Abstract
Spinocerebellar ataxia type 1 (SCA1) is one of nine inherited neurodegenerative diseases caused by the expansion of a CAG trinucleotide repeat encoding a polyglutamine tract. SCA1 patients lose motor coordination and develop slurred speech, spasticity, and cognitive impairments. Difficulty with coordinating swallowing and breathing eventually causes death. Genetic evidence indicates that the disease mutation induces a toxic gain of function in the SCA1 encoded protein ATXN1. The discovery that residues in ATXN1 outside of the polyglutamine tract are crucial for pathogenesis hinted that alterations in the normal function of this protein are linked to its toxicity. Biochemical and genetic studies provide evidence that the polyglutamine expansion enhances interactions that are normally regulated by phosphorylation at Ser776 and a subsequent alteration in its interaction with other cellular proteins. Moreover, the finding that other ATXN1 interactions are decreased in disease suggests that the polyglutamine expansion contributes to disease by both a gain-of-function mechanism and partial loss of function.
Background
As mentioned by all of the contributions to this minireview series, a unique aspect of the human genome is that it contains many short polymorphic repeat elements that are susceptible to expansions causing a series of inherited disorders (1). One class of such disorders is the so-called polyglutamine neurodegenerative diseases, in which the unstable CAG trinucleotide repeat is located within the coding region of the mutant gene. Expansion of this repeat is thought to lead to a polyglutamine-induced gain-of-function mutation in the protein. Presently, there are nine disorders caused by a CAG repeat expansion. These are Huntington disease, spinal bulbar muscular atrophy (Kennedy disease), dentatorubropallidoluysian atrophy, and six dominantly inherited spinocerebellar ataxias. In this review, we focus on molecular mechanistic insights gained from studies of SCA13 and highlight some points that might prove relevant to other polyglutamine disorders. Attention is directed at addressing three general issues common to all of the polyglutamine disorders as well as many of the other neurodegenerative diseases. These are as follow: what underlies the observation that with increasing age comes an increase in risk for disease, what is the molecular basis for the cell specificity of pathology, and how does expansion of the glutamine tract corrupt the affected “host” protein?
SCA1: The Disease
SCA1 is typically a late-onset fatal autosomal dominant neurodegenerative disease that, like all ataxias, is characterized by loss of motor coordination and balance. The clinical features of SCA1 also include slurred speech, swallowing difficulty, spasticity, and cognitive impairments. Most SCAs are characterized by onset of cerebellar atrophy, which is then followed by degeneration of other neural structures. A characteristic feature of SCA1 pathology is the atrophy and loss of Purkinje cells from the cerebellar cortex. Purkinje cells are the major integrative neuron of the cerebellar cortex. Their axons form the sole efferent pathway from the cerebellar cortex, projecting onto the deep cerebellar nuclei (2). As SCA1 progresses, pathology is noted in other regions of the brain, including the deep cerebellar nuclei, especially the dentate nucleus, the inferior olive, the pons, and the red nuclei. Cranial nerve nuclei III, X, and XII can also show signs of pathology.
The SCA1 gene was cloned in 1993 and found to encode a novel protein designated ATXN1 (ataxin-1) (3). Normal SCA1 alleles contain from 6 to 42 CAG repeats, with those greater than 21 being interrupted with one to three CAT trinucleotides. On the other hand, disease alleles are pure CAG tracts ranging from 39 to 82 units (4). Individuals carrying a mutant SCA1 allele can have symptoms starting as early as the first decade. By the sixth decade, disease penetrance is essentially complete. Analysis of mutant SCA1 alleles provides some insight into the basis of the variability of age of onset. The length of the repeat tract is a major contributor to the age of disease onset. Individuals with 70 or more repeat units have a juvenile form of SCA1, whereas those containing mutant alleles with 40–50 repeats have onset in the fourth or fifth decade of life. Thus, the longer the repeat length on the mutant allele, the earlier is the age of onset. The mutant alleles also show germ line instability such that in successive generations. The repeat can expand further, causing earlier onset of symptoms and increasing the severity of the disease in successive generations, a phenomenon known as anticipation.
