Abstract
During seedling establishment, cotyledons of the rain forest tree Hymenaea courbaril mobilize storage cell wall xyloglucan to sustain growth. The polysaccharide is degraded and its products are transported to growing sink tissues. Auxin from the shoot controls the level of xyloglucan hydrolytic enzymes. It is not yet known how important the expression of these genes is for the control of storage xyloglucan degradation. In this work, partial cDNAs of the genes xyloglucan transglycosylase hydrolase (HcXTH1) and β-galactosidase (HcBGAL1), both related to xyloglucan degradation, and two other genes related to sucrose metabolism [alkaline invertase (HcAlkIN1) and sucrose synthase (HcSUS1)], were isolated. The partial sequences were characterized by comparison with sequences available in the literature, and phylogenetic trees were assembled. Gene expression was evaluated at intervals of 6 h during 24 h in cotyledons, hypocotyl, roots, and leaves, using 45-d-old plantlets. HcXTH1 and HcBGAL1 were correlated to xyloglucan degradation and responded to auxin and light, being down-regulated when transport of auxin was prevented by N-1-naphthylphthalamic acid (NPA) and stimulated by constant light. Genes related to sucrose metabolism, HcAlkIN1 and HcSUS1, responded to inhibition of auxin transport in consonance with storage mobilization in the cotyledons. A model is proposed suggesting that auxin and light are involved in the control of the expression of genes related to storage xyloglucan mobilization in seedlings of H. courbaril. It is concluded that gene expression plays a role in the control of the intercommunication system of the source–sink relationship during seeding growth, favouring its establishment in the shaded environment of the rain forest understorey.
Keywords: Auxin, cell wall, β-galactosidase, Hymenaea, invertase, storage mobilization, XTH, xyloglucan
Introduction
The Leguminosae is among the most important families in the Neotropics. Worldwide, around 18 000 species belong to this family and, from studies of the chemical composition of their seeds, it has been estimated that about half of the species contain galactomannan, an endospermic cell wall storage polysaccharide (Buckeridge et al., 2000). However, some species from the Leguminosae have specialized in a different way, relying on the cell wall polysaccharide xyloglucan. Species from the two genera Copaifera and Hymenaea use xyloglucan as the carbon source to establish initial growth, and this feature has been thought to be correlated with the success in the colonization of biomes, such as the Amazon and the savannahs in South America, by these genera (Buckeridge et al., 2000).
The genus Hymenaea is thought to have originated in Africa and subsequently spread and adapted very well in the Neotropical regions of South America, generating many different species (Lee and Langenheim, 1975). Within the genus, the species Hymenaea courbaril is considered as one of the most successful mainly because of its shade and drought tolerance (Gerhardt, 1993; Souza and Valio, 1999; Santos and Buckeridge, 2004). The species stores ∼45% of the seed dry mass in the cotyledons as xyloglucan (Buckeridge and Dietrich, 1990), whose function has been shown to be the support for initial seedling growth and development until autotrophic growth is established (Santos and Buckeridge, 2004).
Xyloglucan, besides being a source of carbon, also plays an important role in defining the structural properties of plant cell walls and the regulation of plant growth and development (Levy et al., 1997). This polymer is composed of a cellulose-like (1→4)-linked β-D-glucan main chain, which is partially substituted by α-D-Xylp side chains at O-6. Depending on the source, the side chain can be β-D-Galp-(1→2)-α-D-Xylp, α-L-Fucp-(1→2)-β-D-Galp-(1→2)-α-Xylp, or even more complex groups (Buckeridge et al., 2000). Xyloglucans are polysaccharides with remarkable structural regularity. Under hydrolysis with cellulase, a persistent pattern of oligosaccharides is produced. These are usually based on a cellotetraose chain in which three out of every four glucoses in the main chain is branched with xylose, forming a unit named XXXG (for the nomenclature of xyloglucans see Fry et al., 1993). In contrast to other plant tissues, in seeds the storage xyloglucan is composed of galactose branches that form XLXG, XXLG, and XLLG, and no fucosylated oligosaccharides are found. Comparative studies of the fine structure of seed storage xyloglucans have shown the similar structural pattern of subunits, but the H. courbaril xyloglucan displays unique structural features, with ∼50% of it being composed of a family of oligosaccharides based on five instead of four glucoses in the main chain (XXXXG) (Buckeridge et al., 1992, 1997; Tiné et al., 2006).
During early seedling growth, xyloglucan mobilization proceeds, leading to sucrose production in the cotyledons. Sucrose is transported to the developing sink tissues (shoot and root) (Santos and Buckeridge, 2004). It has been observed for cotyledons of Tropaelum majus, Copaifera langsdorffii, and H. courbaril that the polysaccharide is attacked by three hydrolases and one transglycosylase (Buckeridge et al., 2000). In all xyloglucan mobilization systems studied to date, it has been shown that polysaccharide degradation is achieved by the coordinated action of at least four cell wall hydrolases. The main chain of the polymer is attacked by xyloglucan endo-transglycosylase hydrolase (XTH) which, depending on the species, may be of the hydrolytic type (XET; xyloglucan endo-transglycosylase) or the transglycosylase type (XTH). The oligosaccharides produced are attacked by β-galactosidose, α-xylosidase, and β-glucosidase, so that free glucose and xylose are produced (Buckeridge et al., 2000).
In H. courbaril as well as in T. majus, auxin is thought to be an important positive regulator of cell wall xyloglucan storage mobilization. It has been proposed that xyloglucan degradation is dependent on the polar transport of auxin in H. courbaril (Santos et al., 2004).
In addition, light also seems to influence directly the rate of xyloglucan mobilization in cotyledons of developing seedlings of H. courbaril by modulating the production of xyloglucan hydrolases (Santos and Buckeridge, 2004; Santos et al., 2004). The mechanisms involved in this light-mediated xyloglucan hydrolysis are uncertain but are possibly related to the photomorphogenetic growth pattern.
This intercommunication system between light, hormones, and sugar metabolism highlights a very efficient mechanism used to store and use carbon during seedling establishment. The possible control related to a switch from skotomorphogenesis and photomorphogenesis programmes might explain the successful development of the seedlings of H. courbaril in the very low light intensity conditions found in the understorey of the rain forests. Lower light intensities lead seedlings to etiolate (skotomorphogenesis) until they cross the litter layer of the forest, after which the photomorphogenesis rules prevail.
Light has also been proposed to stimulate the production of auxin that would be transported to the cotyledons where it would signal for xyloglucan degradation to take place, which would result in sucrose production. The resulting sucrose would then be translocated to the growing seedling and, when the number of leaves is such that net photosynthesis can support autotrophic growth, the shrunken cotyledons would have already degraded most of their reserves and would fall, leaving a young plant ready to face life in the understory of the forest.
It seems, therefore, that the interplay of auxin and light signals plays important roles in the control of storage mobilization in the H. courbaril cotyledons. Mobilization means here degradation of xyloglucan to maintain the synthesis of sucrose that will be translocated to sustain growth of developing sink organs.
In order to obtain further insight into xyloglucan mobilization the question was asked of whether auxin and light act by regulating the expression of key genes involved in the mobilization process. In this work, partial cDNA for XTH and β-galactosidase (xyloglucan degradation), sucrose synthase, and alkaline invertase (sucrose metabolism) were cloned and the expression of the corresponding genes under inhibition of polar transport of auxin and the presence or absence of light during the mobilization process were analysed in different organs and during circadian light changes. The results suggest that regulation of mRNA accumulation is important for xyloglucan mobilization in H. courbaril.
Materials and methods
Plant material and experimental design
Seeds of H. courbaril L. were obtained from trees growing in a gallery forest at Sao João da Boa Vista county (22º00′S; 47º18′W), São Paulo, Brazil. Seeds were scarified manually with sandpaper and allowed to germinate in distilled water in Petri dishes at 25 °C for 15 days, when radicle protrusion occurred. After germination, the seedlings were transferred to pots (0.5 l) containing vermiculite and were allowed to grow in a greenhouse under a 12 h photoperiod at ambient temperature.
Thirty-day-old plants were subjected to shoot excision, and auxin transport inhibition in these plants was induced by application of 200 mM N-1-naphthylphthalamic acid (NPA) in lanolin as described previously (Santos et al., 2004). For light/darkness experiments, developing seedlings were transferred to continuous light (50 μmol of photons m2 s−1) at 28 °C or to darkness at 28 °C for 20 d.
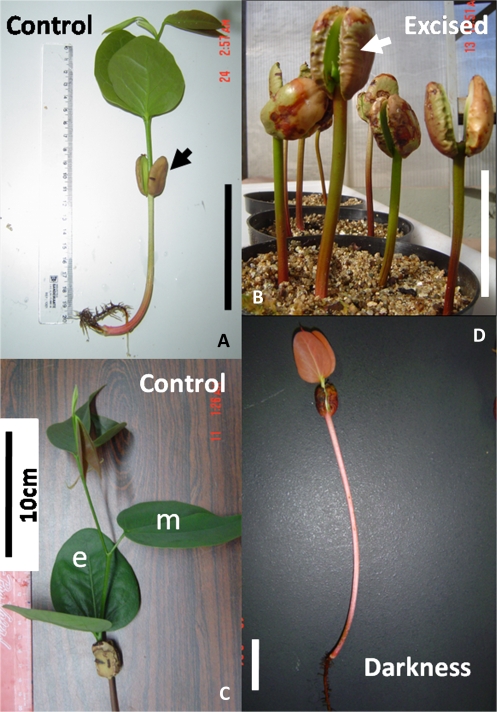
After these treatments, seedlings were allowed to grow until the 45th day, when samples of cotyledons, leaves, hypocotyls, and roots from 15 plants were harvested (Fig. 1). The 45th day was chosen due to the fact that xyloglucan mobilization is approximately half way to being complete and at this point the cotyledons display high level of activities of xyloglucan hydrolases (Tiné et al., 2000; Santos and Buckeridge, 2004).
