Abstract
Introducing structural modifications into biomolecules represents a powerful approach to dissect their functions and roles in biological processes. Bacterial polysaccharides, despite their rich structural information and essential roles in bacterium-host interactions and bacterial virulence, have largely been unexplored for in vivo structural modifications. In this study, we demonstrate the incorporation of a panel of monosaccharide analogs into bacterial polysaccharides in a highly homogenous manner via metabolic engineering of a promiscuous sugar nucleotide biosynthetic pathway. In addition, the bioorthorgonal functional groups metabolically incorporated were exploited for cell surface labeling using in vitro selective chemical ligation reactions. In summary, our study presents a general, facile and effective approach for in vivo generation of novel tailor-made bacterial polysaccharides.
Keywords: chemical remodeling, metabolic engineering, sugar nucleotide biosynthesis, fucose
The surfaces of almost all microbes are decorated with polysaccharides of remarkable structural variations. These polysaccharides play critical roles in interactions between bacteria and their host environments, such as mediation of bacterial adhesion, facilitation of biofilm formation, and conferring resistance to the host immune defense by mimicking host glycoconjugates (1). Therefore, polysaccharides of pathogenic bacteria have long been recognized as a major virulence factor. This property has been exploited to develop polysaccharide-based vaccines against infectious diseases such as pneumonia, meningitis and sepsis (2). Moreover, increasing interests have been paid in recent years to identify small molecule inhibitors to interfere with polysaccharide biosynthesis as novel antibiotics (3).
The ability to introduce structural modifications into biomolecules presents a powerful tool to dissect their functions and roles in biological processes. For example, introduction of nonnatural amino acids into proteins by various chemical and genetic means has permitted the characterization of protein folding, enzyme catalytic mechanisms and ligand-receptor interactions (4). Recently, metabolic introduction of a bioorthorgonal functional group such as an azide into mammalian cell surface carbohydrates on glycoconjugates has become a powerful technology for studying glycosylation in native cellular environments (5). However, compared with these fascinating research accomplishments, the area of introducing modifications into bacterial polysaccharides has largely remained unexplored. To date, only a handful of examples have been demonstrated. Jennings and coworkers chemically converted native N-acetylated Group B meningococcal polysaccharides (homopolymers of α2,8-linked sialic acids) into N-propionylated derivatives, which induced high titers of polysaccharide specific IgG antibodies (6). The Nishimura group chemically synthesized a series of cell wall precursors tagged with a fluorescein and a keto-group. They showed that these precursors could be metabolically incorporated onto the cell wall (7). In another example, Bertozzi, Gibson and coworkers demonstrated the in vivo incorporation of N-acyl modified sialic acid analogs into Haemophilus ducreyi cell surface lipooligosaccharides (8). In these examples, the in vitro chemical conversion has the advantage of achieving high homogeneity, but these modifications are detached from cellular environments. Although the in vivo incorporation of unnatural substrates has the appeal of providing useful information in the cellular context, these modifications are generally substoichiometric, because of the competition of natural and unnatural substrates in the biosynthetic pathway. Therefore, there exists a need to develop a strategy to generate homogenously modified polysaccharides using the cell's in vivo biosynthetic systems. Toward this goal, in this study we demonstrated in Escherichia coli that an exogenous sugar nucleotide salvage pathway not only can functionally replace the corresponding native de novo pathway, but also is promiscuous to metabolize different sugar analogs into polysaccharides, resulting in homogenously modified structures (Fig. 1A). Moreover, the bioorthorgonal chemical handles incorporated allow further chemical elaborations, as demonstrated by direct labeling of bacterial cell surface polysaccharides.
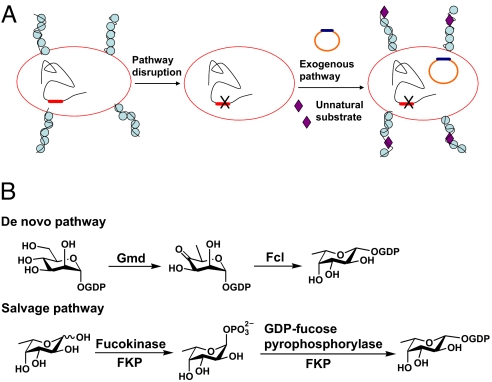
Fig. 1.
