Abstract
Although nanoparticle-protein conjugates have been synthesized for numerous applications, bioconjugation remains a challenge, often resulting in denaturation or loss of protein function. This is partly because the protein–nanoparticle interface is poorly understood, which impedes the use of nanoparticles in nanomedicine. Although the effects of nanoparticle ligand and material on protein structure have been explored, the choice of the labeling site on the protein has not yet been systematically studied. To address this issue, we label cytochrome c site-specifically with a negatively charged Au nanoparticle via a covalent thiol–Au bond. The attachment site is controlled by cysteine mutations of surface residues. The effect of labeling on protein structure is probed by circular dichroism. Protein unfolding is the most severe when the nanoparticle is attached to the N- and C-terminal foldon, the core motif of cytochrome c. Also, when the nanoparticle is attached in the vicinity of charged residues, the amount of structural damage is greater because of salt-dependent electrostatic interactions with charged ligand bis(p-sulfonatophenyl) phenylphosphine on the nanoparticle. Molecular dynamics simulations also elucidate local to global structural perturbation depending on labeling site. These results suggest that the labeling site must be considered as one of the main design criteria for nanoparticle–protein conjugates.
Keywords: bioconjugation, protein folding, circular dichroism, molecular dynamics simulation, nanoscale interface
Nanomaterials hold much promise for biomedical applications such as multiscale imaging (1–3), novel therapeutic approaches (4), and biomolecular sensing (5). The emerging field of nanomedicine is based on exploiting the unique size- and material-dependent properties of nanomaterials. However, the biggest barrier in effectively using them is their biological interface. Historically, biological–inorganic interfaces suffer complications such as surface fouling in medical devices and nonspecific adsorption of proteins on sensors, inhibiting practical use. The problem significantly worsens for nanoscale systems because of their high surface-to-volume ratio. Nanoscale surfaces are physically and chemically different from the bulk, requiring nontraditional tools to effectively probe them. These factors suggest a pressing need for deeper understanding of the “nano-bio” interface.
One particular issue related to the interface that limits the potential of nanoparticles (NPs) is conjugation to a biomolecule. Covalently linking a NP to a protein can affect both its structure and function. Even if protein structure is retained, the NP can sterically hinder substrate binding. NPs are large compared with dye molecules, and their interface with proteins is highly complex. NPs are not hard spheres but crystals with facets, edges, and vertices, and their passivating ligands can change conformation or come on and off the NP. Consequently, there are numerous nonspecific interactions between the NP and protein, where amino acid side chains noncovalently bind to the NP or ligand in undesirable ways. This often leads to protein denaturation, rendering the NP–protein conjugate useless.
Despite these complications, proteins on nanostructures are often assumed to be fully folded, an oversimplification that can severely obfuscate quantitative data in fluorescence resonance energy transfer (FRET) (6) and activity measurements (7), or lead to fundamental obstacles in comprehending biological side effects and cytotoxicity. To date, efforts have focused predominantly on exploiting material properties of NPs (8, 9), but ultimately these endeavors are limited by a poor understanding of the NP–protein interface. To realize the full potential of NPs in biological applications, understanding and ultimately controlling these interactions is critical.
To this end, efforts have begun to elucidate how NP labeling affects protein structure and function. For covalently linked proteins, the structural effects of ligand charge (10, 11) and NP material (12) have been studied. However, a key factor not yet explored is the effect of the labeling site, which may affect the protein conformation and activity, and thus would be crucial for obtaining quantitative information from NP–protein conjugates. Whereas the advances in conjugation chemistry have enabled site-specific labeling, the choice of labeling site must be strategized. Proteins have different domains of varying structural and functional importance, and charge and hydrophobicity are rarely uniform on protein surfaces. Obviously, labeling sites necessary for protein function, such as binding or active sites, must be avoided. Furthermore, each domain on a protein can stabilize the structure differently, so labeling sites must be carefully chosen to minimize destabilization. We have previously shown that protein structure in NP–protein conjugates is dominated by interactions with amino acid side chains in local vicinity of the conjugation site (10, 12). However, to date, there is no study that systematically investigates the effect of labeling site on protein structure, and the choice of labeling site has been largely overlooked.
Here, we conjugate Au NPs to the protein cytochrome c (cyt c) at different sites and study the effect on protein structure. We show that electrostatic interactions of amino acid side chains with the NP ligand are partially responsible for protein behavior, but more importantly, there are key motifs on the protein that must not be labeled to avoid denaturation. Conversely, labeling a protein with a NP can be used to induce local denaturation. Based on this, we propose a general guideline for NP functionalization: To preserve protein structure, flexible and loosely folded motifs should be labeled instead of nucleation centers for folding. Fundamental understanding of the effect of labeling site on protein structure, in combination with material, size, and ligand dependence, will enable strategic labeling of diverse proteins with a variety of biomedical importance.
Results
Choice of Protein and NP.
