Abstract
We have conducted an integrative genomics analysis of serological responses to non-HLA targets after renal transplantation, with the aim of identifying the tissue specificity and types of immunogenic non-HLA antigenic targets after transplantation. Posttransplant antibody responses were measured by paired comparative analysis of pretransplant and posttransplant serum samples from 18 pediatric renal transplant recipients, measured against 5,056 unique protein targets on the ProtoArray platform. The specificity of antibody responses were measured against gene expression levels specific to the kidney, and 2 other randomly selected organs (heart and pancreas), by integrated genomics, employing the mapping of transcription and ProtoArray platform measures, using AILUN. The likelihood of posttransplant non-HLA targets being recognized preferentially in any of 7 microdissected kidney compartments was also examined. In addition to HLA targets, non-HLA immune responses, including anti-MICA antibodies, were detected against kidney compartment-specific antigens, with highest posttransplant recognition for renal pelvis and cortex specific antigens. The compartment specificity of selected antibodies was confirmed by IHC. In conclusion, this study provides an immunogenic and anatomic roadmap of the most likely non-HLA antigens that can generate serological responses after renal transplantation. Correlation of the most significant non-HLA antibody responses with transplant health and dysfunction are currently underway.
Keywords: integrative genomics, kidney compartment, kidney transplantation, non-HLA antigen
Despite advances in immunosuppressive therapies and the resultant reduction in the incidence of acute rejection, declining graft function remains a paramount clinical concern, because recent studies have shown no benefit of the reduction of acute rejection incidence on graft life expectancy (1). This may relate to the heterogeneity of the acute rejection injury (2) (3), but there is extensive evidence that antibodies recognizing and engaging with donor antigens also play a key role in renal allograft outcomes (4).
Antibodies recognizing HLA molecules (major histocompatibility antigens) are the most important group of antibodies for renal transplantation. HLA antibodies can be present before transplantation, because of prior exposure to nonself HLA molecules (after pregnancy, blood transfusion or prior allo-transplantation), or can be produced de novo after transplantation (5). Donor-specific anti-HLA alloantibodies can initiate rejection through complement-mediated and antibody-dependent, cell-mediated cytotoxicity (6, 7). In contrast to these “major” histocompatibility antibodies, “minor” non-HLA antigens have been implicated in renal allograft outcome, and likely have a much stronger role in clinical transplantation than previously thought (8). Antibodies against MICA (MICA = MHC class I polypeptide-related sequence A), a locus related to HLA determining a polymorphic series of antigens similar to HLA, have been associated with decreased graft survival (9, 10). Duffy [a chemokine receptor, the Duffy antigen-receptor for chemokines (DARC)], and Kidd polymorphic blood group antigens, may be associated with chronic renal allograft histological injury (11). Antibodies against Agrin, the most abundant heparin sulfate proteoglycan present in the glomerular basal membrane, have been implicated in transplant glomerulopathy (12), and agonistic antibodies against the Angiotensin II type 1 receptor (AT1R-AA) were described in renal allograft recipients with severe vascular types of rejection and malignant hypertension (13).
It is expected that there are many more unidentified non-HLA non-ABO immune antigens that might evoke specific antibody responses after renal transplantation (8). However, without target antigen identification, antibody screening for specificity is near impossible. The advent of high-density protein microarrays has made screening for serum antibodies against thousands of human proteins more efficient, as seen in recent publications in autoimmune disease (14) and cancer (15).
Here, we use protein arrays to query de novo or augmented postkidney transplantation antibody responses against a selection of HLA and non-HLA targets in 18 transplant recipients. We are able to simultaneously interrogate posttransplant antibody responses to >5,000 individual human proteins, but for these antibody responses to be clinically relevant, it would be important to interrogate if they are directed against the transplanted kidney. Second, if kidney specific antigens are preferentially recognized after transplantation, are some kidney compartments more immunogenic than the others? To address these questions, tissue-specific and microdissected kidney compartment-specific gene expression raw data were downloaded from the public domain [e.g., SYMATLAS (http://symatlas.gnf.org/SymAtlas)], and gene identifiers from cDNA and Affymetrix platforms were mapped to their corresponding protein identifiers on the ProtoArray platform. Our hypothesis was that kidney alloantigens can elicit de novo or augmented antibody recognition after kidney transplantation. Microdissected normal kidney compartment-specific lists of expressed genes, were cross-mapped to protein identifiers on the ProtoArray platform, to determine whether antigens in a specific kidney compartment were being targeted after transplantation.
Results
Identification of de Novo Non-HLA Non-ABO Antibody Formation After Renal Transplantation.
The formation of de novo antibodies after renal transplantation was assessed by comparing 18 posttransplant serum samples with 18 paired pretransplant serum samples. Of the 5,056 proteins present on the ProtoArray, an average of 61% (range across patients 21–96%) had an increased signal after transplantation. A complete list of all 5,056 antigens has been deposited in the Gene Expression Omnibus (GEO) (16).
Mapping Genes That Are Specifically Expressed in Different Compartments of Normal Kidney.