Remodeling bacterial polysaccharides by metabolic pathway engineering. (A) Strategy for in vivo generation of modified polysaccharides. (B) Two natural pathways for the biosynthesis of GDP-fucose.
Results and Discussion
Fucose-Containing Bacterial Polysaccharides as a Model System.
Fucose-containing E. coli O86 O-antigen (lipopolysaccharide, LPS), whose structure was solved by our group (9), was used as a model system. We chose to introduce fucose analogs into this polysaccharide to demonstrate our strategy. Fucose is a common monosaccharide component of bacterial polysaccharides. It has been implicated as a main determinant for bacterium-induced immune reactivity and is often a virulence factor (10). There are 2 pathways in nature for generating GDP-fucose (10), the sugar nucleotide donor for fucosyltransferase-catalyzed incorporation of fucose into oligosaccharides, polysaccharides and glycoconjugates (Fig. 1B). One is called the de novo pathway, in which GDP-fucose is generated from GDP-mannose by GDP-mannose dehydratase (Gmd) and GDP-fucose synthetase (Fcl). The other one is the salvage pathway, in which fucose is first phosphorylated by fucokinase, and then converted to GDP-fucose by GDP-fucose pyrophosphorylase. The de novo pathway is present in both prokaryotes and eukaryotes, whereas the salvage pathway was originally believed to be present only in eukaryotes. Recently, the Comstock group (11) identified the first and so far the only microbial GDP-fucose salvage pathway from human symbionts Bacterioides. Inspired by this result and recent research on metabolic incorporation of unnatural sugars into mammalian glycans, we replaced the E. coli native GDP-fucose de novo pathway with the salvage pathway from Bacterioides fragilis, with which we were able to introduce a panel of fucose analogs into polysaccharides with high efficiency and fidelity (Fig. 2A).
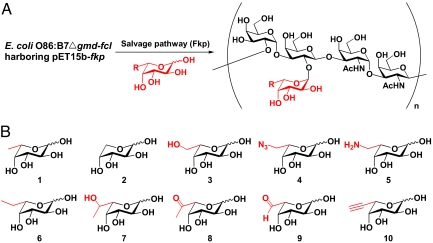
Fig. 2.
Introducing modifications into polysaccharides. (A) Generation of polysaccharides containing fucose analogs (E. coli O86 as a model system). (B) Fucose analogs used in the study. Modifications are located at C-6 position of fucose.
Functional Replacement of GDP-Fucose de Novo Pathway with the Salvage Pathway.
To demonstrate the successful incorporation of the functional salvage pathway, we first disrupted the de novo pathway in E. coli O86:B7 by specifically replacing the gmd-fcl gene sequence with CAT gene as described in ref. 12. Cell surface lipopolysaccharides (LPSs) from Δgmd-fcl mutant and wild-type strain were isolated and visualized with silver staining (13). The wild-type strain produced a ladder-like smooth LPS phenotype (Fig. S1, lane 1). The Δgmd-fcl mutant, however, displayed a semirough phenotype (Fig. S1, lane 2), in which only lipid A-core structure and a single repeating unit were expressed. Therefore, disruption of the GDP-fucose de novo pathway genes depleted the GDP-fucose pool, which presumably rendered the cells unable to produce a complete repeating unit, resulting in the abolishment of polysaccharide formation. The ability of the mutant to produce the smooth LPS phenotype was restored by introducing a recombinant plasmid pTRC99Af containing the gmd-fcl gene (Fig. S1, lane 3). This result showed that the GDP-fucose de novo pathway (gmd-fcl gene) is the only pathway in E. coli O86:B7 to produce GDP-fucose and is essential for the synthesis of polysaccharides containing fucose.
