Abstract
Peroxisomes are ubiquitous eukaryotic organelles housing diverse enzymatic reactions, including several that produce toxic reactive oxygen species. Although understanding of the mechanisms whereby enzymes enter peroxisomes with the help of peroxin (PEX) proteins is increasing, mechanisms by which damaged or obsolete peroxisomal proteins are degraded are not understood. We have exploited unique aspects of plant development to characterize peroxisome-associated protein degradation (PexAD) in Arabidopsis. Oilseed seedlings undergo a developmentally regulated remodeling of peroxisomal matrix protein composition in which the glyoxylate cycle enzymes isocitrate lyase (ICL) and malate synthase (MLS) are replaced by photorespiration enzymes. We found that mutations expected to increase or decrease peroxisomal H2O2 levels accelerated or delayed ICL and MLS disappearance, respectively, suggesting that oxidative damage promotes peroxisomal protein degradation. ICL, MLS, and the β-oxidation enzyme thiolase were stabilized in the pex4–1 pex22–1 double mutant, which is defective in a peroxisome-associated ubiquitin-conjugating enzyme and its membrane tether. Moreover, the stabilized ICL, thiolase, and an ICL-GFP reporter remained peroxisome associated in pex4–1 pex22–1. ICL also was stabilized and peroxisome associated in pex6–1, a mutant defective in a peroxisome-tethered ATPase. ICL and thiolase were mislocalized to the cytosol but only ICL was stabilized in pex5–10, a mutant defective in a matrix protein import receptor, suggesting that peroxisome entry is necessary for degradation of certain matrix proteins. Together, our data reveal new roles for PEX4, PEX5, PEX6, and PEX22 in PexAD of damaged or obsolete matrix proteins in addition to their canonical roles in peroxisome biogenesis.
Keywords: Arabidopsis thaliana, organelle remodeling, peroxisome, protein turnover
Peroxisomes are ubiquitous eukaryotic organelles that characteristically possess H2O2-producing oxidases and H2O2-decomposing catalases. Specific reactions housed in peroxisomes vary by species, developmental stage, and cell type. For example, young seedling peroxisomes contain glyoxylate cycle enzymes, whereas mature leaf peroxisomes contain photorespiration enzymes (1). Peroxisomal proteins are nuclearly encoded and inserted posttranslationally into the peroxisome matrix or membrane with the assistance of peroxins (peroxisome biogenesis proteins) (2, 3). Although matrix protein import into peroxisomes is increasingly understood, mechanisms for recognizing and eliminating damaged or obsolete peroxisomal proteins remain largely obscure. Damage to peroxisomal proteins can occur through interactions with reactive oxygen species (ROS) produced by peroxisomal oxidative reactions. Most peroxisomal ROS are detoxified by catalase and the ascorbate-glutathione cycle (4); however, some ROS inevitably damage peroxisomal proteins (5). Mechanisms for detecting and eliminating damaged peroxisomal proteins have not been described; however, removal of excess or nonfunctional peroxisomes in yeast and mammals can occur through pexophagy, a specialized form of autophagy (6). Although autophagy occurs in plants (7), plant pexophagy has not been reported.
Plant peroxisomes possess at least one unidentified means to eliminate obsolete matrix proteins. Peroxisomes in oilseed plants such as Arabidopsis undergo protein content shifts that coincide with developmental progression. After germination, a crucial function of oilseed peroxisomes is catabolizing fatty acid stores to fuel seedling growth until photoautotrophic growth begins. The glyoxylate cycle enzymes isocitrate lyase (ICL) and malate synthase (MLS) convert acetyl-CoA from fatty acid β-oxidation into gluconeogenesis substrates and are found in peroxisomes of mature seeds and germinated seedlings (8). The disappearance of ICL and MLS a few days after germination coincides with the appearance of peroxisomal photorespiratory enzymes. In cucurbit cotyledons, transitional peroxisomes containing both glyoxylate cycle and photorespiratory enzymes can be visualized using immuno-electron microscopy (9–11). The presence of transitional peroxisomes implies that mature leaf peroxisomes derive from content remodeling of seedling peroxisomes, that is, the degradation of glyoxylate cycle enzymes and import of photorespiratory enzymes, rather than (or in addition to) general destruction and resynthesis of entire organelles (12).
Parallels between certain peroxins (encoded by PEX genes) required for recycling the PEX5 peroxisomal matrix protein receptor and proteins acting in ER-associated protein degradation (ERAD) have been noted in studies of the evolutionary origins of peroxisomes (13, 14). Similar to protein export in ERAD, recycling PEX5 from the yeast peroxisome back to the cytosol after cargo delivery requires a ubiquitin-conjugating enzyme (PEX4), several RING proteins that may act as ubiquitin-protein ligases, and AAA ATPases (PEX1 and PEX6) (13, 14). Moreover, when yeast PEX5 is not efficiently recycled from the peroxisome after import, it is multiubiquitinated and degraded by the proteasome (15–17). It is unknown whether parallels between ERAD components and the PEX5-recycling peroxins are limited to PEX5 degradation or extend to degradation of damaged or obsolete matrix proteins.
The ubiquitin-conjugating enzyme PEX4 is tethered to the cytosolic peroxisomal face by the membrane anchor PEX22 (18). The Arabidopsis pex4–1 mutant (19) displays reduced responsiveness to the proto-auxin indole-3-butyric acid (IBA), which appears to be converted to the active auxin indole-3-acetic acid in peroxisomes (20). pex4–1 peroxisome-deficient phenotypes are enhanced in the pex4–1 pex22–1 double mutant (19). Moreover, ICL disappearance is delayed in pex4–1 pex22–1 (19), suggesting a role for PEX4 in peroxisomal matrix protein degradation. In this study, we characterize peroxisome-associated matrix protein degradation (PexAD) during peroxisome content remodeling in Arabidopsis. In the pex4–1 pex22–1 mutant, the transition from seedling to mature leaf peroxisomes is delayed; obsolete matrix proteins persist despite normal mRNA disappearance and matrix protein import. Degradation can also be delayed by mislocalization of certain peroxisomal proteins to the cytosol in a pex5 mutant, indicating that normal PexAD requires peroxisomal import. Our results suggest that some peroxins function not only in matrix protein import, but also in matrix protein degradation, perhaps by promoting export from the peroxisome.