A pathological hallmark of SCA1 as well as most of the other polyglutamine disorders is the presence of the large inclusions containing the mutant polyglutamine protein. In the case of SCA1, the inclusions are in the nuclei of several different types of neurons. Besides containing mutant ATXN1, the inclusions are positive for ubiquitin and components of the proteasome and chaperone systems (5). From the moment of their identification, the inclusions were viewed as having a central role in polyglutamine-mediated pathogenesis (6). Subsequent studies in SCA1 pathogenesis questioned whether the large inclusions of mutant ATXN1 are themselves pathogenic. For example, transgenic mice expressing a form of mutant ATXN1 that fails to form large inclusions still show signs of disease (7); SCA1–82Q transgenic mice crossed with mice lacking the E3 ligase UBE3A show no inclusions and yet have more pronounced pathology than those with normal levels of UBE3A and inclusions (8). Finally, mice in which an expanded stretch of CAG repeats encoding 154 glutamines was inserted into the endogenous mouse SCA1 gene in place of the typical two CAG units at this locus develop inclusions in many neuronal types (9). Curiously, however, the Purkinje cells of these knock-in mice are the last type of neuron to form inclusions usually after 30 weeks, yet they show the most severe signs of pathology starting by 10 weeks. In contrast, neurons in the cerebral cortex form inclusions by 6 weeks on and show no signs of pathology. Overall, these studies provide strong genetic evidence that disassociates mutant ATXN1-induced pathology from the formation and presence of inclusions. Thus, although the inclusions signal the fact that the mutant protein is different and somehow must resist clearance and/or mount a cellular response that results in its deposition in inclusions, the genetic data suggest that the protein is far more toxic when it is not sequestered in these inclusions. Interestingly, several subsequent studies have confirmed these observations in SCA7 knock-in models (10), in cellular models of toxic huntingtin fragments (11), and most recently in amyotrophic lateral sclerosis (12).
ATXN1 Protein: Function and Relationship to Disease Pathogenesis
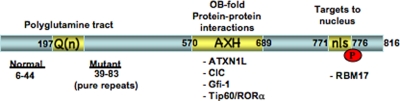
Genetic evidence strongly indicates that SCA1 pathogenesis involves a gain-of-function mechanism, i.e. the phenotype requires the expression of mutant ATXN1 with an expanded repeat tract (13), whereas loss of function of ATXN1 does not cause an SCA1-like phenotype in mice (14). Data revealing that the toxic effects of the disease causing polyglutamine expansion are determined by the context of other domains in the ATXN1 protein suggested that the endogenous normal function and regulation of ATXN1 interactions are critical for pathogenesis. The in vivo studies revealed that in addition to the polyglutamine tract, located toward the N terminus of ATXN1, at least three other regions are important for its function (Fig. 1). These elements include an evolutionarily conserved AXH domain, a nuclear localization sequence, and Ser776 (which undergoes phosphorylation).
FIGURE 1.
Structural elements in ATXN1 that are critical for its function and pathogenesis of SCA1. The functional elements are indicated as yellow boxes. The sequence shown encompasses a full-length form of ATXN1 with 816 amino acids corresponding to a wild-type allele with 30 repeat units (residues 197–226). Wild-type alleles contain from 6 to 44 CAG repeats at this site, whereas mutant pathogenic alleles have a size range spanning 39 to 83 perfect CAG repeats. The AXH domain (residues 570–689) forms an oligonucleotide-binding fold and is important for dimerization of ATXN1 as well as its ability to interact with several proteins. The NLS (nls) spans amino acids 771–776 and targets ATXN1 to the nucleus. Finally, the C-terminal residue of the NLS, Ser776, is a site of phosphorylation in ATXN1. Phosphorylation of Ser776, as well as expansion of the polyglutamine tract, increases the interaction of ATXN1 with RBM17.
