Abstract
Gametophytic self-incompatibility (GSI) in Solanaceae, Rosaceae, and Plantaginaceae is controlled by a multiallelic S-locus. The specificities of pistil and pollen are controlled by separate S-locus genes, S-RNase and SLF/SFB, respectively. Although the S-specificity is determined by the S-locus genes, factors located outside the S-locus are also required for expression of GSI. HT-B is one of the pistil non-S-factors identified in Nicotiana and Solanum, and encodes a small asparagine/aspartate-rich extracellular protein with unknown biochemical function. Here, HT-B was cloned from Petunia and characterized. The structural features and expression pattern of Petunia HT-B were very similar to those of Nicotiana and Solanum. Unlike other solanaceous species, expression of HT-B was also observed in self-compatible Petunia species. RNA interference (RNAi)-mediated suppression of Petunia HT-B resulted in partial breakdown of GSI. Quantitative analysis of the HT-B mRNA accumulation in the transgenics showed that a 100-fold reduction is not sufficient and a >1000-fold reduction is required to achieve partial breakdown of GSI.
Keywords: HT-B, Petunia inflata, pistil, RNAi, self-incompatibility
Introduction
Gametophytic self-incompatibility (GSI) is a genetic system that enables the pistil to reject pollen from genetically related plants, and thus contributes to promotion of outcrossing. In the families Solanaceae, Rosaceae, and Plantaginaceae, this system is controlled by a single polymorphic S-locus. When the S-haplotype of pollen matches one of the two S-haplotypes of a diploid pistil, the pollen is recognized as self and rejected by the pistil (de Nettancourt, 2001). The S-specificities of pistil and pollen of these families are determined by different S-locus genes, S-RNase and SLF/SFB, respectively (Kao and Tsukamoto, 2004; McClure, 2006; McClure and Franklin-Tong, 2006). Despite its apparent simplicity, the S-locus alone is not sufficient to elicit the S-RNase-based GSI reaction. Genetic analyses have suggested that factors located outside the S-locus are required for expression of the GSI mechanism (Ai et al., 1991; Bernatzky et al., 1995; Hosaka and Hanneman, 1998a, b). Furthermore, expression of high enough levels of S-RNases in transgenic self-compatible (SC) Nicotiana tabacum, N. plumbaginifolia, or cultivated tomato (Solanum lycopersicum) did not confer the S-specific pollen rejection function (Murfett et al., 1996; Kondo et al., 2002b). These findings indicate that non-S-factors are required for GSI to occur. However, very limited numbers of non-S-factors have been cloned and characterized so far.
The stylar 120 kDa glycoprotein (120K) is a non-S-factor identified in Nicotiana. 120K is an extracellular arabinogalactan protein capable of binding to the S-RNase (Cruz-Garcia et al., 2005). RNA interference (RNAi)-mediated suppression of 120K resulted in the breakdown of the capability of the pistil to reject self-pollen, suggesting that it is required for GSI function (Hancock et al., 2005). Another non-S-factor gene cloned to date is HT-B of Nicotiana and Solanum. HT-B is a pistil-expressed, extracellular, small asparagine/aspartate (N/D)-rich protein and was first identified in Nicotiana. Antisense suppression of HT-B caused loss of the pistil part function of GSI, although the transgenic plants retained normal levels of S-RNases (McClure et al., 1999). In wild relatives of tomato and potato, the species of Solanum, HT-B genes have also been isolated and characterized. Kondo et al. (2002b) isolated two HT-like genes, HT-B and HT-A, and found that SC cultivated tomato (S. lycopersicum) has defects in both genes. They further extended the analysis of the HT genes to other SC and self-incompatible (SI) wild relatives of tomato, and suggested that the mutation affecting the expression level of HT-B may have been primarily responsible for the evolution of SC in cultivated tomato and its wild relatives (Kondo et al., 2002a). Functions of HT-A and HT-B were examined experimentally in S. chacoense, a wild potato. Antisense suppression of HT-A did not affect the SI phenotype, while RNAi-mediated silencing of HT-B changed the phenotype, suggesting that HT-B but not HT-A is involved in SI (O'Brien et al., 2002).
Although the biochemical function of HT-B is not known at present, immunolocalization has shown that HT-B protein is taken up by the pollen tube, and then localized in the membrane of vacuoles in which S-RNases, which have been taken up, are stored (Goldraij et al., 2006). Based on the finding that degradation of HT-B protein is more prominent in compatible pollination than in incompatible pollination, it was hypothesized that HT-B is involved in destabilization of vacuoles, and interaction between the S-RNase and its cognate SLF specifically inhibits the degradation of HT-B, which results in breakdown of the vacuole and release of the S-RNase into the cytoplasm of the self pollen (Goldraij et al., 2006; McClure, 2006).
In Petunia, a solanaceous species, HT-B has not been identified yet. An attempt to clone HT-B from Petunia resulted in the isolation of a new class of HT-like gene, HTL (Sassa and Hirano, 2006). HTL shared several characteristics with HT genes of Nicotiana and Solanum, i.e. the deduced amino acid sequence including a signal peptide region and conserved cysteine residues near the C-terminus, and style-specific expression. However, the Petunia HTL protein lacked the N/D-rich domain. Furthermore, suppression by RNAi did not affect the SI phenotype of transgenic Petunia, suggesting that the Petunia HTL is not the orthologue of the non-S-factor HT-B of Nicotiana and Solanum, and is a new member of the HT-like gene family (Sassa and Hirano, 2006). Recently, similar HT-like proteins that lack the N/D-rich domain were also identified in Nicotiana and designated as HT-M. RNAi-mediated suppression of HT-M did not affect the SI phenotype of the transgenic Nicotiana (Kondo and McClure, 2008).
In this study, the isolation of the Petunia HT-B gene which encodes a protein with the N/D-rich domain is described. RNAi experiments with this gene caused partial breakdown of S-specific pollen rejection. Quantitative analysis of the HT-B mRNA accumulation in the transgenics showed that a 100-fold reduction is not sufficient and a >1000-fold reduction is required to achieve partial breakdown of GSI.
