Abstract
We report here that 4-dibenzo[a,d]cyclohepten-5-ylidene-1-[4-(2H-tetrazol-5-yl)-butyl]-piperidine (AT-56) is an orally active and selective inhibitor of lipocalin-type prostaglandin (PG) D synthase (L-PGDS). AT-56 inhibited human and mouse L-PGDSs in a concentration (3–250 μm)-dependent manner but did not affect the activities of hematopoietic PGD synthase (H-PGDS), cyclooxygenase-1 and -2, and microsomal PGE synthase-1. AT-56 inhibited the L-PGDS activity in a competitive manner against the substrate PGH2 (Km = 14 μm) with a Ki value of 75 μm but did not inhibit the binding of 13-cis-retinoic acid, a nonsubstrate lipophilic ligand, to L-PGDS. NMR titration analysis revealed that AT-56 occupied the catalytic pocket, but not the retinoid-binding pocket, of L-PGDS. AT-56 inhibited the production of PGD2 by L-PGDS-expressing human TE-671 cells after stimulation with Ca2+ ionophore (5 μm A23187) with an IC50 value of about 3 μm without affecting their production of PGE2 and PGF2α but had no effect on the PGD2 production by H-PGDS-expressing human megakaryocytes. Orally administered AT-56 (<30 mg/kg body weight) decreased the PGD2 production to 40% in the brain of H-PGDS-deficient mice after a stab wound injury in a dose-dependent manner without affecting the production of PGE2 and PGF2α and also suppressed the accumulation of eosinophils and monocytes in the bronco-alveolar lavage fluid from the antigen-induced lung inflammation model of human L-PGDS-transgenic mice.
PGD24 is a lipid mediator involved in sleep (1, 2) and inflammatory responses (3). PGD2 activates two different types of receptors (i.e. DP1 (4) and DP2 (also known as CRTH2 (5))). PGD2 regulates sleep (2, 6) and pain (7) via DP1 receptors in the central nervous system. This prostanoid also causes contraction of airway smooth muscle via DP1 receptors (8) and mediates chemotaxis of eosinophils and basophils into the lung via DP2 receptors (9) in the periphery. Therefore, PGD2 coordinately regulates allergic reactions, especially airway inflammation, via these two receptors (9).
PGD2 is formed by the following sequence of enzyme reactions after cell activation: 1) cytosolic phospholipase A2 is translocated to the endoplasmic reticulum and perinuclear membranes in a Ca2+-dependent manner, where it cleaves arachidonic acid from the membrane phospholipids; 2) arachidonic acid is converted to PGH2, a common precursor of various prostanoids, by the membrane-bound cyclooxygenases (COXs); and 3) PGH2 is further isomerized to PGD2 by PGD synthase (PGDS). There are two distinct types of PGDS, namely lipocalin-type PGDS (L-PGDS) (10–13) and hematopoietic PGDS (H-PGDS) (14–16).
L-PGDS contributes to the production of PGD2 in the central nervous system (10, 17, 18), ocular tissues (19), cardiovascular systems (20), and male genital organs (21) of various mammals and is involved in the regulation of non-rapid eye movement sleep (2, 22), sex determination (23), protection of atherosclerosis (24, 25), and adipogenesis (26). L-PGDS is secreted into the cerebrospinal fluid (CSF) (27, 28), seminal plasma (29, 30), and plasma (20) as β-trace, a major protein in human CSF (31).
L-PGDS is the only enzyme among the members of the lipocalin gene family (11), which is composed of various secretory proteins involved in binding and transporting small lipophilic substances, such as retinoids and thyroids (32). L-PGDS also binds retinoic acid, retinal (33), biliverdin (34), bilirubin (34), gangliosides (35), and amyloid β peptides (36) with high affinities of Kd = 20–200 nm, indicating that L-PGDS may act as a transporter protein of these lipophilic compounds and as an endogenous chaperon to prevent amyloid β aggregation. On the other hand, H-PGDS is the first known vertebrate homolog of the σ class of glutathione S-transferases (37). Because both of these enzymes have evolved from different origins to acquire the same catalytic function, these two enzymes are considered to be a new example of functional convergence (38, 39).
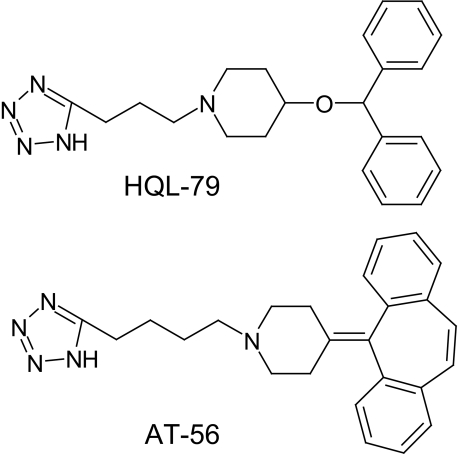
Inorganic quadrivalent selenium (Se4+) compounds are known to be noncompetitive and reversible inhibitors of L-PGDS (40) and to inhibit the sleep of animals in a time- and dose-dependent manner after infusion into the third ventricle of rats or an intraperitoneal injection into mice (2, 41). However, no organic inhibitor of L-PGDS has been reported until now. We recently found that 4-benzhydryloxy-1-(3-(1H-tetrazol-5-yl)-propyl)-piperidine (HQL-79; Fig. 1) is an orally active and selective inhibitor of H-PGDS (42). Because L-PGDS catalyzes the same reaction as H-PGDS, we suspected that some derivative(s) of HQL-79 might have an inhibition activity toward L-PGDS. Among various tetrazol compounds, we found that 4-dibenzo[a,d]cyclohepten-5-ylidene-1-[4-(2H-tetrazol-5-yl)-butyl]-piperidine (AT-56; Fig. 1) is a specific inhibitor of L-PGDS (43), having a higher potency and biological availability than seleno-compounds in vivo.
