Abstract
Although cancer types differ substantially, many cancers share common gene expression signatures. Consistent with this observation, we find convergent and representative distributions and correlation vectors that are distinct in cancer and noncancer ensembles. These differences originate in many genes, but comparatively few genes account for the major differences. We identify genes with different combinatorial regulation in cancer and noncancer as indicated by significant differences in their correlation vectors. Among the identified genes are many established oncogenes and apoptotic genes (such as members of the Bcl-2, the MAPK, and the Ras families) and new candidate oncogenes. Our findings expand and complement the tumorigenic role of up and down regulation of these genes by emphasizing cancer-specific changes in their couplings and correlation patterns at genome-wide level that are independent from their mean levels of expression in cancer cells. Given the central role of these genes in defining the cancerous state it may be worth investigating them and the differences in their combinatorial regulation for developing wide-spectrum anticancer drugs.
Keywords: canonical distributions, combinatorial regulation, correlation vectors
Analysis and clustering of gene expression datasets have identified numerous molecular events accompanying malignant transformation (1, 2). Many of the transformation events are specific to subsets of tissues and cancer types (3, 4). Indeed, gene expression in cancer cell-lines reflects their ostensible tissues of origin (5). Furthermore, gene expression profiles differ significantly in different cancers (6, 7) and help identify and subdivide even cancers previously assigned to the same histopathalogical type (3, 4).
However, many cancer types are believed to share a common gene expression signature (8, 9). If indeed this is true, it suggests some underlying “near-universal” cellular dysfunction that leads to cancer. The cancer signatures that are common to many cancer types might reflect either convergent evolution (due to selection of proliferative and metastatic phenotypes) or a transition to one of many predefined genetic programs [attractor states (10) of the gene regulatory network] found in embryonic developmental processes, which occurring in the improper context bestows a malignant phenotype upon the cell. The latter possibility would imply that the cancerous transformation is not just a random sequence of mutations selected based on their proliferative and metastatic advantages but a regulated process leading to hardwired cellular phenotypes (10). If this hypothesis is correct, the identification and characterization of gene expression signatures common to many cancer types might suggest an approach for effectively altering the malignant phenotype.
The methods developed for identifying such signatures include comparison of gene expression levels in cancer and noncancer, using a variety of techniques such as machine learning and classification approaches, TSP and TSPG (9). Still, features common to many cancer types are neither easily nor reliably detected by classification approaches (11, 12) or direct clustering (13) of expression data. This may be in part due to the techniques becoming swamped by numerous differences (rather than finding the commonalities) among cancer types and the limited number of analyzed datasets (11, 14). Furthermore, differences between cancer types can be incidental (idiopathic) mutations that arise because of the intrinsic genomic instability of cancer cells. Such mutations are highly variable between different cancer types and irrelevant to proliferation and metastasis processes themselves.
To avoid these difficulties, we have studied the pairwise gene-gene correlations (and their organization) computed by averaging across thousands of gene expression datasets representing many cancer types. Such averaging integrates thousands of expression datasets and emphasizes trends common to cancer types while at the same time canceling (averaging out) inconsequential differences and features specific to individual cancer types. To go beyond the simplest pairwise correlations and look for cancer specific correlation signatures, we compared the correlation vectors and the clusters of correlation vectors in separate subensembles of data drawn from cancer and noncancer ensembles. This approach allows us to identify cancer specific correlations (and their organization at multiple scales) that may not be evident in the changes of the expression levels of individual genes.