The AXH domain is a 120-amino acid stretch that is highly homologous to a portion of HBP1 (high mobility group box transcription factor-binding protein 1) (15, 16). The AXH domain folds independently into an oligonucleotide-binding fold, a structural motif found in oligonucleotide-binding proteins, and is able to bind RNA in a manner similar to that reported for full-length ATXN1 (17). In addition, the AXH domain of ATXN1 acts as dimerization domain and has a cluster of charged surface residues suggested to form a second binding surface (16). In fact, a number of proteins that bind to ATXN1 via interactions with its AXH domain were identified (18–21). Among these interacting proteins is the ATXN1 paralog BOAT (brother of ATXN1; ATXN1L), which, like ATXN1, contains an AXH domain (18). In addition, several transcriptional regulators interact with ATXN1 through the AXH domain. Such transcriptional regulators include the corepressor SMRT (silencing mediator of retinoid and thyroid hormone receptors) (18), Drosophila SENS (Senseless) and its mammalian homolog Gfi1 (growth factor-independent 1) (19), the human homolog of the Drosophila repressor CIC (Capicua) (20), and the RORα-Tip60 complex (21). In the case of SENS/Gfi1 and RORα-Tip60, mutant ATXN1 seems to enhance the degradation of these factors, whereas in the case of CIC, ATXN1 seems to be a stabilizing factor, given that loss of ATXN1 leads to a decrease in the steady-state level of CIC. The finding that ATXN1 can be SUMOylated at at least five residues, including sites in its AXH domain, is consistent with the idea that ATXN1 functions in regulating transcription (22) because SUMOylation is a common post-translational modification of nuclear proteins with a role in transcription (23). Interestingly, the SUMOylation of ATXN1 is negatively affected by the polyglutamine tract length and is dependent on ATXN1 being transported to the nucleus.
The importance of the nuclear localization of ATXN1 for toxicity is supported by a genetic study. ATXN1 contains a functional NLS toward its C terminus (7). Inactivation of the NLS by a single amino acid substitution, replacing Lys at position 776 with Thr, disrupting entry into the nucleus, results in an ATXN1 mutant that is no longer pathogenic despite having a long polyglutamine stretch. This result indicates that the ability of mutant ATXN1 to cause disease might be linked to a function of the protein in the nucleus (7). Moreover, this finding supports the conclusion that the toxicity of the expanded SCA1 allele is mediated at the protein rather than RNA level.
Consistent with the theme that residues outside of the polyglutamine stretch may influence the function and pathogenicity of ATXN1, genetic studies provided evidence that phosphorylation of Ser776 is necessary for toxicity of glutamine-expanded ATXN1. Ser776 is a site that is phosphorylated in both wild-type and glutamine-expanded ATXN1. The in vivo studies revealed that substituting Ser776 with Ala, a residue that is not phosphorylated, abrogates the toxicity of polyglutamine-expanded ATXN1 (24). This study again confirmed that pathogenicity is mediated by the protein and not by the RNA, but notably shifted the focus from the polyglutamine tract to other domains in the proteins for toxicity. In other words, the idea that toxicity is simply a result of an expanded toxic polyglutamine tract that might escape the cellular degradation and quality control machinery becomes less likely. Given the evidence that phosphorylation of ATXN1 at Ser776 is required along with polyglutamine expansion for pathogenesis, then one might predict that there are proteins whose interaction with ATXN1 is regulated by both the length of the polyglutamine tract and phosphorylation of Ser776. Phosphorylation of Ser776 does create a binding site for 14-3-3 proteins (25), but this interaction retards the degradation of the mutant protein more so than wild-type ATXN1, although it is not affected by the length of the polyglutamine tract and is not enhanced by a phospho-mimicking aspartic acid substitution (S776D).4 Recently, RBM17 (RNA-binding motif protein 17) was shown to interact with ATXN1 in a manner dependent on polyglutamine expansion as well as phosphorylation of Ser776 (26). ATXN1 and RBM17 can be co-immunoprecipitated from wild-type mouse cerebellar extracts. Yeast two-hybrid and glutathione S-transferase pulldown assays show that the binding of RBM17 to ATXN1 is increased with the expansion of the polyglutamine tract and by the replacement of Ser776 with Asp. Further analysis indicated that ATXN1 and RBM17 are part of a large protein complex in vivo and that the proportion of RBM17 in the complex increases upon polyglutamine expansion. Interestingly, the amount of ATXN1 in another complex containing the CIC protein decreases with polyglutamine expansion (26).