Fig. 1.
Aspect of seedlings of Hymenaea courbaril on the 45th day after the beginning of imbibition. (A) A typical specimen of H. courbaril. Note the investment in the shoot (arrow=cotyledon). (B) Seedlings that had their shoot excised 35 d after imbibition. These will develop new branches in ∼30 d (see Santos et al., 2004) (arrow=cotyledon). (C) Closer view of the shoot, showing the eophylls (e) (first leaf) and metaphylls (m) (second leaf). (D) An etiolated seedling of H. courbaril after 20 days in darkness. Note that the hypocotyl is ∼5 times longer in the dark and eophylls did not develop, characterizing the programme of skotomorphogenesis. Bars represent 10 cm.
Sampling during the 24 h day–night cycle was obtained every 6 h (0, 6, 12, 18, and 24 h). The sampled material was immediately frozen in liquid nitrogen and stored, at most for 3 months, before RNA extraction. The choice to evaluate expression during a whole day was made on the basis of previous observations that xyloglucan hydrolases presented diurnal variations (A Santos and MS Buckeridge, unpublished results). The time in all experiments was recorded in days after the beginning of imbibition. All assays and time points were conducted in triplicate.
Total RNA extraction, primer design, cDNA synthesis and amplification, cloning, and sequencing of PCR products
RNA extraction was performed with different protocols according to the part of the seedling being analysed, i.e. cotyledon, hypocotyl, leaves, or root. For roots and leaves, the method used was the same as that described by Gesteira et al. (2003), and for hypocotyls a Concert Total RNA extraction solution (Invitrogen) was used following the manufacturer's instructions. For cotyledons, a procedure described by Daohong et al. (2004) was used with the following modifications. A 1 g aliquot of cotyledons was ground into a fine powder in liquid nitrogen and then 10 ml of buffer solution (0.5 M TRIS-HCl pH 8, 0.01 M EDTA, 0.25 M LiCl, 2.5% SDS, and 0.1% β-mercaptoethanol) was added. The mixture was vigorously stirred for 1 min and phenol:chloroform:isoamylic alcohol (25:24:1, v/v/v) was added and stirred again for 1 min. The material was centrifuged at 4 °C, 13 000 g, for 15 min. This step was repeated twice. The RNA was then precipitated twice. The first precipitation was performed by adding 0.5 ml of 12 M LiCl and kept for 2 h at –80 °C. After 30 min of centrifugation the pellet was washed with 6 M LiCl. After a further 15 min of centrifugation under the same conditions, the pellet was resuspended in 0.3 ml of diethylpyrocarbonate (DEPC)-treated water and a second precipitation was performed using 1/10 volume of 3 M sodium acetate, pH 5.2 and a 0.5 vol. of absolute ethanol. The mixture was stirred and kept overnight at –80 °C. After 30 min of centrifugation, the pellet was washed with 70% ethanol, resuspended in DEPC-treated water, and stored at –80 °C. RNA integrity was checked by 1% agarose gel electrophoresis with 6% formaldehyde in MOPS buffer (20 M MOPS, 0.6 M sodium acetate, 0.01 M EDTA, pH 8). The RNAs were stored at –80 °C until used for cDNA synthesis.
The reverse transcription of total RNAs to cDNA was performed using Ready-To-Go RT-PCR beads (Amersham Biosciences). The reaction was carried out following the manufacturer's instructions, using 3.5 μg of total RNA samples and 1 mM primer Oligo dT12-18 (Invitrogen). The cDNA was stored at –20 °C until use.
Coding sequences for actin, xyloglucan endo-transglycosylase, β-galactosidase, alkaline invertase, and sucrose synthase from eudicotyledons were obtained from GenBank (NCBI: http://www.ncbi.nlm.nih.gov/Genbank/index.html). Amino acid sequence alignments were performed with Clustal W (http://www.ebi.ac.uk/clustalw) using default parameters. Gaps were removed for phylogenetic analyses which were performed using the Prodist program to calculate distances among sequences, and the Neighbor–Joining tree constructing algorithm (Saitou and Nei, 1987) was used to establish the evolutionary relationships among the homologous sequences.
To amplify partial cDNAs, pairs of nested and degenerated primers were designed from conserved protein domains detected in the sequences from organisms phylogenetically closely related to Hymenaea, i.e. the Leguminosae. The primers synthesized had 17–24 nucleotides (Table S2).
For PCR amplification of cDNAs, the following solution in a total volume of 50 μl was used: 10 μl of cDNA reaction, 5 μl of 10× PCR buffer, 1.5 μM MgCl2 (50 mM), 2.5 μl of dNTP mix (10 μM), 1 mM for the degenerated 3′ primer and 1 mM for the degenerated 5′ primer, and 0.3 μl of Taq DNA polymerase (5 U μl−1;– Invitrogen). The PCR conditions for β-galactosidase, xyloglucan transglycosylase hydrolase, alkaline invertase, and sucrose synthase were: initial denaturation for 2 min at 95 °C followed by 5 min at 72 °C and 36 cycles (94 °C for 5 s, 55 °C for 1 h 30 min, and 72 °C for 45 s), and a final extension of 10 min at 72 °C. Actin was used as an internal control in semi-quantitative RT-PCR experiments.
The amplification products of the expected size were purified after separation by 2% agarose gel electrophoresis using the kit Wizard SV Gel and PCR Clean-Up System (Promega) according to the manufacturer's instructions.
PCR products were cloned into the TA-Cloning vector (Invitrogen) following the manufacturer's instructions. Sequencing was performed using a DNA sequencing Big Dye Kit™ (Applied Biosystems) following the manufacturer's instructions, and analysed with an automatic sequencer ABI PRISM™ 377 (Perkin Elmer).
The sequences that were obtained were then compared with GenBank sequences using the blastn tool (NCBI: http://www.ncbi.nlm.nih.gov/GenBank/index.html) to confirm their identities.
Semi-quantitative RT-PCR and densitometric quantification
The semi-quantitative RT-PCR experiments were performed using pairs of specific primers which were designed from the partial cDNA sequences of the H. courbaril enzymes which were obtained as described above. The primers promoted the amplification of a 360 bp fragment for actin, a 400 bp fragment for β-galactosidase (BGAL), a 330 bp fragment for xyloglucan transglycosylase hydrolase (XTH), a 440 bp fragment for alkaline invertase (AlkIN), and a 410 bp fragment for sucrose synthase (SUS) (Table S2). The number of cycles for PCR amplification was 36 for all genes and 25 for actin. The optical densities of cDNA amplification products were obtained using eagle eye II (Stratagene). All assays were conducted in triplicate, and for each experiment three technical replicates were used. The densitometry analyses of the gels were performed with Gel-Pro Analyser software (V. 3.1 Gel-Pro; Media Cybernetics, Inc.) (data no shown).
Construction of phylogenetic trees
Arabidopsis and rice xyloglucan transglycosylase hydrolase, β-galactosidase, alkaline invertase, and sucrose synthase were identified by using the blast program and the query sequences at TAIR (The Arabidopsis Information Resource) in order to identify genes that encode in the Arabidopsis genome. Orthologues in rice (Oryza sativa) were also searched for using tbastn. Alignment was performed using Clustal W.
Phylogenetic reconstruction was performed from distances calculated from the aligned amino acid sequences using the JTT matrix (Jones et al., 1992), and the tree was inferred by the Neighbor–Joining method (Saitou and Nei, 1987). The numbers of amino acid positions used were 145, 111, 273, and 382 for β-galactosidase, xyloglucan transglycosylase hydrolase, alkaline invertase, and sucrose synthase, respectively. The analyses were carried out using the software MEGA4 (Tamura et al., 2007).
In order to classify the cDNAs obtained from cotyledons of H. courbaril β-galactosidase (HcBGAL1; EU370969), xyloglucan transglycosylase hydrolases (HcXTH1; EU370971), alkaline invertase (HcAlkIN1; EU370968), and sucrose synthase (HcSUS1; EU370970), trees were generated using the representatives of the Leguminosae (see Supplementary Table S1 available at JXB online for accession numbers).