Materials and methods
Plant materials
SI P. inflata lines (S3LS3L and Sk1Sk1) were described previously (Sassa and Hirano, 2006). Taking advantage of its high transformation efficiency, SC P. hybrida cv. Mitchell was used for transformation (Ausubel et al., 1980). The SI near-isogenic line ‘Mitchell’ [NIL Mitchell (S3LS3L, HT-BiHT-Bi)] was bred by introduction of S3L and HT-Bi of P. inflata into ‘Mitchell’ by backcrossing using ‘Mitchell’ as a recurrent parent. S genotypes of segregants were analysed by multiplex PCR. DNA was extracted from the plants as described (Sassa, 2007) and used for PCR with a primer pair FSSR1 and RSSR1 for S3L-RNase (∼400 bp, Sassa and Hirano, 2006) and a pair of primers FSm1 (5′-CAGATGTCTACAGTCAATCAG-3′) and RSmPT15 (5′-CGCGGATCCTCACGGTCGAAACATAATCCC-3′) for Sm-RNase of ‘Mitchell’ (∼320 bp, HS and ARP, unpublished data). HT-B genotypes of backcrossed progeny were analysed by PCR with FHTCtrm1 (5′-ACGCTTCAAAAACAAGGAGG-3′) and RHTBPT15 (5′-CGCGGATCCTAACAACACATGGTTTGGC-3′) that generate fragments of 213 bp (inflata allele, HT-Bi=PiHT-B) and 190 bp (Mitchell allele, HT-Bm=PhHT-B). A BC9F1 plant (S3LSm, HT-BiHT-Bm) was bud pollinated with self-pollen, and a BC9F2 plant of S3LS3L, HT-BiHT-Bi was selected [NIL Mitchell (S3LS3L, HT-BiHT-Bi)]. SI of NIL Mitchell (S3LS3L, HT-BiHT-Bi) was confirmed by self-pollination.
Cloning of cDNA and the genomic fragment
Total RNA was extracted from the pistils of P. inflata (S3LS3L), and reverse transcribed with oligo d(T) RACE-N primer to generate first-strand cDNA as described by Ushijima et al. (2003). The cDNA fragment of PiHT-B was first amplified by FHT-3 (5′-RWTGAAYGAYSCAACACTCC-3′) and HT-C1 [5′-TCCTTTATTCAACCAAT(C/T)TCATATTA-3′, Kondo et al. (2002b)], and the product was then used as the template for nested PCR with FHT-4 (5′-STGTKCASSTTGCAMWTGCC-3′) and RHTB1 (5′-CTAACAACAARCGGYTTKAC-3′). 5’ RACE (rapid amplification of cDNA ends) was conducted by using a specific reverse primer RHTb1 (5′-GTCGTTGTTATCATTATCACC-3′) which was designed based on the RT-PCR fragment for PiHT-B. Based on the 5′ RACE clone sequence, the forward primer FPH5E (5′-ATTCACAAACTAAATATCAACAAAC-3′) was designed and used to amplify the full-length cDNA of PiHT-B by 3′ RACE using a high fidelity DNA polymerase Pyrobest (Takara, Ohtsu, Japan). The PCR products were cloned into a pZErO-2 vector (Invitrogen, Carlsbad, CA, USA) and sequenced. The DDBJ/GenBank/EMBL accession number of PiHT-B is AB191255. The HT-B allele of ‘Mitchell’ was also cloned by RT-PCR (PhHT-B, AB468968).
A bacterial artificial chromosome (BAC) library of P. inflata (S3LSk1) was constructed by using pECBAC1 (Amplicon Express, Pullman, WA, USA). The library consisting of 64 436 clones with an average insert size of 129 kb (∼7.2-fold genome coverage) was screened by the HT-B cDNA clone of S. peruvianum (Kondo et al., 2002b) or by PiHT-B as probe under low and high stringency, respectively.
Isolation and gel blot analysis of DNA and RNA
Isolation of nucleic acids, electrophoresis, blotting, and hybridization with digoxigenin (DIG)-labelled probes were performed as described in Sassa and Hirano (2006).
RNA silencing
The construct for RNA silencing was prepared by using pHANNIBAL (Wesley et al., 2001). The HT-B coding region was amplified from cDNA by Pyrobest polymerase with iFPHT1 (5′-GCTCTAGACTCGAGTTAATTCGTCCAAAATATG-3′) and iRPHT1 (5′-GCATCGATGGTACCAAGATAATCATCGCCATTAC-3′), and was cloned into pHANNIBAL in sense and antisense orientation separated by the Pdk intron (Wesley et al., 2001). The resultant HT-B silencing cassette was excised by SacI and SpeI, and inserted into the SacI–XbaI sites of pBINPLUS (van Engelen et al., 1995) to obtain pBINiPiHT-B. The silencing construct was introduced into Agrobacterium tumefaciens LBA4404 to transform ‘Mitchell’ by the leaf disk method (Horsch et al., 1985). For the analysis of the effect of HT-B suppression on the incompatibility phenotype, transgenic ‘Mitchell’ lines were crossed with SI P. inflata (S3LS3L) or NIL Mitchell (S3LS3L, HT-BiHT-Bi) to introduce a functional S3L allele. The heterozygous hybrid transgenics (S3LSm) were pollinated with pollen from incompatible (S3LS3L) or compatible genotypes (Sk1Sk1 or SmSm) to analyse the effect of the transgene on the S-specific pollen rejection.