Next, we investigated the possibility of functional expression of the salvage pathway in the Δgmd-fcl mutant. The salvage pathway fkp gene was cloned from Bacteroides fragilis strain NCTC9343. The fkp encodes a bifunctional enzyme possessing both fucokinase and pyrophosphorylase activity (11). The fkp was constructed in pET15b vector and expressed in E. coli BL21 (DE3). The protein was purified by Ni2+ affinity chromatography to near homogeneity (Fig. S2). A 50-μL reaction containing ATP, GTP, fucose and Fkp was carried out. GDP-fucose was identified in the reaction mixture by TLC, MS and a capillary electrophoresis (CE) assay (Figs. S3 and S4A). This result confirmed the in vitro enzymatic function of Fkp to produce GDP-fucose from fucose, ATP and GTP. Next, the recombinant plasmid (pET15b-fkp) was introduced into the Δgmd-fcl mutant to form Δgmd-fcl (fkp). When the recombinant strain was grown in LB medium containing fucose, the LPS was similar to that of the wild-type (Fig. S1, lane 6). However, in the medium alone or the one containing glucose, the semirough LPS phenotype did not change (Fig. S1, lane 5). These results suggested that the salvage pathway can functionally replace the de novo pathway in E. coli O86:B7 to generate polysaccharides in the presence of exogenous fucose.
Promiscuity of the Salvage Pathway Toward Fucose Analogs in Vitro and in Vivo.
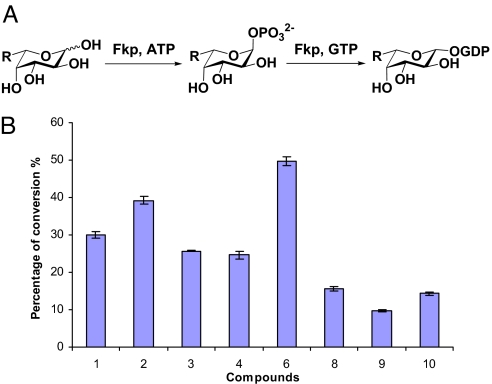
We next investigated the promiscuity of the salvage pathway in vitro. Fucose (coumpound 1, Fig. 2B) and 9 fucose analogs (compounds 2–10, Fig. 2B) with modifications at the C-6 position (see SI Appendix for compound synthesis and characterization) were used in the study. The C-6 position modification of fucose has been demonstrated to be tolerant by the mammalian salvage pathway and fucosyltransferases (14, 15). Reactions containing fucose or a fucose analog were carried out in a total volume of 20 μL, and the formation of the corresponding GDP-sugars was detected and quantified by CE assays (Fig. S4). The relative activities of Fkp toward different compounds were evaluated by the percentage conversions of substrates to products in 15 min (Fig. 3). The percentage conversions rang from ≈10% to as high as 50%, indicating a promiscuous yet differential recognition of Fkp toward different modifications. Compounds 5 and 7 with a aminomethyl and a 1-hydroxylethyl group respectively, did not give corresponding GDP-sugar peaks at the set wavelength (254 nm), thus no information on product conversion was obtained. However, reactions did occur as being verified by the observation of GDP-sugar formation in ESI-MS analysis, and the incorporation into polysaccharides was confirmed by CE-MS (see Fig. S5and discussion below).
Fig. 3.
Analysis of Fkp specificity toward fucose analogs. (A) In vitro enzymatic conversion of fucose to GDP-fucose (salvage pathway); Fkp possesses both fucokinase and pyrophosphorylase activities. (B) Percentage conversions of substrates to products by Fkp in 15 min in vitro reaction.