We obtained from the Stanford Microarray Database (http://med.stanford.edu/jhiggins/Normal_Kidney/download.shtml) a previously reported cDNA microarray gene expression dataset, in which 34 samples were obtained from 7 different renal compartments of normal kidneys: glomerular (n = 4), inner cortex (n = 5), outer cortex (n = 5), inner medulla (n = 5), outer medulla (n = 5), pelvis (n = 5) and papillary tip samples (17). Raw data were downloaded and genes significant for each compartment selected by SAM (18), using an FDR ≤5%. Probes, specific to each kidney compartment, were retained in our analysis based on the published filtered list of 16,293 cDNA probes from ≈42,000. The specific gene lists for each compartment are given in Table S1.
Cross mapping kidney compartment specific gene probes to protein targets on the ProtoArray to select potential kidney compartment specific proteins, Cross mapping of gene IDs on the gene expression microarray and the ProtoArray platforms was conducted using AILUN software (19). The number of compartment specific genes that were also measured on the ProtoArray platform, are shown in Table S2.
Identification of HLA and MICA Antibodies After Kidney Transplantation.
Fifty percent of patients (9/18) had showed positive de novo donor specific antibody responses clinically detected by flow cytometry performed at the Stanford histocompatibility lab, detected at a mean time of 24 months posttransplantation. We compared the ability to detect anti-HLA antibodies by ProtoArray measurements, with clinical measurement of anti-HLA antibodies by flow cytometry. On the ProtoArray, there are only 4 proteins are annotated as major histocompatibility antigens, specific for class I (HLA-B) and class II (HLA-DPA1, HLA-DMA, and HLA-DRA). Despite the paucity of available HLA antigens to interrogate on this platform, 5 of these 9 patients showed de novo anti-HLA antibody generation against either/or HLA-class I and HLA-class II by ProtoArray, with no false-positives. This yields a calculated performance for detecting anti-HLA antibodies by ProtoArray at 56% sensitivity, 100% specificity, 100% positive predictive value, and 70% negative predictive value. Of note, based on the cross mapping of the ProtoArray platform by AILUN, there was no specific renal compartment noted to express HLA antigens selectively. Anti-MICA antibodies, previously described to increase in the posttransplant period in patients after transplantation, and often associated with adverse graft outcomes (9, 10), was at a higher level after transplantation in 72% of patients in this study. There was no correlation between the mean intensity of posttransplant antibody responses and HLA match or mismatch grades.
Integrative cDNA-Protein Microarray Analysis for Compartment-Specific Immunogenicity.
A list of proteins against which each patient developed significant antibody responses was generated for each patient, across a variety of possible threshold levels of immune response signal intensities. Each list was then tested to see whether these antigens were significantly over-enriched by their corresponding genes, expressed in any particular individual renal compartment from the microarray dataset (17), by using the hypergeometric distribution (Table 1). If there was no renal compartment-specific posttransplant antibody targeting, then over-enrichment of compartment-specific antigens would not be expected as the significance threshold of ProtoArray signals were reduced. Contrary to this expectation, we saw significant over-enrichment of compartment-specific antigens in every renal transplant patient. Surprisingly, for 14 of the total 18 patients (78%), we found that the renal pelvis was the anatomic location showing the most significant enrichment for posttransplant antibody immune responses (Fig. 1A). Based on this approach we can rank each of the 7 kidney compartments with regards to their immunogenic potential to mount specific antibody responses to kidney specific targets after transplantation. This ranking can be based on the level of a specific antibody against that compartment, and the mean of all antibody levels targeting that compartment. Regardless of the rank method, the highest antibody levels were consistently noted against the renal pelvis (average rank order of highest antibody = 167), followed by the outer renal cortex (average rank order of highest antibody = 397). When compartment-specific antibody responses were examined overall, the renal pelvis again had the highest average antibody level (564), followed by the outer renal cortex (364) (Fig. 1 A and B). Although other compartments of the kidney were targeted by antibodies at lower levels, it is noteworthy that antibodies to outer medulla were not noted to be significant in any patient (Fig. 1A and 2 and Fig. S1).
Table 1.
Frequency of compartment-specific humoral-antibodies and P value of enrichment across different kidney compartments at multiple ProtoArray thresholds
| ProtoArray threshold | Glomeruli |
Inner Cortex |
Inner Medulla |
Outer Cortex |
Outer Medulla |
Pelvis |
Papillary Tip |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | P | No. | P | No. | P | No. | P | No. | P | No. | P | No. | P | |
| 50 | 1 | 0.26 | 5 | 0.07 | 0 | 0.89 | 5 | 0.18 | 0 | 0.68 | 8 | 0.11 | 2 | 0.28 |
| 100 | 4 | 0.20 | 6 | 0.16 | 0 | 0.79 | 6 | 0.10 | 0 | 0.46 | 17 | 0.04 | 5 | 0.17 |
| 150 | 5 | 0.16 | 7 | 0.15 | 1 | 0.70 | 7 | 0.02 | 1 | 0.37 | 26 | 0.01 | 6 | 0.16 |
| 200 | 8 | 0.14 | 8 | 0.10 | 1 | 0.62 | 11 | 0.03 | 1 | 0.34 | 33 | 0.01 | 8 | 0.14 |
| … | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 1,300 | 48 | 0.04 | 57 | 0.01 | 3 | 0.27 | 104 | 0.02 | 9 | 0.15 | 156 | 0.04 | 57 | 0.07 |
| 1,350 | 50 | 0.04 | 58 | 0.01 | 3 | 0.27 | 106 | 0.02 | 9 | 0.14 | 161 | 0.04 | 58 | 0.07 |
| 1,400 | 54 | 0.05 | 61 | 0.01 | 3 | 0.27 | 112 | 0.02 | 10 | 0.15 | 167 | 0.04 | 62 | 0.06 |
| 1,409 | 54 | 0.05 | 61 | 0.01 | 3 | 0.27 | 112 | 0.02 | 10 | 0.15 | 169 | 0.04 | 62 | 0.06 |
Data are shown for a single representative patient (ID = 15).