Results
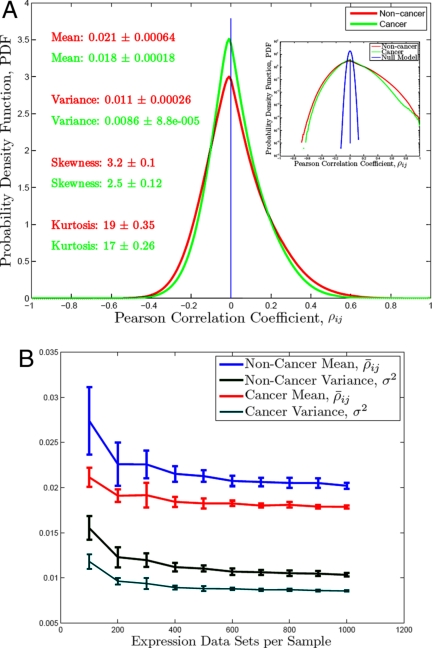
We divided the National Center for Biotechnology Information (NCBI) gene expression profiles from the HG-U133A gene microarray into 2 groups: (i) noncancer, 2,512 expression datasets; (ii) cancer, 2,239 expression datasets. (Details are given in Materials and Methods.) These 2 groups constituted our 2 ensembles over which all subsequent averages were taken. We then calculated pairwise (Pearson) correlations among all (N = 22,283) reported U133A probes* by averaging across cells from many tissue types. For the ith and the jth genes with expression vectors xi and xj the correlation is ρij ≡ (〈xixj〉 − 〈xi〉 〈xj〉)/(σxi σxj); σxi ≡ . Here and throughout the article angular brackets denote arithmetic average, 〈x〉 = (1/M)ΣiM xi, where M is the number of observations across which the averaging is done to compute the correlations. The 2 distributions of pairwise correlations for the 2 ensembles (Fig. 1A) converged to 2 highly reproducible probability density functions. (The convergence process is illustrated in Fig. 1B.) Very similar convergence to these stable distributions is observed when the expression datasets are (randomly) assembled into bootstrap† subensembles (thus allowing overlap between subensembles) and when the expression datasets are subdivided into orthogonal subensembles without overlap. The results shown below are for subensembles containing 1,000 expression datasets. (This size offers a good compromise between convergence, overlap and reproducibility. Smaller samples (500 datasets) give noisier but otherwise very similar results.) Given this convergence, it seems likely these 2 distributions (Fig. 1A) contain canonical information on differences between cancer and noncancer in system-wide gene regulation.‡ Large scale differences between them, if they can be analyzed, would imply some unifying concepts for cancer itself.
Fig. 1.
Distributions of pairwise correlations (ρij) for subensembles from cancer, non-cancer and for fully randomized expression data (A) and convergence of the distributions (B).
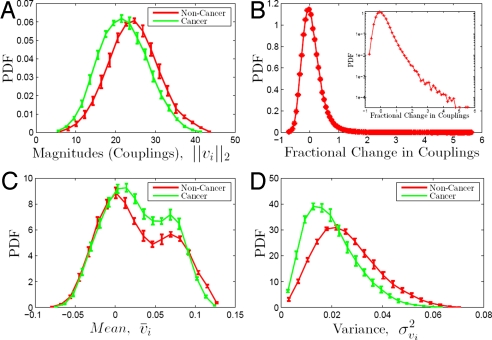
The ordered set of pairwise correlations between the ith and the other N genes (represented on the U133A gene microarray) may also be thought of as a correlation vector vi({j}) ≡ (ρi1, …, ρij, …, ρiN), denoted as vi(n) and vi(c) for noncancer and cancer subensembles respectively. In biological terms, vi captures a combinatorial pattern of covariation between the ith gene and all other genes that may reflect synthetic (synergistic and/or antagonistic) genetic interactions. It transpires that the differences between cancer and noncancer are highlighted more strongly by these correlation vectors, and related quantities. The first such quantity is the length of vi, ‖vi‖2 ≡ , which reflects the overall strength of correlations or couplings of the ith gene to all other genes. In Fig. 2A we see that for noncancer subensembles the distribution (ρ̄c = 0.89 ± 0.01) of ‖vi‖2 is shifted to higher values compared with the distribution for cancer subensembles. (The quantification of the reproducibility of this and all subsequent results is described in Methods.) This shift can indicate either that genes in cancer are less coupled to all other genes or that the cancer types are more variable, for example because of genomic instability. Subsequent results (based on the collinearity and proximity between the ith cancer vectors for different cancer subensembles) suggest that the difference in coupling is likely to be a consequence of gene regulatory couplings in addition to genome instability. To quantify the difference in the coupling of the ith gene in cancer and in noncancer we define the fractional change in coupling: ΔCi = (; here and throughout the article the dot product of vectors x⃗ and y⃗ is: x⃗ · y⃗ ≡ xTy ≡ Σixiyi. The distribution (ρ̄c = 0.84 ± 0.02) of ΔC (Fig. 2B) possesses a long tail to higher values of ΔC. Genes belonging to this tail are coupled much more strongly in noncancer compared with cancer tissues. For example, among the genes with ΔC ≥ 1 (meaning that their coupling to all other genes is at least 2 times stronger in noncancer compared with cancer cells) there is a diverse set of highly over-represented gene ontology (GO) terms,§ that is genes with these GO functions are much more commonly represented than expected in an equal-size, randomly assembled set of genes. Such overrepresented GO terms include multicellular organismal process, cell–cell signaling, response to stimulus, signal transduction, cell proliferation and cell death (Dataset S1). This set of genes includes many receptors (such as epidermal growth factor receptors, insulin-like growth factor receptors, chemokine receptors, tumor necrosis factor receptors, colony stimulating factor receptors) mediating cell growth, differentiation and proliferation signals. Another prominent group of genes in this set are members of the melanoma antigen family and other oncogenes. See SI Appendix for a full list of the genes and the highly enriched GO terms. The correlation vectors, vi = (ρi1, …,ρiN), and their distributions can be analyzed further. For example, the normalized projection of a correlation vector on the sum of unit vectors corresponding to all other genes, v̄i = (1/N)Σj = 1j = Nρij, has an interesting bimodal distribution (ρ̄c = 0.92 ± 0.01). Thus, we find in both the cancer and noncancer subensembles (see Fig. 2C) 2 clearly defined peaks. As demonstrated in the section on vi clusters, the smaller peak corresponds to a large cluster ℂ of highly positively correlated genes whose correlation vectors are close to each other (in the Euclidean sense). The implication is that those genes are correlated to all others in a fairly similar manner. In turn this suggests a large scale universal modular machinery, shared by all cell types, with genes of noncancer cells more strongly correlated to this module. We also find (Fig. 2D) that within the noncancer ensemble many more genes have correlation vectors with higher variances relative to the cancer assemble, suggesting that noncancer cells possess more differentiated and distinctly regulated gene-gene correlations. Again, this might point toward a connection between cancer and forms of system-level disregulation.