Having demonstrated the promiscuity of the salvage pathway in vitro, we turned to investigate whether the fucose analogs can be metabolized into polysaccharides in vivo. The Δgmd-fcl(fkp) strain was grown in LB medium supplemented with 10 compounds respectively. These compounds showed no toxicity to the cells as the cell growth rates were comparable to that of the wild-type. LPSs isolated from these recombinant strains displayed smooth phenotypes (Fig. S6). However, these LPSs had lower degrees of polymerization (ranging from 3 to 12 repeating units) compared with the wild-type (up to 20 resolvable repeating units). This observation may reflect the specificity of Wzy, the enzyme that catalyzes the polymerization of repeating units. Nevertheless, it appeared that the fucose analogs can be used by bacterium engineered with the GDP-fucose salvage pathway to produce polysaccharides, although to a less extent compared with fucose.
Expression of Modified Polysaccharides on Bacterial Cell Surfaces.
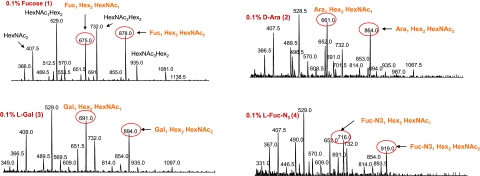
To confirm the successful incorporation of fucose analogs into polysaccharides, we used combined capillary electrophoresis mass spectrometry (CE-MS) technique (16) to determine the structures. Fig. S5 shows the full spectra of O-deacetylated LPSs from wild-type and mutant cells. In each spectrum, lipid-A associated ions were detected at m/z 855.9, 468.6 and 388.5, corresponding to the consecutive loss of a H3PO4, an N-acylated glucosamine and the second H3PO4 group, respectively. These fragment ions were generated within the orifice-skimmer region of the mass spectrometer because of the in-source collision-induced dissociation. The information on the repeating unit could be easily obtained by selecting the ion at m/z 204.2 as a precursor to perform precursor ion scan experiments, which corresponds to the HexNAc oxonium ion. The precursor-treated spectra showed a close-up at repeating units from each strain and the fragmentation patterns. Fig. 4 presents selected examples of precursor-treated spectra (see Fig. S7for the rest spectra). In the spectra, major peaks were identified and the corresponding sugar sequences were assigned. The peaks corresponding to the fragments containing fucose or fucose analogs were circled in red. In the wild-type strain, 2 peaks m/z 675.0 and 878.0 represent fragments Fuc-HexNAc-Hex2 and Fuc-HexNAc2-Hex2, respectively. The presence of a fucose (Mw = 146) can be derived from the difference between ions of m/z 675.0 and 529.0, and that between m/z 878.0 and 732.0. Analysis of the LPSs from mutant cells fed with compound 3 showed 2 characteristic peaks m/z 691.0 and 894.0, corresponding to fragments Gal-HexNAc-Hex2 and Gal-HexNAc2-Hex2, respectively. The presence of Gal (Mw = 162) was also confirmed by the difference between ions of m/z 691.0 and 529.0, and that between m/z 894.0 and 732.0. Spectra from other modified polysaccharides can also be analyzed likewise. It is worth noting that in all of the spectra for mutants, the fucose containing peaks m/z 675.0 and 878.0 were not detected, suggesting that fucose analogs completely replace the native fucose in modified polysaccharides.
Fig. 4.
Selected examples of structural analysis of modified polysaccharides by mass spectrometry. Major peaks in the spectra correspond to different fragments of repeating unit structures. The information was obtained by selecting the ion at m/z 204.2, which corresponds to the HexNAc oxonium ion, as a precursor to perform precursor ion scan experiments. The peaks corresponding to the fragments containing fucose or fucose analogs are circled in red.
Labeling LPS and Bacterial Cell Surfaces with Selective Chemical Reactions in Vitro.