Results
Phylogenetic characterization of partial cDNA for H. courbaril β-galactosidase, xyloglucan transglycosilase hydrolase, alkaline invertase, sucrose synthase, and actin
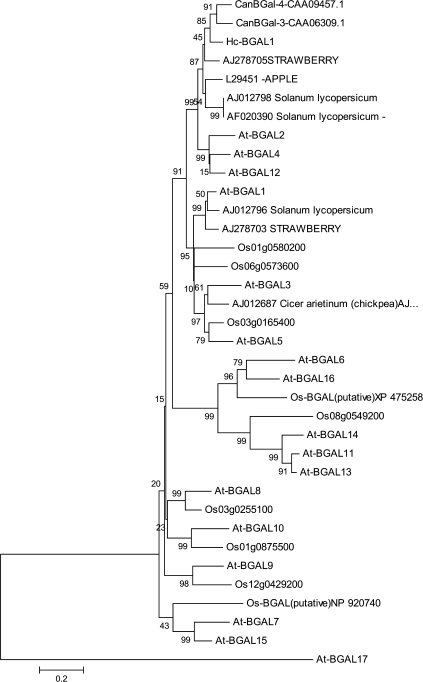
The strategy to obtain partial cDNA sequences encoding H. courbaril relied basically on the use of two pairs of nested degenerated primers corresponding to conserved sequences. The amplification products with the expected length (Table S2) corresponding to 452 bp for actin, 435 bp for β-galactosidase, 333 bp for XTH, 819 bp for alkaline invertase, and 1146 bp for sucrose synthase were cloned and sequenced. Their identity was confirmed according to their similarity to known sequences. The H. courbaril (Hc) amino acid sequences were further characterized by a phylogenetic analysis. As expected, the partial sequences for HcBGAL1, HcXTH1, HcAlkIN1, and HcSUS1 were found to group more strongly with homologous sequences from Leguminosae. This analysis also allowed the more precise classification of the Hymenaea sequences. It was found that HcBGAL1 is more closely related to group I β-galactosidases; HcXTH1 to group I XTHs; HcAlkIN1 to group β alkaline invertase, and HcSUS1 to the SUS1 group of sucrose synthase (Supplementary Table S1 at JXB online). A comparison of HcBGAL1 with other plant β-galactosidases is shown in Fig. 2. This gene is inserted in a group that contains TBG4, an exogalactanase from tomato, and also genes from mung bean and chickpea. HcBGAL1 separated completely from a group that contains several genes from Arabidopsis and rice, except for the genes AtBGAL2, AtBGAL12, and AtBGAL4.
Fig. 2.
Phylogenetic relationship of β-galactosidases of Angiosperms. Distances were calculated on the basis of positions 181–332 in A. thaliana (At-BGAL4). The nucleotide sequence can be accessed at NCBI: EU370969. Accession numbers are as follows: A. thaliana (At-GAL1, AT3G13750; At-GAL2, AT3G52840; At-GAL3, AT4G36360; At-BGAL4, AT5G56870; At-GAL5, AT1G45130; At-BGAL6, AT5G63800; At-GAL7, AT5G20710; At-BGAL8, AT2G28470; At-BGAL9, AT2G32810; At-BGAL10, AT5G63810; At-BGAL11, AT4G35010; At-GAL12, AT4G26140; At-BGAL13, AT2G16730; At-BGAL14, AT4G38590; At-GAL15, AT1G31740, At-BGAL16, AT1G77410; At-BGAL17, AT1G72990; Oryza sativa (10 members), Leguminosae (CanBGAL-4, CAA09457; CanBGAL-3, CAA06309; chickpea, AJ012687), tomato (AJ012796; AF020390; AJ012798), apple (L29451), and strawberry (AJ278705; AJ278703). The values presented represent the bootstraps.
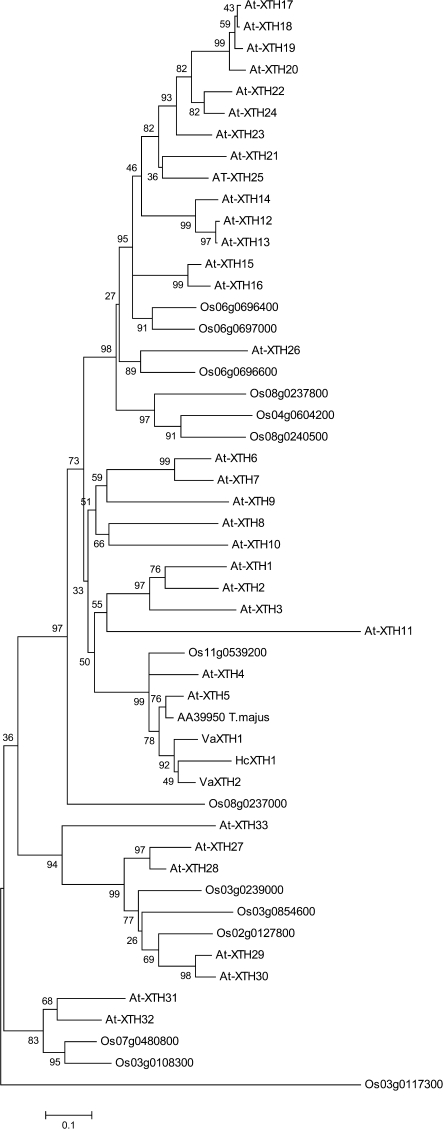
HcXTH1 appears closer to the genes VaXTH1 and VaXTH2 (Fig. 3), both coding for XTHs supposedly related to cell wall expansion in Vigna angulares. It is also noticeable that HcXTH1 belongs to the same possible group of orthologues of an XTH from nasturtium (T. majus) that is the only XTH clearly related to storage xyloglucan metabolism (see below).
Fig. 3.
Phylogenetic relationship of xyloglucan transglycosylase hydrolase (XTH) of Angiosperms. Distances were calculated on the basis of positions 87–197 of A. thaliana (At-XTH5). The nucleotide sequence can be accessed at NCBI: EU370971. Accession numbers are as follows: A. thaliana (At-XTH1, AT4G13080; At-XTH2, AT4G13090; At-XTH3, AT3G25050; At-XTH4, AT2G06850; At-XTH5, AT5G13870; At-XTH6, AT5G65730; At-XTH7, AT4G37800; At-XTH8, AT1G11545; At-XTH9, AT4G03210; At-XTH10, AT2G14620; At-XTH11, AT3G48580; At-XTH12, AT5G57530; At-XTH13, AT5G57540; At-XTH14, AT4G25820; At-XTH15, AT4G14130; At-XTH16, AT3G23730; At-XTH17, AT1G65310; At-XTH18, AT4G30280; At-XTH19, AT4G30290; At-XTH20, AT5G48070; At-XTH21, AT2G18800; At-XTH22, AT5G57560; At-XTH23, AT4G25810; At-XTH24, AT4G30270; At-XTH25, AT5G57550; At-XTH26, AT4G28850; At-XTH27, AT2G01850; At-XTH28, AT1G14720; At-XTH29, AT4G18990; At-XTH30, AT1G32170; At-XTH31, AT3G44990; At-XTH32, AT2G36870), Oryza sativa (14 members), Leguminosae (Va-XTH2, AB086396; VaXTH1, AB086395), andTropaeolum majus (AA39950). The values presented represent the bootstraps.
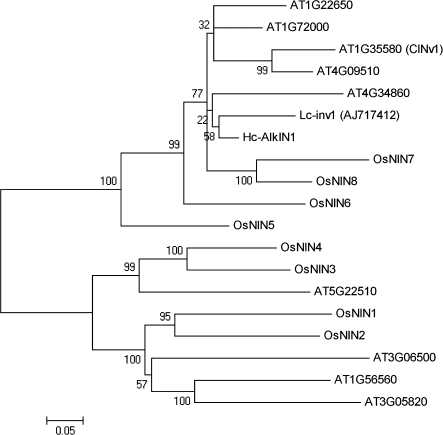
Relatively little is known about the features of genes related to alkaline invertases. HcAlkIN1 was found to be close to a gene from the legume Lotus corniculatus (Lcinv1) (Fig. 4). This class of genes is referred to as neutral alkaline invertase in the literature and its function is poorly understood.
Fig. 4.
Phylogenetic relationship of alkaline/neutral invertase of Angiosperms. Distances were calculated on the basis of positions 278–556 of A. thaliana (AT4G34860). The nucleotide sequence can be accessed at NCBI: EU370968. Accession numbers are as follows: A. thaliana (nine members), Oryza sativa (OsNIN1, AY575558; OsNIN2, AY575559; OsNIN3, AY575560; OsNIN4, AY575561; OsNIN5, AY575562; OsNIN6, AY575563; OsNIN7, AY575564; OsNIN8, AY575565), and Lotus corniculatus (Lc-inv1, AJ717412). The values presented represent the bootstraps.
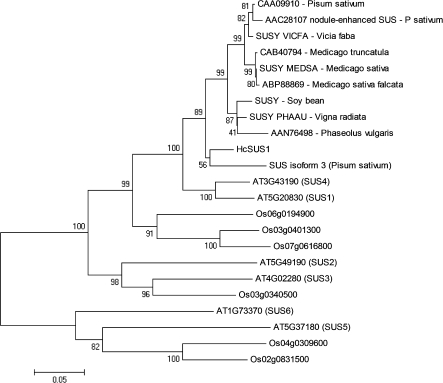
Figure 5 shows the phylogenetic relationship between HcSUS1 and other plants SUS sequences. This gene belongs to a possible group of orthologues including two Arabidopsis (SUS1 and SUS4) and three rice genes (Os06g0194900, Os03g0401300, and Os07g0616800). HcSUS1 clustered together with several legumes sequences, among which the most closely related seems to be one from Pisum sativum (SUS isoform 3).
Fig. 5.
Phylogenetic relationship of sucrose synthase of Angiosperms. Distances were calculated on the basis of positions 429–810 of A. thaliana (SUS4). The nucleotide sequence can be accessed at NCBI: EU370970. Accession numbers are as follows: A. thaliana (At-SUS1, AT5G20830; At-SUS2, AT5G49190; At-SUS3, AT4G02280; At-SUS4, AT3G43190; At-SUS5, AT5G37180; At-SUS6, AT1G73370), Oryza sativa (six members), and legumes (SUSY VICFA, P31926; SUSY MEDSA, O65026; SUSY PHAAU, Q01390; SUS isoform 3, CAC32462). The values presented represent the bootstraps. Dicot=eudicotyledons.