Fig. 2.
Correlation vectors. Error bars correspond to the standard deviations from 10 subensembles. (A) Length (Norm). (B) Fractional difference in coupling, ΔCi. (C) Projection on the body diagonal (mean). (D) Variance.
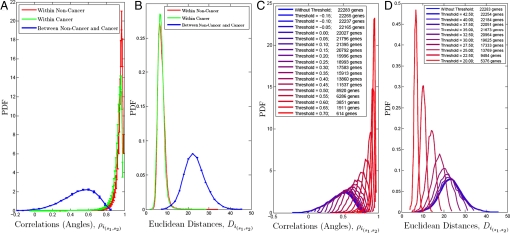
So far we have identified differences between cancer and noncancer by focusing on aggregate statistics (distributions of all correlations, and couplings, projections and variances for the correlation vectors) calculated within the 2 ensembles. To further differentiate the 2 ensembles while emphasizing gene identities, we explore measures that more directly compare corresponding ρij correlations in the cancer and noncancer ensembles. For example, the similarity (or rather dissimilarity) between the ith correlation vectors (and therefore their corresponding correlations) of 2 subensembles (s1 and s2) can be measured by the “correlation angle” ρi(s1,s2) and the Euclidean distance Di(s1,s2) between the correlation vectors, vi(s1) and vi(s2): ρi(s1,s2) ≡ (〈vi(s1)vi(s2)〉 − 〈vi(s1)〉 〈vi(s2)〉)/(σvi(s1) σvi(s2)), and Di(s1,s2) ≡ . Recall that each such vector corresponds to a specific gene (the vector components being its correlations within a given ensemble) and the distance or angle between vectors associated with the same gene (the 2 separate vectors being calculated in the cancer and noncancer ensembles) is therefore a measure of the differences between the correlations (ρij) of this gene in cancer and noncancer. In biological terms, differences between vi(c) and vi(n) (quantified by the angle and the distance between the vectors) reflect different combinatorial regulation of the ith gene in cancer and in noncancer.
In Fig. 3A we see very pronounced difference (as evidenced by a quite different distribution of angles) between cancer and noncancer. Thus, most correlation vectors are collinear within either the cancer or noncancer ensembles, but have different directions when comparing cancer and noncancer subensembles. Indeed, gene-gene correlations are typically different in cancer and noncancer subensembles, pointing toward very different macroscopic behaviors. Furthermore, given that the gene-gene correlations for different subensembles within the cancer ensemble are very similar it seems unlikely that cancer induced genomic instability is the only origin of the difference between cancer and noncancer. That is, if the differences arise from intrinsic tendency of cancer cells to rearrange their genomes, then we might expect different cancers to lead to different rearrangements, and then ρij correlations within different cancer subensembles would also be quite different.
Fig. 3.
Comparisons of correlation vectors. (A and B) Distributions of angles/correlations (A) and Euclidean distances (B) between vi calculated with all genes within and between ensembles. (C and D) Decrease in the angle (C) and distance (D) between vi(n) and vi(c) with the gradual and systematic removal of genes whose vi are most distinct between cancer and noncancer.