Widely Expressed but Selectively Toxic
Although ATXN1 is widely expressed in the central nervous system and elsewhere (27), the most frequent and severe pathology is in cerebellar Purkinje cells. Thus, an important goal of SCA1 research is to understand the molecular basis of this cell specificity of the disease. A simple scenario in which ATXN1 normally serves a function critical for Purkinje cells that is eliminated upon expansion of the polyglutamine seems unlikely because mice lacking Atxn1 do not develop ataxia and Purkinje cell pathology (14) as seen in mice overexpressing mutant ATXN1 (13) or knock-in mice expressing ATXN1 with an expanded polyglutamine tract (9). An alternate scenario is that ATXN1 mediates its cell-specific toxicity by affecting the function of protein partners that are specific to vulnerable neurons. The interaction of ATXN1 with RORα-Tip60 offers possible explanations as to why Purkinje cells are more susceptible in SCA1. RORα is an orphan nuclear receptor that mediates the expression of a group of genes known to have a role in Purkinje cell development and function (28). Loss of Rora results in a congenital form of ataxia and severe cerebellar hypoplasia (29–31). Moreover, mice with a partial loss of Rora are reported to have an age-dependent Purkinje cell atrophy similar to that seen in SCA1 transgenic mice (32). In SCA1 transgenic mice, RORα is decreased in Purkinje cells, and there is a corresponding decrease in the expression of many RORα-regulated genes (21). Although ATXN1 does not interact directly with RORα, there is evidence indicating that it does interact directly with the RORα co-activator Tip60. By some mechanism that is yet to be determined, upon the interaction of mutant ATXN1 with Tip60, it seems that RORα is destabilized, and its function is compromised. It is noteworthy that it is when mutant ATXN1 compromises the RORα pathway during early cerebellar postnatal development that Purkinje cells become particularly susceptible to mutant ATXN1 in adults. The importance of a developmental ATXN1 effect on RORα for the severity of SCA1 in the adult was shown using a conditional mouse model of SCA1. Delay of the postnatal expression of mutant ATXN1 until completion of cerebellar development led to a substantial reduction in disease severity in adults in comparison with disease severity seen in mice with early postnatal SCA1 transgene expression (21). Additional factors that are likely to contribute to Purkinje cell vulnerability are Gfi1 and RBM17, given their relatively high levels in Purkinje cells in comparison with other cerebellar neurons.
The issue of selective neuronal vulnerability is likely to be complex and influenced by a multitude of factors. We propose that among such contributing factors are the relative levels of ATXN1. It is very clear that the levels of mutant ATXN1 are a major contributor to disease severity and Purkinje cell pathology. Therefore, if Purkinje cells express twice as much ATXN1 as other neurons, this could put them at increased risk for degeneration when the protein is mutated. Such subtle differences in the levels of ATXN1, although critical for its toxicity, are unlikely to be uncovered using current immunolabeling assays. Along the same lines, the levels of the interacting partners are likely contributors to the cell-specific vulnerabilities. For example, if RBM17 levels are slightly higher in vulnerable neurons, the toxic gain-of-function effects of ATXN1 would be more pronounced in such neurons. The levels and expression patterns of modifying proteins such as the enzyme that phosphorylates and dephosphorylates ATXN1 and enzymes that SUMOylate or ubiquitinate are also probable contributors to selective vulnerability.