Expression patterns of the four genes in response to auxin polar transport inhibition and different light treatments
On the basis of the fact that the presence of gene expression and enzyme activities of the four genes cloned in this work is associated with most plant organs and different isoforms of the proteins, it was decided to evaluate the levels of expression of these genes in roots, cotyledons, leaves, and stem of developing seedlings of H. courbaril. The results of the experiments with excision of the top shoot and NPA treatment are shown in Fig. 6 and the experiment with continuous light and darkness is shown in Fig. 7.
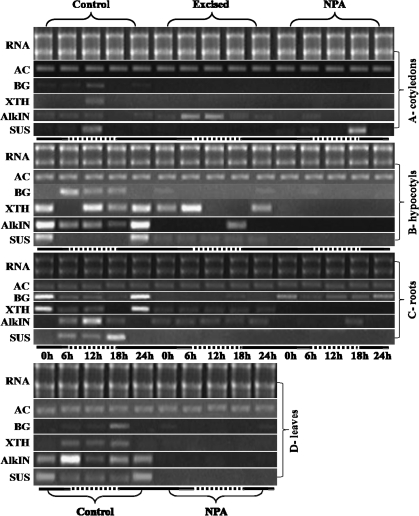
Fig. 6.
Patterns of gene expression of actin (AC), HcBGAL1 (BG), HcXTH1 (XTH), HcAlkIN1 (AlkN), and HcSUS1 (SUS) in different organs of 45-d-old seedlings of H. courbaril. Gene expression was followed using semi-quantitative RT-PCR during 24 h by sampling every 6 h. Solid white bars represent the dark periods and dashed lines the light periods during the experiment. Treatments were excision of the top shoot and inhibition of auxin polar transport by application of NPA. Densitometry was performed for each gel and essentially confirmed the patterns observed in this figure.
Fig. 7.
Patterns of gene expression of actin (AC), HcBGAL1 (BG), HcXTH1 (XTH), HcAlkIN1 (AlkN), and HcSUS1 (SUS) in different organs of 45-d-old seedlings of H. courbaril. Gene expression was followed using semi-quantitative RT-PCR during 24 h by sampling every 6 h. White solid bars represent the dark periods and dashed lines the light periods during the experiment. After germination, developing seedlings were kept under continuous light or darkness for 20 d. Densitometry was performed for each gel and essentially confirmed the patterns observed in this figure.
Cotyledons
Figure 6A presents the patterns of expression of HcBGAL1 (BG), HcXTH1 (XTH), HcAlkIN1 (AlkN), and HcSUS1 (SUS) in cotyledons of 45-d-old seedlings of H. courbaril. Both HcBGAL1 and HcXTH1 presented a peak of mRNA production at midday. HcBGAL1 was expressed in control seedlings with a peak of transcript at 12 h. Transcripts of this gene were found neither under excised conditions (top shoot of the plant excised) nor in NPA-treated seedlings (Fig. 6A). HcXTH1 displayed a very similar pattern of expression to HcBGAL1 (Fig. 6A). Under excision treatment, HcAlkIN1was up-regulated and under NPA treatment it was down-regulated (Fig. 6A). HcSUS1 was expressed during the light period, and under excision treatment no transcript was observed. Under NPA treatment, the HcSUS1 gene was expressed similarly to control seedlings, with a peak of transcript at 18 h (Fig. 6A).
Figure 7A shows the expression patterns of the four cloned genes in cotyledons of H. courbaril under different light conditions. Under constant light, HcBGAL1 was expressed only at 18 h and in darkness it was expressed only at 0 h and 24 h. Under constant light, no transcripts of HcXTH1 were observed and under darkness the gene was up-regulated (Fig. 7A). HcAlkIN1 was strongly up-regulated in constant light whereas in the darkness it was down-regulated (Fig. 7A). Regarding HcSUS1, in control seedlings a high level of mRNA was observed at 12 h. Cotyledons from seedlings growing under constant light for 20 d presented a peak of expression at 6 h. The level of expression of HcSUS1 remained low and constant in seedlings growing in darkness (Fig. 7A).
Hypocotyls
HcBGAL1 was expressed in control seedlings with a peak of transcript at 6 h, and under excision and NPA treatments the gene was down-regulated (Fig. 6B). HcXTH1 presented peaks of transcripts every 12 h. Under excision and NPA treatments, the gene was down-regulated. HcAlkIN1 showed a peak of transcript at 0 h and 24 h, and under excision and NPA treatments the gene was down-regulated (Fig. 6B). HcSUS1 presented a peak of transcription at 0 h and 24 h, and under excision treatment the gene was constitutively expressed. Under treatment with NPA, no transcript was observed (Fig. 6B).
Under constant light, the HcBGAL1 gene was up-regulated and under darkness it was expressed similarly to control seedlings, with peaks of transcripts every 12 h (Fig. 7B). HcXTH1 was down-regulated under constant light and darkness, being constitutively expressed (Fig. 7B). Under constant light, the HcAlkIN1 gene was strongly up-regulated whereas under darkness the gene was down-regulated. HcSUS1 presented a similar pattern to the control hypocotyls, whereas it was completely inhibited in the dark (Fig. 7B).
Roots
HcBGAL1 was expressed in control seedlings with a peak of transcript at 0 h and 24 h (Fig. 6C). Under excised conditions, the gene was down-regulated and under NPA treatment the gene was expressed similarly to control seedlings, with peaks of transcripts at 0 h and 24 h (Fig. 6C). HcXTH1 displayed a peak of transcription in the middle of the night in roots. It was down-regulated (showing a constitutive behaviour) under excised conditions and it was completely inhibited by NPA treatment (Fig. 6C). HcAlkIN1 was expressed in control seedlings with a peak of transcript at 12 h. Under excised conditions, the gene was constitutively expressed and under NPA treatment it has been found to be down-regulated. HcSUS1 was expressed during both light and dark periods, with a peak of transcript at 18 h, and under excision and NPA treatment no transcripts were observed (Fig. 6C).
HcBGAL1 was up-regulated under constant light and down-regulated in the darkness (Fig. 7C). Under constant light, HcXTH1 showed a similar pattern to the control and was down-regulated in darkness (Fig. 7C). HcAlkIN1 was up-regulated in constant light and down-regulated in the dark. Under constant light, HcSUS1 was up-regulated, displaying constitutive expression throughout the day. Under darkness the gene was constitutively expressed from 0 h to 12 h. At 18 h it was down-regulated and transcripts appeared again at 24 h (Fig. 7C).
Leaves
HcBGAL1 was expressed during the light and dark periods, with a peak of transcription at 18 h in control seedlings. Under NPA treatment, the gene was down-regulated. HcXTH1 was expressed with a constitutive peak of transcript at 6, 12, and 18 h. HcAlkIN1 presented a peak of transcription at 6 h, and a peak of transcription of HcSUS1 was observed at 0 h and 24 h. NPA down-regulated all the four genes studied (Fig. 6D).
Under constant light, the HcBGAL1 gene was up-regulated, with peaks of transcripts every 12 h. In darkness, the gene was down-regulated. Under constant light, HcXTH1 of leaves was down-regulated during the day and under darkness the gene was strongly up-regulated (Fig. 7D). Under constant light, HcAlkIN1 behaved similarly to the control, whereas in darkness the gene was down-regulated (Fig. 7D). In constant light HcSUS1 was up-regulated and under darkness the gene was down-regulated (Fig. 7B).
All gels were analysed by densitometry (not shown), confirming the patterns described above.
Discussion
Phylogeny and putative biological roles for the four partial sequences cloned from H. courbaril
The cloning of the partial cDNA sequences of the β-galactosidase, XTH, invertase, and sucrose synthase genes from H. courbaril permitted phylogenetic analyses to be performed with other known genes (Figs 2–5, respectively).
β-Galactosidase
HcBGAL1 is close to TBG4, a gene belonging to the tomato gene family (Smith and Gross, 2000), and also to CanBGal3 from chickpea (Esteban et al., 2003). TBG4 has the same sequence as the lupin exo-galactanase gene, which has been functionally well characterized as a cell wall gene directly related to cell wall storage mobilization (Buckeridge and Reid, 1994). The same is the case for CanBGal3, which has been experimentally associated with pectin degradation. In the case of HcBGAL1, it remains to be elucidated why this association with pectin-related genes occurs if the main storage polysaccharide is xyloglucan. A possible explanation is that this enzyme might be associated with granting access to xyloglucan-degrading enzymes during mobilization. Amaral (2005) found that the primary walls of the cotyledons of H. courbaril are rich in pectins and these walls might have to be modified during mobilization in order to grant access to storage xyloglucan. Alternatively, the preservation of sequence similarity might not be related to enzyme specificity, as in this work the full sequence was not cloned. Indeed, Alcântara et al. (1999, 2006) found unique β-galactosidases in xyloglucan-storing cotyledons of C. langsdorffii and H. courbaril (this enzyme was named hcbetagal), respectively. Sequencing of these proteins along with cloning of the full sequence of HcBGAL1 will probably provide the answer to whether or not hcbetagal (see also Fig. 8) and HcBGAL1 are the same entity.
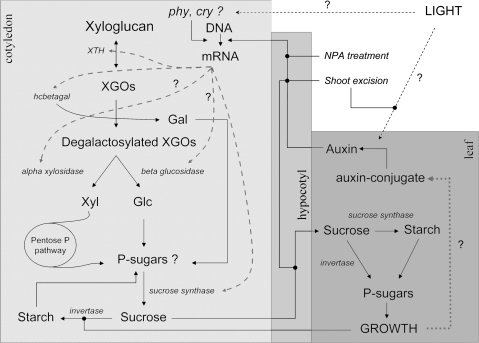
Fig. 8.