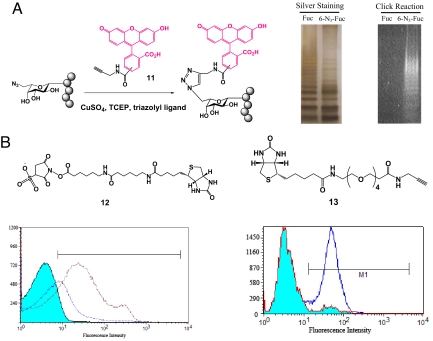
Many chemical functional groups have been used in chemical biology. Common ones include azide, alkyne, ketone, amine and hydroxyl sulfur. Azide is the most viable and versatile one in live cell applications, because of its stability and applicability in selective chemical conjugation with functionalized phosphines or with alkynes under physiological conditions (5). The azido group incorporated into polysaccharides using compound 4 can thus be further derivatized in vitro using chemical reactions (17). We isolated LPSs from E. coli O86:B7 Δgmd-fcl(fkp) strains cultured in medium supplemented with fucose and compound 4, respectively, and performed a Cu(I)-catalyzed [3+2] cycloaddition reaction with fluorescein conjugated alkyne reagent 11. The result showed that fluorescence was readily detected in the LPSs of E. coli O86:B7 Δgmd-fcl(fkp) fed with 6-azido-fucose 4, whereas no fluorescence was seen in the LPSs from E. coli O86:B7 Δgmd-fcl(fkp) fed with fucose (Fig. 5A). Comparison with various controls (Fig. S8), demonstrated that this reaction was highly specific.
Fig. 5.
Further derivatization of modified polysaccharides using in vitro chemical reactions. (A) Fluorescence labeling of azido-containing LPS by Cu(I)-mediated [3+2] cycloaddition. (B) Bacterial cell surface labeling with biotinylated activated ester 12 and biotinylated alkyne 13. Fluorescence signal was analyzed by flow cytometry.
Functionalized polysaccharides obtained by metabolic pathway engineering also provide the possibility for direct cell surface labeling by chemical reactions. Two different functionalized polysaccharides with amino (compound 5) and azido (compound 4) groups, respectively, were used as examples to demonstrate cell surface labeling. Amino groups mentabolically engineered to the polysaccharides on cell surface can react with biotinylated sulfosuccinimidyl-activated ester (compound 12) in PBS buffer to form stable amide bonds. The biotin-coated bacterial cells can be probed by staining with FITC conjugated streptavidin and the resulted fluorescence signal can be analyzed by flow cytometry. As shown in Fig. 5B, E. coli O86:B7 Δgmd-fcl(fkp) cells fed with compound 5 showed significantly increased fluorescence intensity compared with control samples. One of the controls, cells fed with fucose and underwent the same chemical reaction and FITC staining showed increased fluorescence signal compared with the one without any in vitro treatment. The possible explanation is that the amino group in both LPS core structures and membrane proteins can react with sulfo-NHS conjugated biotin to give positive fluorescence signal. However, cells expressing azido group displayed superior labeling efficiency and specificity with extremely low background fluorescence (Fig. 5B). These results indicate that azido group is more suitable than amino group for specific cell surface labeling.
Conclusion
In this study, we demonstrate a general, facile and effective approach to introduce modifications into polysaccharides in vivo. The key to the success of this approach lies in 2 points: (i) the functional equivalence of the salvage pathway to the de novo pathway in producing GDP-Fuc; and (ii) the promiscuity of the salvage pathway in metabolizing unnatural fucose derivatives to produce the corresponding sugar nucleotides. The functional expression of the salvage pathway in the mutant strain depleted of the de novo pathway has permitted the cell to directly salvage the exogenous sugars from the medium to synthesize sugar nucleotides. Thus, modifications on sugar analogs are generally preserved during conversion by the salvage pathway. In addition, since the only native de novo pathway is disrupted, the generation of natural sugar substrates is efficiently depleted. In this case, the expression of cell surface polysaccharides exclusively relies on the sugar nucleotides produced by the salvage pathway. Therefore, highly homogenous modifications on polysaccharides can be achieved. Among the chemical modifications incorporated are groups suitable for bioorthorgonal conjugation, such as an azido group. They serve as useful handles for in vitro chemical labeling of polysaccharides and bacterial cell surface. This labeling technique has potential applications in bacterial cell imaging, prodrug conjugation and directed drug delivery. In our study, even though the proof-of-principle experiments have been demonstrated with fucose analogs, in principle, this strategy can be applied to other monosaccharide components, such as galactose (Gal), N-acetylgalactosamine (GalNAc), N-acetylglucosamine (GlcNAc) and sialic acid (Neu5Ac). Study is being carried out in our lab to demonstrate engineering of bacterial polysaccharides with analogs of different monosaccharides.