FIGURE 1.
Chemical structures of HQL-79 and AT-56.
In the present study, we characterized AT-56 and found it to be a competitive inhibitor of L-PGDS against PGH2 by kinetic analysis and also used NMR analysis to determine the binding mode of AT-56 to L-PGDS. In addition we found that AT-56 inhibited the production of PGD2 by L-PGDS-expressing cultured cells, H-PGDS gene knock-out (KO) mice, and human L-PGDS overexpressing transgenic (TG) mice.
EXPERIMENTAL PROCEDURES
Chemicals—AT-56 was a generous gift from TAIHO Pharmaceutical Company (Saitama, Japan). AT-56 was dissolved in DMSO for in vitro experiments and in 0.5% methylcellulose for oral administration to mice, due to its high lipophilicity. In aqueous solution, the maximum solubility of AT-56 was determined to be 90 μm in saline containing 1% DMSO and 260 μm in 10% DMSO, as determined by UV absorption at 287 nm. Other reagents were purchased from Sigma or Wako (Osaka, Japan), unless otherwise specified.
Purification of Human CSF L-PGDS (β-Trace) and Recombinant Mouse L-PGDS—Human CSF L-PGDS (β-trace) was purified from human CSF, which was donated by Dr. M. Mase (Nagoya City University Hospital), as reported (36).
The full-length cDNA for mouse L-PGDS, which is composed of 189 amino acid residues (GenBank™ accession number X89222), was ligated into the BamHI-EcoRI site of the expression vector pGEX-2T plasmid (GE Healthcare). The N-terminal 22-amino acid residues of the signal peptide were deleted, and the C89A/C186A- and W54A-substituted recombinant enzymes were expressed in Escherichia coli DH5α (TOYOBO, Osaka, Japan). Site-directed mutagenesis was performed by using a QuikChange® site-directed mutagenesis kit (Stratagene, La Jolla, CA). The recombinant enzymes retained PGDS activity comparable with that of the wild-type enzyme and were stable for long term use. The mutated L-PGDS was expressed as a GSH transferase fusion protein, purified by column chromatography with GSH-Sepharose 4B (GE Healthcare), and eluted from the column by incubation with thrombin (100 units/100 μl of resin), as reported previously (44). The recombinant protein was further purified to apparent homogeneity by gel filtration chromatography with Superdex 75 in 5 mm Tris/HCl (pH 8.0).
Fluorescence Quenching Assays—13-cis-Retinoic acid and AT-56 were dissolved in DMSO to give a 2 mm stock solution. Various concentrations of 13-cis-retinoic acid and AT-56 in 10 μl of DMSO were added to the L-PGDS solution in 990 μl of 5 mm Tris/HCl (pH 8.0). After incubation at room temperature for 60 min, the intrinsic tryptophan fluorescence was detected with an FP-750 spectrofluorometer (JASCO, Tokyo, Japan) at an excitation wavelength at 282 nm and an emission wavelength at 338 nm, as reported previously (44).
Enzyme Activity Assays—Enzyme activities of L-PGDS (45), H-PGDS (46), and microsomal PGE synthase (m-PGES-1 (47)) were measured with 10 μm [1-14C]PGH2 as a substrate in 100 mm Tris-HCl, pH 8.0, in the presence of 1 mm GSH, 0.1 mg/ml IgG, and 10% DMSO. The activities of COX-1 and COX-2 were measured as described earlier (48) with 50 μm [1-14C]arachidonic acid (PerkinElmer) used as a substrate in 100 mm Tris-HCl, pH 8.0, containing 2 μm hematin, 5 mm l-tryptophan, 0.1 mg/ml IgG, and 10% DMSO. The products were separated by thin layer chromatography. The conversion rate from 14C-labeled substrate to 14C-labeled products was calculated by using an imaging plate system (Fuji Film, Tokyo, Japan). The kinetic constants were determined by Lineweaver-Burk plots prepared with SigmaPlot software (version 10.0 for Windows; Systat Software, Inc.).
NMR Titration Experiment—The NMR samples of mouse L-PGDS were prepared in a mixture of 50 mm sodium phosphate in 75% H2O, 15% D2O, 10% DMSO at pH 7.0, as reported previously (49). The protein concentration was adjusted to ∼1 mm in a 5-mm microcell NMR tube (Shigemi, Tokyo, Japan) for NMR experiments. NMR experiments were performed at 27.5 °C by using an INOVA600 spectrometer (Varian, Palo Alto, CA) equipped with shielded gradient triple resonance probes (49).
The binding of L-PGDS to AT-56 was monitored by NMR titration of 15N-labeled L-PGDS with unlabeled ligands by using 1H-15N heteronuclear single quantum correlation experiments. The combined 1H and 15N chemical shift changes over the range of the titration from 0 to 2.0 equivalents of AT-56 were plotted. The composite chemical shift differences were calculated according to the empirical equation Δδtot = {(ΔδHN × WHN)2 + (ΔδN × WN)2}½, where ΔδHN and ΔδN are the chemical shift changes of 1H and 15N, respectively. The weighting factors used were WHN = 1 and WN = 0.2.