On the contrary, the similarity within cancer and differences between cancer and noncancer subensembles suggests the possibility that there is a large-scale system-level difference in gene-gene correlations differentiating cancer and noncancer phenotypes.¶ Very similar points may be made by use of Euclidean distances between the correlation vectors, rather than the angles. Results are presented in Fig. 3B. It is interesting to explore how widely distributed throughout the system these underlying differences between cancer and noncancer really are. That is, we seek to understand how many genes contribute to the large distance and angle between vi(c) and vi(n). One way to approach this question is to remove genes for which vi(c) and vi(n) are most different (thereby potentially most relevant in defining cancer noncancer differences) and recalculate the correlation vectors for the remaining genes until the angles and distances approach the ones for vectors coming from the same ensemble (Fig. 3 C and D). Such systematic removal of genes leads to a surprising conclusion. Distinctions between vi(c) and vi(n) persist until the subensembles contain only several thousand genes. Evidently, large number of genes contribute to the angle and the distance between vi(c) and vi(n). Therefore, at a macroscopic level the differences between cancer and noncancer are system (and genome) wide. This conclusion is also supported by the high participation ratio‖ of the difference vectors, di ≡ vi(n) − vi(c), 6,819 ± 2,044 compared with 122 ± 2 for the null model. (The null model is for completely randomized expression data.)
There is an interesting subtlety here; despite the observation of reproducible system-wide macroscopic differences, the correlations of some genes (from the long tails of the distributions of ρ(s1,s2) and D(s1,s2)) consistently account for the largest differences between cancer and noncancer. Again, it is found that certain biological functions and processes are overrepresented by these sets of genes. Such overrepresented (the probability of observing such enrichment for the corresponding GO terms by chance alone is smaller than 10−18) processes include apoptosis, development, generation of precursor metabolites and energy, protein synthesis, regulatory processes and biopolymer/macromolecular metabolic process. Highlighted groups of genes include caspases and many members of the Bcl-2, Ras, MAPK and TNF families. The up and down regulation of these genes has been shown to affect strongly the proliferation and survival of cancer cells and now we demonstrate cancer specific changes in their combinatorial regulation that are independent from their mean levels (average up or down regulation) in the cancer ensemble. In addition, we find very significant changes in the combinatorial regulation of ribosomal genes (from the 47 genes with the largest Di(s1,s2), 17 correspond to ribosomal proteins) and enzymes from the central biosynthetic and energy generating metabolism that likely reflect the high energy and biosynthetic demands of aggressively proliferating cancers. Among the metabolic enzymes, 42 are dehydrogenases, which is a very likely indication of disregulation of the redox state of cancer cells. This conclusion is bolstered by the fact that among the genes with Di(s1,s2) > 30, 12 genes are cytochrome c oxidases and reductases. Such changes in the combinatorial regulation of key metabolic and oxidation/reduction enzymes likely points to the molecular origins of aerobic glycolysis (the so called Warburg Effect) that is one of the hallmarks of cancer (15). For a full list of the genes and the over-represented GO terms (Dataset S1).
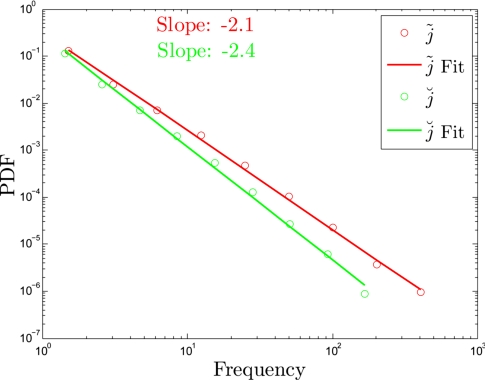
From the distributions in Fig. 3 we see that most genes participate a little in the differences between cancer and noncancer, with a few genes contributing the most to that difference. To explore further the distribution of participation, let us define, j̃i and j̆i as the genes (that is the “dimensions”) along which vi(c) and vi(n) are respectively most** positively and negatively separated when the 2 vectors are plotted in the space of genes. Then, considering the 2 ensembles, the frequency of j̃ is equal to the number of vi(c) − vi(n) pairs that are farthest apart along the dimension of j̃. We plot the distributions of j̃i and j̆i in Fig. 4, and these show quantitatively that a few genes maximize the separation between vi(c) and vi(n) for many genes, whereas many genes maximize the separation between vi(c) and vi(n) for a few genes. A small number of genes are very frequent (and therefore often participate in the largest difference between correlation vectors in cancer and noncancer) whereas most genes (79%) have zero frequencies (Fig. 4). For reasons not yet clear to us, the frequency distributions of j̃ and j̃ appear to follow power laws with exponents of −2.1 and −2.4 respectively (Fig. 4). In other words, if we think of the most distinct correlations between cancer and noncancer as edges (links) in a functional network and the genes as vertices (nodes), the degree distribution of this network follows an almost perfect power law with a relatively limited dynamical range.