Quantitative RT-PCR
Total RNA preparations were treated with DNase I twice, and the absence of genomic DNA in the RNA was confirmed by PCR. First-strand cDNA was synthesized from 1 μg of the DNase-treated RNA with ReveTra Ace reverse transcriptase (Toyobo, Osaka, Japan) and random hexamer primer. Quantitative RT-PCR (qRT-PCR) was performed with SYBR Premix Ex Taq II (TaKaRa, Ohtsu, Japan) on a MyiQ Single-Color Real-Time PCR Detection System (Bio-Rad) according to the manufacturer's instruction. Each reaction was performed with 4 μl of a 1:20 (v/v) dilution of the synthesized cDNA with 0.4 μM of each primer in a 20 μl reaction volume. The cycling conditions were as follows: 95 °C for 2 min followed by 45 cycles of denaturation at 95 C for 5 s, annealing at 55 °C for 10 s, and extension at 72 °C for 15 s. The specificity of the PCR amplification was verified by a dissociation curve analysis. Ser/Thr protein phosphatase 2A (PP2A), which was found to be very stably expressed in Arabidopsis (Czechowski et al., 2005) and tomato (K Ushijima et al., unpublished data), was used as an internal control to calculate the efficiency of the cDNA synthesis. The primers used are as follows; PP2A (PhPP2AQtF, 5′-AGCTTGGTGCTCTATGCATGC-3′ and PhPP2AQtR, 5′-TTCCTCAGCAAGGCGCTTAAC-3′), PiHT-B (PiHTBLQtF, 5′-CTAAATATCAACAAACTTCTATAAG-3′ and PiHTBLQtR, 5′-CTTGTTTTTGAAGCGTAG-3′), and PiHTL (PiHTLsQtF, 5′-GGGATCTACAAATATCAACATC-3′ and PiHTLsQtR, 5′-CCTAGCAGCAACCTCTGA-3′). The transcript levels of PiHT-B and PiHTL were normalized to that of PP2A.
Pollination phenotypes
Emasculated flowers were pollinated at anthesis. After 48 h they were harvested and their pistils were fixed in acetic acid:70% ethanol (1:3) for 1 d. Pistils were then treated with 1 N NaOH for 3 h and stained with aniline blue (0.1%) for 24 h. Pollen tube growth was observed by fluorescence microscopy. For fruit set analysis, pollination was performed as described above and fruits were detached from the plants and photographed 2 weeks after pollination. All plants were crossed at least three times with each pollen donor for each type of experiment.
Results
Identification of Petunia HT-B
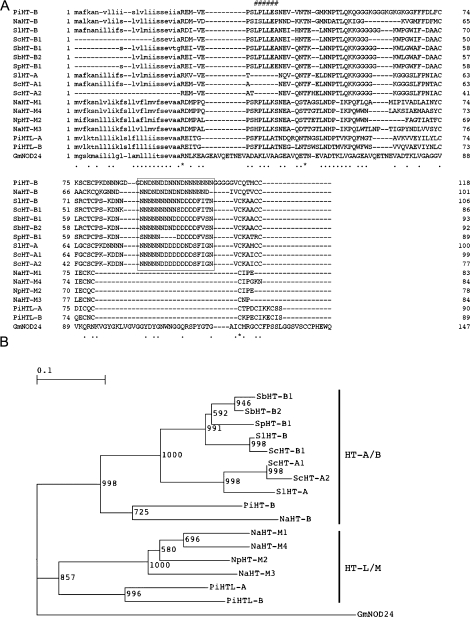
PiHT-B was isolated from pistils of P. inflata by RT-PCR using degenerate primers designed according to HT sequences of Nicotiana and Solanum. Figure 1A shows the deduced amino acid sequence of PiHT-B and other related proteins. PiHT-B encodes a 119 residue amino acid protein with a predicted cleavage site before Arg24 (SignalP, Nielsen et al., 1997). The pI of the mature protein is calculated to be 4.42. PiHT-B, unlike the previously described PiHTL (Sassa and Hirano, 2006) and like other HT-B proteins, contains a C-terminal N/D-rich domain (23 residues) with three cysteine residues at each side. HT-A proteins have a characteristic deletion of 5–7 residues near the N-terminal region of mature proteins (Kondo et al., 2002b; O'Brien et al., 2002). The corresponding region of PiHT-B was much closer to that of solanaceous HT-Bs and Petunia HTL than to HT-As (Fig. 1A). Phylogenetic analysis showed that the HT-like proteins were first classified into two groups, the HT-A/B group and the HT-L/M group (Fig. 1B). PiHT-B was categorized in the HT-A/B group, and was closely related to HT-B of Nicotiana. Identities between PiHT-B and the other solanaceous proteins ranged between 34.9% and 58.0%.
Fig. 1.
Amino acid sequence alignment and phylogenetic tree of HT-like proteins. (A) The amino acid sequences of HT-like proteins were aligned by CLUSTAL W (Thompson et al., 1994). Putative signal peptides are shown in lower case. A chacteristic deletion for HT-A is denoted with #. The N/D-rich domain was boxed. (B) A Neighbor–Joining tree (Saitou and Nei, 1987) of HT-like proteins. NOD24 protein of soybean (GmNOD24; M10595) was chosen as an outgroup for rooting, because it is a member of the recently proposed expanded HT family, the HT/NOD-24 family, and is distantly related to solanaceous HT-like proteins (Kondo and McClure, 2008). Numbers denote bootstrap values with 1000 replicates.
A P. inflata BAC library was screened with the S. peruvianum HT-B probe (Kondo et al., 2002b) and the PiHT-B cDNA under low and high stringency conditions, respectively. Two positive clones, 22C21 and 21I9, were obtained. Southern blotting analysis of these two clones revealed that they exhibited very similar restriction patterns, suggesting that they are overlapping with each other (data not shown). No other positive clones were recovered from the library that is ∼7.2-fold genome coverage, suggesting that there are no HT-B-like sequences in the genome of P. inflata other than PiHT-B. This is consistent with the result of genomic Southern blot analysis (data not shown).