Polyglutamine Corruption of ATXN1: A Complex Balance
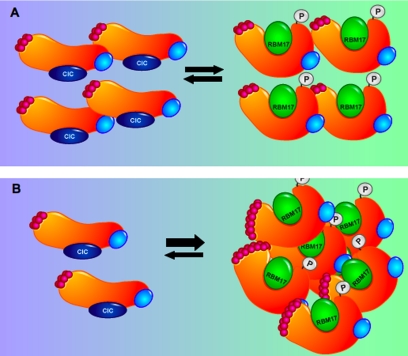
Data gathered from a multitude of biochemical and genetic in vivo studies are finally revealing the mechanism of pathogenesis of SCA1. The finding that protein domains outside of the polyglutamine tract are necessary for the toxicity of polyglutamine-expanded ATXN1 provided the first clue that the polyglutamine expansion might exert its toxicity by altering the function or interactions of other domains in ATXN1. The discovery that RBM17 interacts preferentially with the phosphorylated form of ATXN1 and with the polyglutamine-expanded form (both required for the toxicity of ATXN1) provided insight about how the polyglutamine expansion might influence other regions of ATXN1. In this particular case, perhaps the expansion enhances interactions (ATXN1/RBM17) that are normally tightly regulated by phosphorylation. The finding that some interactions are enhanced or gained (ATXN1/RBM17) while others are relatively decreased (ATXN1/CIC) revealed a new insight about SCA1 pathogenesis. These findings suggest a model whereby both a gain-of-function mechanism and a loss-of-function mechanism contribute to SCA1 pathology (Fig. 2). It is also quite likely that alterations in additional interactions, again some lost and some gained, contribute to disease. Given, however, that total loss of ATXN1 does not cause SCA1 pathology (14), it is likely that the gain-of-function mechanism is the main contributor to the disease and that the partial loss-of-function mechanism is a minor contributor or is simply a contributor in a background sensitized by the toxic gain-of-function effects.
FIGURE 2.
Model of SCA1 in which expansion of the polyglutamine tract in ATXN1 shifts the balance of protein complexes. A, wild-type ATXN1 normally exists in at least two large complexes, with one containing ATXN1 that is not phosphorylated at Ser776 and CIC. The other complex contains wild-type ATXN1 phosphorylated at Ser776 associated with RBM17. B, with expansion of the polyglutamine stretch into the mutant range, the balance of these two complexes is shifted such that less ATXN1 is associated with CIC and more ATXN1 is in a complex with RBM17. The small red circles depict glutamine residues.
Perspective
Late-onset neurodegenerative disorders, including polyglutamine disorders, Alzheimer disease, amyotrophic lateral sclerosis, and Parkinson disease, share many common features: selective neuronal vulnerability in the face of widely expressed causative genes, accumulation of the mutant proteins in neurons, and possible toxicity of overexpression of wild-type protein (at least true for ATXN1 (33), α-synuclein (34), and amyloid precursor protein (35)). The majority of the genetic data in humans and mice supports a gain-of-function mechanism. Thus, given these similarities, what lessons learned from extensive studies of SCA1 are worthy of consideration when thinking about the broader class of neurodegenerative diseases? A few come to mind. First, focusing on the full-length protein is critical. Although polyglutamine tracts are toxic in their own right if overexpressed in cells and animals, the insight we have learned about the regions that are necessary for the polyglutamine tract to mediate its toxicity (for example, Ser776) would have been missed if we had not focused our genetic studies on full-length ATXN1. Second, so far, the biochemical and protein interaction data point to alterations in native interactions rather than “novel” toxic interactions of the mutant protein. For the polyglutamine disorders, an implication of this concept is that although an expanded polyglutamine tract is necessary for pathogenesis, it is not sufficient. Besides the examples provided here for ATXN1 and SCA1, there are studies with spinal bulbar muscular atrophy and Huntington disease indicating that residues outside of the polyglutamine tract impact the severity of the disease (36–38), supporting the idea that is likely to be a principle broadly applicable to the neurodegenerative disorders. Thus, extensive characterization of the normal function and interaction of the disease-causing protein will likely pay off. Third, although the genetic data in humans and mice unequivocally support a gain-of-function mechanism, the biochemical and animal studies reveal a subtle but clinically relevant role for a partial loss-of-function mechanism (26). Fourth, paralogs are likely to provide insight about protein functions and interactions but also might be exploited as modifiers that may be good targets to manipulate to alter the disease course (39). Thus, extensive studies of paralogs and their interactions and modifications might pay off in this class of disorders. Finally, the finding that mouse models expressing inducible forms of mutant ATXN1 (40), mutant Tau (41), or a mutant fragment of huntingtin (42) functionally recover when the mutant transgene is turned off is very exciting and provides hope that if we can identify drugs that subdue gained interactions (for example, by decreasing Ser776 phosphorylation in ATXN1), individuals who are already symptomatic might be able to regain some functional recovery.