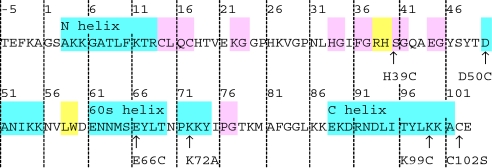
Cyt c is ideal for this study because its structure (13, 14) and folding have been extensively characterized, both experimentally (15, 16) and computationally (17, 18). Cyt c from Saccharomyces cerevisiae was used because its unique exposed Cys at position 102 can be linked covalently to Au NPs. Two other Cys at positions 14 and 17 are covalently bound to the heme and unavailable for labeling. Site-directed mutagenesis has inserted Cys mutations distributed throughout different motifs [39 = loop B; 50 = loop C; 66 = 60's helix; 99 and 102 = C-terminal helix; Fig. 1a, Table 1 (19, 20)]. We used 1.5-nm diameter Au NPs coated with BPS ligands, which partially unfold cyt c when covalently attached at C102 (10). Although a neutral ligand such as PEG would be better at keeping the conjugated protein folded, it would not be able to discriminate optimum labeling sites. K99C was tested in addition to C102 because C102 is buried in a hydrophobic pocket, whereas K99 is exposed to the solvent. Previous work has shown that flipping out C102 to form disulfides in cyt c dimerization perturbs protein structure slightly (10, 21).
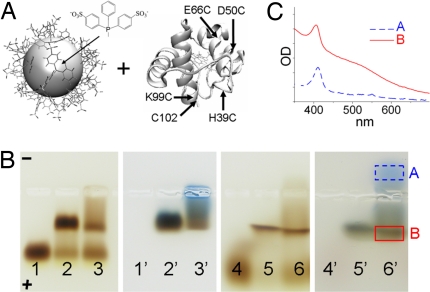
Fig. 1.
NP–protein conjugates. (A) Labeling yeast cyt c (Right) with 1.5-nm Au NP with BPS capping groups (Left). Sizes are to scale. Positions of labeled Cys in mutants and WT are marked by arrows. (B) Agarose gel electrophoresis. NP alone (lane 1), NP incubated with WT yeast cyt c (lane 2), or horse cyt c (lane 3) in 0.5× Tris-borate-EDTA buffer (TBE). Lanes 4–6: the same as lanes 1–3, except with 1 M NaCl. Lanes 1′–6′: corresponding blue-stained gel. (C) Absorption spectra of samples extracted from lane 6′ (marked by boxes). Absorption of NP–cyt c conjugates (red solid line) is the sum of a Soret peak at ≈410 nm due to cyt c and a broad band due to NPs.
Table 1.
Sequences of WT cyt c and mutants
Cys mutations indicated by arrows. Residue numbers follow that of PDB ID code 2BCN. Cyan, α-helices; pink, hydrogen bonded turn; yellow, β-bridge.
Site-Specific NP Labeling of cyt c on Surface Cys.
To check whether the NP was site-specifically and covalently labeled and not nonspecifically adsorbed onto the protein, we compared the conjugation of NPs with yeast cyt c and horse cyt c. Horse cyt c has similar sequence and structure but lacks the surface Cys. The positively charged horse cyt c attaches to negatively charged NPs by Coulombic interactions. We determined conditions under which yeast cyt c form conjugates to the NPs with higher yields than horse cyt c. Gel electrophoresis probed conjugation (Fig. 1B). Lane 1 contains free NPs that run to the anode. Lane 2 has NP incubated with yeast cyt c and lane 3 with horse cyt c. Both proteins attach to the NPs, as evidenced by lower mobility bands in both lanes. NP–cyt c conjugates have reduced gel mobility compared with free NPs because of larger size and lower net charge. Blue staining confirms the presence of protein in lower-mobility bands and shows that all cyt c are NP-labeled, because no band runs to the anode (lanes 2′ and 3′). In 1 M NaCl (Fig. 1B, lanes 4–6), the efficiency of horse cyt c labeling is reduced, likely because of salt screening. This is illustrated by a higher intensity of the NP band (lane 6 compared with lane 5), and a protein band that runs to the cathode (lane 6′). The absorption spectrum of this band has a Soret peak (410 nm), indicative of unlabeled horse cyt c (Fig. 1C). In contrast, salt does not affect the NP labeling efficiency of yeast cyt c, because no band due to free protein appears in lane 5′. This supports that C102 forms a covalent thiol–Au bond with the NP.
Structure of NP-Labeled Mutants Probed by CD.
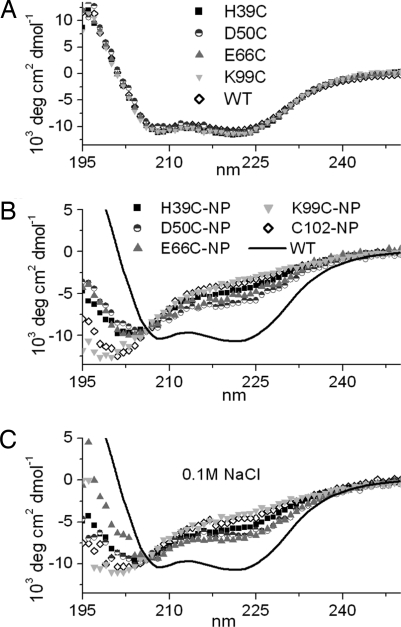
Far-UV CD spectroscopy confirmed that all unlabeled mutants were fully folded (Fig. 2A) (20). CD spectra of each of the mutants and wild type (WT) conjugated to NPs in 10 mM NaPi, pH 7.3, with (Fig. 2B) and without (Fig. 2C) 0.1 M NaCl, are shown. Compared with WT (solid line), NP conjugation results in structural disturbance for all of the conjugates, because molar ellipticity at 222 nm, θ222, increases and α-helical content decreases (Table 2). The protein is more disturbed when the NP is attached to the C-terminal helix (K99C-NP and C102-NP). Without salt, D50C-NP retains its α-helicity the most. With 0.1 M NaCl, folding improved in all conjugates to different degrees. Improvement was minimal for D50C, K99C, and C102, whereas H39C and E66C improved the most (Table 2). Soret and Q-band peaks show retained redox activity in several conjugates [supporting information (SI) Fig. S1].