Results and Discussion
Certain Peroxisomal Proteins Undergo Developmentally Coordinated Changes in Abundance.
The Arabidopsis glyoxylate cycle proteins ICL and MLS are model substrates for studying peroxisome-associated protein degradation. ICL and MLS transcripts are present only in very young seedlings (8, 21); this programmed transcriptional cessation allows monitoring of matrix protein turnover using immunoblotting. Moreover, both ICL and MLS are expressed in Arabidopsis cotyledons (22, 23), which grow by cell expansion without division following germination (24). Thus, the decline in cotyledon ICL and MLS protein abundance after ICL and MLS transcriptional cessation results from protein degradation and not dilution among growing tissues.
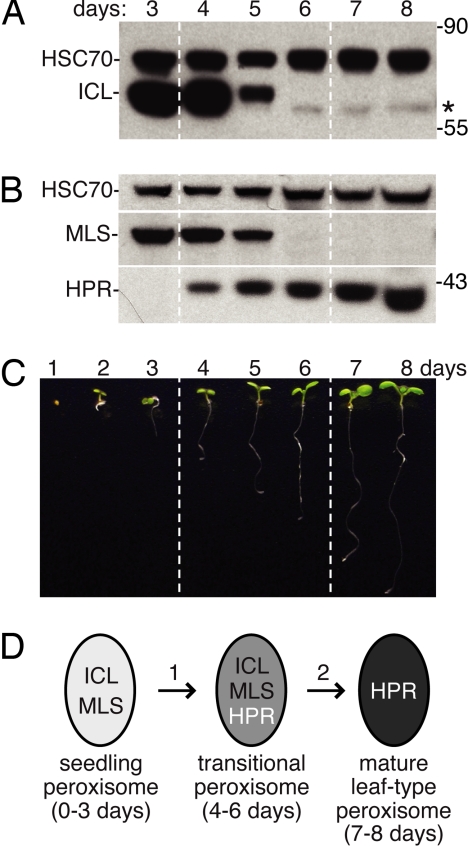
We analyzed wild-type cotyledons and found that ICL and MLS were present 3 and 4 days after sowing, had reduced abundance on day 5, and were no longer detected after day 6 (Fig. 1 A and B). By contrast, the photorespiratory enzyme hydroxypyruvate reductase (HPR) was first detected on day 4 and remained present throughout the 8-day time course (Fig. 1B). The timing of ICL and MLS disappearance and HPR appearance in Arabidopsis cotyledons resembles changes in glyoxylate cycle and photorespiratory enzyme activities and mRNA levels in Arabidopsis and other oilseed seedlings (8, 9, 21, 25, 26). Furthermore, the 2- to 3-day window in which both glyoxylate cycle and photorespiratory enzymes are present supports the possibility that transitional peroxisomes containing both enzyme types exist in Arabidopsis (Fig. 1D), as in other plants (9–11).
Fig. 1.
Levels of the peroxisomal enzymes isocitrate lyase (ICL), malate synthase (MLS), and hydroxypyruvate reductase (HPR) are developmentally regulated. (A and B) Immunoblots of protein extracts from wild-type cotyledons (12 per lane) from 3- to 8-day-old seedlings were probed with antibodies to ICL, the cytosolic loading control HSC70, MLS, and HPR. Asterisk marks a cross-reacting band detected by the α-ICL antibodies. Positions of molecular weight markers (in kDa) are indicated on the right. (C) Photograph of light-grown 1- to 8-day-old wild-type (Col-0) seedlings. (D) Diagram of early postgerminative enzyme composition changes in Arabidopsis peroxisomes.
Varying Peroxisomal H2O2 Alters ICL and MLS Stability.
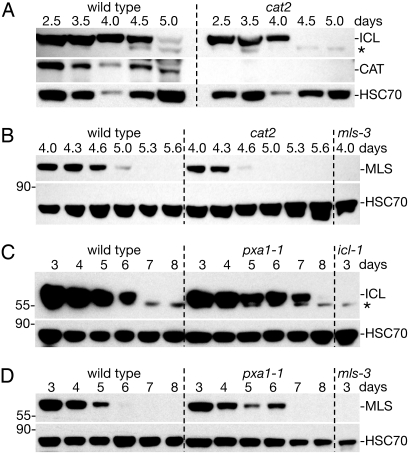
H2O2, a toxic byproduct of peroxisomal oxidative reactions such as fatty acid β-oxidation, likely damages peroxisomal proteins (4). This damage is normally minimized by peroxisomal catalase (CAT), which decomposes H2O2. In an Arabidopsis cat2 antisense line, ICL and MLS enzyme activities are decreased (27), presumably because these enzymes are damaged by excess H2O2. To determine whether oxidative damage increases matrix protein turnover, we examined ICL and MLS stability in a cat2 insertion mutant that lacks seedling CAT immunoreactivity (Fig. 2A). We found that both ICL and MLS disappeared 8 to 12 hours faster in cat2 cotyledons than in wild-type cotyledons (Fig. 2 A and B). Blocking β-oxidation by preventing fatty acid entry into peroxisomes, as in the pxa1–1 mutant (28), decreases seedling H2O2 levels (27). We found that both ICL and MLS persisted ≈1 day longer in pxa1–1 cotyledons than in wild-type cotyledons (Fig. 2 C and D). Our demonstration that seedlings modulate ICL and MLS turnover in response to genetic alterations expected to vary peroxisomal H2O2 levels suggests that some form of PexAD disposes of H2O2-damaged peroxisome matrix proteins.
Fig. 2.
Mutants varying peroxisome H2O2 production exhibit altered ICL and MLS degradation kinetics. (A and B) Immunoblots of protein extracts from wild-type and cat2 cotyledons (12 per lane) were probed with α-ICL, α-catalase (CAT), α-HSC70, and α-MLS antibodies. (C and D) Immunoblots of total protein extracts from wild-type and pxa1–1 cotyledons (12 per lane) were sequentially probed with α-ICL and α-HSC70 (C) or α-MLS and α-HSC70 (D) antibodies. Null mls-3 and icl-1 (8) mutants provide controls for the MLS (B, D) and ICL (C) antibodies, respectively. Asterisk marks a cross-reacting band detected by the α-ICL antibodies that remains present in the icl-1 null mutant, and positions of molecular weight markers (in kDa) are indicated on the left.