PG Production by Cultured Cells—L-PGDS-expressing human medulloblastoma TE-671 cells and H-PGDS-possessing human megakaryoblastic MEG-01S cells were purchased from American Type Culture Collection. TE-671 (1 × 106 cells/well) and MEG-01S (5 × 105/well) cells were seeded into multiplates and cultured in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum, 4 mm l-glutamine, 4.5 g/liter glucose, 100 units/ml penicillin, and 100 μg/ml streptomycin sulfate under a 5% CO2 atmosphere at 37 °C. MEG-01S cells were caused to differentiate by treatment with 12-O-tetradecanoylphorbol-13-acetate to express H-PGDS and COX-1, as described previously (50).
After the cells had been cultured for 1 day, AT-56 was added to them at different doses ranging from 0 to 100 μm, and the cells were then incubated at 37 °C for 10 min. Thereafter, they were stimulated with a calcium ionophore, A23187 (5 μm), at 37 °C for 15 min. The culture media were harvested and centrifuged at 10,000 × g for 5 min to remove the cells, and the supernatant was removed and stored at –80 °C until the measurements of PGs could be made. In some experiments, these cells were prelabeled with [1-14C]arachidonic acid (3.7 kBq/well) for 12 h before the assay. PGD2, PGE2, and PGF2α in the culture medium were quantified as described below.
PG Production by Stab Wound Brain Injury—The protocols used for all animal experiments in this study were approved by the Animal Research Committee of the Osaka Bioscience Institute.
H-PGDS KO mice (14–16-week-old males or females weighing 25–30 g, C57BL/6 strain) (2, 51) containing only L-PGDS were used for the stab injury model of the brain. AT-56 of various doses (1, 3, 10, and 30 mg/kg body weight) was administered orally to H-PGDS KO mice 1 h before the stab wound injury. Under pentobarbital (50 mg/kg) anesthesia, a 25-gauge needle was inserted into the frontal cortex of the brain of H-PGDS KO mice at a position 2 mm caudal to the bregma, 2 mm lateral to the sagittal suture, and 1 mm deep. After the needle had been withdrawn, the brains were harvested, immediately frozen in liquid nitrogen, and stored in a deep freezer (–80 °C) until the measurements of PG contents could be made.
Measurement of PG Contents—The amounts of PGs in the cell culture media and brain tissues were determined as described previously (22). In brief, the cell culture media and the frozen brain tissues were homogenized in ethanol containing 0.02% HCl at pH 2.0 and [3H]PGD2, [3H]PGE2, and [3H]PGF2α (PerkinElmer Life Sciences) as tracers to estimate the recovery during the purification procedure. After centrifugation at 500 × g for 20 min, the ethanol extracts were applied onto Sep-Pak C18 cartridges (Waters Associates, Milford, MA), washed with hexane, eluted with ethyl acetate, and fractionated by reverse-phase high performance liquid chromatography. The amounts of PGD2, PGE2, and PGF2α were measured by using their respective enzyme immunoassay kits (Cayman Chemical, Ann Arbor, MI), according to the manufacturer's instructions.
L-PGDS-mediated Allergic Airway Inflammation—Human L-PGDS-overexpressing TG mice (14–16-week-old males weighing 25–30 g, FVB strain (22)) were actively sensitized by an intraperitoneal injection of 10 μg of ovalbumin (Sigma) in 0.2 ml of aluminum hydroxide gel (Serva, Heidelberg, Germany) on days 0 and 14 and then exposed to aerosolized ovalbumin (50 mg/ml in sterile saline) for 20 min on day 21 (52). At 2 days after the ovalbumin challenge, the bronchoalveolar lavage fluid was collected as reported previously (52). Total and differential cell counts (500 cells) were obtained based on standard morphologic criteria after the cells had been cytospun onto glass slides and stained with Diff-Quik (Dade Diagnostics).
Pharmacokinetics of AT-56—Male C57BL/6 mice (7 weeks old, weighing 22–26 g; Japan SLC, Shizuoka, Japan) were given a single oral dose of 10 mg/kg AT-56 or a single intravenous dose of 2 mg/kg AT-56. Plasma was collected after euthanasia from 3 mice/group at 30 min and 1, 2, 4, 8, and 24 h for oral administration and 5 and 30 min and 1, 2, 4, 8, 12, and 24 h after intravenous dosing. Whole blood sample was collected into a heparinized syringe at each time point. Plasma was obtained by centrifugation and was stored at –30 °C until analysis.
Concentrations of AT-56 in plasma were determined by high performance liquid chromatography coupled to mass spectrometry. Samples were separated by chromatography with a Cadenza HS-C18 column (2 mm × 150 mm; Imtact Corp., Kyoto, Japan) at a column temperature of 33 °C with a 5–90% acetonitrile gradient in 0.02% formic acid at a flow rate of 0.2 ml/min for 12 min. Injection volume was 10 μl. AT-56 was detected by mass spectrometry with the system (Waters Associates) composed of the 2695 separation module, 996 photodiode array detector, and ZQ-4000 mass spectrometry detector with an electrospray ionization source. The electrospray ionization interface was operated at a source temperature of 115 °C and a desolvation temperature of 350 °C. Cone gas and desolvation gas flow were 124 and 606 liters/h, respectively. Cone voltage for AT-56 was positive 33 V for m/z = 398. Pharmacokinetic parameters were calculated using noncompartmental analysis with WinNonlin, version 5.0.1 (Pharsight, Mountain View, CA).