Fig. 1.
Antigenic targets enrichment and signal intensity for 7 kidney compartments. (A) Enrichment of humoral responses to antigenic targets for 7 kidney compartments across 18 kidney transplant patients. Each patient is represented by a similarly-colored circle. The location of the circles (displayed on the nephron structure) indicates the specificity of the antigenic response to a particular compartment of the kidney. The larger filled circle indicates the highest antibody response; the smaller filled circle indicates the next highest antibody response. Large empty circles indicate over-enrichment at lower levels. Most patients had over-enrichment with highest level antibodies targeting the renal pelvis, with outer cortex as the next highest compartment. Background image is from the 20th U.S. edition of Gray's Anatomy of the Human Body and is in the public domain (41). (B) Rank order and antibody signal intensity for posttransplant serological responses across the 7 kidney compartments. The top 5 antigenic targets are listed for each compartment. (Left) Rank order (x axis) for posttransplant serological responses across the 7 kidney compartments (y axis). The highest detection of antibody immune response is ranked for each of the 7 compartments. Each double solid circle indicates that the signal rank was detected as a significant enrichment level across all 18 patient samples. The dashed line with an arrow indicates that the span of ProtoArray targets until antibody detection. (Right) The average signal intensity for antibody immune responses for each of the 7 compartments is shown across all 18 patient samples. Each bar represents the average ± standard error of immune response signal intensity. The targets were selected by meeting the following criteria: (i) antibody immune response signal intensity was positive for at least 70% samples and (ii) coefficient of variation across all 18 samples was <1.7. The top 5 targets are listed next to the corresponding kidney compartment. Only 3 targets met these criteria for the inner medulla. ARHGEF6 and STMN3 were further selected for validation studies for compartmental localization of the protein in the kidney by immunohistochemistry.
Spearman correlation coefficient analysis between the intensity of posttransplant antibody responses to 5,056 antigenic targets on the ProtoArray and time posttransplantation, shows that although overall many of the antibody responses are associated with posttransplant sample time (r = 0.69, P = 0.0016), this is independent of the discovery of antibodies generated after transplantation to kidney compartment specific protein targets.
Specificity of Antibody Responses for Renal Antigens.
To assess whether the antibodies detected after renal transplantation were indeed specific against targets only expressed in kidney tissue, we used tissue-specific datasets from other organs for comparison. Tissue-specific gene expression data were used from a published study by Su et al. (20), on 79 human and 61 mouse tissues, hybridized on Affymetrix GeneChip Human Genome U133 Arrays. From this dataset, we arbitrarily selected organ-specific gene expression profiles for heart and pancreas, and found 122 genes expressed significantly in heart tissue, and 26 in pancreatic tissue. There was no significant enrichment by hypergeometric analysis of serological responses against heart or pancreatic tissue-specific antigens in the sera of any of the 18 kidney transplant recipients. It thus appears that kidney compartment-specific non-HLA antigenic targets are specifically recognized and can mount significant antibody responses after kidney transplantation (Fig. 2).
Fig. 2.
Enrichment of posttransplant serological responses, specific to kidney, heart, and pancreas. This graph displays the −log P values of representative posttransplant antibody responses by hypergeometric analysis (patient ID = 15) (y axis) against each of the 7 kidney compartments and heart and pancreas as arbitrary control tissues. Values were plotted against a series of ranked ProtoArray antibody intensities, at every 50 consecutive measurements (x axis); −log P = 1.3 (a solid horizontal brown line) indicates P = 0.05. The top 100 antibodies by intensity are significantly over-enriched with pelvis specific targets (purple line rises above horizontal). For other patients, see Fig. S1.
Immunohistochemistry (IHC) to Confirm Compartment Specific Localization of Antigens in the Kidney.
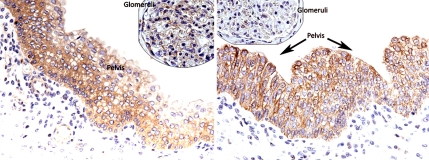
To confirm that compartment-specific serological responses are mounted against specific kidney compartment antigens, we sought to demonstrate whether the antigenic localization, predicted by the integrated genomics approach used, could be replicated by immunohistochemical localization of the same antigen in the predicted renal compartment. Two antibodies were selected for IHC. One antibody, anti-ARHGEF6, had high posttransplant signals in 100% (18/18) patients and was predicted to be specifically expressed in 2 compartments of the kidney: the renal pelvis and the glomerulus. IHC confirmed accurate localization of this antigen with positive cytoplasmic staining in the pelvic urothelium, and the glomerulus (Fig. 3Left). The second antibody, anti-STMN3, had increased posttransplant signal intensity in 83% (15/18) patients with a prediction for strong expression in the renal pelvis, which was confirmed by IHC (Fig. 3 Right). Thus, it appears that the integrated genomics approach to predict specific tissue localization of genes and proteins in human tissues is accurate and can be validated.