ICL and MLS Are Stabilized in the Arabidopsis pex4–1 pex22–1 Mutant.
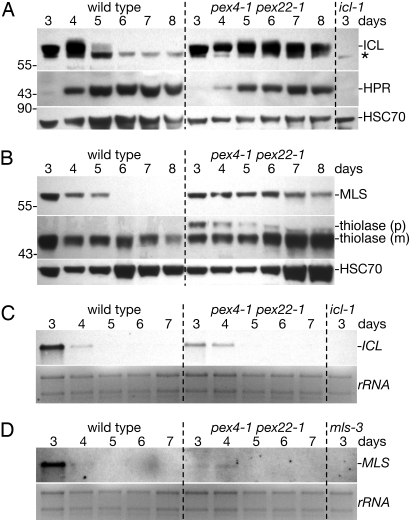
We previously found that ICL persists in pex4–1 pex22–1 seedlings after it disappears from wild-type seedlings (19), suggesting that the PEX4 ubiquitin-conjugating enzyme might promote ICL degradation. To determine whether this apparent stabilization could be explained by protein dilution as seedlings matured, we examined developmental persistence of peroxisomal matrix proteins using extracts from equal numbers of cotyledons, which grow by cell expansion but not division after germination (24). We detected ICL for at least 3 days longer in pex4–1 pex22–1 cotyledons than in wild-type cotyledons (Fig. 3A), confirming a role for PEX4 in ICL turnover. Similarly, we found MLS for several days longer in pex4–1 pex22–1 cotyledons than in wild-type cotyledons (Fig. 3B), indicating that MLS turnover was also PEX4 dependent. We also monitored thiolase, a fatty acid β-oxidation protein with a type 2 peroxisome targeting signal (PTS2) that is cleaved after import into the peroxisome matrix (29), which was most abundant at 3 days and gradually declined to a lower basal level as wild-type seedling matured (21) (Fig. 3B). We found that pex4–1 pex22–1 thiolase levels remained relatively constant throughout the time course (Fig. 3B), suggesting that PTS2 proteins also could be subject to PEX4-dependent turnover. Notably, thiolase in pex4–1 pex22–1 was predominantly the mature form (Fig. 3B), demonstrating that stabilization occurred after peroxisomal import and subsequent PTS2 removal.
Fig. 3.
ICL and MLS degradation is delayed in pex4–1 pex22–1. (A and B) Immunoblots of protein extracts from wild-type and pex4–1 pex22–1 cotyledons (12 per lane) from 3- to 8-day-old seedlings were probed with α-ICL, α-HPR, α-HSC70, α-MLS, and α-thiolase antibodies. The α-thiolase antibody recognizes precursor (p) and mature (m) thiolase polypeptides. Asterisk marks a cross-reacting band detected by the α-ICL antibodies, and positions of molecular weight markers (in kDa) are indicated on the left. (C and D) ICL and MLS mRNA levels in wild-type and pex4–1 pex22–1 seedlings. RNA gel blots of 4 μg total RNA from 3- to 7-day-old seedlings were analyzed with DNA probes to ICL (C) or MLS (D). Fluorescence from ethidium bromide-stained ribosomal RNA (rRNA) is shown as a loading control.
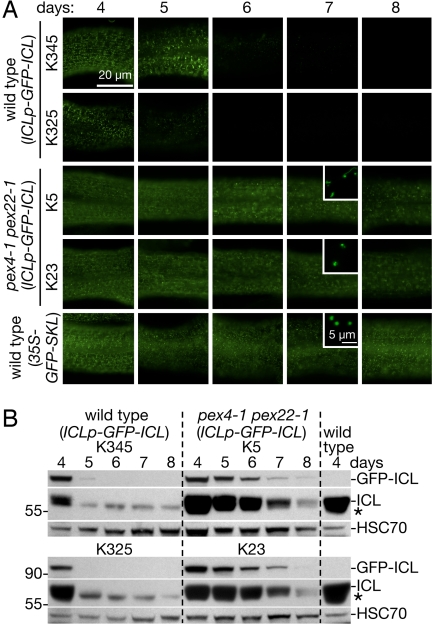
To examine ICL stability in living tissues, we examined wild-type and pex4–1 pex22–1 seedlings transformed with a GFP-ICL construct driven by the native ICL 5′ sequence (ICLp-GFP-ICL). We observed that GFP-ICL fluorescence in wild-type hypocotyls (Fig. 4A) and GFP-ICL protein in wild-type cotyledons (Fig. 4B) both disappeared between 5 and 6 days after sowing, consistent with kinetics of native ICL disappearance from wild-type cotyledons (Figs. 3A and 4B). By contrast, we observed GFP-ICL fluorescence (Fig. 4A) and protein (Fig. 4B) through 8 days in pex4–1 pex22–1 lines expressing the ICLp-GFP-ICL transgene. The persistent GFP-ICL fluorescence in pex4–1 pex22–1 (ICLp-GFP-ICL) resembled GFP-SKL fluorescence in wild-type hypocotyls expressing 35S-GFP-SKL, a peroxisomal GFP variant (30) driven by the strong viral 35S promoter (Fig. 4A). The punctate GFP-ICL subcellular localization (Fig. 4A Insets) and the partial thiolase processing (Fig. 3B) (19) confirm that peroxisomal import functions in pex4–1 pex22–1.
Fig. 4.