Statistics—Comparisons were analyzed for statistical significance by Dunnett's multicomparison test using SigmaPlot (Systat Software, CA). p < 0.05 was considered significant.
RESULTS
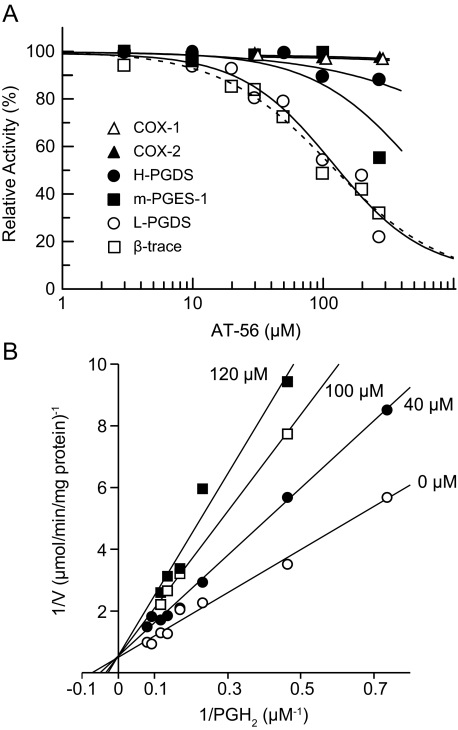
Inhibition of the L-PGDS Activity by AT-56—AT-56 inhibited the PGDS activity of both β-trace purified from human CSF and purified recombinant mouse L-PGDS C89A/C186A mutant in a concentration (10–250 μm)-dependent manner. The L-PGDS activity of both preparations was inhibited to 30% with 250 μm AT-56, and the IC50 value was calculated to be 95 μm. However, the activities of the purified COX-1, COX-2, m-PGES-1, or H-PGDS in the arachidonate cascade were not significantly affected by AT-56 up to 250 μm (Fig. 2A).
FIGURE 2.
Specific inhibition of L-PGDS by AT-56. A, human CSF L-PGDS (β-trace), the recombinant mouse L-PGDS C89A/C186A mutant, the purified m-PGES-1, and H-PGDS were incubated with 10 μm PGH2 and 1 mm GSH at 25 °C for 30 s in the presence of 0–250 μm AT-56 in 10% DMSO. The purified COX-1 and COX-2 were incubated with 50 μm arachidonic acid in the presence of 0–250 μm AT-56. B, Lineweaver-Burk plot for L-PGDS. Recombinant mouse L-PGDS C89A/C186A mutant was incubated with various concentrations of PGH2 (3–20 μm), 100 mm Tris-HCl (pH 8), and 1 mm dithiothreitol in the presence of 0 (○), 40 (•), 100 (□), or 120 μm (▪) AT-56 in 10% DMSO.
Kinetic experiments revealed that AT-56 inhibited the recombinant L-PGDS in a competitive manner against the substrate PGH2, as shown in Lineweaver-Burk plots (Fig. 2B), in which the Vmax value remained unchanged but the Km value increased when the AT-56 concentration was increased (0–120 μm). The apparent Ki value of AT-56 was calculated to be 75 μm, which was 5.3-fold higher than the Km value of the L-PGDS activity for PGH2 (14 μm) in the presence of 10% DMSO.
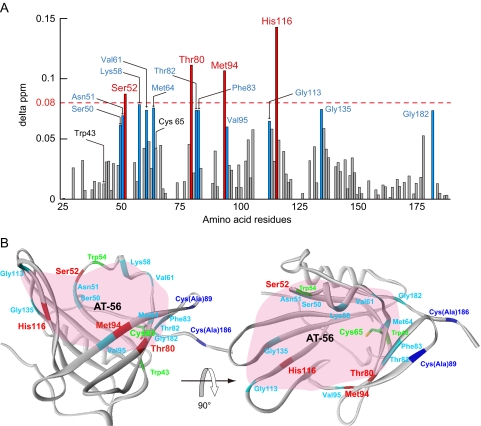
Binding Interaction between L-PGDS and AT-56 Analyzed by NMR—We have previously determined the NMR structure of the mouse L-PGDS C89A/C186A mutant (49). The interaction sites of L-PGDS with AT-56 were determined by the chemical shift perturbation method (53) based on the two-dimensional 1H-15N heteronuclear single quantum correlation NMR spectra of L-PGDS after the successive addition of AT-56 (Fig. 3). Upon AT-56 binding, large changes (>0.08 ppm) of chemical shift were observed at several amino acid residues of L-PGDS (i.e. Ser52, Thr80, Met94, and His116) (Fig. 3A). These residues are located at the upper part of the central cavity of the β-barrel structure of the L-PGDS molecule (Protein Data Bank code 2E4J; Fig. 3B), in which the PGH2-binding site containing the catalytic center of Cys65 is located (49). However, the AT-56 binding did not cause any significant chemical shift in the regions of the retinoic acid-binding pocket at the lower part of the central cavity of L-PGDS (49).
FIGURE 3.
Interactions of L-PGDS with AT-56 as examined by NMR. A, composite 1H and 15N chemical shift differences (delta ppm) versus the amino acid sequence of recombinant mouse L-PGDS C89A/C186A mutant. B, overall structure of L-PGDS after AT-56 binding in a ribbon representation. In both panels, the residues with relatively large changes in chemical shift (≥0.08) are represented in red, whereas residues with shifts in the middle range (0.06 ≤ Δ ppm < 0.08) are shown in sky blue. In B, the AT-56-binding site predicted from NMR signal perturbation is shaded in pink.