Fig. 2.
Secondary structure of conjugates. CD spectra of unlabeled (A) and NP-labeled proteins (B and C), with no salt (B), with 0.1 M NaCl (C).
Table 2.
α-Helical content of NP-cyt c and change in α-helicity in 0.1 M NaCl
| Conjugate | No salt, % | 0.1 M NaCl, % | Δα-helicity, % |
|---|---|---|---|
| H39C-NP | 7.3 | 15.6 | 8.3 |
| D50C-NP | 16.2 | 17.5 | 1.3 |
| E66C-NP | 7.7 | 20.7 | 13.0 |
| K99C-NP | 4.8 | 6.1 | 1.3 |
| C102-NP | 5.6 | 6.9* | 1.3 |
Molecular Dynamics Simulations.
To provide atomistic insights into the structure and stability of NP-cyt c conjugates, we performed MD simulations. The NP was modeled as a 1.5-nm diameter dummy atom with 21 BPS ligands distributed on its surface. Inductively Coupled Plasma Optical Emission Spectrometry (ICP-OES) measured an S molar concentration 2.2× lower than Au, confirming a BPS coverage comparable to that reported for triphenylphosphine on 1.5-nm Au NPs, 21 BPS/NP (22). We postulated that the protein displaces one BPS when forming a thiol bond with Au (16).
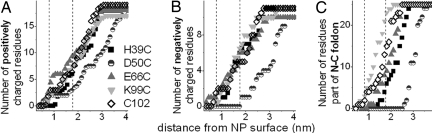
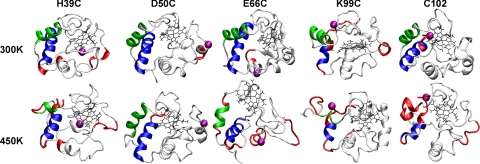
After building the NP–cyt c conjugates and performing initial minimization, we calculated distances of charged residues from the NP surface (Fig. 3 A and B). Compared with other conjugates, D50C-NP has significantly fewer charged residues near the NP surface. Other conjugates have a similar distribution of negatively charged residues. E66C-NP has slightly more positive charges in the vicinity of BPS. During MD simulations at 300 K, slight structural disturbance was observed. Because unfolding of proteins on NPs can take up to hours (23), we raised the temperature to 450 K to accelerate unfolding (24). We chose 450 K as unlabeled yeast cyt c remained folded over the course of the simulation. Snapshots of protein conformations at the end of the simulations at 300 K and 450 K (NP removed for clarity; Fig. 4) show that side chains of positively charged residues interact with BPS ligands (e.g., Fig. 5), and the secondary structures are perturbed differently depending on NP attachment site (Fig. 4).
Fig. 3.
Distribution of positive (A) and negative charges (B), and residues (C) part of the N-/C-terminal helices around the NP. Distances are between Au surface and amino acid charged end groups (A and B) or Cα atoms (C), before MD when the protein structure is folded. Dashed lines: Average distance of BPS O atoms from NP surface (8 Å) and one Debye length (9.6 Å) away from BPS O at 0.1 M NaCl.
Fig. 4.
Snapshots of protein structure at the end of 8-ns MD simulations at 300 K and 450 K. Green/blue: N-/C-terminal helices; red, loss of >50% α-helicity from WT structure (compare Fig. S2); purple sphere, NP attachment site. The program VMD was used for visualization (44).
Fig. 5.
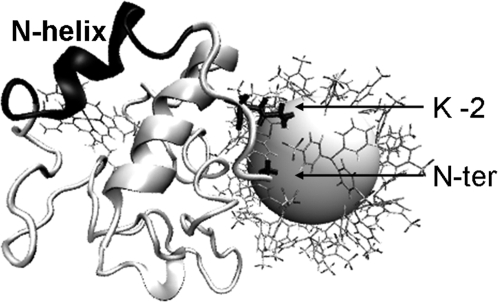
Interaction of positive charges of K-2 and the N terminus with BPS. The snapshot (in “perspective” view) is from MD simulation of E66C-NP at 450 K.
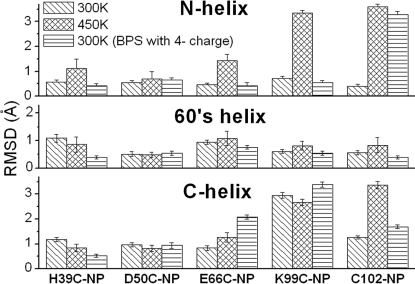
Structural disturbances observed in MD simulations agree with secondary structure changes observed experimentally. Root-mean-square deviation (rmsd) of Cα atoms in the WT conformation (Fig. 6) reveals that NP labeling of residue 66 perturbs the 60's helix, whereas labeling residues 99 and 102 perturbs both the N- and C-terminal helices that are in contact. Labeling residue 39 perturbs the β-bridge at positions 38–39 and 58–59 (Fig. 4). This shows that the NP generally disturbs the structure to which it is attached. These results based on rmsd are also supported by analysis of α-helicity of each of the mutant conjugates and the WT (Figs. S2–S4).