Fig. 3.
IHC staining for ARHGEF6 and STMN3 on control kidney tissue. Cytoplasmic staining is observed in the pelvic urothelium with ARHGEF6 and STMN3. Glomerular staining is also observed for ARHGEF6. (Left) ARHGEF6 shows positive staining in renal pelvis and glomerulus. Faint staining is seen in proximal tubules, podocytes and epithelial cells. (Right) STMN3 shows positive staining exclusively in the pelvis. Mild staining was seen in proximal tubules, podocytes, and epithelial cells.
Discussion
This is the first study to explore the use of high-density protein microarrays to study non-HLA serological responses after kidney transplantation. We demonstrate that ProtoArray measurements reveal increased serological responses that can be recognized in all patients after renal transplantation, across 61% of the targets interrogated on the ProtoArray. To ascertain that these serological responses are specific to the transplanted organ, analysis for over-enrichment was performed using a method of integration of these serum antibody measurements with previously published compartment-specific normal kidney gene expression measurements. These studies demonstrated that the posttransplant serological responses detected were in fact selectively recognizing kidney antigens; furthermore these antibody responses were recognizing relevant kidney-compartment specific antigens. We discovered that these posttransplant serological responses were specific to the kidney, and were not noted randomly to other organs such as the heart and pancreas; thus suggesting that these serological responses may in fact be specific to the kidney transplant. The 7 different renal compartments studied were found to vary in immunologic potential after transplantation, with the renal pelvis generating the highest levels of compartment-specific antibody responses. Immunohistochemistry based localization for 2 selected antigens confirmed the predicted tissue localization of the antigens, derived from the integrated genomics approach in this study.
Integrative genomics has been defined as the study of complex interactions between genes, organisms and environment of biological data. Methods in integrative genomics have been used to find genes associated with rare diseases, such as Leighs Syndrome (21), polygenic disorders such as obesity (22). We are using integrative genomics, using publicly-available histopathological gene expression measurements as a kind of lens, to focus the set of antibody level changes into a specific set relevant to kidney transplantation. The advantage of using an integrative genomics method is that, although each patient may have immunogeneic antigens in the same compartment of the kidney, these specific antigens may not be the same antigens across every patient. Only by considering the measurements anatomically does one find a consistent pattern across all patients, seen in Fig. 1A.
We find that all 18 patients demonstrate an increase in antibodies against Rac/Cdc42 guanine nucleotide exchange factor 6 (ARHGEF6), a protein we show is expressed in the renal pelvis and glomerulus. ARHGEF6 has been shown to be expressed at a moderate level in human kidney (23). ARHGEF6 is activated by phosphatidylinositol 3-kinase (24), known to regulate PTEN (25), and has been shown to be required for chemoattractant-induced recruitment of neutrophils and activation of cell-cycle component Cdc42 in the mouse (26). Although an ARHGEF6−/− has been created and shows no gross defects (26), no kidney ischemic-reperfusion injury phenotype has yet to be reported for this model. ARHFEG6 is currently known to have at least 3 missense single nucleotide polymorphisms, one of which has an average minor allele frequency as high as 0.43, suggesting there are large prevalent differences in the structure of this protein across populations.
We also show that 15 patients demonstrate an increase in antibodies against Stathmin-like 3 (STMN3), a protein we show is expressed in the renal pelvis. STMN3 has been shown to be expressed at a moderate level in cells from the human kidney (27). Pig models of kidney ischemic-reperfusion injury have shown that expression amounts of a related gene, STMN1, is correlated with reduction of ischemia (28). Mouse models of kidney ischemic-reperfusion injury have shown that STMN1 has increased expression in renal tubules and is necessary for the recovery phase (29). Although a STMN3−/− has been created (Lexicon Genetics), there is no kidney ischemic-reperfusion injury phenotype for this model.
We are unaware of other studies on the presence, immunogenicity, and significance of pelvis epithelial antigens in human transplants, and the extent of pelvis specific antibody responses may depended on the avidity of the antibody, and the dose of the antigen presented. The findings that the pelvis compartment of the kidney shows the greatest intensity of de novo posttransplant antibodies, that ARHGEF6 and STMN3 are confirmed to localize in the renal pelvis, and both mount robust antibody responses after transplantation of the kidney, all enable us to consider a common mechanism that links all of these findings together: the role of peri-operative ischemia and reperfusion injury, which may expose specific antigenic injury targets in the pelvis early after transplantation, and ongoing (and as yet unexplained) posttransplant injury triggers for the renal pelvis and parenchyma. Peri-operative factors (brain death, surgery, cold storage, reperfusion) are known to lead to ischemic injury in the renal pelvis, and in very rare but extreme conditions, have been shown to lead to even pelvic necrosis (30). Reperfusion injury is known to lead to STMN1 up-regulation, and leads to neutrophil recruitment (31). During this process, it is plausible that ARHGEF6 protein is also up-regulated in these neutrophils. This speculative model could be tested in a mouse model with measures of specific protein increases in STMN3 and ARHGEF6, in combination with escalating doses of FK506 or other immunosuppressive agents, which are known to reduce peri-operative ischemia and reperfusion injury.