Style-specific expression of PiHT-B
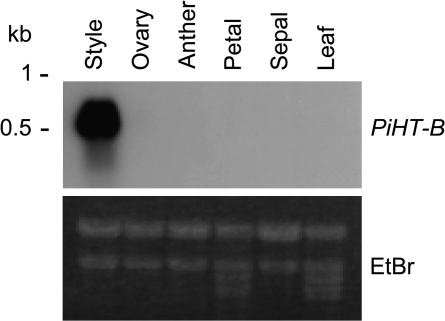
RNA blot analysis was performed with RNA from style-stigma (hereafter style), ovary, anther, sepal, petal, and leaf of P. inflata, and the results are shown in Fig. 2. PiHT-B expression was restricted to the style as is that of other solanaceous HT-B genes.
Fig. 2.
RNA blot analysis of organ-specific expression of PiHT-B in P. inflata. A 10 μg aliquot of total RNA was loaded in each lane, blotted, and hybridized with a PiHT-B probe. The ethidium bromide-stained gel is shown to ascertain equal loading conditions.
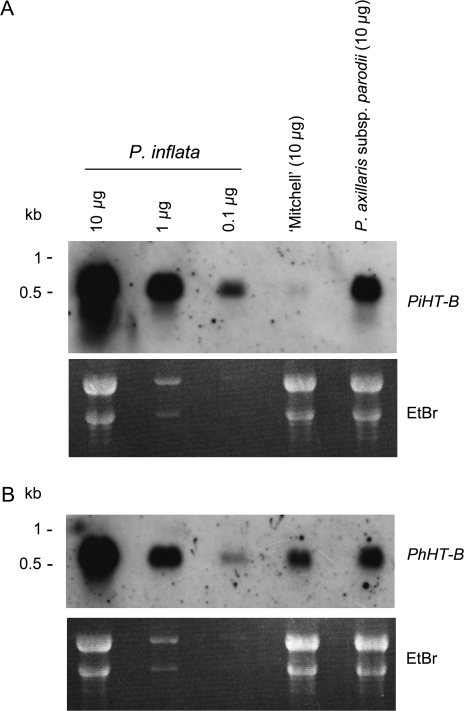
HT-B expression was also compared between P. inflata (SI), P. axillaris subsp. parodii (SC), and ‘Mitchell’ (SC). The HT-B allele of ‘Mitchell’, PhHT-B, was cloned by RT-PCR and also used as a probe. Nucleotide and deduced amino acid identities between the two Petunia genes/proteins were 91.6% and 90.7%, respectively. As shown in Fig. 3, P. inflata presented a very strong signal when the blots were hybridized with either PiHT-B or PhHT-B. In contrast, ‘Mitchell’ showed almost no signal when the blot was hybridized with PiHT-B and a moderate one when hybridized with PhHT-B. Petunia axillaris subsp. parodii had a moderate signal with both probes, although a slightly stronger intensity is observed with PiHT-B.
Fig. 3.
RNA blot analysis of PiHT-B and PhHT-B in styles of P. inflata, ‘Mitchell’ and P. axillaris subsp. parodii. A 10 μg aliquot of total RNA was loaded in each lane, blotted, and hybridized with P. inflata (A, PiHT-B) and ‘Mitchell’ HT-B (B, PhHT-B) probes. Aliquots of 1 μg and 0.1 μg of P. inflata RNA were also loaded in separate lanes to allow for comparison of the level of expression. The ethidium bromide-stained gels are shown to ascertain equal loading conditions.
RNAi-mediated suppression of PiHT-B
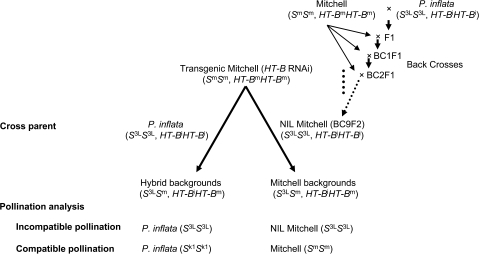
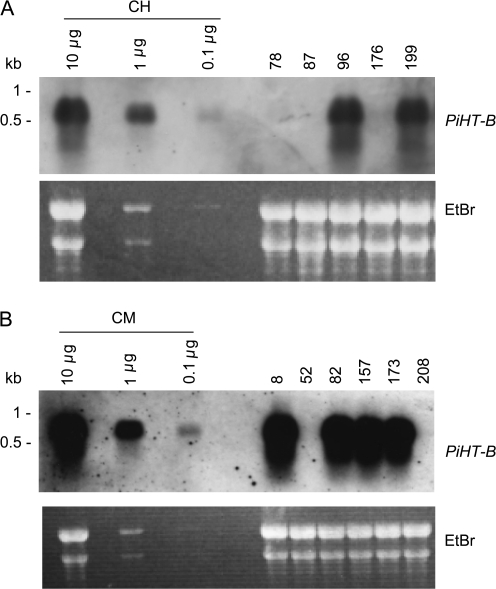
Taking advantage of its high transformation efficiency, SC line ‘Mitchell’ was used to introduce the RNAi construct for PiHT-B. The transgenics were crossed with SI lines P. inflata (S3LS3L) or NIL Mitchell (S3LS3L, HT-BiHT-Bi) to produce ‘hybrid background’ and ‘Mitchell background’ transgenics, respectively (Fig. 4). Independent ‘Mitchell’ transgenics were used to produce ‘hybrid backgrounds’ and ‘Mitchell backgrounds’. The two types of heterozygous transgenics (S3LSm, HT-BiHT-Bm), ‘hybrid backgrounds’ and ‘Mitchell backgrounds’, were used to analyse the effect of PiHT-B suppression on the S-specific pollen rejection function. Similarly, an HTL-suppressed P. inflata plant (Sassa and Hirano, 2006) was crossed with ‘Mitchell’ HT-B RNAi transgenic plants to produce double transformants. Transgenics were analysed by Southern blotting to confirm the presence of transgenes (data not shown). Five independent ‘Mitchell’×P. inflata (S3LS3L) transformants (hybrid backgrounds; S3LSm, HT-BiHT-Bm) were subjected to RNA blot analysis (Fig. 5A). Suppression levels varied from almost no suppression to approximately a >100-fold reduction in comparison with the untransformed control. Likewise, six independent ‘Mitchell’×NIL Mitchell (S3LS3L, HT-BiHT-Bi) transgenics (Mitchell backgrounds; S3LSm, HT-BiHT-Bm) showed different levels of suppression (Fig. 5B). Those S-heterozygous transgenics were subjected to pollination with S-homozygous lines for phenotype analyses.