Fig. 6.
rmsd of Cα atoms from the WT conformation averaged over the last 500 ps of MD. Error bar: standard deviation over the time period. Fig. S4 shows rmsd trajectories.
To study the role of Coulombic interactions, the charge on each BPS molecule was increased from −2e to −4e (e = 1.6 × 10−19 C). This had a pronounced effect on the C102-NP conjugate, causing the N helix to unfold at 300 K within the 8-ns simulation time (Fig. 6).
Discussion
We find that the choice of labeling site for the NP can have a profound influence on protein structure. This conclusion was based on conformations of cyt c site-specifically labeled with NP at various positions, which were probed by using CD spectroscopy and further analyzed by MD simulations.
Electrostatic Effects.
Confirming earlier observations (10), electrostatic interactions between the protein and the NP ligand play a prominent role in the protein stability. Addition of 0.1 M NaCl results in an improvement of folding for all labeled mutants (Fig. 2C, Table 2). Thus, the degree of denaturation can be tuned by salt, especially for H39C or E66C. Addition of 0.1 M NaCl increases charge screening, and enables the protein to retain structure. However, the amount of structural recovery for each conjugate is different, with Δα ranging from 1.3 to 13.0% (Table 2). The Debye length at 0.1 M NaCl is 0.96 nm. A large portion of charged residues are screened at 0.1 M NaCl, because they are located further than 0.96 nm from BPS O atoms (Fig. 3 A and B). Results of MD simulations are consistent with the secondary structural measurements from CD, and yield insight into molecular interactions involved. Interaction of the amino acid side chains with BPS molecules is observed (Fig. 5), underscoring the importance of modeling the NP ligand explicitly.
D50C-NP has significantly fewer charges near BPS molecules (Fig. 3 A and B). This conjugate is the most folded both experimentally and in simulations, as evidenced by the least α-helical disturbance (Figs. 4 and 6). The present analysis confirms local electrostatic effects previously observed in NP labeling of WT yeast cyt c at position 102 (10). It also suggests that based on electrostatics alone, the labeling of sites with the least number of charged residues in the vicinity results in the least structural perturbation.
NP Destabilizes the Whole Protein when Attached on Structures Essential for Folding.
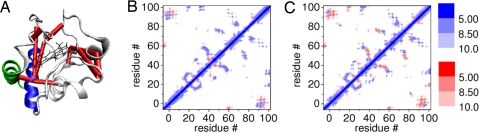
However, the different amounts of denaturation and recovery on charge screening cannot be explained solely by electrostatics. In both CD and MD, NP labeling of K99C and C102 results in strong denaturation that is barely improved by salt addition (Δα = +1.3%). This contrasts with E66C-NP, which also has the surface Cys on an α-helical motif with a similar number of charged residues as K99C, but retains significantly more of its structure in presence of salt (7.7% → 20.7%). Thus, local electrostatics are not the only factor influencing protein structure. When the NP is on positions 99 or 102 in the C-terminal α-helix, both the N- and C-terminal helices are substantially distorted (Figs. 4 and 6). This indicates that the mere presence of the NP is a large enough perturbation to the folding of these helices. Folding studies of cyt c have shown that these positions are on a motif critical for folding. Hydrogen exchange experiments on cyt c have shown that the N- and C-terminal helices, known as the N–C foldon, fold first (15) and bind together by hydrophobic residues at the interface. Four residues at the N–C interface, Gly-6, Phe-10, Leu-94, and Tyr-97, are the only non-heme-binding residues conserved among known cyt c sequences (25), underscoring the importance of this interface. Once the N–C foldon is locked in place, it guides the folding of the rest of the protein. Simulations also support this sequential stepwise folding pathway (17, 18). The N–C foldon is crucial for the stability of the whole protein and mutations in either the N- or C-terminal helix significantly affects folding (26, 27). It thus acts as a “safety pin” for the entire structure, while other motifs such as the 60's helix, are less stable and thus can denature more locally. We monitor N–C foldon conformation in MD simulations via the solvent-accessible surface area (ASA) of its hydrophobic cluster (28). Although no significant change is observed for simulations with normal charges on BPS, ASA increases for K99C-NP and C102-NP when charges on BPS are enhanced (Fig. S5). Indeed, our contact analysis reveals that the N–C helices form a “hub” that is linked to other multiple domains that constitute additional foldons of cyt c (Fig. 7A) (29). The level of denaturation can also be characterized by changes in the contact map (Fig. 7 B and C and Fig. S6). Thus, the NP prohibits not only formation of the C-terminal helix but also the N–C helix contact, affecting the stability of the entire protein.
Fig. 7.
Contact map of cyt c. (A) WT contact network. Centers of mass of 2 domains are connected by a red cylinder if there are residues with Cα atoms closer than 8.5 Å. Domains connected to the N–C foldon are known to fold before the network on the right (β-bridge and 50's helix) (45). Contact maps of D50C-NP (B) and K99C-NP (C), averaged over the last 500 ps of MD at 450 K (blue) on top of the WT contact map (red). Scale corresponds to distances between Cα atoms (Å). Wider red area in C indicates more denaturation. (See also Fig. S6.)