Fig. 4.
Frequency of genes maximizing the separation between vi(n) and vi(c).
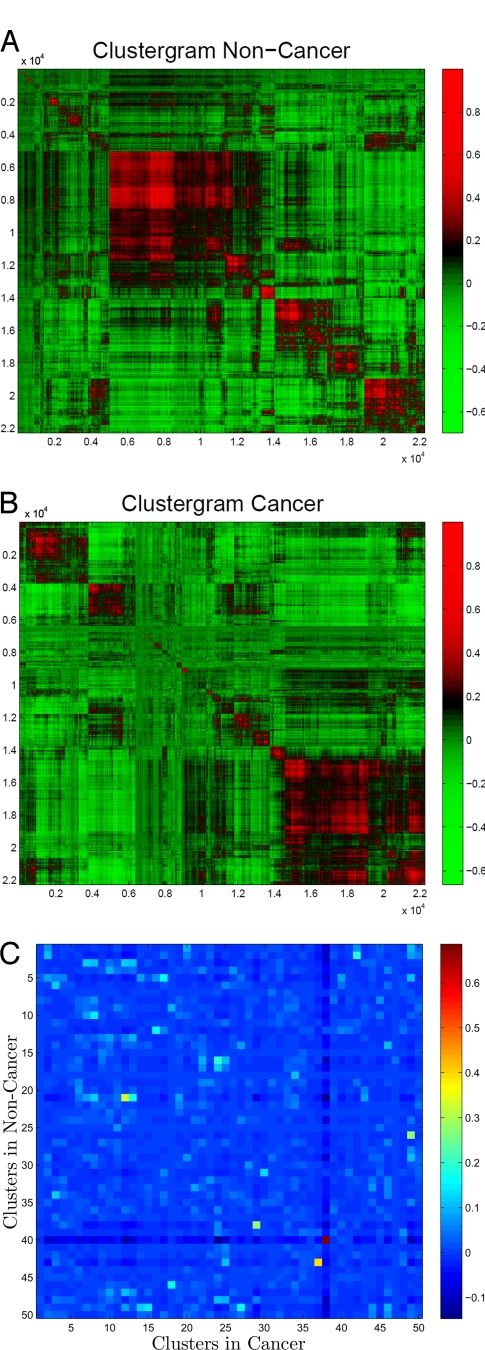
So far we have analyzed the differences between vi(c) and vi(n) (the correlation vectors corresponding to the same gene in different ensembles) by comparing the gene-gene correlations and their vectors in cancer and noncancer. However, this tells us little about the underlying cooperative biological processes in cells. To identify such cooperative units, we now seek to select sets of genes that are strongly coupled to each other, and that are therefore expected to operate in a coherent manner. Thus, we seek to group correlation vectors into clusters (we used hierarchal clustering based on group average Euclidean distance between correlation vectors) for each of the 2 ensembles (Fig. 5). The arrays have as their axes the full list of gene array probes, and the strength of the correlation between genes is illustrated in colors (red - high positive and green - high negative). The gene array probe labels are then permuted so that gene correlation vectors that are most similar to each other are placed adjacent to each other. The outcome is coherent patches of gene array labels corresponding to groups or clusters of genes that are correlated to all other genes in a rather similar manner to each other (Fig. 5 A and B). Most clusters of correlation vectors (cones of closely grouped vectors) are large, well-defined, and reproducible between bootstrap subensembles of an ensemble.
Fig. 5.
Clusters of correlation vectors. (A) Clustered correlation matrix for noncancer. (B) Clustered correlation matrix for cancer. (C) Overlap among clusters of correlation vectors between cancer and noncancer.
We now compare the composition of the clusters in cancer and noncancer to further understand the biologically cooperating modules that distinguish these 2 ensembles. Each cluster in each ensemble was assigned a N dimensional indicator vector, each element corresponding to a gene probe on the array: if the x cluster contains the ith gene, xi = 1 otherwise xi = 0. We now consider the the overlap between the indicator vectors (x and y) for 2 clusters in the 2 ensembles by calculating the Pearson correlation between their corresponding vectors, ρovp. (The overlap estimated by the ratio (rxy) of common to total genes in clusters x and y results in similar estimate, rxy = (x · y)/Σixi + yi.) From the 50 best defined clusters, 49 are non overlapping (contain different genes) in cancer and noncancer (Fig. 5C). Only the largest cluster ℂ (the red square in Fig. 5C) contains many genes common to both cancer and noncancer subensembles, ≈4,200 common genes out of ≈5,800 genes for each cluster. All other clusters in cancer and noncancer have rather small overlap (Fig. 5C), suggesting that the cooperating clusters of genes in the 2 ensembles are very different.