Fig. 4.
Transgenic plant experiments testing the role of PiHT-B in the S-specific pollen rejection. The RNAi construct for PiHT-B was first introduced into the SC line ‘Mitchell’. The transgenics were crossed with SI lines P. inflata (S3LS3L, HT-BiHT-Bi) or NIL Mitchell (S3LS3L, HT-BiHT-Bi) to produce ‘hybrid background’ or ‘Mitchell background’ transgenics, respectively, and used for analysis of the S-specific pollen rejection.
Fig. 5.
RNA blot analysis of PiHT-B in styles of RNAi transgenic plants. (A) Hybrid background transgenic lines 78, 87, 96, 176, and 199 [transgenic ‘Mitchell’×P. inflata (S3LS3L)]. (B) Mitchell background transgenic lines 8, 52, 157, 173, and 208 [transgenic ‘Mitchell’×NIL Mitchell (S3LS3L, HT-BiHT-Bi)]. A 10 μg aliquot of pistil total RNA was loaded in each lane. Aliquots of 1 μg and 0.1 μg of RNA of the untransformed plants were also loaded in separate lanes to assess the level of suppression. CH and CM are untransformed controls of hybrid and Mitchell backgrounds, respectively. The ethidium bromide-stained gel is shown to ascertain equal loading conditions.
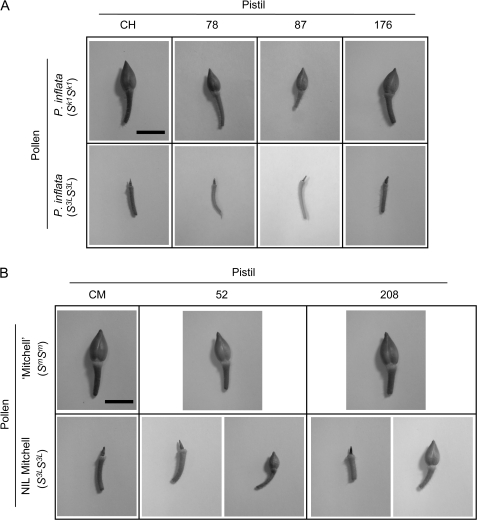
Fruit set analysis revealed that all hybrid background transformants (including the double transformant) presented the same phenotype as that of the control plant regardless of the level of suppression (Table 1). The use of compatible pollen from P. inflata (Sk1Sk1) resulted in the development of large fruits (Fig. 6A). Conversely, the use of incompatible pollen from P. inflata (S3LS3L) led to no fruit formation.
Table 1.
Fruit sets of RNAi transgenics for HT-B
| Pistil | Pollen | |
| Hybrid background (S3LSm) | P. inflata (Sk1Sk1) | P. inflata (S3LS3L) |
| CH | 3/3 | 0/3 |
| 78 | 3/3 | 0/3 |
| 87 | 3/3 | 0/3 |
| 96 | 3/3 | 0/3 |
| 176 | 3/3 | 0/3 |
| 199 | 3/3 | 0/3 |
| 94 (DT) | 3/3 | 0/3 |
| Mitchell background (S3LSm) | ‘Mitchell’ (SmSm) | NIL Mitchell (S3LS3L) |
| CM | 3/3 | 0/3 |
| 8 | 3/3 | 0/3 |
| 52 | 3/3 | 4/11* |
| 82 | 3/3 | 0/3 |
| 157 | 3/3 | 0/3 |
| 173 | 3/3 | 0/3 |
| 208 | 3/3 | 4/17* |
Hybrid background transgenics [transgenic ‘Mitchell’×P. inflata (S3LS3L)] were pollinated with pollen from P. inflata Sk1Sk1 or P. inflata S3LS3L, while Mitchell background transgenics [transgenic ‘Mitchell’×NIL Mitchell (S3LS3L, HT-BiHT-Bi)] were pollinated with ‘Mitchell’ or NIL Mitchell S3LS3L. Data are presented as the number of fruits over pollinations. CH and CM are untransformed controls of hybrid background and Mitchell background, respectively. DT denotes a double transformant.
Fruit sizes were smaller than those obtained by compatible pollination. See text and Fig. 6B.
Fig. 6.
Fruit development in RNAi lines for HT-B. (A) Hybrid background transgenics [transgenic ‘Mitchell’×P. inflata (S3LS3L)]. (B) Mitchell background transgenics [transgenic ‘Mitchell’×NIL Mitchell (S3LS3L, HT-BiHT-Bi)]. Pistils were pollinated, detached from the plants after 2 weeks, and photographed. CH and CM are untransformed controls of hybrid and Mitchell backgrounds, respectively. For plants 52 and 208, two types of responses after incompatible pollination [NIL Mitchell (S3LS3L)] are presented; aborted (left) and developed fruits (right). Bar=1 cm.
The Mitchell background transgenic plants behaved in a similar manner except for those which presented a severe HT-B suppression, lines 52 and 208. These plants produced fruits upon pollination with S3L pollen in some of the crosses (Table 1). However, as shown in Fig. 6B, these fruits were smaller than those produced in crosses between control Mitchell background (CM; S3LSm, HT-BiHT-Bm) and ‘Mitchell’. The average fruit length (±SD) of the CMבMitchell’ crosses was 1.2±0.06 cm, while for plants 52 and 208, when crossed with NIL Mitchell (S3LS3L), the average fruit length was 0.6±0.05 cm and 0.8±0.14 cm, respectively.