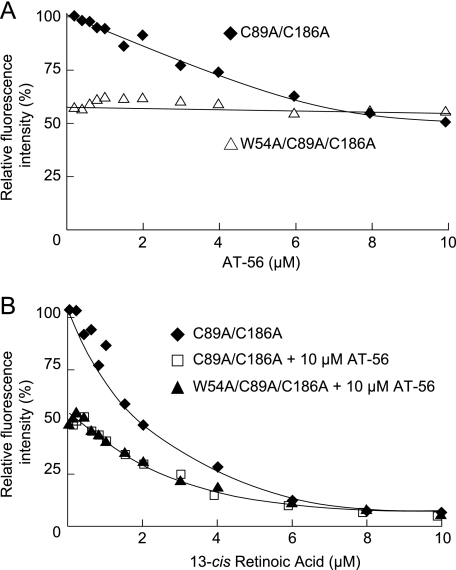
Fluorescence Quenching Study on L-PGDS after Binding of AT-56—To confirm the mode of binding of AT-56 to L-PGDS, we examined the fluorescence quenching of intrinsic Trp residues of mouse L-PGDS after incubation with AT-56. Mouse L-PGDS contains two tryptophan residues, at positions 43 and 54, the former of which is located at the bottom of barrel and the latter in the AB-loop at the entrance of the central cavity of L-PGDS (49). The C89A/C186A mutant of mouse L-PGDS showed fluorescence quenching by the addition of AT-56 (Fig. 4A) in a concentration-dependent manner, and the fluorescence intensity was decreased to about 60% in the presence of 10 μm AT-56. On the other hand, the W54A/C89A/C186A mutant showed about 60% of the tryptophan fluorescence of the C89A/C186A mutant in the absence of AT-56 and did not show the fluorescence quenching in the presence of 10 μm AT-56 (Fig. 4A). The C89A/C186A mutant showed fluorescence quenching by the addition of 13-cis-retinoic acid in a concentration-dependent manner, with the fluorescence intensity being decreased to about 8% in the presence of 10 μm 13-cis-retinoic acid (Fig. 4B). The C89A/C186A mutant in the presence of 10 μm AT-56 and the W54A/C89A/C186A mutant showed about 60% of the fluorescence intensity of the C89A/C186A mutant in the absence of 10 μm AT-56. In both cases, the fluorescence intensity was decreased by the addition of 13-cis-retinoic acid in a concentration-dependent manner to <10% in the presence of 10 μm 13-cis-retinoic acid, giving the same fluorescence quenching curves even in the presence of 10 μm AT-56. These results indicate that AT-56 binds near Trp54 in the AB-loop of L-PGDS but not to the Trp43 residue in the retinoid-binding pocket, being consistent with the results obtained by NMR titration analysis.
FIGURE 4.
Tryptophan fluorescence quenching by AT-56. A, fluorescence quenching of intrinsic tryptophan of C89A/C186A mutant (♦) and W54A/C89A/C186A mutant (▴) of mouse L-PGDS by incubation with AT-56. B, fluorescence quenching of intrinsic tryptophan of the mouse L-PGDS C89A/C186A mutant in the absence (♦) or presence (□) of 10 μm AT-56 and that for the W54A/C89A/C186A mutant in the presence of 10 μm AT-56 (▴) by incubation with 13-cis-retinoic acid.
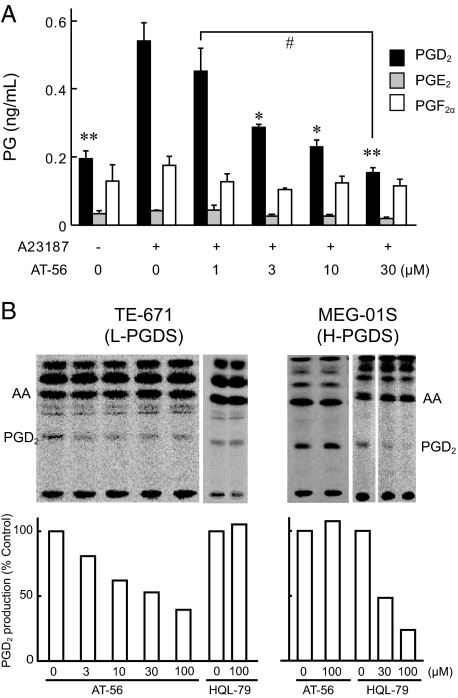
Inhibition of PGD2 Production in L-PGDS-expressing Human Medulloblastoma TE-671 Cells by AT-56—Human medulloblastoma TE-671 cells constitutively express L-PGDS (54). We pretreated TE-671 cells for 10 min with 0 to 30 μm AT-56, stimulated them with calcium ionophore A23187 (5 μm) at 37 °C for 15 min, and then determined the production of PGD2, PGE2, and PGF2α by enzyme immunoassay to investigate the effects of AT-56 on PG production by these cells. AT-56 dose-dependently inhibited the production of PGD2 without affecting the production of PGE2 and PGF2α (Fig. 5A).
FIGURE 5.