GFP-ICL driven from the ICL promoter reports native ICL localization and levels during seedling development. (A) GFP fluorescence in hypocotyls of light-grown seedlings expressing ICLp-GFP-ICL or 35S-GFPSKL constructs at 4–8 days after sowing. Peroxisomal ICLp-GFP-ICL fluorescence diminishes after 5 days in wild-type (lines K325 and K345), whereas fluorescence persists for at least 8 days in pex4–1 pex22–1 (lines K5 and K23). GFP fluorescence from 35S-GFPSKL is constitutive. Exposure times in each row were normalized to the 4-day-old seedling. Bar, 20 μm. Insets are confocal optical slices showing individual peroxisomes in hypocotyl cells; inset bar, 5 μm. (B) Immunoblot of protein extracts (32 cotyledons per lane) from wild-type and pex4–1 pex22–1 plants expressing ICLp-GFP-ICL. Blots were probed with α-ICL and α-HSC70 antibodies. The α-ICL antibodies detect GFP-ICL (absent in untransformed wild-type, right lane), native ICL, and a cross-reacting band marked with an asterisk. Positions of molecular weight markers (in kDa) are indicated on the left.
The persistence of ICL and MLS in pex4–1 pex22–1 seedlings could be explained by (i) delay in onset of transitional peroxisomes, marked by synthesis of photorespiratory enzymes and cessation of ICL and MLS transcription (Fig. 1D, step 1), (ii) delay in conversion of transitional peroxisomes into leaf peroxisomes, marked by degradation of ICL and MLS proteins (Fig. 1D, step 2), or (iii) delays in both conversions. To assess whether transitional peroxisome onset was delayed in pex4–1 pex22–1, we monitored appearance of the photorespiratory enzyme HPR. In both wild-type and pex4–1 pex22–1 cotyledons, HPR appeared between 3 and 4 days (Fig. 3A). Furthermore, we found that ICL and MLS mRNAs disappeared between 4 and 5 days after sowing in both wild-type and pex4–1 pex22–1 seedlings (Fig. 3 C and D), suggesting that transcriptional cessation of these genes occurred normally in the mutant. In fact, ICL and MLS transcript levels were reduced in 3-day-old pex4–1 pex22–1 seedlings compared with wild-type seedlings, an observation that, together with the increased ICL and MLS protein levels in pex4–1 pex22–1, supports our conclusion that ICL and MLS degradation is slowed in pex4–1 pex22–1.
We concluded that the switch from seedling to transitional peroxisomes, marked by HPR appearance (Fig. 3A) and the cessation of ICL and MLS transcription (Fig. 3 C and D), occurred with normal timing in pex4–1 pex22–1, whereas the change from transitional to mature leaf-type peroxisomes, marked by ICL and MLS degradation, was substantially delayed.
ICL and MLS Are Stabilized in a pex5 and a pex6 Mutant.
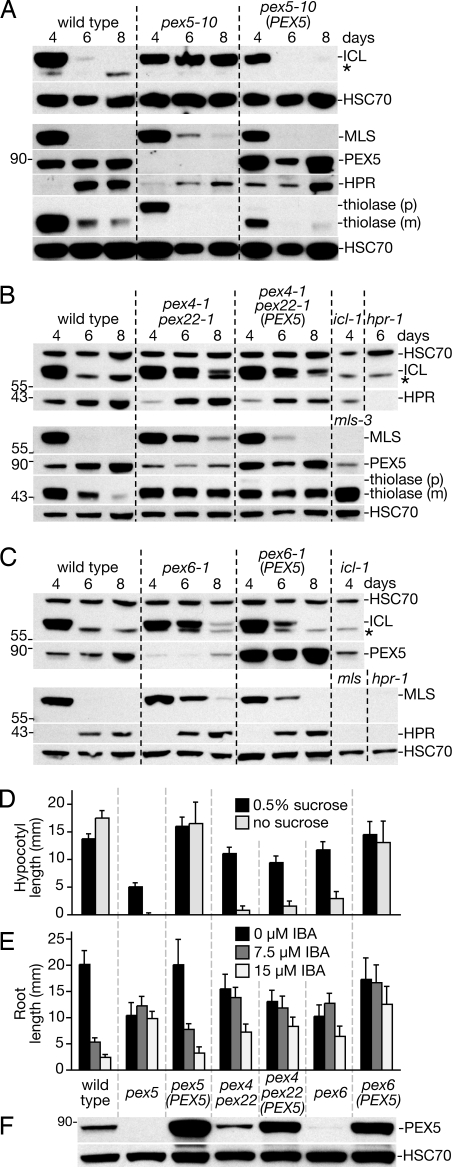
To explore the necessity of peroxisomal import for PexAD, we analyzed matrix protein stability in pex5–10 (19). PEX5 is the receptor that delivers PTS1-containing proteins, including ICL, MLS, and HPR, to the peroxisome matrix. Because the PEX7 PTS2 receptor binds PEX5, import of PTS2 proteins, including thiolase, is also PEX5 dependent in plants (31, 32). We found that ICL was stabilized through at least 8 days in pex5–10 seedlings (Fig. 5A). In contrast to the dramatic ICL stabilization, thiolase and MLS were still degraded in pex5–10, although MLS was slightly stabilized (Fig. 5A). As expected, a construct driving a PEX5 cDNA from the 35S promoter (35S-PEX5) restored PEX5 protein levels (Fig. 5F), ICL and MLS instability (Fig. 5A), thiolase processing (Fig. 5A), sucrose independence (Fig. 5D), and IBA responsiveness (Fig. 5E) to pex5–10. Because PEX5 is needed for matrix protein import, we concluded that peroxisomal import is required for efficient ICL degradation, whereas thiolase and MLS appear to be targeted for degradation even when mislocalized to the cytosol, presumably via a PexAD-independent mechanism.
Fig. 5.
ICL is stabilized in pex5–10 and pex6–1. (A-C) Immunoblots of cotyledon (12 per lane) protein extracts from the indicated lines probed with α-ICL, α-MLS, α-PEX5, α-HPR, α-thiolase, and α-HSC70 antibodies. (D) Sucrose-dependent hypocotyl elongation of 35S-PEX5 lines. Seeds from lines in A-C were plated on the indicated medium and incubated overnight under white light, and were then incubated for 5 days in the dark. (E) IBA-resistant root growth of 35S-PEX5 lines. Seeds from A-C were plated on media containing 0.5% sucrose and the indicated IBA concentration and grown under yellow-filtered light for 7 days. (F) Immunoblot of protein extracts from whole sucrose-grown seedlings in D (eight 6-day-old dark-grown seedlings per lane) probed with α-PEX5 and α-HSC70 antibodies.