Acknowledgments
We thank Yvonne Klisch for drawing Fig. 2.
This work was supported, in whole or in part, by National Institutes of Health Grants NS22920 and NS45667 (to H. T. O.) and NS27699 and HD24064 (to H. Y. Z.). This is the fourth of five articles in the Unstable Nucleotide Repeat Minireview Series. This minireview will be reprinted in the 2009 Minireview Compendium, which will be available in January, 2010.
Footnotes
The abbreviations used are: SCA1, spinocerebellar ataxia type 1; RORα, retinoic acid receptor-related orphan receptor α; NLS, nuclear localization sequence.
H. Y. Zoghbi and H. T. Orr, unpublished data.
References
- 1.Orr, H. T., and Zoghbi, H. Y. (2007) Annu. Rev. Neurosci. 30 575–621 [DOI] [PubMed] [Google Scholar]
- 2.Voogd, J., and Glickstein, M. (1998) Trends Neurosci. 21 370–375 [DOI] [PubMed] [Google Scholar]
- 3.Orr, H. T., Chung, M. Y., Banfi, S., Kwiatkowski, T. J., Jr., Servadio, A., Beaudet, A. L., McCall, A. E., Duvick, L. A., Ranum, L. P., and Zoghbi, H. Y. (1993) Nat. Genet. 4 221–226 [DOI] [PubMed] [Google Scholar]
- 4.Chung, M. Y., Ranum, L. P., Duvick, L. A., Servadio, A., Zoghbi, H. Y., and Orr, H. T. (1993) Nat. Genet. 5 254–258 [DOI] [PubMed] [Google Scholar]
- 5.Cummings, C. J., Mancini, M. A., Antalffy, B., DeFranco, D. B., Orr, H. T., and Zoghbi, H. Y. (1998) Nat. Genet. 19 148–154 [DOI] [PubMed] [Google Scholar]
- 6.Taroni, F., and DiDonato, S. (2004) Nat. Rev. Neurosci. 5 641–655 [DOI] [PubMed] [Google Scholar]
- 7.Klement, I. A., Skinner, P. J., Kaytor, M. D., Yi, H., Hersch, S. M., Clark, H. B., Zoghbi, H. Y., and Orr, H. T. (1998) Cell 95 41–53 [DOI] [PubMed] [Google Scholar]
- 8.Cummings, C. J., Reinstein, E., Sun, Y., Antalffy, B., Jiang, Y.-h., Ciechanover, A., Orr, H. T., Beaudet, A. L., and Zoghbi, H. Y. (1999) Neuron 24 879–892 [DOI] [PubMed] [Google Scholar]
- 9.Watase, K., Weeber, E. J., Xu, B., Antalffy, B., Yuva-Paylor, L., Hashimoto, K., Kano, M., Atkinson, D. L., Sweatt, J. D., Orr, H. T., Paylor, R., and Zoghbi, H. Y. (2002) Neuron 34 905–919 [DOI] [PubMed] [Google Scholar]
- 10.Yoo, S. Y., Pennesi, M. E., Weeber, E. J., Xu, B., Atkinson, R., Chen, S., Armstrong, D. L., Wu, S. M., Sweatt, J. D., and Zoghbi, H. Y. (2003) Neuron 37 383–401 [DOI] [PubMed] [Google Scholar]
- 11.Arrasate, M., Mitra, S., Schweitzer, E. S., Segal, M. R., and Finkbeiner, S. (2004) Nature 431 805–810 [DOI] [PubMed] [Google Scholar]
- 12.Proescher, J. B., Son, M., Elliott, J. L., and Culotta, V. C. (2008) Hum. Mol. Genet. 17 1728–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burright, E. N., Clark, H. B., Servadio, A., Matilla, T., Feddersen, R. M., Yunis, W. S., Duvick, L. A., Zoghbi, H. Y., and Orr, H. T. (1995) Cell 82 937–948 [DOI] [PubMed] [Google Scholar]