Inhibition of L-PGDS activity by AT-56 in TE-671 cells but not in MEG-01S cells. TE-671 cells expressing L-PGDS were pretreated with various concentrations of AT-56 for 15 min and then incubated with or without 5 μm A23187 for 10 min to measure prompt PGD2 release. A, the amount of PGD2 released from TE-671 cells was measured by enzyme immunoassay. AT-56 dose-dependently inhibits PGD2 production from L-PGDS-expressing TE-671. Data are presented as the mean ± S.E. *, p < 0.05; **, p < 0.01 as compared with the value in the absence of AT-56 and in the presence of A23187. #, p < 0.05 as compared with the value in the presence of 1 μm AT-56 (Dunnett's test). B, selective inhibition by AT-56 of [14C]PGD2 production in TE-671 cells and in H-PGDS-expressing MEG-01S cells. TE-671 and MEG-01S cells were prelabeled with [1-14C]arachidonic acid and stimulated with 5 μm A23187 for 15 min in the presence of various concentrations of AT-56 (3–100 μm). Radiolabeled arachidonic acid and its metabolites were extracted from the culture medium, separated by thin layer chromatography, and analyzed by autoradiography. AA, arachidonic acid.
Dose-dependent inhibition of the A23187-induced PGD2 production by AT-56 (3–100 μm) was also confirmed by using [1-14C]arachidonic acid-prelabeled TE-671 cells (Fig. 5B). However, the production of other 14C-labeled metabolites was not inhibited by AT-56 up to 100 μm. Moreover, AT-56 had no effect on the production of PGD2 by human H-PGDS-expressing MEG-01S cells (50). H-PGDS-specific inhibitor, HQL-79, inhibited PGD2 production from MEG-01S cells but not from TE-671 cells. These results indicate that AT-56 selectively inhibits PGD2 production catalyzed by L-PGDS without affecting the production of other prostanoids.
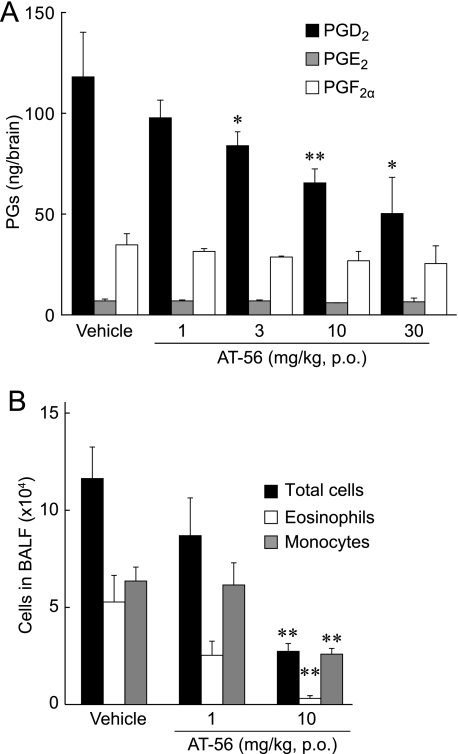
Suppression of PGD2 Production in the Stab-wounded Brain by Oral Administration of AT-56—To investigate the effect of AT-56 in vivo, we used H-PGDS KO mice, which express only L-PGDS in all of their organs (51). In a stab wound brain injury model, the PGD2 content in the brain of the wounded H-PGDS KO mice (118 ± 22 ng/brain) was significantly increased as compared with that of the control mice without injury (0.23 ± 0.04 ng/brain). When various doses of AT-56 were administered orally 1 h before the injury, the total amount of PGD2 in the brain decreased dose-dependently to 40% with 30 mg/kg AT-56 (Fig. 6A). The amounts of PGE2 and PGF2α in the brain were not significantly changed in any conditions (Fig. 6A). These results indicate that orally administered AT-56 inhibited the L-PGDS reaction in the brain.
FIGURE 6.
Inhibitory effect of AT-56 on PGD2 production by stab wounding in H-PGDS KO mice and therapeutic effect on antigen-induced lung inflammation in human L-PGDS TG mice. A, AT-56 was orally administered 1 h before the stab wounding, and brains were collected 10 min after the wounding. B, AT-56 was orally administered 1 h before and 24 h after the antigen exposure. The accumulated cells in the bronchoalveolar lavage fluid were collected 48 h after the antigen exposure. Data are presented as the mean ± S.E. *, p < 0.05; **, p < 0.01 as compared with the vehicle-treated group (Dunnett's test). p.o., per os. BALF, bronchoalveolar lavage fluid.
Suppression of L-PGDS-mediated Allergic Airway Inflammation by AT-56—We next evaluated the therapeutic effect of AT-56 on PGD2-mediated lung inflammation. Human L-PGDS TG mice were used in an OVA-induced lung inflammation model (52). The numbers of total cells and infiltrating eosinophils and monocytes in the bronchoalveolar lavage fluid of the L-PGDS TG mice were dose-dependently decreased to 75, 50, and 96% (actual numbers: 8.7 ± 1.9, 2.53 ± 0.73, and 6.16 ± 1.14 × 104 cells/ml for total cells, eosinophils, and monocytes, respectively) in 1 mg/kg AT-56-treated mice and to 23, 6, and 41% (2.7 ± 0.4, 0.31 ± 0.14, and 2.59 ± 0.3 × 104 cells/ml, respectively) in 10 mg/kg AT-56-treated ones compared with the numbers for the vehicle-administered mice (11.6 ± 1.6, 5.28 ± 1.37, and 6.36 ± 0.72 × 104 cells/ml, respectively) (Fig. 6B). These results demonstrate that AT-56 prevented the eosinophil infiltration by inhibiting transgened human L-PGDS in vivo.