We investigated further the origin of the intriguing ℂ cluster, the only cluster conserved between cancer and noncancer and whose presence is also manifested in the smaller peak of the distribution of vi projections on the main body diagonal (Fig. 2C). One possibility is that the genes in ℂ have very similar correlation vectors in the 2 ensembles. Another possibility is that the correlation vectors of these genes are different between ensembles but consistently similar within an ensemble. To distinguish between those 2 possibilities, we compared the distributions of correlation angles ρi(s1,s2) and the Euclidean distances Di(s1,s2) for ℂ genes and for genes outside of the ℂ cluster. We find that for ℂ genes vi(c) and vi(n) are separated by only slightly smaller angles and distances than for genes not belonging to ℂ. Therefore, the genes in ℂ form a remarkably conserved module present both in cancer and in noncancer but correlated very differently to the rest of the genes.
As before, it is interesting to ask which GO term functions are associated with ℂ genes more frequently than expected by chance. Among the most over-represented functions are development, cell–cell signaling, second messenger signaling, cell differentiation and regulation (see Dataset S1). The genes corresponding to these function are coregulated both in cancer and in noncancer cells. Still, even though these genes preserve their cohesiveness as a module, their vi are coupled differently to genes that do not belong to ℂ. This finding lends further support to the hypothesis that the proliferative and migratory phenotypes associated with cancer result from distinct regulation (as reflected by the cancer-specific state of ℂ genes associated with development and regulation) rather than random mutations alone.
Up to now, we have reported similarities within each ensemble and differences between the 2 ensembles (cancer and noncancer) at many levels, from distributions of pairwise correlations to clusters of vi. It is interesting to ask whether we can find such distinctive features between the 2 ensembles by using more conventional methods. For example, how do our results on clustering vi compare to clustering directly all (M = 4,751) gene expression datasets? To address this question, we tried to group cell types and physiological conditions by applying the same agglomerative hierarchal clustering algorithms to the expression datasets (rather than the correlation vectors). Each cluster identified by the agglomerative clustering algorithm contained comparable fractions of cancer and noncancer datasets. Thus, clustering the physiological conditions based on their gene expression levels fails to distinguish cancer form noncancer reliably. This result is consistent with previous reports (5) and in stark contrast to the the reproducible clustering of the cancer correlation vectors. Therefore, the phenomena we have observed are not merely due to the unusually large dataset we have analyzed. Rather, the clustering of correlation vectors outperforms the clustering of gene expression data as a means of identifying distinctive cancer signatures.
Discussion
One possible explanation of these observations is as follows. Let us conceive, for the moment, of the cell as a nonlinear dynamical system whose variables x⃗ are the concentrations of biomolecules (including mRNAs) and whose global attractors (or macroscopic basins) correspond to different differentiated states or cell types. Each cell type thus represents a distinct (kth) macroscopic state characterized by a basin-specific set of gene-gene correlations, 〈xixj〉k. This conjecture is supported by the clustering of gene expression data (5, 3, 10). Evidently, extended regions of these macroscopic basins could have common features with differences manifested only in smaller numbers of directions in the high dimensional space. We find strong evidence for such common features between basins as many 〈xixj〉k correlations appear to be shared between different cell types. Because we calculate correlations by averaging gene expression levels across cell types, the correlations analyzed in this article are consequently a superposition (weighted by the number of datasets, nk, from the kth basin) of all 〈xixj〉k correlations††: 〈xixj〉 = (Σknk 〈xixj〉k)/Σknk. Therefore, if 〈xixj〉k changes sign and magnitude between basins, 〈xixj〉 would be rather small. This outcome is in stark contrast with the large number of strong pairwise correlations in Fig. 1, reflected also in the large magnitudes of the correlation vectors (Fig. 2); these strong correlations must have the same sign and sufficiently large magnitudes in most tissue types. It therefore seems likely that the strong 〈xixj〉 correlations arise from those (regions of dynamical space in which) gene-gene correlations that are conserved across macroscopic states.