Methods
Cloning, expression, purification and characterization of Fkp. The fkp gene was PCR amplified from B. fragilis 9343 (ATCC 25285) chromosome and constructed into NdeI and BamHI sites of pET15b vector (Novagen), and subsequently transformed into E. coli BL21 (DE3) for expression. For expression, the clones were cultured in LB broth with 100 μg/mL of ampicillin and were induced for protein expression using a final concentration of 0.5 mM IPTG and allowed to proceed at 25 °C for 12 h. Bacterial cell lysates were cleared on HiTrap Chelating HP column (5 mL) (GE Healthcare). Unbound proteins were washed off with a buffer containing 20 mM Tris·HCl (pH 7.5), 500 mM NaCl and 50 mM imidazole. Target proteins were eluted using the same buffer but with 500 mM imidazole. The proteins from different purification steps were analyzed by SDS/PAGE. The purified protein concentration in the final solution was determined to be 2.2 mg/mL. For in vitro assay of Fkp activity, 100-μL enzymatic reaction was performed in 50 mM Tris·HCl buffer (pH 7.5) containing 5 mM MgSO4, 10 mM ATP, 10 mM GTP, 10 mM l-fucose, 1 unit of inorganic pyrophosphatase, and 22 μg of purified Fkp. The reaction was incubated at room temperature for 30 min and quenched in boiling water for 1 min. The formation of GDP-fucose was analyzed by TLC (TLC) and further confirmed by ESI-MS.
Determination of Fkp Activity Toward Fucose Analogs Using Capillary Electrophoresis.
Enzymatic assays were carried out in duplicates in a total volume of 20 μL in Hepes buffer (200 mM, pH 7.0) containing 5 mM MgCl2, 1 mM ATP, 1 mM GTP, 1 mM substrate and 0.6 μg of recombinant Fkp. Reactions were allowed to proceed for 15 min at 37 °C and quenched by heating at 95 °C for 2 min. The reaction mixture was then diluted 10-fold and kept on ice. Samples were analyzed by a P/ACE MDQ Capillary Electrophoresis System equipped with a UV detector (Beckman Coulter). CE conditions were as follows: 75 μm i.d. capillary, 25 KV/80 μÅ, 5-s vacuum injections, monitored at 254 nm, running buffer was 25 mM sodium tetraborate, pH 10.0.
Functional Replacement of GDP-Fucose de Novo Pathway with Salvage Pathway.
The gmd-fcl gene was replaced by a chloramphenicol acetyltransferase (CAT) gene using the RED recombination system of phage lambda (12). The CAT gene was amplified from plasmid pKK232–8 using primers binding to the 5′ and 3′ ends of the gene, with each primer carrying 50 bp that flanks gmd-fcl gene. The PCR product was then transformed into E. coli O86:B7 strain carrying pKD20, and chloramphenicol-resistant transformants were selected after induction of the RED genes. To complement the gmd-fcl deficient mutants, the gmd-fcl gene was amplified from E. coli O86:B7 genomic DNA and then cloned into pTRC99A vector. The recombinant vector was then transformed into the Δgmd-fcl strain for transcomplementation. To transform fucose salvage pathway, recombinant vector pET15b-fkp was transformed into the Δgmd-fcl strain. The transformants were grown in LB medium alone or supplemented with 0.1% (final concentration) of various sugars. LPSs were extracted according to the published proteinase K digestion protocol, and analyzed by SDS/PAGE coupled with silver staining.
Analysis of LPS by Mass Spectrometry.