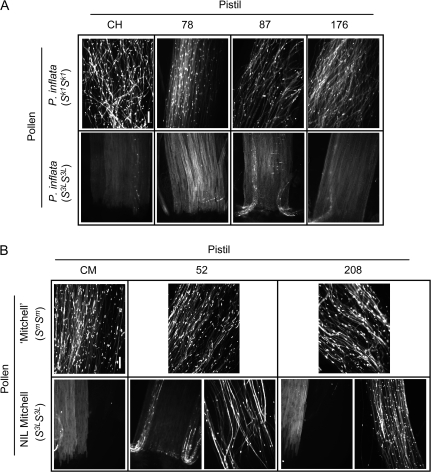
Pollen tube observation confirmed the phenotypes assessed by fruit set (Table 2; Fig 7). All hybrid background transgenics, including the double transformant, presented a characteristic incompatible phenotype when pollinated with S3L pollen, i.e. strong deposition of callose in the pollen tube walls, uneven distribution of callose plugs, irregular direction and a marked decrease in the number of pollen tubes in the middle of the style, with none of them reaching the lower style. Conversely, crosses using Sk1 pollen produced the typical compatible phenotype, in which an overwhelming number of pollen tubes grow harmoniously, with callose plugs regularly spaced, through the style to be visible in great quantities towards the ovary. Mitchell background transgenic plants showed the same phenotype when crossed with ‘Mitchell’ pollen (Table 2; Fig. 7B). Rejection of NIL Mitchell (S3LS3L, HT-BiHT-Bi) pollen occurred, as expected, with the control plant (CM) and with transformants which were slightly or not HT-B suppressed (8, 82, 157, and 173). However, severely suppressed plants (52 and 208) showed a partial SC response upon pollination with S3L pollen. In some crosses, the typical incompatible rejection phenotype was observed. However, other crosses showed partially or fully compatible phenotypes (Fig. 7B).
Table 2.
Pollen tube growth in pistils of RNAi transgenics for HT-B
| Pistil | Pollen | |
| Hybrid background (S3LSm) | P. inflata (Sk1Sk1) | P. inflata (S3LS3L) |
| CH | 4/4 | 0/4 |
| 78 | 4/4 | 0/4 |
| 87 | 3/3 | 0/3 |
| 96 | 3/3 | 0/3 |
| 176 | 3/3 | 0/15 |
| 199 | 3/3 | 0/3 |
| 94 (DT) | 3/3 | 0/8 |
| Mitchell background (S3LSm) | ‘Mitchell’ (SmSm) | NIL Mitchell (S3LS3L) |
| CM | 3/3 | 0/5 |
| 8 | 3/3 | 0/3 |
| 52 | 5/5 | 4/8 |
| 82 | 3/3 | 0/3 |
| 157 | 3/3 | 0/3 |
| 173 | 3/3 | 0/3 |
| 208 | 3/3 | 4/5 |
Data are presented as the number of compatible pollinations over the total pollinations attempted. Crosses were considered as compatible when one or more pollen tubes were observed at the lower part of the style. CH and CM are untransformed controls of hybrid background and Mitchell background, respectively. DT denotes a double transformant.
Fig. 7.
Pollen tube phenotypes in RNAi lines for HT-B. (A) Hybrid background transgenics [transgenic ‘Mitchell’×P. inflata (S3LS3L)]. (B) Mitchell background transgenics [transgenic ‘Mitchell’×NIL Mitchell (S3LS3L, HT-BiHT-Bi)]. CH and CM are untransformed controls of hybrid and Mitchell backgrounds, respectively. For plants 52 and 208, two types of phenotypes in incompatible pollination [NIL Mitchell (S3LS3L)] are presented; incompatibility (left) and partial compatibility (right). Styles were collected 48 h after pollination, stained with aniline blue, and observed under an epifluorescence microscope. All photographs correspond to the lower part of the style. Bar=100 μm.
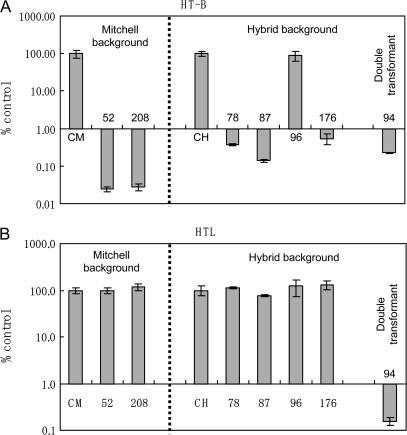
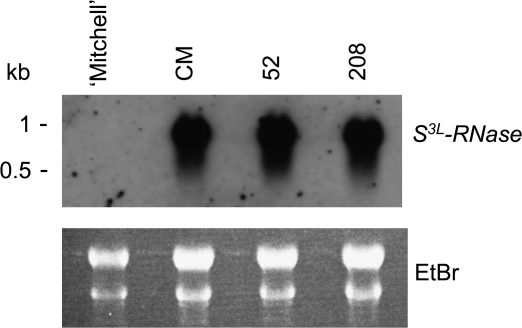
Real-time PCR was performed in selected plants to quantitate the level of reduction of HT-B gene expression. As shown in Fig. 8A, PiHT-B expression in the partial SC ‘Mitchell’ background plants 52 and 208 was reduced by >1000 times in comparison with the control (CM). Suppression in hybrid background plants 78, 87, and 176 was >100 times. The double transformant (plant 94) presented a suppression of >100 times for both genes (Fig. 8A, B). Unaffected expression of S3L-RNase in the partial SC plants 52 and 208 was confirmed through RNA blot analysis including an untransformed plant (CM) and ‘Mitchell’ as positive and negative controls, respectively (Fig. 9).
Fig. 8.
Quantitative analysis of the transcripts of HT-B and HTL in RNAi transgenic plants. Levels of transcripts for HT-B (A) and HTL (B) were determined by quantitative real-time PCR. Values from three independent experiments are expressed as a percentage of controls (±SD). CH and CM are untransformed controls of hybrid and Mitchell backgrounds, respectively.
Fig. 9.