More moderate unfolding is observed when the NP is attached further from the N–C foldon (Figs. 2 and 3C). MD simulations of H39C-NP and E66C-NP show that the N-terminal amino group and Lys-2 (compare Table 1) are close enough to the BPS molecules in these conjugates to form salt bridges, which eventually lead to the disruption of the N helix (Fig. 5). In these conjugates, most of the N–C foldon is away from NP negative charges by more than the Debye length at 0.1 M NaCl (Fig. 3C), which explains the considerable improvement in folding by salt. In contrast, NP labeling at position 50 does not affect the folding of the rest of the protein as dramatically, consistent with CD results. The N–C foldon is still maintained in this conjugate and contacts are more stable (Figs. 4, 6, and 7). Thus, induced disorder is more localized in D50C-NP.
To verify our simulations performed in an implicit solvent environment (Materials and Methods), for D50C-NP and K99C-NP, we performed explicit water MD simulations, where each production run lasted 4.5 ns at 450 K. A stronger structural disturbance in K99C-NP was again observed from the beginning of the simulation, albeit deformation occurred more slowly because of solvent viscosity (Figs. S7–S10).
Ramifications for NP-Labeling Strategies.
The result here indicates that choice of labeling site is crucial for protein structure. It has been suggested that the network of contacts among amino acids in protein structures exhibits the “small-world” feature, in which there are a few key residues that play the role of hubs in the network, stabilizing the structure (30). Labeling motifs that contain these key residues result in the greatest amount of protein denaturation. A very large family of proteins has the N- and C-terminal motifs in contact, which has been suggested to control initial folding, native state stability, and turnover (29). Incidentally, protein termini are the most common NP labeling sites. His tags are predominantly put at N or C termini because of convenience in cloning, labeling chemistry, and conjugate purification. However, if the protein is part of the large family that possesses the N–C contact (29), it could potentially be the worst site for labeling, resulting in denaturation that cannot be rescued by salt screening, and ultimately loss of biological function. For these systems, labeling of low-stability motifs would be optimal. In addition to choosing the “least stable” motifs for labeling, sites should also have minimally charged residues in the vicinity to reduce electrostatic interactions. We find that the level of denaturation correlates with the number of charged amino acids in the local vicinity of the NP labeling site.
Furthermore, by inducing localized perturbations with a careful choice of labeling sites, there is potential for exploiting NP labeling as a tool for studying protein structure and folding dynamics. Obviously this effect strongly depends on NP size, because the amount of interaction with the NP ligand will vary with NP curvature (31, 32); 1.5-nm NPs are small enough to introduce local denaturation effects, because only residues close to the attachment site will interact with the NP and ligands. For larger NPs, disorder in the protein might not be so localized. Dependence on NP size and the effect on redox potential are the subject of future studies, in addition to application to other proteins.
Materials and Methods
Gold Nanoparticle Synthesis.
The 1.5-nm diameter Au NPs were synthesized in toluene with triphenylphosphine ligands (22). Negatively charged BPS (Strem Chemicals) was used to passivate NPs to permit water solubility. Two-phase ligand exchange was used. NPs were drawn into aqueous phase in the presence of BPS. BPS-NPs were purified from excess BPS by size-exclusion HPLC (Varian ProStar) with TSK-gel G4000PW column (TOSOH Bioscience). TEM showed a d = 1.5 ± 0.2 nm (n = 298). Optical absorption showed no plasmon peak, confirming a d <2 nm and no aggregation. NP concentrations were determined by using ε420 = 110.0 mM−1 cm−1 (33). BPS and NP molar ratios were measured by ICP-OES (SPECTRO model Ciros-Vision) assuming 100 atoms/NP.
Expression of Cyt c Mutants.
WT cyt c from S. cerevisiae was purchased (Sigma). Mutant plasmids were obtained from the groups of Harry Gray and Jay Winkler (19) (Table 1, sequences with mutations in bold). C102S mutation is for removal of the WT Cys. Because S. cerevisiae cyt c expressed in Escherichia coli lacks the trimethylation at Lys-72 found in the native protein, a K72A mutation was necessary for preventing the heme from binding to Lys-72 (34). Pletneva et al. (20) determined that the mutations (H39C/K72A/C102S, D50C/K72A/C102S, E66C/K72A/C102S and K72A/K99C/C102S) do not significantly perturb folding.
Plasmids were sequenced (MIT Biopolymers Facility) to verify mutations, and cloned into E. coli BL21DE3 Star cells (Invitrogen). To increase yield, cyt c maturation gene cassette pEC86 with Chloramphenicol (Chl) resistance (35) and cyt c plasmid with Ampicillin (Amp) resistance were cotransformed. Colonies from LB agar plates with 100 μg/mL Amp and 34 μg/mL Chl were screened for best cyt c expression in 5 mL of TB medium containing 100 μg/mL Amp and 34 μg/mL Chl grown for 24 h shaking at 37 °C. When possible, colonies were selected by the pink color of the cell pellet (due to presence of cyt c) and saved for frozen stocks.
Cells from frozen stocks were used to inoculate 10 mL of TB medium containing 100 μg/mL Amp and 34 μg/mL of Chl. After overnight growth, large TB cultures (≈5–10 L) containing 100 μg/mL Amp and 34 μg/mL of Chl were inoculated. Cells were grown for 36 h at 37 °C, harvested by centrifugation, and resuspended in 10 mM NaPi buffer (pH 7.0).