Because PEX5 overexpression can overcome some yeast pex4 defects (33), we examined whether PEX5 overexpression would have an impact on matrix protein degradation in pex4–1 pex22–1. We found that pex4–1 pex22–1 (35S-PEX5) plants, despite elevated PEX5 levels (Fig. 5F), remained sucrose dependent (Fig. 5D) and IBA resistant (Fig. 5E). ICL and thiolase remained stabilized in pex4–1 pex22–1 (35S-PEX5) (Fig. 5B), suggesting that ICL degradation defects did not stem from insufficient PEX5. However, MLS instability was partially restored in pex4–1 pex22–1 (35S-PEX5), suggesting that PEX5 may contribute to MLS degradation in pex4–1 pex22–1.
The ATPase PEX6 is a peroxin thought to act after PEX4 during receptor recycling (18). To determine whether PEX6 also participates in matrix protein turnover, we examined pex6–1, a mutant with decreased PEX5 levels, and pex6–1 (35S-PEX5), which has elevated PEX5 levels (30). As shown previously (30), PEX5 overexpression rescued pex6–1 sucrose dependence (Fig. 5D) and root elongation defects (Fig. 5E) without restoring IBA sensitivity (Fig. 5E). ICL and MLS were both stabilized in pex6–1 and the instability of both proteins was partially restored by PEX5 overexpression (Fig. 5C), consistent with roles for PEX6 and PEX5 in eliminating obsolete peroxisome matrix proteins.
Stabilized ICL Is Peroxisome Associated in pex4–1 pex22–1 and pex6–1 and Cytosolic in pex5–10.
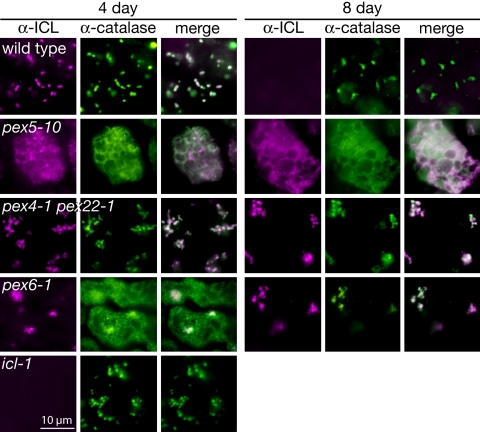
Studies in various organisms have revealed that most null pex alleles, including pex4, pex5, pex6, and pex22, fail to deliver matrix proteins from their site of synthesis in the cytosol to the peroxisome matrix (3). Indeed, the Arabidopsis pex5–10 mutant displays severe matrix protein import defects (19, 34). However, the Arabidopsis pex4–1 missense allele displays only minor defects in thiolase and PTS-tagged GFP import into peroxisomes, despite severe physiological phenotypes suggesting impaired peroxisome function (19). To determine whether ICL stabilization in the pex mutants was accompanied by failed peroxisomal import, we examined ICL subcellular localization via immunofluorescence microscopy. In 4-day-old wild-type cotyledon cells, we detected ICL in punctate structures that co-localized with peroxisomal catalase (Fig. 6). Lack of detectable fluorescence from α-ICL antibodies in the icl-1 null mutant (8) verified the specificity of the antibody, and the absence of ICL signal in 8-day-old wild-type cells (Fig. 6) was consistent with the timing of ICL degradation (Fig. 1A).
Fig. 6.
Stabilized ICL is peroxisome associated in pex4–1 pex22–1 and pex6 and cytosolic in pex5–10. Cotyledons from 4- and 8-day-old light-grown Arabidopsis seedlings were dually immunolabeled with rabbit α-ICL (magenta, left columns) and mouse α-catalase (green, center columns) antibodies and were visualized via fluorescence microscopy. Overlays of ICL and catalase images (white, right columns) show co-localizations. The icl-1 null mutant exhibits background fluorescence from the α-ICL antibody. Bar, 10 μm.
In cotyledon cells from the PTS1 receptor mutant pex5–10 (19), both ICL and catalase were mislocalized to the cytosol at 4 and 8 days (Fig. 6), verifying that our immunolabeling detects cytosolic localization and that the pex5–10 allele is severely disrupted in peroxisomal matrix protein delivery. The stabilization of cytosolic ICL in pex5–10 confirms that peroxisome entry is required for ICL targeting for degradation by the PexAD machinery. In contrast to pex5–10, ICL was peroxisome-associated in pex4–1 pex22–1 cotyledon cells at both 4 and 8 days (Fig. 6), confirming the GFP-ICL localization (Fig. 4A), and suggesting that ICL stabilization in pex4–1 pex22–1 does not result from a failure of ICL to enter peroxisomes but, rather, from an inability of the peroxisome to target ICL for degradation. Similarly, ICL was peroxisome associated in pex6–1 at both 4 and 8 days. Interestingly, catalase localized to peroxisomes at 8 days but not 4 days in pex6–1, suggesting that matrix proteins differentially require PEX6 function for peroxisome entry.
Conclusions
Protein degradation is complicated in eukaryotes by the necessity to eliminate proteins from multiple subcellular compartments. Although oxidative damage from ROS and developmentally induced peroxisome content remodeling necessitate turnover of peroxisomal matrix proteins, little is known about peroxisome-associated protein degradation in any system. We have demonstrated that ICL and MLS are degraded during seedling development (Figs. 1 and 3) and that the rate of this degradation can be accelerated or reduced by changing peroxisome metabolism to increase or decrease peroxisomal H2O2 levels (Fig. 2). Furthermore, we showed that ICL and MLS degradation is dependent upon the peroxins PEX4, PEX5, PEX6, and PEX22 (Figs. 3 and 5) and that the abnormal ICL and MLS persistence in pex4–1 pex22–1 results from ICL and MLS protein stabilization, rather than extended ICL and MLS mRNA persistence (Fig. 3). Blocking ICL import into peroxisomes was sufficient to prevent normal ICL degradation in the pex5–10 PTS1 receptor mutant, and stabilized ICL remained peroxisome-associated in pex4–1 pex22–1 and pex6–1 (Fig. 6). PEX5 overexpression partially restored ICL and MLS degradation in pex6–1 and MLS degradation in pex4–1 pex22–1 but had no effect on ICL degradation in pex4–1 pex22–1 (Fig. 5).