Pathways of storage xyloglucan catabolism in the cotyledon of a developing seedling of Hymenaea courbaril. These steps are considered together as ‘xyloglucan degradation’. The figure also shows some of the events that occur in the hypocotyl and developing leaves. The entire process that includes xyloglucan degradation, sucrose production, and transport through the hypocotyl towards the leaves is termed xyloglucan mobilization. The names of the enzymes are shown in italics. Hypothetically the following correlations between enzymes and genes exist: XTH=HcXTH1, hcbetagal=HcBGAL1, invertase=HcAlkIN1, and sucrose synthase=HcSUS1. Note that there are several points at which feedback control is present. The figure illustrates the points which are thought to control xyloglucan mobilization according to the experiments with application of NPA, top shoot excision, and presence or absence of light. Question marks were added to indicate hypothetical pathways that need further experiments. phy, phytochrome; cry, cryptochrome; Gal, galactose; Glc, glucose; Xyl, xylose.
XTH
The HcXTH1 gene displays closer similarity to nasturtium NXET1, a gene associated with endotransglycosylation, than to the NXG1, one of the first XET genes cloned from cotyledons of T. majus (De Silva et al., 1993) that has been associated with a xyloglucan-storing system analogous to that of H. courbaril. Indeed, later, Rose et al. (1998) found that XET1 was expressed in all vegetative tissues (root, epicotyl, stem, and leaf) except for germinating cotyledons of T. majus. Although the xyloglucan degradation system present in cotyledons of legumes (especially H. courbaril) has been associated with that present in nasturtium (Buckeridge et al., 2000), it is becoming clear that many of the features of the former systems are quite different. Tiné et al. (2000) have already demonstrated that the principal activity of XET in H. courbaril needs a supply of oligosaccharides in order to hydrolyse its own storage xyloglucan, therefore being a true XTH. Alcântara (2000) isolated an XTH from cotyledons of H. courbaril and found that it is indeed an enzyme of the transglycosylase type and not an XET of the hydrolytic type as found by Fanutti et al. (1993) for T. majus. Thus, the finding of closer similarity of HcXTH1 to genes such as VaXTH1 and VaXTH2 is not surprising, especially if we consider that their expression is associated with the action of auxin, as demonstrated in this work. The detection of HcXTH1 in several organs of the seedling of H. courbaril cannot be considered as specifically marking a single gene, as the primers for PCR most probably allow amplification of cDNA of different closely related genes. The sequence DEIDFEFLGNRTG, according to Okazawa et al. (1993), is a conserved sequence present in most XETs found in nature. Therefore, the present detection of this conserved sequence in HcXTH1, along with expression of the gene in several organs of H. courbaril, suggests that the primer used is apparently not specific, but reflects the occurrence of expression of several isoforms of legume-specific XTHs. On the other hand, due to the fact that the main event taking place in the cotyledons of H. courbaril at the 45th day during seedling development is xyloglucan degradation (Santos and Buckeridge, 2004; Santos et al., 2004), one can expect that the expression patterns observed, at least in this organ, reflect important events related to xyloglucan mobilization.
Alkaline invertase
In cotyledons of H. courbaril, relatively high concentrations of sucrose, glucose, and fructose were detected by Santos and Buckeridge (2004) during xyloglucan degradation. A search in the databases using HcAlkIN1 resulted in many sequences with considerable similarity, but there were only a few publications that could indicate a possible physiological role for this invertase. Because it is an alkaline/neutral invertase, it is possible that the product of the gene is active in the cytosol. In this context, its biochemical role might indeed be associated with sucrose degradation. However, it is not possible at this point to understand the role played by HcAlkIN1 in the process of storage mobilization, and further experiments will have to be performed to define the precise function of HcAlkIN1.
Sucrose synthase
One of the genes found to be closely related to HcSUS1 is MtSucS1 which, in Medicago truncatula, was found to be associated with the vascular systems in several parts of the plant as well as in developing seedlings (Hohnjec et al., 1999). These authors conclude that the pattern of gene expression observed indicated an involvement of MtSucS1 with the generation of sink strength. This adds support to the hypothesis that HcSUS1 might be associated with sink strength in developing seedlings of H. courbaril.
Expression of sucrose synthase genes has been shown to be cell specific, developmentally regulated, or regulated by tissue carbohydrate status (Koch et al., 1992; Ruan et al., 1997). Most of the carbon produced during photosynthesis is channelled through the synthesis of sucrose, which is central to plant growth and development. Synthesis of sucrose certainly occurs in cotyledons of H. courbaril, as the products of xyloglucan degradation are thought to be rapidly metabolized into sucrose (Santos and Buckeridge, 2004).
Besides its role in determining accumulation of sucrose in certain tissues, sucrose synthase has also been related to synthesis of cell wall polysaccharides (Amor et al., 1995; Buckeridge et al., 1999). In these cases, the enzyme is thought to be associated with plasma or Golgi membranes and to control the production of UDP-glucose that is used as substrate for synthesis of cellulose, callose, and the mixed linkage β-glucan in grasses. In the present experiments involving excision of the shoot or application of NPA, in all cases of sink or transport organs (leaves, roots, and hypocotyl, respectively) inhibition of expression of HcSUS1 was observed. Thus, it is possible that the lower level of expression of this gene is associated with lower use of sugars to produce cell walls, as the growth rate decreases due to the lack of sugars coming from the storage mobilization organ (i.e. the cotyledon).
Auxin-induced expression patterns for cell wall-related genes
The importance of auxin in plant development has been highlighted by several authors (Romano et al., 1995; Gray et al., 1998; Reed, 2001; Rampey et al., 2004). Auxin signalling has also been implicated as the principal regulatory factor for xyloglucan metabolism during cell wall loosening following development in several plant species (see, for example, Cosgrove, 1993; Catalá et al., 1997; Nishitani, 1997). The regulation of gene expression by auxin has been extensively studied for decades (Theologis and Ray, 1982) and several families of auxin-regulated genes [e.g. members of the SAUR (small auxin-up-regulated) genes, the Aux/IAA gene, and GH3 families] (Catalá et al., 2000; Nakazawa et al., 2001; Reed, 2001; Liscum and Reed, 2002) have been identified. These rapidly induced, auxin-responsive mRNAs are under transcriptional control involving regulatory sequences in the promoter region (Abel and Theologis, 1996; Reed, 2001). Some of these genes controlled by auxin are expressed in distinct spatial and temporal patterns, thus underscoring the diversity of auxin responses in different plant tissues and organs (Abel and Theologies, 1996). However, the patterns of gene expression related to cell wall storage polysaccharides have not been described to date.
Genes that encode XTHs have been implicated by several studies in auxin-mediated regulation of xyloglucan metabolism (Nishitani and Masuda, 1981; Nishitani, 1995; Schindler et al., 1995; Wu et al., 1996; Xu et al., 1996; Catalá et al., 1997, 2001; Davies et al., 1997; Akamatsu et al., 1999; Yokoyama and Nishitani, 2000, 2001; Hyodo et al., 2003; Nakamura et al., 2003; Goda et al., 2004; Yokoyama et al., 2004). The application of indole acetic acid (IAA) on intact seedlings of Arabidopsis thaliana up-regulated expression of XTH members (Xu et al., 1995, 1996; Yokoyama and Nishitani, 2001; Goda et al., 2004; Vissenberg et al., 2005). Catalá et al. (2000) characterized regulation of Cel7 by auxin during fruit development in tomato. Nakamura et al. (2003) analysed two azuki bean XTH genes and concluded that they are up-regulated by auxin but with different responses. Cui et al. (2005) observed up-regulation of XET in rice leaf. Osato et al. (2006) analysed some members of XTH in roots of A. thaliana and concluded that AtXTH19 was up-regulated by auxin.
Considering these observations, gene expression of two of the hydrolases (HcBGAL1 and HcXTH1) which are directly associated with xyloglucan storage degradation were analysed. Expression of genes related to sucrose metabolism in seedlings, i.e. invertase (HcAlkIN1) and sucrose synthase (HcSUS1), was also followed. Due to the observation of changes in the concentrations of sucrose, fructose, and glucose in xyloglucan-degrading cotyledons (Santos and Buckeridge, 2004), these two enzymes can be thought of as key for sucrose metabolism on the 45th day of development and consequently have important roles in the establishment of the seedlings of H. courbaril.
The pattern of expression observed in the control treatments of cotyledons suggests that the genes related to xyloglucan degradation (HcBGAL1 and HcXTH1) in seedlings of H. courbaril are dominantly expressed during the day time (peaks observed at 12 h) and sucrose metabolism genes (HcAlkIN1 and HcSUS1) are expressed during both the light and dark periods, but with peaks observed in the middle of the day (Figs 6A, 7A).
The fact that activity and protein related to β-galactosidase are found during the night (LIV Amaral and MS Buckeridge, unpublished) suggests that the gap between mRNA accumulation and enzyme production/activity in cotyledons of H. courbaril is of ∼6 h. Auxin transport is apparently crucial for the xyloglucan degradation process in the cotyledons of H. courbaril as the present observations showed that no expression is detectable in these organs when auxin polar transport is inhibited by NPA and also when the sink organs (developing leaves) are excised (Fig. 6). Because these new leaves are thought to produce the auxin that is transported to the cotyledon and induces the activity of the enzymes (Santos et al., 2004), it cannot be concluded from the shoot excision treatment that absence of sink strength inhibits expression of HcBGAL1 and HcXTH1. Thus, it remains to be seen whether sink strength or auxin production (or both) are the main factors that control HcXTH1 and HcBGAL1 expression.