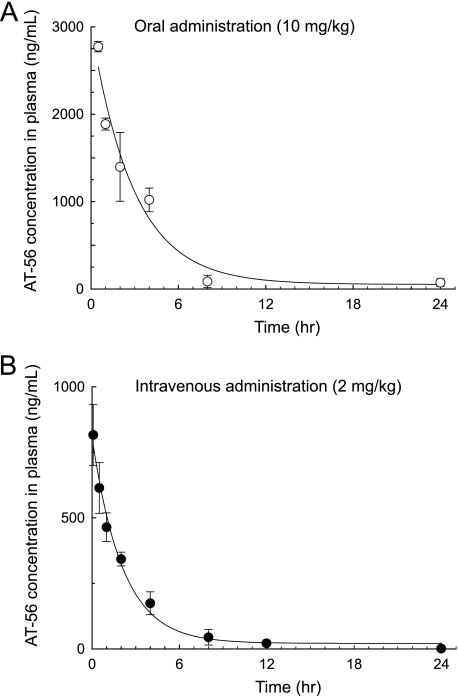
Pharmacokinetic Properties of AT-56—We also determined pharmacokinetic properties of AT-56. Fig. 7 shows the time courses of the plasma concentration of AT-56 after an oral administration to mice at a dose of 10 mg/kg and an intravenous bolus injection of 2 mg/kg. Table 1 summarizes pharmacokinetic parameters of AT-56. After the oral administration, the plasma level of AT-56 reached the maximum (2.15 μg/ml) within 30 min and decreased with time to be lower than the detection limit (0.4 ng/ml) at 12 h after the administration. The area under the concentration-time curve was calculated to be 2.18 and 8.95 μg/ml × h after the intravenous administration of 2 mg/kg and the oral administration of 10 mg/kg, respectively. Based on these data, bioavailability of AT-56 was calculated to be 82%, indicating that orally administered AT-56 was well absorbed in mice.
FIGURE 7.
Time courses of plasma concentrations of AT-56 after oral (10 mg/kg) (A) or intravenous (2 mg/kg) (B) administration of AT-56 to C57BL/6 mice. Data are presented as the mean ± S.E., n = 3/time point.
TABLE 1.
Pharmacokinetic parameters of AT-56
| Route | Dose | AUCa | Cmaxb | C0c | tmaxd | t½e | BAf |
|---|---|---|---|---|---|---|---|
| mg/kg | μg·h/ml | μg/ml | μg/ml | h | h | % | |
| Intravenous | 2 | 1.28 | 0.86 | 0.08 | 2.35 | ||
| Per os | 10 | 8.95 | 2.15 | 0.50 | 1.71 | 82.0 |
AUC, area under the concentration versus time curve from 0 to the last quantifiable time point.
Cmax, maximal concentration.
C0, initial concentration.
tmax, time to Cmax.
t½, half-life.
BA, bioavailability = (AUC p.o. × dose i.v.)/(AUC i.v. × dose p.o.) × 100, where p.o. represents per os and i.v. represents intravenous.
DISCUSSION
In the present study, we demonstrated that AT-56 is an orally active inhibitor specific for L-PGDS with a high bioavailability. The results of the kinetic analyses indicated that AT-56 was a competitive inhibitor against the substrate, PGH2. NMR titration analysis revealed that AT-56 bound to the catalytic pocket of L-PGDS, in which the catalytic center Cys65 is located (49), being consistent with the competitive inhibition of L-PGDS by AT-56. The NMR solution structure is also in good agreement with the results of the fluorescence quenching analysis of L-PGDS, in which AT-56 interacted with the upper part of L-PGDS containing Trp54 but did not affect the fluorescence of Trp43 located in the retinoic acid-binding pocket at the bottom of the hydrophobic cavity of the enzyme.
Our data demonstrate that AT-56 inhibited the isomerization reaction of PGH2 to PGD2 catalyzed by L-PGDS in a competitive manner against PGH2 with a Ki value of 75 μm (Fig. 2), which was 5.3-fold higher than the Km value of the L-PGDS activity for PGH2 (14 μm) in the presence of 10% DMSO. Therefore, in the enzyme assay, AT-56 inhibited the enzyme activity clearly in a high concentration range in the presence of PGH2. On the other hand, in the absence of PGH2, AT-56 binds to the enzyme and induces the fluorescence quenching even in the low concentration range. Various inorganic selenium compounds are also known to inhibit both the purified L-PGDS and the enzyme in the crude brain supernatant in a noncompetitive manner with a Ki value of 10 μm (40). The Ki value of AT-56 was 7.5-fold higher than that of SeCl4. However, the inhibition mode of AT-56 was quite different from that of selenium; i.e. this is the first competitive inhibitor of L-PGDS against PGH2 to be reported. When the same dose of various seleno-compounds was used in cell cultures, such compounds reduced PGD2 production; however, their efficacy cannot be attributed solely to the effects on the PGD2 pathway. Seleno-compounds also inhibited the release of arachidonic acid and the production of all PGs due to the lack of specificity of action. In contrast, when human TE-671 cells were stimulated with the Ca2+ ionophore A23187 in the presence of 10 μm AT-56, only a base level of PGD2 biosynthesis was observed without changing the arachidonic acid release (Fig. 5B).