RNA blot analysis of S3L-RNase in styles of RNAi lines for HT-B. Severely HT-B-suppressed Mitchell background plants 52 and 208 were analysed. ‘Mitchell’ and untransformed Mitchell background (CM) are negative and positive controls, respectively. A 10 μg aliquot of style total RNA was loaded in each lane, blotted, and hybridized with the P. inflata S3L-RNase probe. The ethidium bromide-stained gel is shown to ascertain equal loading conditions.
Discussion
A previous attempt to clone HT-B of Petunia resulted in isolation of PiHTL, which was similar to HT-B but lacked the N/D-rich domain at the C-terminus, representing a new class of HT-like gene. The finding that silencing of PiHTL did not affect GSI suggested that Petunia has other genuine HT-B genes (Sassa and Hirano, 2006). In this study, a new HT-like gene, PiHT-B, was isolated from Petunia and characterized. PiHT-B has characteristics which are typical of HT-B genes, i.e. structures including an N/D-rich domain and style-specific expression. Furthermore, silencing of PiHT-B changed the GSI phenotype, indicating that it is the Petunia orthologue of HT-B. In addition to HT-B, another isoform, HT-A, has been identified in Solanum species (Kondo et al., 2002a, b; O'Brien et al., 2002), while HT-A is not known in Nicotiana (McClure et al., 1999). In Petunia, only HT-B was isolated, and HT-A-like sequences were not identified by the BAC library screening. This may reflect phylogenetic relationships between those species; subfamily Petunioideae, that includes Petunia, is more closely related to Nicotianoideae (Nicotiana) than it is to Solanoideae (Solanum) (Olmstead et al., 1999).
HT-B is the first non-S-factor characterized at the molecular level, and was discovered in a differential screen as being highly expressed in N. alata style but not in the SC species N. plumbaginifolia (McClure et al., 1999). Kondo et al. (2002a, b) isolated HT-B genes from SI and SC relatives of tomato, and showed that high HT-B expression levels were exclusively observed in SI species. Based on the HT data, it was hypothesized that a mutation causing reduced transcription of HT-B in an ancestral species was central to the loss of GSI in tomato relatives (Kondo et al., 2002a). In Petunia, in contrast, HT-B expression was also observed in SC lines, P. axillaris subsp. parodii and P. hybrida ‘Mitchell’, though the levels were lower than that in SI P. inflata. Considering that a >1000-fold reduction of HT-B transcript is required to achieve partial breakdown of GSI, the low but detectable levels of HT-B transcript are unlikely to be related to SC of those Petunia lines. This may suggest that different courses are possible for the evolution of SC in different taxa of Solanaceae.
RNAi-mediated silencing was conducted to analyse the function of PiHT-B. Among the transgenic lines with different levels of suppression of the PiHT-B gene, the most severely affected plants only showed partial breakdown of S-specific pollen rejection function. qPCR analysis showed that the accumulation of PiHT-B mRNA in the partial SC transgenics was reduced by >1000-fold. RNAi lines with >100-fold reduction of PiHT-B showed no change of GSI phenotype, suggesting that a threshold level of PiHT-B expression is required for GSI, and the threshold is very low at <1 % of the wild-type level. In the functional analysis of HT-B of S. chacoense, RNAi-mediated silencing also achieved partial breakdown of GSI, although the level of suppression was not quantitatively assessed (O'Brien et al., 2002). The requirement for threshold level expression is also known for another highly abundant pistil GSI factor, the S-RNase. In contrast to HT-B, the threshold level of the S-RNase is high, and strong expression is required to confer S-specific pollen rejection function to the pistil (Lee et al., 1994; Murfett et al., 1996; Qin et al., 2006). The different threshold levels of these pistil factors may reflect the difference in biochemical function between them. Although the biochemical function of HT-B has not been clarified yet, based on the findings of the probable association of HT-B with the vacuole membrane of pollen tubes and its differential stability between compatible and incompatible pollination, it is hypothesized that HT-B is involved in destabilization of the vacuole of the pollen tube (Goldraij et al., 2006; McClure, 2006). A small amount of HT-B protein may be sufficient for its function on the vacuole membrane. Another possibility is that HT-B plays an indirect role that influences the strength of GSI expression. Analysis of deletion-type or insertional disruption- type mutations of HT-B would clarify if complete abolishment of the protein results in complete breakdown of GSI, and if the small amount of HT-B protein is essential. Identification of interaction partner(s) of HT-B protein would also be necessary to understand the biochemical role of the protein and the mechanism of the S-RNase-based GSI system.
Acknowledgments
We thank Professor Kowyama of Mie University for providing us with the cDNA clone for HT-B of S. peruvianum. We also thank S Kikuchi, H Kakui, and D Heang for comments. This work was supported in part by the Grants-in-Aid for Scientific Research (B, 20380003) from the Ministry of Education, Science, Sports and Culture of Japan to HS.