It is noteworthy that our analysis identified oncogenes that are frequently overexpressed in many cancers and whose overexpression triggers or enhances tumorigenesis in animal cancer models. Yet, our findings are not only a reiteration of established knowledge; rather, our findings extend and complement the role of mere overexpression of those oncogenes by revealing cancer-specific changes in their couplings and correlation patterns to the whole genome. For example, if the only cancer related abnormality of Ras members were their overexpression (increased mean level of expression in cancers) their Pearson correlations to the rest of the genes would not change because the Pearson correlation is not influenced by the mean of the correlated variables (the mean is subtracted). Thus, the change in couplings and the correlation pattern reveal different regulation rather than simply overexpression. More specifically, we find that in cancer some genes (including many growth factor receptors and tumor necrosis factor receptors) are significantly less coupled (compared with noncancer) to all other genes as indicated by the long tail of ΔC toward high values (Fig. 2B). Furthermore, not only is the overall strength of the couplings different but also the pattern of correlations is altered very significantly as demonstrated by the large angles between the cancer and noncancer correlation vectors of many genes, including members of the Bcl-2 and Ras families. These findings point to cancer-specific regulatory programs for oncogenes like RAS. Such programs may vary significantly across different cell and cancer types but they clearly share much in common as demonstrated by the collinearity of correlation vectors in different cancer subensembles. Our analysis provides the stepping stones to understanding the cancer-specific regulatory programs by revealing their characteristic correlation patterns.
The described correlation vector analysis can be generalized to identify common features among different physiological conditions and tissue types. This approach is particularly suitable for integrating and analyzing large datasets, exploring common topological structure in different basins of attraction of the cellular network and emphasizing distinct topological structures of correlations. The main strength of our approach is in characterizing the macroscopic states of the cellular network and thus paving the way for more in depth microscopic characterization of the attractor states and dynamics of living cells.
We may speculate that there would be practical applications of the ideas discussed in this article. For example, one important result is that the differences between cancer and noncancer are system wide. The implications could be significant. Conventional anti-cancer therapies target 1 or a few biomolecules, and thereby may affect only limited parts of the system. Such therapies can be successful if the gene regulatory network has paths of directed edges (biochemical reactions and regulatory interactions) from the targeted molecules to all other genes whose levels and correlations have to change for transitioning from one basin to another. If such paths do not exist and cancers are indeed separate attractor basins, however, one suspects that it would be important to push the regulatory network away from a cancerous basin through a high dimensional separatrix toward a healthy, nonproliferative basin. In turn this casts doubt on the efficacy of drugs affecting limited targets, and points more toward therapies that target highly specific groups of genes in a cooperative manner that can restore the system to normal functioning. The key to triggering such transitions may then be the identification of the phase-space trajectories that are most suitable to take the network away from cancer toward its normal, noncancerous basin. As a first step in this direction, we have identified the genes that contribute the most to the macroscopic differences between cancer and noncancer–those are the genes whose couplings decrease the most (Fig. 2B and Datset S1) and whose correlation vectors differ the most in cancer and noncancer (Fig. 3 and Dataset S1). Future work should identify the regulatory mechanisms at the microscopic level, and thus provide the mechanistic understanding for rational cancer therapies.
Materials and Methods
Data Sampling and Bootstrapping.
All datasets (4,751) were downloaded as raw data (Affymetrix CEl files) from the GEO of NCBI (www.ncbi.nlm.nih.gov/geo) and converted into mRNAs levels using the Affymetrix MAS5 algorithm. Datasets were classified as cancers if their source description contained any of the words: neuroblastoma, pheochromocytoma, adenocarcinoma, leukemia, sarcoma, myeloma, melanoma, hepatoma, carcinoma, lymphoma, cancer, and tumor. The remaining datasets (2,512) were classified as noncancers. Orthogonal bootstrap subensembles (samples) were assembled by choosing datasets (with equal probability) without replacement. This method has the advantage of not including a dataset in 2 independent bootstrap samples but allows limited resamplings for a given sample size. To overcome this limitation, we also resampled datasets (again with equal probability) with replacement, thus allowing for unlimited number of resamplings at the expense of some overlap between subensembles.
Reproducibility and Cross Correlations.
The reproducibility of distributions is quantified by the standard deviations (plotted as error bars) of the distribution frequencies. The standard deviation σu for the u frequency is calculated across the bootstrap subensembles, σu ≡ . Although σu measures the reproducibility of distributions, it does not quantify the reproducibility of the results for individual genes. To quantify how similar is the the result for the ith gene in all bootstrap subensembles, we used cross correlations, ρ̄c.
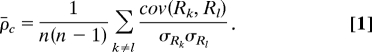 |
Here, n is the number of bootstrap subensembles and Rk and Rl are the vectors with results for all genes from the kth and the lth bootstrap subensembles, Rk, Rl ∈ ℝN. The averaging in computing the covariances and the standard deviations is across all (N = 22,283) gene probes on the arrays.
Supplementary Material
Acknowledgments.