- 14.Matilla, A., Roberson, E. D., Banfi, S., Morales, J., Armstrong, D. L., Burright, E. N., Orr, H. T., Sweatt, J. D., Zoghbi, H. Y., and Matzuk, M. (1998) J. Neurosci. 18 5508–5516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Chiara, C., Giannini, C., Adinolfi, S., de Boer, J., Guida, S., Ramos, A., Jodice, C., Kioussis, D., and Pastore, A. (2003) FEBS Lett. 551 107–112 [DOI] [PubMed] [Google Scholar]
- 16.Chen, Y. W., Allen, M. D., Veprintsev, D. B., Löwe, J., and Bycroft, M. (2004) J. Biol. Chem. 279 3758–3765 [DOI] [PubMed] [Google Scholar]
- 17.Yue, S., Serra, H., Zoghbi, H. Y., and Orr, H. T. (2001) Hum. Mol. Genet. 10 25–30 [DOI] [PubMed] [Google Scholar]
- 18.Mitzutani, A., Wang, L., Rajan, H., Vig, P. J., Alaynick, W. A., Thaler, J. P., and Tsai, C. C. (2005) EMBO J. 24 3339–3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsuda, H., Jafar-Nejad, H., Patel, A. J., Sun, Y., Chen, H.-K., Rose, M. F., Venken, K. J. T., Botas, J., Orr, H. T., Bellen, H. J., and Zoghbi, H. Y. (2005) Cell 122 633–644 [DOI] [PubMed] [Google Scholar]
- 20.Lam, Y. C., Bowman, A. B., Jafar-Nejad, P., Lim, J., Richman, R., Fryer, J. D., Hyun, E., Duvick, L. A., Orr, H. T., Botas, J., and Zoghbi, H. Y. (2006) Cell 127 1335–1347 [DOI] [PubMed] [Google Scholar]
- 21.Serra, H. G., Duvick, L., Zu, T., Carlson, K., Stevens, S., Jorgensen, N., Lysholm, A., Burright, E., Zoghbi, H. Y., Clark, H. B., Andresen, J. M., and Orr, H. T. (2006) Cell 127 697–708 [DOI] [PubMed] [Google Scholar]
- 22.Riley, B. E., Zoghbi, H. Y., and Orr, H. T. (2005) J. Biol. Chem. 280 21942–21948 [DOI] [PubMed] [Google Scholar]
- 23.Muller, S., Ledl, A., and Schmidt, D. (2004) Oncogene 23 1998–2008 [DOI] [PubMed] [Google Scholar]
- 24.Emamian, E. S., Kaytor, M. D., Duvick, L. A., Zu, T., Tousey, S. K., Zoghbi, H. Y., Clark, H. B., and Orr, H. T. (2003) Neuron 38 375–387 [DOI] [PubMed] [Google Scholar]
- 25.Chen, H.-K., Fernandez-Funez, P., Acevedo, S. F., Lam, Y. C., Kaytor, M. D., Fernandez, M. H., Aitken, A., Skoulakis, E. M. C., Orr, H. T., Botas, J., and Zoghbi, H. Y. (2003) Cell 113 457–468 [DOI] [PubMed] [Google Scholar]
- 26.Lim, J., Crespo-Barreto, J., Jafar-Nejad, P., Bowman, A. B., Richman, R., Hill, D. E., Orr, H. T., and Zoghbi, H. Y. (2008) Nature 452 713–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Servadio, A., Koshy, B., Armstrong, D., Antalffy, B., Orr, H. T., and Zoghbi, H. Y. (1995) Nat. Genet. 10 94–98 [DOI] [PubMed] [Google Scholar]
- 28.Gold, D. A., Baek, S. H., Schork, N. J., Rose, D. W., Larsen, D. D., Sachs, B. D., Rosenfeld, M. G., and Hamilton, B. A. (2003) Neuron 40 1119–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamilton, B. A., Frankel, W. N., Kerrebrock, A. W., Hawkins, T. L., Fitz-Hugh, W., Kusumi, K., Russell, L. B., Mueller, K. L., van Berkel, V., Birren, B., Kruglyak, L., and Lander E. S. (1996) Nature 379 736–739 [DOI] [PubMed] [Google Scholar]