In leaves of H. courbaril, HcSUS1 presented a constant level of expression during the whole day (Figs 6D, 7D). The observation that NPA inhibits expression of all genes is intriguing. It might possibly be attributed to the fact that there is no production and transport of sucrose from cotyledons to developing leaves, i.e. the maintenance of the source–sink relationship between the two organs is important to supply sucrose for developing organs.
The hypocotyl can be viewed as the means through which cotyledons (the source) transport synthesized sucrose towards the developing shoot (the main sink). The hypocotyl is also the way through which auxin produced in the top shoot reaches the storage-mobilizing cotyledons. In hypocotyls, HcBGAL1 and HcXTH1 are genes probably associated with cell elongation, one of the main events taking place in this organ. It may be speculated that the sucrose metabolism-related genes HcAlkIN1 and HcSUS1 are probably associated with the supply of energy for hypocotyl tissues. The expression levels of these two genes might also be associated (or at least correlated) with the intensity of transport of sucrose. Therefore, they may be used as an indication of the status of the source–sink relationship between growing tissues and storage degradation. The fact that all four genes had their expression levels severely decreased under the effect of NPA adds a new dimension to the interpretation of the effect observed by Santos et al. (2004). Auxin, produced in the shoot and transported to the cotyledons, not only seems to control expression of the genes related to xyloglucan degradation, but also seems to keep the sucrose metabolism in hypocotyls functional. A similar result was obtained when the top shoot was excised. However, HcXTH1 still displayed a high expression level after excision, suggesting that there are independent wall-related mechanisms different from the control by auxin.
Roots seem to be the organ with the lowest correlation with storage mobilization in seedlings of H. courbaril, as a lower proportion (∼30%) of the xyloglucan degradation products end up in this organ during the whole period of development (Santos and Buckeridge, 2004). Even so, some strong effects of inhibition of polar transport of auxin were observed. Expression of HcBGAL1 was shut down after excision of the shoot, but not under treatment with NPA (Fig. 6C). In the case of HcXTH1, the reverse was observed, with maintenance of expression after excision of the shoots, but complete inhibition under treatment with NPA. These results reflect changes in the wall of roots that are apparently related to the shoot growth. As according to Santos and Buckeridge (2004) the developing leaves display a limited capacity to carry out photosynthesis at this stage of development, it is possible that in the present experiments roots were already becoming dependent on photosynthesis. This hypothesis is supported by the observation that HcSUS1, a gene that is possibly related to sucrose synthesis, was inhibited by both shoot excision and NPA treatment. Thus, the absence of auxin coming from the shoot eliminated expression of an important gene (sucrose synthase) thought to be related to the sink function of the organ (Fig. 6C).
Gene expression and light
Plants are sensitive to a number of abiotic environmental stimuli, including light, and related genes change their expression level in response to light. The existence of distinct gene sets that respond to different stimuli suggests that specific receptors and signal transduction pathways are utilized in response to alterations in light to drive distinct gene expression changes (IIiev et al., 2002).
When a seed germinates in the forest understorey and a seedling starts to develop, a genetic programme named skotomorphogenesis is thought to be active (Alabbadí et al., 2004). This genetic programme maintains hypocotyl elongation and inhibits expression of genes related to photosynthesis. When this seedling reaches a higher light intensity, photosynthesis establishes concomitantly with a rapid increase in leaf area, and hypocotyls cease to elongate.
Seedlings of H. courbaril can face microenvironmental conditions in the understorey of the tropical rain forests that are compatible with the maintenance of skotomorphogenesis. Seedlings of H. courbaril are usually quite large (30–40 cm tall; Fig. 1) but the light conditions in the understorey where they naturally develop can be as low as 22 μmol photons m2 s−1 (Santos and Buckeridge, 2004). According to these authors, the lower the average light intensity, the slower will be the rate of storage mobilization and the corresponding relative growth rate. Furthermore, these authors reported that cotyledons are proportionally more important when light intensity decreases. Yet an important observation regarding the control of storage mobilization in seedlings of H. courbaril by light is that the importance of reserves in the less illuminated environment is directly related to the capacity of the newly developed leaves to establish photosynthesis, i.e. when in higher light intensity, seedlings become autotrophic earlier, becoming independent of the cotyledons sooner, which may induce an earlier fall of cotyledons.
That light interferes with xyloglucan mobilization in cotyledons of H. courbaril during seedling development is certain (Santos and Buckeridge, 2004). However, it is not yet known what type of light is involved and by what mechanism it regulates xyloglucan degradation. Although Santos and Buckeridge (2004) observed that a very low red/far-red ratio (0.5) did not increase seedling height significantly in relation to other red/far-red ratios (1.2 and 1.4), in a separate experiment, Santos et al. (2004) observed that when cotyledons were kept in the darkness (covered with aluminium foil during mobilization), xyloglucan hydrolase activities were significantly inhibited. These observations raise the hypothesis that light may have a somewhat direct control on xyloglucan mobilization at the cotyledon level, but at the same time is indirectly controlled by light through auxin production and transport in leaves.
To evaluate this hypothesis, in the present work, seedlings were kept in constant light and darkness during the storage mobilization period (from the 30th to the 60th day after the beginning of imbibition), and on the 45th day (the maximal rate of degradation of xyloglucan) gene expression was followed for 24 h.
In the literature, physiological experiments indicate that auxin is the major plant hormone closely connected with light transduction signals (Neff et al., 1999; Steindler et al., 1999). The expression of the AtHB2 gene is regulated by phytochrome, and this photoreceptor may regulate the transport of auxin through AtHB2 gene expression (Carabelli et al., 1996). Reed (2001) suggests that light can also regulate Aux/IAA proteins by stabilizing them in A. thaliana. Vandenbussche et al. (2003) observed the induction of auxin-up-regulated genes in plants growing in lower light intensities. Braam and Davis (1990) and Xu et al. (1995) also observed that TCH4 encodes an XET that is also up-regulated by darkness and auxin. How these several stimuli lead to the common molecular response of TCH4 regulation is unknown.
The present experiments were not designed to test the involvement of phytochrome in the control of xyloglucan degradation, but the observations, together with previous observations by Santos and Buckeridge (2004) and Santos et al. (2004), suggest that this is a hypothesis worthwhile testing in future experiments. In general, the present results show that constant light or darkness had a profound effect on the expression of the four genes related to storage xyloglucan mobilization in cotyledons of H. courbaril. In cotyledons, where xyloglucan degradation takes place (Tiné et al., 2000), expression of HcBGAL1 was inhibited by light, whereas the expression of HcXTH1 was induced in the dark. The latter observation is consistent with the literature mentioned above. On the other hand, in control seedlings, which were kept under a 12 h photoperiod, the expression of these two genes was apparently synchronized, with both peaking in the middle of the day. When constant light or darkness was imposed on seedlings during the period of storage mobilization, the expression of HcBGAL1 and HcXTH1 lost synchrony. The observation that the expression of these two genes becomes out of phase in constant light or darkness, according to the model proposed by Tiné et al. (2000), would disconnect the processes of degalactosylation from the process of transglycosylation, therefore halting the production of oligosaccharides that are the substrates for α-xylosidase and β-glucosidase. The disruption of the chain of biochemical events would then halt xyloglucan degradation. Indeed, cotyledons of seedlings grown in the dark were not capable of hydrolysing reserves, as they did not present the characteristic symptoms (i.e. shrunken cotyledons). Although the effect of different light intensities on gene expression was not tested, it is reasonable to speculate that the desynchronization between galactosylation and transglycosylation is likely to be at least part of the explanation for the delay in xyloglucan degradation in low light intensities observed by Santos and Buckeridge (2004). The lower production of auxin due to lower light intensity (LIV Amaral, HP Santos, and MS Buckeridge, unpublished results) might be the other part of the explanation for these observations. However, more experiments taking into consideration the addition of exogenous auxin to the excised seedling to replace the lack of the top shoot are necessary to clarify how different control systems interact.
Santos et al. (2004) proposed a model for storage xyloglucan mobilization in seedlings of H. courbaril in which a complex cross-talking network would in fact modulate the source–sink relationship between cotyledons and parts of the developing seedling. The present report extends this model for genes related to xyloglucan mobilization and sucrose metabolism (Fig. 8), showing for the first time that control at the transcription level is also important for xyloglucan storage mobilization. Auxin produced in leaves induces expression of hydrolases in the cotyledons. Hypothetically, auxin production could be related to growth as well as hydrolysis of its conjugated form (Woodward and Bartel, 2005). Regarding the effect of light, it is not possible to know for sure exactly where signalling occurs. Two hypotheses are possible, one being through transformation of conjugated to free IAA and the other related to action through phytochromes and/or cryptocrome. Further investigation will be necessary to test these hypotheses.
In conclusion, at least the four genes studied in this work have their expression controlled by auxin and light, which are both integrated in a communication network that seedlings of this important rain forest tree use to maximize physiological performance (i.e. carbon use efficiency) in the understorey of the very competitive rain forests such as the Atlantic and the Amazon forests.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Material
Acknowledgments
The authors acknowledge financial support from the Ministry of Science and Technology of Brazil (project MCT-FUNDEPAG 2005.0188), FAPESP (grants 2004/10159-8 and 2007/59708-1) and CNPq, to ADB and TMQ for help in harvesting seedlings. AD Brandão was a doctoral student at Cell Biology Department, UNICAMP.