Small scale isolation of LPSs from wild-type and mutants was carried out as follows: Bacterial cells were collected from 1 mL overnight culture supplemented with various sugars. The cells were washed with 1 mL of H2O and resuspended in 0.5 mL of H2O in a 1.5-mL centrifuge tube. An equal volume of liquefied phenol was added and the tube was heated to 65 °C for 40 min, with vigorous votexing every 10 min. The mixture was centrifuged at 13,000 rpm (Beckman Coulter; JA-20 Fixed-Angle Rotor) for 30 min. The Top aqueous phase was transferred to a new tube and the organic phase was reextracted with 0.5 mL of H2O. The combined aqueous phase was concentrated to 100 μL before subjected to precipitation using 150 μL of 100% ethanol. The LPS was collected by centrifugation for 45 min. This LPS sample needs to be O-deacetylated to make it more amenable to MS analysis. To do that, dried LPS sample was dissolved in 100 μL of anhydrous hydrazine at 37 °C for 45 min. Then chilled acetone (400 μL) was added to remove the excess hydrazine. The solution was kept at −20 °C for overnight, or at −80 °C for 1 h, before centrifugation to pellet the LPS. Finally, the O-deacetylated LPS was dissolved in 20 μL of H2O and subjected to drop dialysis. The sample after dialysis was ready for CE-MS analysis.
Labeling Polysaccharides Via in Vitro Cu(I) Mediated [3+2] Cycloaddition.
Extraction of LPS from bacterial cells was carried out as described. Briefly, 20 mL overnight bacterial culture was centrifuged at 6,000 × g to harvest the cells. The cells were then washed with 30 mM Tris·HCl (pH = 8.0), and resuspended in buffer A containing 30 mM Tris·HCl, pH 8.0, 20% sucrose, and 10 mM EDTA. The cells were lysed by sonication, followed by centrifugation (10,000 × g). The supernatant was collected and subjected to ultracentrifugation (110,000 × g, 4 °C, 2 h). The precipitated gels were resuspended in distilled water for labeling reaction. The cycloaddition conditions were followed as described with TBTA as the triazole ligand (18). The reaction was allowed to proceed at room temperature for 2 h. The labeled product was harvested by ultracentrifugation and washed to remove excess reagents. After processed with proteinase K, the label LPSs were electrophoresed and detected with UV light.
Labeling Live Bacterial Cells via Chemical Reactions with Amine and Sulfo-NHS-Activated Ester.
Overnight culture of E. coli O86:B7 Δgmd-fcl(fkp) (200 μL) in LB medium supplemented with compound 5 was collected, washed 3 times with PBS buffer, and subsequently resuspended in 0.8 mL of PBS. A 10 mM solution of the biotinylated activated ester 12 was prepared by dissolving 2.0 mg of reagent in 300 μL of ultrapure water. A 200 μL of the biotin solution was added to cell resuspension and the reaction mixture was incubated at room temperature for 1 h. The reaction was centrifuged to discard the reaction supernatant. Labeled cells were washed extensively with PBS to remove the unreacted reagent. Cells were then resuspended in 0.5 mL of staining buffer, to which 1 μL of FITC conjugated streptavidin (0.5 mg/mL) was added. The resuspension was incubated at 4 °C with mild agitation for 45 min. Cells were again washed with PBS extensively before subjected to flow cytometry analysis.
Cu(I) Mediated [3+2] Cycloaddition (Click Chemistry).
Mutants expressing compound 4 were subjected to cycloaddition reaction as described in the labeling of LPS. Labeled cells were washed extensively with PBS, stained with FITC conjugated streptavidin, and subsequently subjected to flow cytometry analysis.
Supplementary Material
Acknowledgments.
We thank Cindy Coleman for her administrative assistance and Bob Woodward for proofreading of the manuscript. This work was supported by an endowed Ohio Eminent Scholar Professorship on Macromolecular Structure and Function in the Department of Biochemistry at The Ohio State University (to P.G.W.). X.C. is a Beckman Young Investigator and an Alfred P. Sloan Fellow.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812432106/DCSupplemental.