Our observations that certain peroxins are necessary for the developmental elimination of matrix proteins suggest a model in which PexAD parallels certain features of ERAD. In particular, it is possible that obsolete or damaged proteins destined for degradation are recognized in the peroxisomal matrix and retrotranslocated through the peroxisomal membrane with the assistance of the PEX4 ubiquitin-conjugating enzyme and the PEX6 ATPase. Because both PTS1- and PTS2-containing proteins can be stabilized in pex4–1 pex22–1, it will be interesting to learn whether there are specific PexAD degradation signals or whether all peroxisome matrix proteins undergo general turnover. It also remains to be determined whether developmentally-induced and damage-induced PexAD use the same targeting mechanisms, and whether PexAD defects contribute to the physiological defects of the Arabidopsis pex4–1 and pex6–1 mutants. Future experiments will be required to explore these models, to isolate additional components, and to determine whether substrates in other organisms and in additional peroxisome remodeling transitions are subject to PexAD via mechanisms similar to those uncovered here.
Materials and Methods
Plant Material and Growth Conditions.
All mutants were in the Arabidopsis thaliana Col-0 accession. icl-1 (8), pex4–1 pex22–1 (19), pex5–10 (19), pex6–1 (30), and pxa1–1 (28) were described previously. The cat2 (SALK_144919), mls-3 (SALK_002289), and hpr-1 (SALK_143584) mutants were from the Salk Institute sequence-indexed insertion collection (35). mls-3 has a T-DNA inserted in the second MLS exon and is phenotypically indistinguishable from the previously described mls-2 allele (25).
Before immunoblotting, wild-type and mutant seedlings were grown under continuous white light (experiments using pex5–10 and pxa1–1) or for 1 day in the light, 2 days in the dark, and up to 5 additional days under continuous white light (all other experiments) on plant nutrient medium (36) solidified with 0.6% agar and supplemented with 0.5% sucrose. For phenotypic assays, seedlings were grown for 1 day in the light and 5 days in the dark (sucrose dependence) or for 7 days under yellow-filtered light (IBA resistance).
Plasmid Preparation.
The 35S-GFP-SKL and 35S-PEX5 constructs were described previously (30). The ICLp-GFP-ICL clone was constructed by polymerase chain reaction amplification of a 3.5-kb fragment 5′ of the ICL start site from the P1 clone MSD21, the coding sequence of a cytosolic enhanced GFP (eGFP) from pEGAD (37), and the ICL cDNA from clone U24828 (38), and subcloning each amplicon into separate pCR2.1 vectors (Invitrogen, Carlsbad, CA). The three fragments were combined in pCR2.1 using introduced junctional restriction sites to create an ≈6 kb ICLp-GFP-ICL sequence that was subcloned into pBIN19 (39) and transformed into Agrobacterium tumefaciens strain GV3101 (40) using electroporation. After transformation (41) into wild-type and pex4–1 pex22–1 plants, transgenic (T1) seedlings were selected after growth on kanamycin-containing medium. GFP-ICL localization was analyzed in T2 seedlings via fluorescence microscopy.
Immunoblot Analysis.
Samples were processed for immunoblotting as previously described (30) except that frozen cotyledon homogenates were suspended in 8 μl (3- to 6-day-old seedlings) or 12 μl (7- and 8-day-old seedlings) protein loading buffer, and 8 μl was loaded in each well. After transfer, filters were air dried. Primary antibodies were diluted in blocking buffer as follows: 1:2,000 α-ICL (42), 1:25,000 α-MLS (43), 1:1,000 α-HPR (44), 1:4,000 α-catalase (45), 1:200 α-PEX5 (30), 1:2,500 α-thiolase (prepared from rabbits immunized with an Escherichia coli-produced fusion protein including the C-terminal 350 aa of Arabidopsis PED1/At2g33150), 1:300 α-GFP (632377; Clontech, Mountain View, CA) and 1:8,000 α-HSC70 (SPA-817; StressGen Biotechnologies, Victoria, BC, Canada).
RNA Analysis.
Wild-type, pex4–1 pex22–1, icl-1, and mls-3 seedlings were grown under continuous white light on filter paper-covered medium containing 0.5% sucrose. Seedlings were harvested on days 3–7, immersed in liquid nitrogen, and stored at −80 °C. Frozen tissue was ground using a mortar and pestle, and RNA was isolated using RNeasy Mini kits (Qiagen, Valencia, CA). Total RNA (≈4 μg) was electrophoresed and processed for Northern blotting as previously described (46).
Digoxigenin-labeled ICL and MLS probes were amplified using a PCR DIG Probe Synthesis kit (Roche, Indianapolis, IN), according to the manufacturer's instructions, from a wild-type genomic DNA template.
Microscopy.
Cotyledons were prepared for immunofluorescence microscopy as described (47) with the following modifications. Cell walls in 4-day-old cotyledons were permeabilized for 1.5 hours in 3% (wt/vol) pectinase (Sigma) and 3% (wt/vol) cellulase (Karlan, Cottonwood, AZ) at 37 °C. Cell walls in 8-day-old cotyledons were permeabilized for 1 hour in 2% (wt/vol) pectinase and 2% (wt/vol) cellulase. Cotyledons were dually labeled with rabbit α-ICL (1:1,500) (42) and mouse α-catalase (undiluted hybridoma media from cell line SABP 3B6–7, Princeton Monoclonal Antibody Facility, Princeton, NJ) for 4 hours at 37 °C. Secondary antibodies were goat α-rabbit Alexa 594 (1:1,500) and goat α-mouse Alexa 488 (1:500) (Invitrogen). To delay photobleaching, 1 mg/ml n-propyl gallate (Alfa Aesar, Ward Hill, MA) in 90% glycerol, and phosphate-buffered saline solution was added before adding coverslips.