References
- Abel S, Theologis A. Early genes and auxin action. Plant Physiology. 1996;111:9–17. doi: 10.1104/pp.111.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akamatsu T, Hanzawa Y, Ohtake Y, Takahashi T, Nishitani K, Komeda YY. Expression of endoxyloglucan transferase genes in acaulis mutants of Arabidopsis. Plant Physiology. 1999;121:715–722. doi: 10.1104/pp.121.3.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabbadí D, Gil J, Blázquez MA, García-Martínez JL. Gibberellins repress photomorphogenesis in darkness. Plant Physiology. 2004;134:1050–1057. doi: 10.1104/pp.103.035451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcântara PHN. Federal University of São Paulo (UNIFESP): São Paulo, Brazil; 2000. Isolamento e caracterização das enzimas xiloglucano endotransglicosilase e β-galactosidase do catabolismo do xiloglucano de reserva dos cotilédones de Hymenaea courbaril L. (Leguminosae-Caesalpinioideae). PhD thesis. [Google Scholar]
- Alcântara PHN, Dietrich SMC, Buckeridge MS. Xyloglucan mobilisation and purification of a (XLLG/XLXG) specific β-galactosidase from cotyledons of Copaifera langsdorffii Desf. (Leguminosae) Plant Physiology and Biochemistry. 1999;37:1–11. [Google Scholar]
- Alcântara PHN, Martin L, Silva CO, Dietrich SMC, Buckeridge MS. Purification of a β-galactosidase from cotyledons of Hymenaea courbaril L. (Leguminosae). Enzyme properties and biological function. Plant Physiology and Biochemistry. 2006;44:619–627. doi: 10.1016/j.plaphy.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Amaral LIV. PhD Thesis University of São Paulo, Department of Botany: São Paulo, Brazil; 2005. Metabolismo de carboidratos estruturais e de reserva em cotilédones de Hymenaea courbaril L var. stilbocarpa (Jatobá) [Google Scholar]
- Amor Y, Haigler CH, Johnson S, Wainscott M, Delmer DP. A membrane-associated form of SuSy and its potential role in synthesis of cellulose and callose in plants. Proceedings of the National Academy of Sciences, USA. 1995;92:9353–9357. doi: 10.1073/pnas.92.20.9353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braam J, Davis RW. Rain-, wind-, and touch-induced expression of calmodulin and calmodulin-related genes in Arabidopsis. Cell. 1990;60:357–364. doi: 10.1016/0092-8674(90)90587-5. [DOI] [PubMed] [Google Scholar]
- Buckeridge MS, Carpita NC, Vergara C. The mechanism of synthesis of a cereal mixed-linkage (1,3),(1,4)-β-d-glucan: evidence for multiple sites of glucosyl transfer in the synthase complex. Plant Physiology. 1999;120:1105–1116. doi: 10.1104/pp.120.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckeridge MS, Crombie HJ, Mendes CJM, Reid JSG, Gidley MJ, Vieira CJ. A new family of xyloglucan oligosaccharides from cotyledons of Hymenaea courbaril: structure determination of the oligosaccharide XXXXG by enzymatic sequencing. Carbohydrate Research. 1997;303:233–237. doi: 10.1016/s0008-6215(97)00161-4. [DOI] [PubMed] [Google Scholar]
- Buckeridge MS, Dietrich SMC. Galactomannans from Brazilian legume seeds. Revista Brasileira de Botanica. 1990;13:109–112. [Google Scholar]
- Buckeridge MS, Reid JSG. Purification and properties of a novel β-galactosidase or exo-β-(1,4)-galactanase from the cotyledons of germinated Lupinus angustifolius L. seeds. Planta. 1994;192:502–511. doi: 10.1007/BF00203588. [DOI] [PubMed] [Google Scholar]
- Buckeridge MS, Rocha DC, Reid JSG, Dietrich SMC. Xyloglucan structure and post-germinative metabolism in seeds of Copaifera langsdorffii from savannah and forest populations. Physiologia Plantarum. 1992;86:145–151. [Google Scholar]
- Buckeridge MS, Tiné MAS, Lima DU, Santos HP. Mobilisation of storage cell wall polysaccharides in seeds. Plant Physiology and Biochemistry. 2000;38:141–156. [Google Scholar]
- Carabelli M, Morelli G, Whitelam G, Ruberti I. Twilight-zone and canopy shade induction of the ATHB-2 homeobox gene in green plants. Proceedings of the National Academy of Sciences, USA. 1996;93:3530–3535. doi: 10.1073/pnas.93.8.3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalá C, Rose JKC, Bennett AB. Auxin regulation and spatial localization of an endo-1,4-β-d-glucanase and a xyloglucan endotransglycosylase in expanding tomato hypocotyls. The Plant Journal. 1997;12:417–426. doi: 10.1046/j.1365-313x.1997.12020417.x. [DOI] [PubMed] [Google Scholar]
- Catalá C, Rose JKC, Bennett AB. Auxin-regulated genes encoding cell wall modifying proteins are expressed during early tomato fruit growth. Plant Physiology. 2000;122:527–534. doi: 10.1104/pp.122.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalá C, Rose JKC, York WS, Albersheim P, Darvill AG, Bennett AB. Characterization of a tomato xyloglucan endotransglycosylase gene that is down-regulated by auxin in etiolated hypocotyls. Plant Physiology. 2001;127:1180–1192. doi: 10.1104/pp.010481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. Wall extensibility: its nature, measurement and relationship to plant cell growth. New Phytologist. 1993;124:1–23. doi: 10.1111/j.1469-8137.1993.tb03795.x. [DOI] [PubMed] [Google Scholar]
- Cui D, Neill SJ, Tang Z, Cai W. Gibberellin-regulated XET is differentially induced by auxin in rice leaf sheath bases during gravitropic bending. Journal of Experimental Botany. 2005;56:1327–1334. doi: 10.1093/jxb/eri133. [DOI] [PubMed] [Google Scholar]
- Daohong W, Bochu W, Biao L, Chuanren D, Jin Z. Extraction of total RNA from Chrysanthemum containing hight levels of phenolic and carbohydrates. Colloids and Surfaces. B, Biosurfaces. 2004;36:111–114. doi: 10.1016/j.colsurfb.2004.05.014. [DOI] [PubMed] [Google Scholar]
- Davies C, Boss PK, Robinson SP. Treatment of grape berries, a nonclimacteric fruit, with a synthetic auxin retards ripening and alters the expression of developmentally regulated genes. Plant Physiology. 1997;115:1155–1161. doi: 10.1104/pp.115.3.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva J, Jarman CD, Arrowsmith DA, Stronach MS, Sidebottom C, Reid JSG. Molecular characterisation of a xyloglucan specific endo 1,4-β-d glucanase (xyloglucan endo-transglycosylase) from nasturtium seeds. The Plant Journal. 1993;3:701–711. [PubMed] [Google Scholar]
- Esteban R, Dopico B, Muñoz FJ, Romo S, Martín I, Labrador E. Cloning of a Cicer arietinum β-galactosidase with pectin-degrading function. Plant and Cell Physiology. 2003;44:718–725. doi: 10.1093/pcp/pcg087. [DOI] [PubMed] [Google Scholar]
- Fanutti C, Gidley MJ, Reid JSG. Action of a pure xyloglucan endo-transglycosylase (formerly called xyloglucan-specific endo-(1,4)-β-d-glucanase) from the cotyledons of germinated nasturtion seeds. The Plant Journal. 1993;3:691–700. doi: 10.1046/j.1365-313x.1993.03050691.x. [DOI] [PubMed] [Google Scholar]
- Fry SC, York WS, Albersheim P, et al. An unambiguous nomenclature for xyloglucan-derived oligosaccharides. Plant Physiology. 1993;89:1–3. [Google Scholar]
- Gerhardt K. Tree seedling development in tropical dry abandoned pasture and secondary forest in Costa Rica. Journal of Vegetation Science. 1993;4:95–102. [Google Scholar]
- Gesteira AS, Micheli F, Ferreira CF, Cascardo CM. Isolation and purification of functional total RNA from different organs of cacao tree during its interaction with the pathogen Crinipellis perniciosa. Biotechnology. 2003;35:494–500. doi: 10.2144/03353st02. [DOI] [PubMed] [Google Scholar]
- Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S. Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiology. 2004;134:1–19. doi: 10.1104/pp.103.034736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Ostin A, Sandberg G, Romano CP, Estelle M. High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 1998;95:7197–7202. doi: 10.1073/pnas.95.12.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohnjec N, Becker JD, Puhler A, Perlick AM, Kuster H. Genomic organization and expression properties of the MtSucS1 gene, which encodes a nodule-enhanced sucrose synthase in the model legume Medicago truncatula. Molecular Genetics and Genomics. 1999;261:514–522. doi: 10.1007/s004380050995. [DOI] [PubMed] [Google Scholar]
- Hyodo H, Yamakawa S, Takeda Y, Tsuduki M, Yokota A, Nishitani K, Kohchi T. Active gene expression of a xyloglucan endotransglucosylase/hydrolase gene, XTH9, in inflorescence apices is related to cell elongation in Arabidopsis thaliana. Plant Molecular Biology. 2003;54:473–482. doi: 10.1023/a:1023904217641. [DOI] [PubMed] [Google Scholar]