The outer cortex is also a critical source of immunogenicity, as demonstrated in this study. This is not surprising as functionally significant injury in the kidney transplant is scored and recognized in the renal cortex (32) and the glomerulus. Peri-and posttransplant triggers for cortical and glomerular injury include acute rejection, infection, hypertension, and pharmaceutical agents, including the immunosuppressive drugs used for maintenance therapy in these patients. It is speculated that these cumulative injuries may result in the recognition of compartment specific antigenic targets after transplantation, with generation of de novo non-HLA antibodies.
It is still not clear why these intracellular antigens are targeted by an immune response, many of these responses being potentially against kidney alloantigens; these proteins are not known to be expressed at the cell surface. Our proposed alloantigens here are not the first intracellular peptides seen as autoantibodies (33). One possibility is that immune exposure of these antigens happens secondarily to primary events such as tissue damage from ischemia or damage from reactive oxygen species after reperfusion, which could release normally intracellular peptides for immune presentation. A second possibility is that there are increased levels or unusual forms of these proteins in renal tubule, infiltrative neutrophils, and other cells in response to transplantation. A third possibility is that under conditions of physiological stress, proteins may be expressed and targeted to the cell surface (33).
The next step in this study is to look at a targeted group of antibodies to these minor non-HLA targets and examine them for their correlation with clinical graft outcomes. Because the samples examined in this study do not have clinical graft dysfunction categories, correlation of these antibodies with decline in renal graft function or graft survival could not be performed. Further studies are necessary to determine how these antibody levels, as measured by protein microarrays, correspond to clinical differences, particularly examining their impact on chronic graft injury, and how they change longitudinally. If clinically significant, these levels could be followed to titer pharmacological immunosuppression, or could be studied as a target for depletion. Additional work needs to be done to explain why these antibodies are formed, and whether DNA variants are present in the genes coding for these proteins between donor and recipient are present. The role of the renal pelvis as an immunogenic compartment needs to be explored, especially as a function of varying surgical and medical techniques to limit ischemia and reperfusion injury.
In summary, the utility of high-density protein microarrays to study posttransplantation responses is clear, and the techniques of integrative proteo-genomics can now be extended to this measurement modality to successfully and statistically filter measured responses to just those associated with a particular anatomical compartment. Putting together our high-density protein microarray data with publicly-available gene expression microarray data has yielded more than just the sum of the parts, and more specific questions and hypotheses to target in renal transplantation.
Methods
Patients and Samples.
Thirty-six paired pretransplant and posttransplant serum samples were examined from 18 pediatric kidney allograft recipients on a steroid-free immunosuppressive protocol (34) (Table 2). The mean calculated creatinine clearance (35) was 99.0 ± 26.8 mL/min/1.73m2 at the time of posttransplant sample collection. The study was approved by the Institutional Review Board of Stanford University.
Table 2.
Patient Demographic information for 36 paired samples (pre- and post-transplant) from 18 kidney transplant recipients (ID).
| ID | Sex | Age, years | Race* | ESRD‡ | Sample date pre-txp, mos | Sample date post-txp, mos | Calculated Crcl at sample date, mL/min | Donor (living vs. deceased) | Donor age, years | Donor sex | HLA match |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 7 | 1 | 4 | 0.1 | 12 | 92 | LD | 43 | F | 2 |
| 2 | M | 9 | 1 | 6 | 0.6 | 6 | 56 | DEC | 9 | F | 1 |
| 3 | M | 10 | 5 | 3 | 0.0 | 48 | 99 | LD | 45 | M | 1 |
| 4 | F | 12 | 1 | 2 | 1.0 | 28 | 124 | DEC | 45 | F | 0 |
| 5 | M | 14 | 1 | 2 | 0.8 | 3 | 107 | LD | 44 | F | 4 |
| 6 | M | 19 | 2 | 1 | 0.0 | 30 | 141 | DEC | 25 | M | 1 |
| 7 | F | 9 | 4 | 4 | 0.6 | 72 | 67 | LD | 24 | F | 3 |
| 8 | M | 4 | 2 | 5 | 0.9 | 4 | 75 | DEC | 15 | M | 0 |
| 9 | M | 5 | 5 | 1 | 0.8 | 6 | 99 | DEC | 4 | N/A | 3 |
| 10 | M | 16 | 1 | 4 | 2.7 | 23 | 117 | LD | 19 | M | 2 |
| 11 | F | 19 | 1 | 3 | 0.7 | 9 | 58 | LD | 39 | M | 2 |
| 12 | M | 18 | 1 | 4 | 1.5 | 18 | 113 | DEC | 45 | M | 3 |
| 13 | M | 4 | 4 | 2 | 0.1 | 38 | 117 | LD | 31 | F | 6 |
| 14 | M | 17 | 1 | 3 | 0.1 | 12 | 86 | LD | 47 | F | 2 |
| 15 | F | 1 | 2 | 1 | 0.2 | 6 | 107 | LD | 29 | M | 6 |
| 16 | M | 9 | 5 | 4 | 0.1 | 24 | 138 | LD | 30 | F | 3 |
| 17 | M | 13 | 3 | 1 | 0.0 | 61 | 77 | LD | 37 | F | 2 |
| 18 | M | 14 | 4 | 3 | 0.1 | 47 | 136 | LD | 45 | M | 0 |
Mean age of the patients at transplantation was 11.0 ± 5.5 (range 1–19 years). Twenty-two percent of patients were female, and 67% patients received a kidney from a living donor. The pre-transplant serum samples were collected between August 2001 and April 2006, at 0.6 ± 0.7 (range 0–2.7) months prior to the time of transplantation. The post-transplant serum samples were collected between February 2004 and November 2006, at 24.8 ± 20.8 (range 3–72 months) months after transplantation, as part of the routine follow-up after transplantation. Numerical classification for race and cause of end stage renal disease (ESRD) is shown below. The sample date is shown in months (mos) and the calculated creatinine clearance (CrCl) is based on the Schwartz formula (35).