References
- 1.Westphal O, Jann K, Himmelspach K. Chemistry and immunochemistry of bacterial lipopolysaccharides as cell wall antigens and endotoxins. Prog Allergy. 1983;33:9–39. [PubMed] [Google Scholar]
- 2.Jones C. Vaccines based on the cell surface carbohydrates of pathogenic bacteria. An Acad Bras Cienc. 2005;77:293–324. doi: 10.1590/s0001-37652005000200009. [DOI] [PubMed] [Google Scholar]
- 3.Ostash B, Walker S. Bacterial transglycosylase inhibitors. Curr Opin Chem Biol. 2005;9:459–466. doi: 10.1016/j.cbpa.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 4.Wang L, Xie J, Schultz PG. Expanding the genetic code. Annu Rev Biophys Biomol Struct. 2006;35:225–249. doi: 10.1146/annurev.biophys.35.101105.121507. [DOI] [PubMed] [Google Scholar]
- 5.Prescher JA, Bertozzi CR. Chemical technologies for probing glycans. Cell. 2006;126:851–854. doi: 10.1016/j.cell.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 6.Jennings HJ, Gamian A, Ashton FE. N-Propionylated group B meningococcal polysaccharide mimics a unique epitope on group B Neisseria meningitidis. J Exp Med. 1987;165:1207–1211. doi: 10.1084/jem.165.4.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sadamoto R, et al. Cell-wall engineering of living bacteria. J Am Chem Soc. 2002;124:9018–9019. doi: 10.1021/ja026133x. [DOI] [PubMed] [Google Scholar]
- 8.Goon S, et al. Metabolic incorporation of unnatural sialic acids into Haemophilus ducreyi lipooligosaccharides. Proc Natl Acad Sci USA. 2003;100:3089–3094. doi: 10.1073/pnas.0437851100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yi W, et al. Two different O-polysaccharides from Escherichia coli O86 are produced by different polymerization of the same O-repeating unit. Carbohydr Res. 2005;341:100–108. doi: 10.1016/j.carres.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 10.Ma B, Simala-Grant JL, Taylor DE. Fucosylation in prokaryotes and eukaryotes. Glycobiology. 2006;16:158R–184R. doi: 10.1093/glycob/cwl040. [DOI] [PubMed] [Google Scholar]
- 11.Coyne MJ, et al. Human symbionts use a host-like pathway for surface fucosylation. Science. 2005;307:1778–1781. doi: 10.1126/science.1106469. [DOI] [PubMed] [Google Scholar]
- 12.Yu D, et al. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo H, et al. Molecular analysis of the O-antigen gene cluster of Escherichia coli O86:B7 and characterization of the chain length determinant gene (wzz) Appl Environ Microbiol. 2005;71:7995–8001. doi: 10.1128/AEM.71.12.7995-8001.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rabuka D, et al. A chemical reporter strategy to probe glycoprotein fucosylation. J Am Chem Soc. 2006;128:12078–12079. doi: 10.1021/ja064619y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sawa M, et al. Glycoproteomic probes for fluorescent imaging of fucosylated glycans in vivo. Proc Natl Acad Sci USA. 2006;103:12371–12376. doi: 10.1073/pnas.0605418103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Wang Z, Altman E. In-source fragmentation and analysis of polysaccharides by capillary electrophoresis/mass spectrometry. Rapid Commun Mass Spectrom. 2005;19:1305–1314. doi: 10.1002/rcm.1927. [DOI] [PubMed] [Google Scholar]
- 17.Prescher JA, Bertozzi CR. Chemistry in living systems. Nat Chem Biol. 2005;1:13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- 18.Link AJ, Tirrell DA. Cell surface labeling of Escherichia coli via copper(I)-catalyzed [3+2] cycloaddition. J Am Chem Soc. 2003;125:11164–11165. doi: 10.1021/ja036765z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.