.1.1. [DOI] [PubMed] [Google Scholar]
- 18.Allawi HT, SantaLucia J., Jr Thermodynamics and NMR of internal G. T mismatches in DNA. Biochemistry. 1997;36:10581–10594. doi: 10.1021/bi962590c. [DOI] [PubMed] [Google Scholar]
- 19.Hunter CA. Sequence-dependent DNA structure. The role of base stacking interactions. J Mol Biol. 1993;230:1025–1054. doi: 10.1006/jmbi.1993.1217. [DOI] [PubMed] [Google Scholar]
- 20.Yakovchuk P, Protozanova E, Frank-Kamenetskii MD. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res. 2006;34:564–574. doi: 10.1093/nar/gkj454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones M, Wagner R, Radman M. Repair of a mismatch is influenced by the base composition of the surrounding nucleotide sequence. Genetics. 1987;115:605–610. doi: 10.1093/genetics/115.4.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marsischky GT, Kolodner RD. Biochemical characterization of the interaction between the Saccharomyces cerevisiae MSH2-MSH6 complex and mispaired bases in DNA. J Biol Chem. 1999;274:26668–26682. doi: 10.1074/jbc.274.38.26668. [DOI] [PubMed] [Google Scholar]
- 23.Wang H, Lawrence CW, Li GM, Hays JB. Specific binding of human MSH2. MSH6 mismatch-repair protein heterodimers to DNA incorporating thymine- or uracil-containing UV light photoproducts opposite mismatched bases. J Biol Chem. 1999;274:16894–16900. doi: 10.1074/jbc.274.24.16894. [DOI] [PubMed] [Google Scholar]
- 24.Kolodner RD, Mendillo ML, Putnam CD. Coupling distant sites in DNA during DNA mismatch repair. Proc Natl Acad Sci USA. 2007;104:12953–12954. doi: 10.1073/pnas.0705698104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holmes J, Jr, Clark S, Modrich P. Strand-specific mismatch correction in nuclear extracts of human and Drosophila melanogaster cell lines. Proc Natl Acad Sci USA. 1990;87:5837–5841. doi: 10.1073/pnas.87.15.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas DC, Roberts JD, Kunkel TA. Heteroduplex repair in extracts of human HeLa cells. J Biol Chem. 1991;266:3744–3751. [PubMed] [Google Scholar]
- 27.Constantin N, Dzantiev L, Kadyrov FA, Modrich P. Human mismatch repair: Reconstitution of a nick-directed bidirectional reaction. J Biol Chem. 2005;280:39752–39761. doi: 10.1074/jbc.M509701200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, et al. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell. 2005;122:693–705. doi: 10.1016/j.cell.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 29.Gradia S, et al. hMSH2-hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol Cell. 1999;3:255–261. doi: 10.1016/s1097-2765(00)80316-0. [DOI] [PubMed] [Google Scholar]
- 30.Acharya S, Foster PL, Brooks P, Fishel R. The coordinated functions of the E. coli MutS and MutL proteins in mimsatch repair. Mol Cell. 2003;12:233–246. doi: 10.1016/s1097-2765(03)00219-3. [DOI] [PubMed] [Google Scholar]
- 31.Mendillo ML, Mazur DJ, Kolodner RD. Analysis of the interaction between the Saccharomyces cerevisiae MSH2-MSH6 and MLH1-PMS1 complexes with DNA using a reversible DNA end-blocking system. J Biol Chem. 2005;280:22245–22257. doi: 10.1074/jbc.M407545200. [DOI] [PubMed] [Google Scholar]
- 32.Fang WH, Modrich P. Human strand-specific mismatch repair occurs by a bidirectional mechanism similar to that of the bacterial reaction. J Biol Chem. 1993;268:11838–11844. [PubMed] [Google Scholar]