We thank Mario A. Blanco for insightful discussions and useful advice. This work was supported by Irish Research Council for Science Engineering and Technology, Science Foundation Ireland Research Frontiers Programme (Arrested Matter), and European Union Marie Curie Research Training Network Grant MRTN-CT-2003-504712.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810803106/DCSupplemental.
Even though several microarray probes can correspond to one gene, we use gene and gene probe interchangeably. Because the sub-ensembles used in this article contain hundreds of expression datasets, all genes had sufficiently large mean and variance for computing meaningful correlations. Using only the genes having variance above the median variance gives very similar results.
To increase statistical confidence, we use bootstrapping throughout the article. That is a statistical technique based on multiple resamplings and recalculations of the quantities of interest to test convergence and establish confidence intervals. For more details see, Materials and Methods.
A trivial explanation of those differences between cancer and noncancer (and to all of the differences described in the following analysis) can be systematic experimental errors (such as batch effects) that are very common in one ensemble and much more rare or absent from the other one. Given the large number of different experimental groups contributing the experimental Affymetrix data, however, such ensemble specific biases are very unlikely, which is a noteworthy advantage of out analysis. Furthermore, any systematic errors and biases (if present) in the data of experimental groups contributing both cancer and noncancer datasets are likely to be found in both ensembles rather than be ensemble specific.
For many GO terms, the probability of observing such overrepresentation by chance alone is <10−10. This estimate is Bonferroni corrected for multiple hypothesis testing and based on the the hypergeometric distribution.
We can again establish convergence of the quantities by studying sub-ensembles (s1 and s2) containing different expression datasets only from cancer or only from noncancer. The errors implied by the sub-ensemble fluctuations are reflected in the error bars in Fig. 3A. The reproducibility of the results for individual genes is also excellent, ρ̄c = 0.91± 0.03.
The number of principle components (Npc) of a vector x⃗ is the inverse of the vector's inverse participation ratio (IPR), Npc = 1/IPR; IPR = (1/(x · x)2)Σixi4.
Only the maximum distance was used for defining j̃i and j̆i, because the lower ranks (e.g. the next gene along which vi(c) and vi(n) are farthest apart) are less reproducible between bootstrap subensembles.
This expression is exactly correct only for nonnormalized correlations and has to be corrected with the standard deviations and the means to hold for the Pearson correlations used in the article. However, the overall trend and significance are likely to be the same.
References
- 1.Getz G, Gal H, Kela I, Notterman DA, Domany E. Coupled two-way clustering analysis of breast cancer and colon cancer gene expression data. Bioinformatics. 2003;19:1079–1089. doi: 10.1093/bioinformatics/btf876. [DOI] [PubMed] [Google Scholar]
- 2.Chang HY, et al. Gene expression signature of fibroblast serum response predicts human cancer progression: Similarities between tumors and wounds. PLoS biology. 2004;2:E7. doi: 10.1371/journal.pbio.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bild AH, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 4.Godard S, et al. Classification of Human Astrocytic Gliomas on the Basis of Gene Expression: A Correlated Group of Genes with Angiogenic Activity Emerges As a Strong Predictor of Subtypes. Cancer Res. 2003;63:6613–6625. [PubMed] [Google Scholar]
- 5.Ross DT, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 6.Alizadeh A, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 7.Lapointe J, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci USA. 2004;101:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nature genetics. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 9.Xu L, Geman D, Winslow RL. Large-scale integration of cancer microarray data identifies a robust common cancer signature. BMC Bioinformatics. 2007;8:275–288. doi: 10.1186/1471-2105-8-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang S, Ingber D. A non-genetic basis for cancer progression and metastasis: Self-organizing attractors in cell regulatory networks. Breast Disease. 2007;26:27–54. doi: 10.3233/bd-2007-26104. [DOI] [PubMed] [Google Scholar]
- 11.Ein-Dor L, Zuk O, Domany E. Thousands of samples are needed to generate a robust gene list for predicting outcome in cancer. Proc Natl Acad Sci USA. 2006;103:5923–5928. doi: 10.1073/pnas.0601231103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stefan Michiels SK, Hill C. Prediction of cancer outcome with microarrays: A multiple random validation strategy. Lancet. 2005;365:488–492. doi: 10.1016/S0140-6736(05)17866-0. [DOI] [PubMed] [Google Scholar]
- 13.Bertucci F, et al. Gene expression profiles of poor-prognosis primary breast cancer correlate with survival. Hum Mol Genet. 2002;11:863–872. doi: 10.1093/hmg/11.8.863. [DOI] [PubMed] [Google Scholar]
- 14.Hsu P, Sabatini D. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 15.Hu Z, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96–108. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.