A Zeiss Axioplan 2 (Thornwood, NY) equipped with Narrow Band GFP and Texas Red filters (Chroma, Rochingham, VT) was used for imaging immunolabeled and GFP-expressing tissues. Confocal images were acquired using a Zeiss LSM 5 (488 nm excitation; BP 500–550 nm). Images were cropped and colorized using National Instititutes of Health ImageJ and adjusted for brightness in Adobe Photoshop CS (San Jose, CA).
Acknowledgments.
We thank Masayoshi Maeshima (Nagoya University, Nagoya, Japan), John Harada (University of California, Davis, CA), Richard Trelease (Arizona State University, Phoenix), and Douglas Randall (University of Missouri, Columbia, MO) for the ICL, MLS, CAT, and HPR antibodies, respectively. We thank C. Robertson McClung (Dartmouth College, Hanover, NH) for cat2 seeds, Ian Graham (University of York, York, United Kingdom) for icl seeds, the Arabidopsis Biological Resource Center at Ohio State University for DNA clones (MSD21 and U24828) and seeds from Salk Institute insertion lines, and Naxhiely Martinez, Sarah Ratzel, and Lucia Strader for critical comments on the manuscript. This research was supported by the National Science Foundation (IOS-0315596 and MCB-0745122), the Robert A. Welch Foundation (C-1309), and a postdoctoral fellowship to M.J.L. (USDA 2008–20659).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Michels PAM, et al. Peroxisomes, glyoxysomes and glycosomes. Mol Membr Biol. 2005;22:133–145. doi: 10.1080/09687860400024186. [DOI] [PubMed] [Google Scholar]
- 2.Leon S, Goodman JM, Subramani S. Uniqueness of the mechanism of protein import into the peroxisome matrix: Transport of folded, co-factor-bound and oligomeric proteins by shuttling receptors. Biochim Biophys Acta, Mol Cell Res. 2006;1763:1552–1564. doi: 10.1016/j.bbamcr.2006.08.037. [DOI] [PubMed] [Google Scholar]
- 3.Eckert JH, Erdmann R. Peroxisome biogenesis. Rev Physiol Biochem Pharmacol. 2003;147:75–121. doi: 10.1007/s10254-003-0007-z. [DOI] [PubMed] [Google Scholar]
- 4.del Río LA, Sandalio LM, Corpas FJ, Palma JM, Barroso JB. Reactive oxygen species and reactive nitrogen species in peroxisomes. Production, scavenging, and role in cell signaling. Plant Physiol. 2006;141:330–335. doi: 10.1104/pp.106.078204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yanik T, Donaldson RP. A protective association between catalase and isocitrate lyase in peroxisomes. Arch Biochem Biophys. 2005;435:243–252. doi: 10.1016/j.abb.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 6.Sakai Y, Oku M, van der Klei IJ, Kiel JA. Pexophagy: Autophagic degradation of peroxisomes. Biochim Biophys Acta. 2006;1763:1767–1775. doi: 10.1016/j.bbamcr.2006.08.023. [DOI] [PubMed] [Google Scholar]
- 7.Thompson AR, Vierstra RD. Autophagic recycling: Lessons from yeast help define the process in plants. Curr Opin Plant Biol. 2005;8:165–173. doi: 10.1016/j.pbi.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 8.Eastmond PJ, et al. Postgerminative growth and lipid catabolism in oilseeds lacking the glyoxylate cycle. Proc Natl Acad Sci USA. 2000;97:5669–5674. doi: 10.1073/pnas.97.10.5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Titus DE, Becker WM. Investigation of the glyoxysome-peroxisome transition in germinating cucumber cotyledons using double-label immunoelectron microscopy. J Cell Biol. 1985;101:1288–1299. doi: 10.1083/jcb.101.4.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishimura M, Yamaguchi J, Mori H, Akazawa T, Yokota S. Immunochemical analysis shows that glyoxysomes are directly transformed to leaf peroxisomes during greening of pumpkin cotyledons. Plant Physiol. 1986;81:313–316. doi: 10.1104/pp.81.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sautter C. Microbody transition in greening watermelon cotyledons. Double immunocytochemical labeling of isocitrate lyase and hydroxypyruvate reductase. Planta. 1986;167:491–503. doi: 10.1007/BF00391225. [DOI] [PubMed] [Google Scholar]
- 12.Nishimura M, Hayashi M, Kato A, Yamaguchi K, Mano S. Functional transformation of microbodies in higher plant cells. Cell Struct Funct. 1996;21:387–393. doi: 10.1247/csf.21.387. [DOI] [PubMed] [Google Scholar]
- 13.Gabaldon T, et al. Origin and evolution of the peroxisomal proteome. Biol Direct. 2006;1:8. doi: 10.1186/1745-6150-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schluter A, et al. The evolutionary origin of peroxisomes: An ER-peroxisome connection. Mol Biol Evol. 2006;23:838–845. doi: 10.1093/molbev/msj103. [DOI] [PubMed] [Google Scholar]
- 15.Kragt A, Brouwer TV, van den Berg M, Distel B. The Saccharomyces cerevisiae peroxisomal import receptor Pex5p is monoubiquitinated in wild type cells. J Biol Chem. 2005;280:7867–7874. doi: 10.1074/jbc.M413553200. [DOI] [PubMed] [Google Scholar]
- 16.Kiel JAKW, Otzen M, Veenhuis M, van der Klei IJ. Obstruction of polyubiquitination affects PTS1 peroxisomal matrix protein import. Biochim Biophys Acta, Mol Cell Res. 2005;1745:176–186. doi: 10.1016/j.bbamcr.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 17.Kiel JAKW, Emmrich K, Meyer HE, Kunau WH. Ubiquitination of the peroxisomal targeting signal type 1 receptor, Pex5p, suggests the presence of a quality control mechanism during peroxisomal matrix protein import. J Biol Chem. 2005;280:1921–1930. doi: 10.1074/jbc.M403632200. [DOI] [PubMed] [Google Scholar]
- 18.Thoms S, Erdmann R. Peroxisomal matrix protein receptor ubiquitination and recycling. Biochim Biophys Acta. 2006;1763:1620–1628. doi: 10.1016/j.bbamcr.2006.08.046. [DOI] [PubMed] [Google Scholar]