- 33.Allawi HT, SantaLucia J., Jr Nearest-neighbor thermodynamics of internal A.C mismatches in DNA: Sequence dependence and pH effects. Biochemistry. 1998;37:9435–9444. doi: 10.1021/bi9803729. [DOI] [PubMed] [Google Scholar]
- 34.Allawi HT, SantaLucia J., Jr Nearest neighbor thermodynamic parameters for internal G. A mismatches in DNA. Biochemistry. 1998;37:2170–2179. doi: 10.1021/bi9724873. [DOI] [PubMed] [Google Scholar]
- 35.Allawi HT, SantaLucia J., Jr Thermodynamics of internal C. T mismatches in DNA. Nucleic Acids Res. 1998;26:2694–2701. doi: 10.1093/nar/26.11.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wuthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley; 1986. [Google Scholar]
- 37.Gorenstein DG. Conformational dynamics of DNA and protein-DNA complexes by 31P NMR. Chem Rev. 1994;94:1315–1338. [Google Scholar]
- 38.Aramini JM, Cleaver SH, Pon RT, Cunningham RP, Germann MW. Solution structure of a DNA duplex containing an alpha-anomeric adenosine: Insights into substrate recognition by endonuclease IV. J Mol Biol. 2004;338:77–91. doi: 10.1016/j.jmb.2004.02.035. [DOI] [PubMed] [Google Scholar]
- 39.Fazakerley, et al. Structures of mismatched base pairs in DNA and their recognition by the Escherichia coli mismatch repair system. EMBO J. 1986;5:3697–3703. doi: 10.1002/j.1460-2075.1986.tb04702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patel DJ, Kozlowski SA, Ikuta S, Itakura K. Dynamics of DNA duplexes containing internal G-T, G-A, A-C, and T-C pairs: Hydrogen exchange at and adjacent to mismatch sites. Fed Proc. 1984;43:2663–2670. [PubMed] [Google Scholar]
- 41.Gueron M, Leroy JL. Studies of base pair kinetics by NMR measurement of proton exchange. Methods Enzymol. 1995;261:383–413. doi: 10.1016/s0076-6879(95)61018-9. [DOI] [PubMed] [Google Scholar]
- 42.Moe JG, Russu IM. Kinetics and energetics of base-pair opening in 5′-d(CGCGAATTCGCG)-3′ and a substituted dodecamer containing G. T mismatches. Biochemistry. 1992;31:8421–8428. doi: 10.1021/bi00151a005. [DOI] [PubMed] [Google Scholar]
- 43.Yang W. Poor base stacking at DNA lesions may initiate recognition by many repair proteins. DNA Repair. 2006;5:654–666. doi: 10.1016/j.dnarep.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 44.SantaLucia J., Jr A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc Natl Acad Sci USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Packer MJ, Dauncey MP, Hunter CA. Sequence-dependent DNA structure: Tetranucleotide conformational maps. J Mol Biol. 2000;295:85–103. doi: 10.1006/jmbi.1999.3237. [DOI] [PubMed] [Google Scholar]
- 46.Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 47.Mazurek A, Berardini M, Fishel R. Activation of human MutS homologs by 8-oxo-guanine DNA damage. J Biol Chem. 2002;277:8260–8266. doi: 10.1074/jbc.M111269200. [DOI] [PubMed] [Google Scholar]
- 48.Aramini JM, Kalisch BW, Pon RT, van de Sande JH, Germann MW. Structure of a DNA duplex that contains alpha-anomeric nucleotides and 3′-3′ and 5′-5′ phosphodiester linkages: Coexistence of parallel and antiparallel DNA. Biochemistry. 1996;35:9355–9365. doi: 10.1021/bi960612p. [DOI] [PubMed] [Google Scholar]
- 49.Eisenthal R, Danson MJ, Hough DW. Catalytic efficiency and kcat/Km: A useful comparator? Trends Biotechnol. 2007;25:247–249. doi: 10.1016/j.tibtech.2007.03.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.