- 30.Steinmayr, M., Andre, E., Conquet, F., Rondi-Reig, L., Delhaye-Bouchaud, N., Auclair, N., Daniel, H., Crepel, F., Mariani, J., Sotelo, C., and Becker-Andre, M. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 3960–3965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dussalt, I., Fawcett, D., Matthyssen, A., Badr, J.-A., and Giuère, V. (1998) Mech. Dev. 70 147–153 [DOI] [PubMed] [Google Scholar]
- 32.Hadj-Sahraoui, N., Frederic, F., Zanjani, H., Delhaye-Bouchaud, N., Herrup, K., and Mariani, J. (2001) Dev. Brain Res. 126 201–209 [DOI] [PubMed] [Google Scholar]
- 33.Fernandez-Funez, P., Nino-Rosales, M. L., de Gouyon, B., She, W.-C., Luchack, J., Martinez, P., Turiegano, E., Benito, J., Capovilla, M., Skinner, P., McCal, A., Canal, I., Orr, H. T., Zoghbi, H. Y., and Botas, J. (2000) Nature 408 101–106 [DOI] [PubMed] [Google Scholar]
- 34.Singleton, A. B., Farrer, M., Johnson, J., Singleton, A., Hague, S., Kachergus, J., Hulihan, M., Peuralinna, T., Dutra, A., Nussbaum, R., Lincoln, S., Crawley, A., Hanson, M., Maraganore, D., Adler, C., Cookson, M. R., Muenter, M., Baptista, M., Miller, D., Blancato, J., Hardy, J., and Gwinn-Hardy, K. (2003) Science 302 841. [DOI] [PubMed] [Google Scholar]
- 35.Salehi, A., Delcroix, J.-D., Belichenko, P. V., Zhan, K., Wu, C., Valletta, J. S., Takimoto-Kimura, R., Kleschevnikov, A. M., Sambamurti, K., Chung, P. P., Xia, W., Villar, A., Campbell, W. A., Kulnane, L. S., Nixon, R. A., Lamb, B. T., Epstein, C. J., Stokin, G. B., Goldstein, L. S. B., and Mobley, W. C. (2006) Neuron 51 29–42 [DOI] [PubMed] [Google Scholar]
- 36.Katsuno, M., Adachi, H., Kume, A., Li, M., Nakagomi, Y., Niwa, H., Sang, C., Kobayashi, Y., Inukai, A., and Sobue, G. (2002) Neuron 35 843–854 [DOI] [PubMed] [Google Scholar]
- 37.Chevalier-Larson, E. S., O'Brien, C. J., Wang, H., Jenkins, S. C., Holder, L., Lieberman, A. P., and Merry, D. E. (2004) J. Neurosci. 24 4778–4786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Graham, R. K., Deng, Y., Slow, E. J., Haigh, B., Bissada, N., Lu, G., Pearson, J., Shehadeh, J., Bertram, L., Murphy, Z., Warby, S. C., Doty, C. N., Roy, S., Wellington, C. L., Leavitt, B. R., Raymod, L. A., Nicholson, D. W., and Hayden, M. R. (2006) Cell 125 1179–1191 [DOI] [PubMed] [Google Scholar]
- 39.Bowman, A. B., Lam, Y. C., Jafar-Nejad, P., Chen, H.-K., Richman, R., Samaco, R. C., Fryer, J. D., Kahle, J. J., Orr, H. T., and Zoghbi, H. Y. (2007) Nat. Genet. 39 373–379 [DOI] [PubMed] [Google Scholar]
- 40.Zu, T., Duvick, L. A., Kaytor, M. D., Berlinger, M., Zoghbi, H. Y., Clark, H. B., and Orr, H. T. (2004) J. Neurosci. 24 8853–8861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Santacruz, K., Lewis, J., Spires, T., Paulson, J., Kotilinek, L., Ingelsson, M., Guimaraes, A., DeTure, M., Ramsden, M., McGowan, E., Forster, C., Yue, M., Orne, J., Janus, C., Mariash, A., Kuskowski, M., Hyman, B., Hutton, M., and Ashe, K. H. (2005) Science 309 476–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamamoto, A., Lucas, J. J., and Hen, R. (2000) Cell 101 57–66 [DOI] [PubMed] [Google Scholar]