- 19.Zolman BK, Monroe-Augustus M, Silva ID, Bartel B. Identification and functional characterization of Arabidopsis PEROXIN4 and the interacting protein PEROXIN22. Plant Cell. 2005;17:3422–3435. doi: 10.1105/tpc.105.035691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zolman BK, Yoder A, Bartel B. Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics. 2000;156:1323–1337. doi: 10.1093/genetics/156.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rylott EL, Hooks MA, Graham IA. Co-ordinate regulation of genes involved in storage lipid mobilization in Arabidopsis thaliana. Biochem Soc Trans. 2001;29:283–287. doi: 10.1042/0300-5127:0290283. [DOI] [PubMed] [Google Scholar]
- 22.Charlton WL, Johnson B, Graham IA, Baker A. Non-coordinate expression of peroxisome biogenesis, beta-oxidation and glyoxylate cycle genes in mature Arabidopsis plants. Plant Cell Rep. 2005;23:647–653. doi: 10.1007/s00299-004-0879-7. [DOI] [PubMed] [Google Scholar]
- 23.Graham IA, Smith LM, Leaver CJ, Smith SM. Developmental regulation of expression of the malate synthase gene in transgenic plants. Plant Mol Biol. 1990;15:539–549. doi: 10.1007/BF00017829. [DOI] [PubMed] [Google Scholar]
- 24.Mansfield SG, Briarty LG. The dynamics of seedling and cotyledon cell development in Arabidopsis thaliana during reserve mobilization. Int J Plant Sci. 1996;157:280–295. [Google Scholar]
- 25.Cornah JE, Germain V, Ward JL, Beale MH, Smith SM. Lipid utilization, gluconeogenesis, and seedling growth in Arabidopsis mutants lacking the glyoxylate cycle enzyme malate synthase. J Biol Chem. 2004;279:42916–42923. doi: 10.1074/jbc.M407380200. [DOI] [PubMed] [Google Scholar]
- 26.Trelease RN, Becker WM, Gruber PJ, Newcommb EH. Microbodies (glyoxysomes and peroxisomes) in cucumber cotyledons—correlative biochemical and ultrastructural study in light- and dark-grown seedlings. Plant Physiol. 1971;48:461. doi: 10.1104/pp.48.4.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eastmond PJ. Monodehyroascorbate reductase4 is required for seed storage oil hydrolysis and postgerminative growth in Arabidopsis. Plant Cell. 2007;19:1376–1387. doi: 10.1105/tpc.106.043992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zolman BK, Silva ID, Bartel B. The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid β-oxidation. Plant Physiol. 2001;127:1266–1278. [PMC free article] [PubMed] [Google Scholar]
- 29.Helm M, et al. Dual specificities of the glyoxysomal/peroxisomal processing protease Deg15 in higher plants. Proc Natl Acad Sci USA. 2007;104:11501–11506. doi: 10.1073/pnas.0704733104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zolman BK, Bartel B. An Arabidopsis indole-3-butyric acid-response mutant defective in PEROXIN6, an apparent ATPase implicated in peroxisomal function. Proc Natl Acad Sci USA. 2004;101:1786–1791. doi: 10.1073/pnas.0304368101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woodward AW, Bartel B. The Arabidopsis peroxisomal targeting signal type 2 receptor PEX7 is necessary for peroxisome function and dependent on PEX5. Mol Biol Cell. 2005;16:573–583. doi: 10.1091/mbc.E04-05-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayashi M, Yagi M, Nito K, Kamada T, Nishimura M. Differential contribution of two peroxisomal protein receptors to the maintenance of peroxisomal functions in Arabidopsis. J Biol Chem. 2005;280:14829–14835. doi: 10.1074/jbc.M411005200. [DOI] [PubMed] [Google Scholar]
- 33.van der Klei IJ, et al. The ubiquitin-conjugating enzyme Pex4p of Hansenula polymorpha is required for efficient functioning of the PTS1 import machinery. EMBO J. 1998;17:3608–3618. doi: 10.1093/emboj/17.13.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee JR, et al. Cloning of two splice variants of the rice PTS1 receptor, OsPex5pL and OsPex5pS, and their functional characterization using pex5-deficient yeast and Arabidopsis. Plant J. 2006;47:457–466. doi: 10.1111/j.1365-313X.2006.02797.x. [DOI] [PubMed] [Google Scholar]
- 35.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 36.Haughn GW, Somerville C. Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol Gen Genet. 1986;204:430–434. [Google Scholar]
- 37.Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR. Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA. 2000;97:3718–3723. doi: 10.1073/pnas.97.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamada K, et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- 39.Bevan M. Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 1984;12:8711–8721. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koncz C, Schell J. The promoter of the TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet. 1986;204:383–396. [Google Scholar]
- 41.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 42.Maeshima M, Yokoi H, Asahi T. Evidence for no proteolytic processing during transport of isocitrate lyase into glyoxysomes in castor bean endosperm. Plant Cell Physiol. 1988;29:381–384. [Google Scholar]
- 43.Olsen LJ, et al. Targeting of glyoxysomal proteins to peroxisomes in leaves and roots of a higher plant. Plant Cell. 1993;5:941–952. doi: 10.1105/tpc.5.8.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kleczkowski LA, Randall DD. Purification and characterization of a novel NADPH(NADH)-dependent hydroxypyruvate reductase from spinach leaves—comparison of immunological properties of leaf hydroxypyruvate reductases. Biochem J. 1988;250:145–152. doi: 10.1042/bj2500145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kunce CM, Trelease RN, Turley RB. Purification and biosynthesis of cottonseed (Gossypium hirsutum L) catalase. Biochem J. 1988;251:147–155. doi: 10.1042/bj2510147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dugas DV, Bartel B. Sucrose induction of Arabidopsis miR398 represses two Cu/Zn superoxide dismutases. Plant Mol Biol. 2008;67:403–417. doi: 10.1007/s11103-008-9329-1. [DOI] [PubMed] [Google Scholar]
- 47.Sauer M, Paciorek T, Benkova E, Friml J. Immunocytochemical techniques for whole-mount in situ protein localization in plants. Nat Protoc. 2006;1:98–103. doi: 10.1038/nprot.2006.15. [DOI] [PubMed] [Google Scholar]