*1, Caucasian; 2, Hispanic; 3, Asian; 4, African American; 5, other.
†1, glomerulonephritis; 2, polycystic kidney disease; 3, dysplasia; 4, reflux nephropathy; 5, obstructive uropathy; 6, other.
Plasma Profiling Using the Protein Microarray.
Serum antibodies were profiled using the Invitrogen Human ProtoArray v3.0 (Invitrogen), containing 5,056 nonredundant human proteins expressed in a baculovirus system, purified from insect cells, and printed in duplicate onto a nitrocellulose-coated glass slide. Pearson correlation coefficients and standard deviations between duplicated spots across all proteins were calculated (r > 0.87 for all patients) (36) (15). Antibody signal intensity was Signalused = SignalAb − Signalbackground (Fig. 4, step1). De novo antibodies were identified as (Fig. 4, step2) Immune ResponseAb = Signalused posttransplant − Signalused pretransplant. For details on ProtoArray methods, please refer to SI Methods.
Fig. 4.
Integrative genomics flow chart. Work flow (step 1–10) for identifying antigenic target.
Determining Kidney Compartment and Control Organ Specific Gene Expression.
Compartmental gene expression measurements from 7 normal kidney tissues (17) (Fig. 4, step 3) were downloaded from Stanford Microarray Database for 16,293 significant cDNA probes. SAM (18) 2-unpaired class and multiclass analyses were performed. Genes were selected by (i) FDR <5% by 2-class and multiclass, (ii) fold change >1 between the target compartment versus all other compartments, and (iii) 2-unpaired class analysis was used to identify up-regulated compartment-specific targets (Fig. 4, step 4). Previously published control non-kidney tissue sample were obtained from GEO (accession no. GSE1133) (20). A total of 3,539 genes from the Affymetrix datasets overlapped with the ProtoArray platform. The t test with Bonferroni-Dunn correction for multiple testing was performed by randomly selecting 2 control tissues (heart and pancreas) to generate their tissue specific genes (FDR <5%). To overcome the problem of persistently evolving genome and transcriptome annotations, we used AILUN (19) to reannotate all probe IDs to the most recent National Center for Biotechnology Information Entrez Gene ID (Fig. 4, step 5 and step 9). Across 2 platforms, 3,835 genes/proteins were identified, which are considered as the population pool in this study.
Integrated Bioinformatics and Statistical Analysis.
Each compartment's expression levels were statistically significantly different from each patient's antibody profiles (Kolmogorov–Smirnov 2-sample test P < 0.001, and nonsignificant Spearman correlation coefficients). For analysis details please see SI Methods. In summary, ProtoArray measurements were conducted serially across multiple antibody level thresholds, and arbitrarily at each 50 consecutive interval. Serum antibodies were tested for over-enrichment of kidney-compartment specific genes, using the hypergeometric test (39, 40). (Fig. 4, step 6, and Table 1). Using these counts, we then calculated whether there was an over-enrichment of a compartment within a patient's antibody list at that threshold, using the hypergeometric distribution using the following equation and as described in refs. 37–40. A P < 0.05 was considered significant enrichment for that particular anatomic location at that threshold antibody level. Although we acknowledge that these P values are not controlled for multiple hypotheses testing, we only used the relative ordering of these values for compartment ranking and discovery.
 |
k = Frequency of observed hits at a certain threshold; n = Population pool, 3,835 in this study; m = Threshold at each 50 interval; n = Expected observation, 161 for glomeruli, 201 for inner cortex, 9 for inner medulla, 336 for outer cortex, 29 for outer medulla, 466 for pelvis, and 167 for papillary tip.
Immunohistochemistry Staining.
Immunohistochemistry staining was performed on paraffin embedded, formalin fixed normal kidney tissue sampled from a radical nephrectomy performed for renal cell carcinoma. Rabbit anti-human antibodies (ATLAS, Inc.; Protein Tech Group, Inc.) were used, at a dilution of 1:50. Endogenous peroxidase was blocked and the DAKO Envision system (DAKO) was used for detection. Staining was optimized using appropriate positive and negative controls performed on normal tissue microarray.