To isolate the protein, cells were lysed by French Press at 28 PSI (Constant Systems Basic Z Model). 20 nM DNase, 1 mM phenylmethanesulfonylfluoride (PMSF), and 1 mM DTT were added and the suspension was centrifuged at 6000 rpm for 30 min and the supernatant was collected. To purify the protein, the solution was loaded onto a Fast Flow CM Sepharose Econo-column equilibrated with 20 mM NaPi buffer with 1 mM DTT and 1 mM PMSF (pH 7.0). Cyt c was eluted with a stepwise salt gradient (0–0.5 M NaCl). Protein was further purified by ion-exchange chromatography with a Mono S Sepharose column by using a FPLC system with the same buffer, and then by size exclusion chromatography with TSK-gel G4000PW column on a HPLC system (VarianProStar). Protein purity was confirmed by SDS/PAGE. Cyt c concentrations were determined by using ε410 = 106.1 mM−1 cm−1.
Conjugation of Cyt c with NPs.
Cyt c were washed from excess DTT with 5-kDa centrifugal filters at 4 °C and resuspended in 10 mM NaPi buffer at pH 7.3 with or without 0.1 M NaCl. NPs were attached to cyt c by incubation overnight in the same buffer at 4 °C and then studied by CD and absorption spectroscopy. Cyt c/NP incubation ratio was 1:1.5 to ensure that there is at most 1 protein per NP so as to remove effects of protein–protein interactions on the NP surface that may influence structure (12). To show site-specific labeling, WT yeast cyt c and horse cyt c (Sigma) were incubated with NPs in 0.5× TBE with and without 1 M NaCl for 1 h and loaded in a 1% agarose gel for electrophoresis in 0.5× TBE with a field strength of 10 V/cm. Gels were blue stained for protein (SimplyBlue SafeStain, Invitrogen). Samples were extracted from an identical gel for absorption spectroscopy with 0.2-μm centrifugal filters (Nanosep, Pall) at 104 × g.
Spectroscopic Analysis.
Absorption spectra were obtained (Varian Cary 50 Spectrophotometer) at room temperature with a 1-cm pathlength cuvette. Scan rate was 5 nm/s. Far-UV CD spectra of proteins, NPs, and conjugates were obtained (Aviv model 202) at 25 °C in a 1-mm-pathlength cuvette. NP contribution to CD signal was subtracted. An isodichroic point at θ206 was found for heat-induced cyt c denaturation, as reported for glycerol-induced denaturation (36). Because cyt c concentration in NP:cyt c solutions could not be computed with high precision from absorption spectra because of NP absorbance in the Soret region, CD spectra of NP-cyt c conjugates were normalized to θ206 = −9.5 × 103 deg cm2 dmol−1. We used the CDSSTR algorithm (37) to deconvolve the spectra into % α-helicity.
Molecular Dynamics Simulations.
MD simulations were performed by using CHARMM version 32 (38), with the generalized Born with simple switching (GBSW) implicit solvent (39). Although we did not retain 4 internal water molecules in the crystal, WT remained folded at 300K in simulations with GBSW. A 20-Å cutoff was applied for nonbonded interactions with a switch function between 16 Å and 20 Å. To verify results from simulations by using GBSW, explicit water simulations were performed for D50C-NP and K99C-NP (see SI Text and Figs. S7–S10). The param22 all-atom force field (40) was used and modified to include NP parameters. The NP core was modeled as one dummy atom with Lennard–Jones parameters (ε = 0.02 and rmin/2 = 14 Å). Each BPS ligand was modeled explicitly with partial charges estimated from known parameters for the benzene ring and methyl sulfate. The BPS molecule was constructed from coordinates of a triphenylphosphine molecule linked to an Au atom (41). Twenty-one BPS molecules were uniformly distributed on the NP surface. Cyt c were constructed from Protein Data Bank (PDB) ID code 2BCN (42) and mutated accordingly. The heme was covalently linked to Cys 14 and 17. Covalent bonds were maintained between the heme iron and Met-80 and His-18, which are assumed to remain formed during the timescale of our simulations (17). Cyt c was linked to the NP surface by replacing one BPS ligand by the labeled Cys. To prevent formation of close contacts, this was done by approaching the protein sequentially in steps of 1 Å, and performing an energy minimization between each step until the S of the thiol reached the target coordinate. During energy minimization and dynamics, the position of the S of the labeled Cys and the P atoms of BPS ligands were fixed at 10 Å from the NP center, based on reported P–Au and S–Au bond lengths (41, 43).
For MD, the leap-frog method was used with a 2-fs time step and the SHAKE algorithm to constrain bonds involving hydrogen atoms. The system was heated and equilibrated from 48 K to 300 K for 20 ps followed by a production run at 300 K for 8 ns, or from 48 K to 450 K for 40 ps and maintained at 450 K for 8 ns. During production runs, temperature fluctuated by 1.3%, and no energy or temperature drift was observed. rmsd of Cα atoms from energy-minimized WT configuration were calculated for the N, C, and 60's helices.
Supplementary Material
Acknowledgments.
We thank Harry Gray, Jay Winkler, and Jennifer Lee for generously providing plasmids and helpful advice on mutant expression and purification; Center for Materials Science and Engineering for the use of the transmission electron microscope; Computational and Systems Biology Initiative (CSBi) Biophysical Instrumentation Facility for CD spectrophotometer; the Van Vliet group for UV-vis spectrophotometer; MIT Nuclear Reactor Laboratory for Inductively Coupled Plasma Optical Emission Spectrometry; the CSBi Computational facility; the Thomas Schwartz lab for use of French Press; Ky Lowenhaupt for FPLC purification; and Viviana Serra and Sisi Zhu for help with experiments.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807299106/DCSupplemental.