- IIiev EA, Xu W, Polisensky DH, OH M, Torisky RS, Clouse SD, Braam J. Transcriptional and posttranscriptional regulation of Arabidopsis TCH4 expression by diverse stimuli. Roles of cis regions and brassinosteroids. Plant Physiology. 2002;130:1–14. doi: 10.1104/pp.008680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Computer Applications in the Biosciences. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Koch KE, Nolte KD, Duke ER, McCarty DR, Avigne WT. Sugar levels modulate differential expression of maize sucrose synthase genes. The Plant Cell. 1992;4:59–69. doi: 10.1105/tpc.4.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YT, Langenheim JH. A systematic revision of the genus Hymenaea (Leguminosae; Caesalpinioideae; Detarieae) University of California Publications in Botany. 1975;69:1–109. [Google Scholar]
- Levy S, Maclachlan G, Staehelin LA. Xyloglucan side chains modulate binding to cellulose during in vitro binding assays as predicted by conformational dynamics simulations. The Plant Journal. 1997;11:373–386. doi: 10.1046/j.1365-313x.1997.11030373.x. [DOI] [PubMed] [Google Scholar]
- Liscum E, Reed JW. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Molecular Biology. 2002;49:387–400. [PubMed] [Google Scholar]
- Nakamura T, Yokoyama R, Tomita E, Nishitani K. Two azuki bean XTH genes, VaXTH1 and VaXTH2, with similar tissue-specific expression profiles are differently regulated by auxin. Plant and Cell Physiology. 2003;44:16–24. doi: 10.1093/pcp/pcg002. [DOI] [PubMed] [Google Scholar]
- Nakazawa M, Yabe N, Ichikawa T, Yamamoto YY, Yoshizumi T, Hasunuma K, Matsui M. DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. The Plant Journal. 2001;25:213–221. doi: 10.1046/j.1365-313x.2001.00957.x. [DOI] [PubMed] [Google Scholar]
- Neff MM, Nguyen SM, Malancharuvil EJ, et al. BAS1: a gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 1999;96:15316–15323. doi: 10.1073/pnas.96.26.15316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani K. The role of endoxyloglucan transferase in the organization of plant cell walls. International Review of Cytology. 1997;173:157–206. doi: 10.1016/s0074-7696(08)62477-8. [DOI] [PubMed] [Google Scholar]
- Nishitani K. Endo-xyloglucan transferase, a new class of transferase involved in cell wall construction. Journal of Plant Research. 1995;108:137–148. [Google Scholar]
- Nishitani K, Masuda Y. Auxin-induced changes in the cell wall structure: changes in the sugar compositions, intrinsic viscosity and molecular weight distributions of matrix polysaccharides of the epicotyl cell wall of Vigna angularis. Physiologia Plantarum. 1981;52:482–494. [Google Scholar]
- Okazawa K, Sato Y, Nakagawa T, Asada K, Kato I, Tomita E, Nishitani K. Molecular cloning and cDNA sequencing of endoxyloglucan transferase, a novel class of glycosyltransferase that mediates molecular grafting between matrix polysaccharides in plant cell walls. Journal of Biological Chemistey. 1993;268:25364–25368. [PubMed] [Google Scholar]
- Osato Y, Yokoyama R, Nishitani K. A principal role for AtXTH18 in Arabidopsis thaliana root growth: a functional analysis using RNAi plants. Journal of Plant Research. 2006;119:153–162. doi: 10.1007/s10265-006-0262-6. [DOI] [PubMed] [Google Scholar]
- Rampey RA, LeClere S, Kowalczyk M, Ljung K, Sandberg G, Bartel B. A family of auxin-conjugate hydrolases that contributes to free indole-3-acetic acid levels during Arabidopsis germination. Plant Physiology. 2004;135:978–988. doi: 10.1104/pp.104.039677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW. Roles and activities of Aux/IAA proteins in Arabidopsis. Trends in Plant Science. 2001;6:420–425. doi: 10.1016/s1360-1385(01)02042-8. [DOI] [PubMed] [Google Scholar]
- Romano CP, Robson PRH, Smith H, Estelle M, Klee H. Transgene-mediated auxin overproduction in Arabidopsis: hypocotyl elongation phenotype and interactions with the hy6-1 hypocotyl elongation and axr1 auxin-resistant mutants. Plant Molecular Biology. 1995;27:1071–1083. doi: 10.1007/BF00020881. [DOI] [PubMed] [Google Scholar]
- Rose JK, Brummell DA, Bennett AB. Two divergent xyloglucan endotransglycosylases exhibit mutually exclusive patterns of expression in nasturtium. Plant Physiology. 1998;110:493–499. doi: 10.1104/pp.110.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan YL, Chourey PS, Delmer DP, Perez-Grau L. The differential expression of sucrose synthase in relation to diverse patterns of carbon partitioning in developing cotton seed. Plant Physiology. 1997;115:375–385. doi: 10.1104/pp.115.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Santos HP, Buckeridge MS. The role of the storage carbon of cotyledons in the establishment of seedlings of Hymenaea courbaril L. under different light conditions. Annals of Botany. 2004;94:819–830. doi: 10.1093/aob/mch209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos HP, Purgatto E, Mercier H, Buckeridge MS. The control of storage xyloglucan mobilisation in cotyledons of Hymenaea courbaril L. Plant Physiology. 2004;135:287–299. doi: 10.1104/pp.104.040220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler T, Bergfeld R, Schopfer P. Arabinogalactan proteins in maize coleoptiles: developmental relationship to cell death during xylem differentiation but not to extension growth. The Plant Journal. 1995;7:25–36. doi: 10.1046/j.1365-313x.1995.07010025.x. [DOI] [PubMed] [Google Scholar]
- Smith DL, Gross KC. A family of at least seven β-galactosidase genes is expressed during tomato fruit development. Plant Physiology. 2000;123:1173–1183. doi: 10.1104/pp.123.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza RP, Valio IFM. Carbon translocation as affected by shade in saplings of shade tolerant and intolerant species. Biologia Plantarum. 1999;42:631–636. [Google Scholar]
- Steindler C, Matteucci A, Sessa G, Weimar T, Ohgishi M, Aoyama T, Morelli G, Ruberti I. Shade avoidance responses are mediated by the ATHB-2 HD-Zip protein, a negative regulator of gene expression. Development. 1999;126:4235–4245. doi: 10.1242/dev.126.19.4235. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24:1496–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Theologis A, Ray P. Early auxin-regulated polyadenylylated mRNA sequences in pea stem tissue. Proceedings of the National Academy Sciences, USA. 1982;79:418–421. doi: 10.1073/pnas.79.2.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiné MAS, Cortelazzo AL, Buckeridge MS. Xyloglucan mobilisation in cotyledons of developing plantlets of Hymenaea courbaril L. (Leguminosae-Caesalpinoideae) Plant Science. 2000;154:117–126. doi: 10.1016/s0168-9452(99)00245-9. [DOI] [PubMed] [Google Scholar]
- Tiné MAS, Silva CO, Lima DU, Carpita NC, Buckeridge MS. Fine structure of a mixed-oligomer storage xyloglucan from seeds of Hymenaea courbaril. Carbohydrate Polymers. 2006;66:444–454. [Google Scholar]
- Vandenbussche F, Vriezen WH, Smalle J, Laarhoven LJJ, Harren FJM, Straeten DVD. Ethylene and auxin control the Arabidopsis response to decreased light intensity. Plant Physiology. 2003;133:517–527. doi: 10.1104/pp.103.022665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissenberg K, Oyama M, Osato Y, Yokoyama R, Verbelen JP, Nishitani K. Differential expression of AtXTH17, AtXTH18, AtXTH19 and AtXTH20 genes in Arabidopsis roots. Physiological roles in specification in cell wall construction. Plant and Cell Physiology. 2005;46:192–200. doi: 10.1093/pcp/pci013. [DOI] [PubMed] [Google Scholar]
- Woodward A, Bartel B. Auxin: regulation, action and interaction. Annals of Botany. 2005;95:707–735. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SC, Blumer JM, Darvill AG, Albersheim P. Characterization of an endo-1,4-glucanase gene induced by auxin in elongating pea epicotyls. Plant Physiology. 1996;110:163–170. doi: 10.1104/pp.110.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Campbell P, Vargheese AK, Braam J. The Arabidopsis XET-related gene family: environmental and hormonal regulation of expression. The Plant Journal. 1996;9:879–889. doi: 10.1046/j.1365-313x.1996.9060879.x. [DOI] [PubMed] [Google Scholar]
- Xu W, Purugganan MM, Polisensky DH, Antosiewicz DM, Fry SC, Braam J. Arabidopsis TCH4, regulated by hormones and the environment, encodes a xyloglucan endotransglycosylase. The Plant Cell. 1995;7:1555–1567. doi: 10.1105/tpc.7.10.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama R, Nishitani K. Functional diversity of xyloglucan-related proteins and its implications in the cell wall dynamics in plants. Plant Biology. 2000;2:598–604. [Google Scholar]
- Yokoyama R, Nishitani K. A comprehensive expression analysis of all members of a gene family encoding cell-wall enzymes allowed us to predict cis-regulatory regions involved in cell-wall construction in specific organs of Arabidopsis. Plant and Cell Physiology. 2001;42:1025–1033. doi: 10.1093/pcp/pce154. [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Rose JKC, Nishitani K. A surprising diversity and abundance of XTHs (xyloglucan endotransglucosylase/hydrolases) in rice, classification and expression analysis. Plant Physiology. 2004;134:1088–1099. doi: 10.1104/pp.103.035261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.