Supplementary Material
Acknowledgments.
We thank the members of the M.M.S. and A.J.B. laboratories for thier help. This work was supported by National Institute of Allergy and Infectious Diseases Grant R01 AI61739 (to M.S., L.L., and T.K.S.) and National Library of Medicine Grant K22 LM008261 (to A.J.B.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE10452).
This article contains supporting information online at www.pnas.org/cgi/content/full/0900563106/DCSupplemental.
References
- 1.Pascual M, Theruvath T, Kawai T, Tolkoff-Rubin N, Cosimi AB. Strategies to improve long-term outcomes after renal transplantation. N Engl J Med. 2002;346:580–590. doi: 10.1056/NEJMra011295. [DOI] [PubMed] [Google Scholar]
- 2.Sarwal M, et al. Molecular heterogeneity in acute renal allograft rejection identified by DNA microarray profiling. N Engl J Med. 2003;349:125–138. doi: 10.1056/NEJMoa035588. [DOI] [PubMed] [Google Scholar]
- 3.Kirk AD, et al. CTLA4-Ig and anti-CD40 ligand prevent renal allograft rejection in primates. Proc Natl Acad Sci USA. 1997;94:8789–8794. doi: 10.1073/pnas.94.16.8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Terasaki P, Mizutani K. Antibody mediated rejection: Update 2006. Clin J Am Soc Nephrol. 2006;1:400–403. doi: 10.2215/CJN.02311205. [DOI] [PubMed] [Google Scholar]
- 5.Akalin E, Pascual M. Sensitization after kidney transplantation. Clin J Am Soc Nephrol. 2006;1:433–440. doi: 10.2215/CJN.01751105. [DOI] [PubMed] [Google Scholar]
- 6.Vongwiwatana A, Tasanarong A, Hidalgo LG, Halloran PF. The role of B cells and alloantibody in the host response to human organ allografts. Immunol Rev. 2003;196:197–218. doi: 10.1046/j.1600-065x.2003.00093.x. [DOI] [PubMed] [Google Scholar]
- 7.Mauiyyedi S, Colvin RB. Humoral rejection in kidney transplantation: New concepts in diagnosis and treatment. Curr Opin Nephrol Hypertens. 2002;11:609–618. doi: 10.1097/00041552-200211000-00007. [DOI] [PubMed] [Google Scholar]
- 8.Opelz G. Non-HLA transplantation immunity revealed by lymphocytotoxic antibodies. Lancet. 2005;365:1570–1576. doi: 10.1016/S0140-6736(05)66458-6. [DOI] [PubMed] [Google Scholar]
- 9.Terasaki PI, Ozawa M, Castro R. Four-year follow-up of a prospective trial of HLA and MICA antibodies on kidney graft survival. Am J Transplant. 2007;7:408–415. doi: 10.1111/j.1600-6143.2006.01644.x. [DOI] [PubMed] [Google Scholar]
- 10.Zou Y, Stastny P, Susal C, Dohler B, Opelz G. Antibodies against MICA antigens and kidney-transplant rejection. N Engl J Med. 2007;357:1293–1300. doi: 10.1056/NEJMoa067160. [DOI] [PubMed] [Google Scholar]
- 11.Lerut E, et al. Duffy and Kidd blood group antigens: Minor histocompatibility antigens involved in renal allograft rejection? Transfusion. 2007;47:28–40. doi: 10.1111/j.1537-2995.2007.01060.x. [DOI] [PubMed] [Google Scholar]
- 12.Joosten SA, et al. Antibody response against the glomerular basement membrane protein agrin in patients with transplant glomerulopathy. Am J Transplant. 2005;5:383–393. doi: 10.1111/j.1600-6143.2005.00690.x. [DOI] [PubMed] [Google Scholar]
- 13.Dragun D, et al. Angiotensin II type 1-receptor activating antibodies in renal-allograft rejection. N Engl J Med. 2005;352:558–569. doi: 10.1056/NEJMoa035717. [DOI] [PubMed] [Google Scholar]
- 14.Leitner WW, et al. Alphavirus-based DNA vaccine breaks immunological tolerance by activating innate antiviral pathways. Nat Med. 2003;9:33–39. doi: 10.1038/nmxx. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hudson ME, Pozdnyakova I, Haines K, Mor G, Snyder M. Identification of differentially expressed proteins in ovarian cancer using high-density protein microarrays. Proc Natl Acad Sci USA. 2007;104:17494–17499. doi: 10.1073/pnas.0708572104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrett T, et al. NCBI GEO: Mining tens of millions of expression profiles–database and tools update. Nucleic Acids Res. 2007;35:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higgins JP, et al. Gene expression in the normal adult human kidney assessed by complementary DNA microarray. Mol Biol Cell. 2004;15:649–656. doi: 10.1091/mbc.E03-06-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen R, Li L, Butte AJ. AILUN: Reannotating gene expression data automatically. Nat Methods. 2007;4:879. doi: 10.1038/nmeth1107-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mootha VK, et al. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc Natl Acad Sci USA. 2003;100:605–610. doi: 10.1073/pnas.242716699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.English SB, Butte AJ. Evaluation and integration of 49 genome-wide experiments and the prediction of previously unknown obesity-related genes. Bioinformatics. 2007;23:2910–2917. doi: 10.1093/bioinformatics/btm483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kutsche K, Gal A. The mouse Arhgef6 gene: cDNA sequence, expression analysis, and chromosome assignment. Cytogenet Cell Genet. 2001;95(3–4):196–201. doi: 10.1159/000059346. [DOI] [PubMed] [Google Scholar]