References
- 1.Liu W, et al. Compact biocompatible quantum dots functionalized for cellular imaging. J Am Chem Soc. 2008;130:1274–1284. doi: 10.1021/ja076069p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruchez M, et al. Semiconductor nanocrystals as fluorescent biological labels. Science. 1998;281:2013–2016. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- 3.Gao X, et al. In vivo cancer targeting and imaging with semiconductor quantum dots. Nat Biotechnol. 2004;22:969–976. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 4.Neuberger T, et al. Superparamagnetic nanoparticles for biomedical applications: Possibilities and limitations of a new drug delivery system. J Magn Magn Mater. 2005;293:483–496. [Google Scholar]
- 5.Medintz IL, et al. A fluorescence resonance energy transfer-derived structure of a quantum dot-protein bioconjugate nanoassembly. Proc Natl Acad Sci USA. 2004;101:9612–9617. doi: 10.1073/pnas.0403343101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pons T, et al. Solution-phase single quantum dot fluorescence resonance energy transfer. J Am Chem Soc. 2006;128:15324–15331. doi: 10.1021/ja0657253. [DOI] [PubMed] [Google Scholar]
- 7.Montesano-Roditis L, Glitz DG, Traut RR, Stewart PL. Cryo-electron microscopic localization of protein L7/L12 within the Escherichia coli 70 S ribosome by difference mapping and nanogold labeling. J Biol Chem. 2001;276:14117–14123. doi: 10.1074/jbc.M008430200. [DOI] [PubMed] [Google Scholar]
- 8.Salata O. Applications of nanoparticles in biology and medicine. J Nanobiotechnol. 2004;2:3. doi: 10.1186/1477-3155-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roco MC. Nanotechnology: Convergence with modern biology and medicine. Curr Opin Biotech. 2003;14:337–346. doi: 10.1016/s0958-1669(03)00068-5. [DOI] [PubMed] [Google Scholar]
- 10.Aubin-Tam ME, Hamad-Schifferli K. Gold nanoparticle cytochrome c complexes: The effect of nanoparticle ligand charge on protein structure. Langmuir. 2005;21:12080–12084. doi: 10.1021/la052102e. [DOI] [PubMed] [Google Scholar]
- 11.Verma A, Nakad H, Simard JM, Rotello VM. Recognition and stabilization of peptide α-helices using templatable nanoparticle receptors. J Am Chem Soc. 2004;126:10806–10807. doi: 10.1021/ja047719h. [DOI] [PubMed] [Google Scholar]
- 12.Aubin-Tam M-E, Zhou H, Hamad-Schifferli K. Structure of cytochrome c at the interface with magnetic CoFe2O4 nanoparticles. Soft Matter. 2008;4:554–559. doi: 10.1039/b711937b. [DOI] [PubMed] [Google Scholar]
- 13.Louie GV, Hutcheon WLB, Brayer GD. Yeast iso-1-cytochrome c—A 2.8A resolution three-dimensional structure determination. J Mol Biol. 1988;199:205–314. doi: 10.1016/0022-2836(88)90315-4. [DOI] [PubMed] [Google Scholar]
- 14.Louie GV, Brayer GD. High-resolution refinement of yeast iso-1-cytochrome c and comparisons with other eukaryotic cytochromes c. J Mol Biol. 1990;214:527–555. doi: 10.1016/0022-2836(90)90197-T. [DOI] [PubMed] [Google Scholar]
- 15.Maity H, Maity M, Englander SW. How cytochrome c folds, and why: Submolecular foldon units and their stepwise sequential stabilization. J Mol Biol. 2004;343:223–233. doi: 10.1016/j.jmb.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 16.Hostetter DR, et al. Partially formed native tertiary interactions in the A-state of cytochrome c. J Mol Biol. 1999;289:639–644. doi: 10.1006/jmbi.1999.2764. [DOI] [PubMed] [Google Scholar]
- 17.Roccatano D, et al. Selective excitation of native fluctuations during thermal unfolding simulations: Horse heart cytochrome c as a case study. Biophys J. 2003;84:1876–1883. doi: 10.1016/S0006-3495(03)74995-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.García AE, Hummer G. Conformational dynamics of cytochrome c: Correlation to hydrogen exchange. Proteins Struct Funct Genet. 1999;36:175–191. [PubMed] [Google Scholar]
- 19.Pletneva EV, Gray HB, Winkler JR. Many faces of the unfolded state: Conformational heterogeneity in denatured yeast cytochrome c. J Mol Biol. 2005;345:855–867. doi: 10.1016/j.jmb.2004.10.085. [DOI] [PubMed] [Google Scholar]
- 20.Pletneva EV, Gray HB, Winkler JR. Snapshots of cytochrome c folding. Proc Natl Acad Sci USA. 2005;102:18397–18402. doi: 10.1073/pnas.0509076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moench SJ, Satterlee JD. Proton NMR Comparison of the Saccharomyces cerevisiae ferricytochrome c isozyme-1 monomer and covalent disulfide dimer. J Biol Chem. 1989;264:9923–9931. [PubMed] [Google Scholar]
- 22.Weare WW, Reed SM, Warner MG, Hutchison JE. Improved synthesis of small (dcore 1.5 nm) phosphine-stabilized gold nanoparticles. J Am Chem Soc. 2000;122:12890–12891. [Google Scholar]