References
- Ai Y, Kron E, Kao T-H. S-alleles are retained and expressed in a self-compatible cultivar of Petunia hybrida. Molecular and General Genetics. 1991;230:353–358. doi: 10.1007/BF00280291. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Bahnsen K, Hanson M, Mitchell A, Smith HJ. Cell and tissue culture of haploid and diploid Petunia ‘Mitchell’. Plant Molecular Biology Newsletter. 1980;1:26–31. [Google Scholar]
- Bernatzky R, Glaven RH, Rivers BA. S-related protein can be recombined with self-compatibility in interspecific derivatives of Lycopersicon (tomato) Biochemical Genetics. 1995;33:215–225. [PubMed] [Google Scholar]
- Cruz-Garcia F, Hancock N, Kim D, McClure B. Stylar glycoproteins bind to S-RNase in vitro. The Plant Journal. 2005;42:295–304. doi: 10.1111/j.1365-313X.2005.02375.x. [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nettancourt D. Incompatibility and incongruity in wild and cultivated plants. Berlin: Springer; 2001. [Google Scholar]
- Goldraij A, Kondo K, Lee CB, et al. Compartmentalization of S-RNase and HT-B degradation in self-incompatible Nicotiana. Nature. 2006;439:805–810. doi: 10.1038/nature04491. [DOI] [PubMed] [Google Scholar]
- Hancock CN, Kent L, McClure B. The 120 kDa glycoprotein is required for S-specific pollen rejection in Nicotiana. The Plant Journal. 2005;43:716–723. doi: 10.1111/j.1365-313X.2005.02490.x. [DOI] [PubMed] [Google Scholar]
- Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rgers SG, Fraley RT. A simple and general method for transferring genes into plants. Science. 1985;227:1229–1231. doi: 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
- Hosaka K, Hanneman RE. Genetics of self-compatibility in a self-incompatible wild diploid potato species Solanum chacoense. 1. Detection of an S locus inhibitor (Sli) gene. Euphytica. 1998a;99:191–197. [Google Scholar]
- Hosaka K, Hanneman RE. Genetics of self-compatibility in a self-incompatible wild diploid potato species Solanum chacoense. 2. Localization of an S locus inhibitor (Sli) gene on the potato genome using DNA markers. Euphytica. 1998b;103:265–271. [Google Scholar]
- Kao T, Tsukamoto T. The molecular and genetic bases of S-RNase-based self-incompatibility. The Plant Cell. 2004;16:S72–S83. doi: 10.1105/tpc.016154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo K, McClure B. New microsome-associated HT-family proteins from Nicotiana respond to pollination and define an HT/NOD-24 protein family. Molecular Plant. 2008;1:634–644. doi: 10.1093/mp/ssn018. [DOI] [PubMed] [Google Scholar]
- Kondo K, Yamamoto M, Itahashi R, Sato T, Egashira H, Hattori T, Kowyama Y. Insights into the evolution of self-compatibility in Lycopersicon from a study of stylar factors. The Plant Journal. 2002a;30:143–153. doi: 10.1046/j.1365-313x.2002.01275.x. [DOI] [PubMed] [Google Scholar]
- Kondo K, Yamamoto M, Matton DP, Sato T, Hirai M, Norioka S, Hattori T, Kowyama Y. Cultivated tomato has defects in both S-RNase and HT genes required for stylar function of self-incompatibility. The Plant Journal. 2002b;29:627–636. doi: 10.1046/j.0960-7412.2001.01245.x. [DOI] [PubMed] [Google Scholar]
- Lee H-S, Huang S, Kao T-H. S proteins control rejection of incompatible pollen in Petunia inflata. Nature. 1994;367:560–563. doi: 10.1038/367560a0. [DOI] [PubMed] [Google Scholar]
- McClure B. New views of S-RNase-based self-incompatibility. Current Opinion in Plant Biology. 2006;9:639–646. doi: 10.1016/j.pbi.2006.09.004. [DOI] [PubMed] [Google Scholar]
- McClure B, Franklin-Tong V. Gametophytic self-incompatibility: understanding the cellular mechanisms involved in ‘self’ pollen tube inhibition. Planta. 2006;224:233–245. doi: 10.1007/s00425-006-0284-2. [DOI] [PubMed] [Google Scholar]
- McClure B, Mou B, Canevascini S, Bernatzky R. A small asparagine-rich protein required for S-allele-specific pollen rejection in Nicotiana. Proceedings of the National Academy of Sciences, USA. 1999;96:13548–13553. doi: 10.1073/pnas.96.23.13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murfett JM, Strabala TJ, Zurek DM, Mou B, Beecher B, McClure BA. S RNase and interspecific pollen rejection in the genus Nicotiana: multiple pollen rejection pathways contribute to unilateral incompatibility between self-incompatible and self-compatible species. The Plant Cell. 1996;8:943–958. doi: 10.1105/tpc.8.6.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Engineering. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- O'Brien M, Kapfer C, Major G, Laurin M, Bertrand C, Kondo K, Kowyama Y, Matton DP. Molecular analysis of the stylar-expressed Solanum chacoense small asparagine-rich protein family related to the HT modifier of gametophytic self-incompatibility in Nicotiana. The Plant Journal. 2002;32:985–996. doi: 10.1046/j.1365-313x.2002.01486.x. [DOI] [PubMed] [Google Scholar]
- Olmstead RG, Sweere JA, Spangler RE, Bohs L, Palmer JD. Phylogenetic and provisional classification of the Solanaceae based on chloroplast DNA. In: Nee M, Symon DE, Lester PN, Jessop JP, editors. Solanaceae IV. Kew: Royal Botanic Gardens; 1999. pp. 111–137. [Google Scholar]
- Qin X, Bolin L, Soulard J, Morse D, Cappadocia M. Style-by-style analysis of two sporadic self-compatible Solanum chacoense lines supports a primary role for S-RNases in determining pollen rejection thresholds. Journal of Experimental Botany. 2006;9:2001–2013. doi: 10.1093/jxb/erj147. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor–joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sassa H. A technique to isolate DNA from woody and herbaceous plants by using a silica-based plasmid extraction column. Analytical Biochemistry. 2007;363:166–167. doi: 10.1016/j.ab.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Sassa H, Hirano H. Identification of a new class of pistil-specific proteins of Petunia inflata that is structurally similar to, but functionally distinct form, the self-incompatibility factor HT. Molecular Genetics and Genomics. 2006;275:97–104. doi: 10.1007/s00438-005-0067-7. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushijima K, Sassa H, Dandekar AM, Gradziel TM, Tao R, Hirano H. Structural and transcriptional analysis of the self-incompatibility locus of almond: identification of a pollen-expressed F-box gene with haplotype-specific polymorphism. The Plant Cell. 2003;15:771–781. doi: 10.1105/tpc.009290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Engelen FA, Molthoff JW, Conner AJ, Nap J-P, Pereira A, Stiekema WJ. pBINPLUS: an improved plant transformation vector based on pBIN19. Transgenic Research. 1995;4:288–290. doi: 10.1007/BF01969123. [DOI] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, et al. Construct design for efficient, effective and high-throughput gene silencing in plants. The Plant Journal. 2001;27:581–590. doi: 10.1046/j.1365-313x.2001.01105.x. [DOI] [PubMed] [Google Scholar]