- 24.Yoshii S, et al. alphaPIX nucleotide exchange factor is activated by interaction with phosphatidylinositol 3-kinase. Oncogene. 1999;18:5680–5690. doi: 10.1038/sj.onc.1202936. [DOI] [PubMed] [Google Scholar]
- 25.Li Z, et al. Regulation of PTEN by Rho small GTPases. Nat Cell Biol. 2005;7:399–404. doi: 10.1038/ncb1236. [DOI] [PubMed] [Google Scholar]
- 26.Li Z, et al. Directional sensing requires G beta gamma-mediated PAK1 and PIX alpha-dependent activation of Cdc42. Cell. 2003;114:215–227. doi: 10.1016/s0092-8674(03)00559-2. [DOI] [PubMed] [Google Scholar]
- 27.Bieche I, et al. Expression of stathmin family genes in human tissues: Non-neural-restricted expression for SCLIP. Genomics. 2003;81:400–410. doi: 10.1016/s0888-7543(03)00031-4. [DOI] [PubMed] [Google Scholar]
- 28.Jayle C, et al. Comparison of protective effects of trimetazidine against experimental warm ischemia of different durations: Early and long-term effects in a pig kidney model. Am J Physiol Renal Physiol. 2007;292:F1082–1093. doi: 10.1152/ajprenal.00338.2006. [DOI] [PubMed] [Google Scholar]
- 29.Zahedi K, et al. Stathmin-deficient mice develop fibrosis and show delayed recovery from ischemic-reperfusion injury. Am J Physiol Renal Physiol. 2006;290:F1559–1567. doi: 10.1152/ajprenal.00424.2005. [DOI] [PubMed] [Google Scholar]
- 30.Hidalgo G, Vyas S, Hong J, Singh A, Baqi N. Complete necrosis of the renal pelvis and ureter after cadaveric renal transplantation. Pediatr Transplant. 2000;4:60–62. doi: 10.1034/j.1399-3046.2000.00088.x. [DOI] [PubMed] [Google Scholar]
- 31.Koo DD, Welsh KI, Roake JA, Morris PJ, Fuggle SV. Ischemia/reperfusion injury in human kidney transplantation: An immunohistochemical analysis of changes after reperfusion. Am J Pathol. 1998;153:557–566. doi: 10.1016/S0002-9440(10)65598-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Racusen LC, et al. The Banff 97 working classification of renal allograft pathology. Kidney Int. 1999;55:713–723. doi: 10.1046/j.1523-1755.1999.00299.x. [DOI] [PubMed] [Google Scholar]
- 33.Jordan P, Kubler D. Autoimmune diseases: Nuclear autoantigens can be found at the cell-surface. Mol Biol Rep. 1995;22:63–66. doi: 10.1007/BF00996307. [DOI] [PubMed] [Google Scholar]
- 34.Sarwal MM, et al. Continued superior outcomes with modification and lengthened follow-up of a steroid-avoidance pilot with extended daclizumab induction in pediatric renal transplantation. Transplantation. 2003;76:1331–1339. doi: 10.1097/01.TP.0000092950.54184.67. [DOI] [PubMed] [Google Scholar]
- 35.Schwartz GJ, Haycock GB, Edelmann CM, Jr, Spitzer A. A simple estimate of glomerular filtration rate in children derived from body length and plasma creatinine. Pediatrics. 1976;58:259–263. [PubMed] [Google Scholar]
- 36.Zhu X, Gerstein M, Snyder M. ProCAT: A data analysis approach for protein microarrays. Genome Biol. 2006;7:R110. doi: 10.1186/gb-2006-7-11-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spellman PT, Rubin GM. Evidence for large domains of similarly expressed genes in the Drosophila genome. J Biol. 2002;1:5. doi: 10.1186/1475-4924-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fury W, Batliwalla F, Gregersen PK, Li W. Overlapping probabilities of top ranking gene lists, hypergeometric distribution, and stringency of gene selection criterion. Conf Proc IEEE Eng Med Biol Soc. 2006;1:5531–5534. doi: 10.1109/IEMBS.2006.260828. [DOI] [PubMed] [Google Scholar]
- 39.Tavazoie S, Hughes JD, Campbell MJ, Cho RJ, Church GM. Systematic determination of genetic network architecture. Nat Genet. 1999;22:281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- 40.Curtis RK, Oresic M, Vidal-Puig A. Pathways to the analysis of microarray data. Trends Biotechnol. 2005;23:429–435. doi: 10.1016/j.tibtech.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 41.Gray H. Anatomy of the Human Body. 20th Ed. Philadelphia: Lea & Febiger; 1918. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.