- 23.You C-C, De M, Han G, Rotello VM. Tunable inhibition and denaturation of α-chymotrypsin with amino acid-functionalized gold nanoparticles. J Am Chem Soc. 2005;127:12873–12881. doi: 10.1021/ja0512881. [DOI] [PubMed] [Google Scholar]
- 24.Day R, Bennion BJ, Ham S, Daggett V. Increasing temperature accelerates protein unfolding without changing the pathway of unfolding. J Mol Biol. 2002;322:189–203. doi: 10.1016/s0022-2836(02)00672-1. [DOI] [PubMed] [Google Scholar]
- 25.Ptitsyn OB. Protein folding and protein evolution: Common folding nucleus in different subfamilies of c-type cytochromes? J Mol Biol. 1998;278:655–666. doi: 10.1006/jmbi.1997.1620. [DOI] [PubMed] [Google Scholar]
- 26.Colón W, et al. Side chain packing of the N- and C-terminal helices plays a critical role in the kinetics of cytochrome c folding. Biochemistry. 1996;35:5538–5549. doi: 10.1021/bi960052u. [DOI] [PubMed] [Google Scholar]
- 27.Hampsey DM, Das G, Sherman F. Amino acid replacements in yeast bo-1-cytochrome c. J Biol Chem. 1986;261:3259–3271. [PubMed] [Google Scholar]
- 28.Hwang W, Zhang S, Kamm RD, Karplus M. Kinetic control of dimer structure formation in amyloid fibrillogenesis. Proc Natl Acad Sci USA. 2004;101:12916–12921. doi: 10.1073/pnas.0402634101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krishna MMG, Englander SW. The N-terminal to C-terminal motif in protein folding and function. Proc Natl Acad Sci USA. 2005;102:1053–1058. doi: 10.1073/pnas.0409114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vendruscolo M, Dokholyan NV, Pac E, Karplus M. Small-world view of the amino acids that play a key role in protein folding. Phys Rev E. 2002;65 doi: 10.1103/PhysRevE.65.061910. 061910. [DOI] [PubMed] [Google Scholar]
- 31.Shang W, Nuffer JH, Dordick JS, Siegel RW. Unfolding of ribonuclease A on silica nanoparticle surfaces. Nano Lett. 2007;7:1991–1995. doi: 10.1021/nl070777r. [DOI] [PubMed] [Google Scholar]
- 32.Vertegel AA, Siegel RW, Dordick JS. Silica nanoparticle size influences the structure and enzymatic activity of adsorbed lysozyme. Langmuir. 2004;20:6800–6807. doi: 10.1021/la0497200. [DOI] [PubMed] [Google Scholar]
- 33.Hainfeld JF, Furuya F. A 1.4nm gold cluster covalently attached to antibodies improve immunolabeling. J Histochem Cytochem. 1992;40:177–184. doi: 10.1177/40.2.1552162. [DOI] [PubMed] [Google Scholar]
- 34.Pollock WBR, et al. Bacterial expression of a mitochondrial cytochrome c Trimethylation of Lys72 in yeast iso-1-cytochrome c and the alkaline conformational transition. Biochemistry. 1998;37:6124–6131. doi: 10.1021/bi972188d. [DOI] [PubMed] [Google Scholar]
- 35.Thöny-Meyer L, Künzler P, Hennecke H. Requirements for maturation of Bradyrhizobium japonicum cytochrome c550 in Escherichia coli. Eur J Biochem. 1996;235:754–761. doi: 10.1111/j.1432-1033.1996.00754.x. [DOI] [PubMed] [Google Scholar]
- 36.Bongiovanni C, Sinibaldi F, Ferri T, Santucci R. Glycerol-induced formation of the molten globule from acid-denatured cytochrome c: Implication for hierarchical folding. J Protein Chem. 2002;21:35–41. doi: 10.1023/a:1014179031881. [DOI] [PubMed] [Google Scholar]
- 37.Sreerama N, Woody RW. Computation and analysis of protein circular dichroism spectra. Method Enzymol. 2004;383:318–351. doi: 10.1016/S0076-6879(04)83013-1. [DOI] [PubMed] [Google Scholar]
- 38.Brooks BR, et al. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 39.Im W, Lee MS, Brooks CL., III Generalized born model with a simple smoothing function. J Comput Chem. 2003;24:1691–1702. doi: 10.1002/jcc.10321. [DOI] [PubMed] [Google Scholar]
- 40.MacKerell AD, Jr, Feig M, Brooks CL., III Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 41.Wang J-C, Wang Y. Structure of [Au(PPh3)2][C(CN)3] Acta Crystallogr C. 1993;49:131–132. [Google Scholar]
- 42.Kang SA, Hoke KR, Crane BR. Solvent isotope effects on interfacial protein electron transfer in crystals and electrode films. J Am Chem Soc. 2006;128:2346–2355. doi: 10.1021/ja0557482. [DOI] [PubMed] [Google Scholar]
- 43.Majumder C, Kulshreshtha SK. Structural and electronic properties of Aun (n=2–10) clusters and their interactions with single S atoms: Ab initio molecular dynamics simulations. Phys Rev B. 2006;73:155427–155437. [Google Scholar]
- 44.Humphrey W, Dalke A, Schulten K. VMD—Visual molecular dynamics. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 45.Maity H, et al. Protein folding: The stepwise assembly of foldon units. Proc Natl Acad Sci USA. 2005;102:4741–4746. doi: 10.1073/pnas.0501043102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.