At present, the reason why the IC50 value in the cell was lower than the IC50 and Ki values for purified L-PGDS is unknown. L-PGDS requires sulfhydryl compounds, such as reduced GSH, dithiothreitol, or β-mercaptoethanol, for its catalytic reaction, although it catalyzes the isomerization of the substrate PGH2 to the product PGD2. The endogenous sulfhydryl compounds have not been identified in its reaction, and GSH or dithiothreitol have been used as a reductant in in vitro experiments. In this present study, we used GSH for the enzyme inhibition assay of AT-56. Thus, it is possible that the reductant for the L-PGDS reaction is different from the endogenous one. It might be also possible that L-PGDS requires some other cofactor for its catalysis. AT-56 might effectively inhibit the L-PGDS reaction in the presence of such endogenous cofactor.
Recently, we demonstrated that L-PGDS produced PGD2 coupled with COX-2 in TE-671 cells (43). L-PGDS coupled to COX-2 may be more sensitive to AT-56 than L-PGDS itself. Since AT-56 is a relatively lipophilic compound, its local concentration around the L-PGDS-COX-2 complex within endoplasmic reticulum may be high enough to efficiently inhibit the production of PGD2 within the cell.
Although both L-PGDS and H-PGDS became evolutionarily diversified from quite different ancestor gene families (38, 39), these enzymes can catalyze the same isomerization reaction. HQL-79 recently has been identified as an H-PGDS inhibitor. In this study, we found that AT-56, which is an HQL-79-derivative compound, also has an inhibitory activity against the L-PGDS reaction both in vitro and in vivo, suggesting that the active site architecture for the substrate binding and the catalytic reaction mechanism of L-PGDS could be similar to those of H-PGDS.
Here, we demonstrated pharmacologically and biochemically that AT-56 is an orally effective inhibitor selective for L-PGDS. Especially, it should be noted that AT-56 specifically inhibited the production of PGD2 catalyzed by L-PGDS but only marginally affected the production of other prostanoids. In this sense, AT-56 is an even better PGD2-blocking compound than inorganic selenium compounds. Earlier we demonstrated that PGD2 produced by L-PGDS regulates physiological sleep (1) and pain (7) and also that L-PGDS acts as an extracellular transporter for various useful or harmful hydrophobic compounds (34–36). Thus, AT-56 may be predicted to selectively suppress the drowsiness or pain reaction mediated by L-PGDS-catalyzed PGD2 without showing the various side effects caused by the suppression of cytoprotective and anti-inflammatory PGs.
We did not detect any acute toxic effects of AT-56 after its oral administration even at a dose of 100 mg/kg. AT-56 possesses a direct and previously unknown inhibitory effect on L-PGDS in vitro and in vivo, suggesting that AT-56 might be a useful prototypic molecule to develop selective and/or nonselective inhibitors for L-PGDS and H-PGDS, which may act as anti-somnolence and anti-inflammatory drugs, respectively. The development of such selective and nonselective inhibitors of both enzymes would be helpful to determine the role of PGD2 in animal models. Such inhibitors would be drug candidates or actual drugs.
Acknowledgments
We thank O. Hayaishi (Osaka Bioscience Institute) for generous support of this study. We also thank Dr. K. Shigeno and Dr. K. Tanaka (TAIHO Pharmaceutical Company) for chemical synthesis and pharmacokinetics analysis of AT-56, respectively; T. Tsurumura and M. Sakata (Osaka Bioscience Institute) for assistance in enzyme assays and in vivo experiments, respectively; and Dr. M. Mase (Nagoya City University Hospital) for providing human CSF.
Author's Choice—Final version full access.
This work was supported in part by a grant from the Japan Foundation for Applied Enzymology (to K. A.), Grant-in Aid for Scientific Research of MEXT 19590094 (to K. A.), and Osaka City. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations and trivial names used are: PGD2, PGE2, PGF2α, and PGH2, prostaglandin D2,E2,F2α, and H2, respectively; PG, prostaglandin; AT-56, 4-dibenzo[a,d]cyclohepten-5-ylidene-1-[4-(2H-tetrazol-5-yl)-butyl]-piperidine; PGDS, prostaglandin D synthase; L-PGDS, lipocalin-type prostaglandin D synthase; H-PGDS, hematopoietic prostaglandin D synthase; CSF, cerebrospinal fluid; COX, cyclooxygenase; KO, knockout; TG, transgenic; HQL-79, 4-benzhydryloxy-1-(3-(1H-tetrazol-5-yl)-propyl)-piperidine.
References
- 1.Ueno, R., Ishikawa, Y., Nakayama, T., and Hayaishi, O. (1982) Biochem. Biophys. Res. Commun. 109 576–582 [DOI] [PubMed] [Google Scholar]
- 2.Qu, W. M., Huang, Z. L., Xu, X. H., Aritake, K., Eguchi, N., Nambu, F., Narumiya, S., Urade, Y., and Hayaishi, O. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 17949–17954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewis, R. A., Soter, N. A., Diamond, P. T., Austen, K. F., Oates, J. A., and Roberts, L. J., 2nd. (1982) J. Immunol. 129 1627–1631 [PubMed] [Google Scholar]
- 4.Hirata, M., Kakizuka, A., Aizawa, M., Ushikubi, F., and Narumiya, S. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 11192–11196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagata, K., Hirai, H., Tanaka, K., Ogawa, K., Aso, T., Sugamura, K., Nakamura, M., and Takano, S. (1999) FEBS Lett. 459 195–199 [DOI] [PubMed] [Google Scholar]
- 6.Mizoguchi, A., Eguchi, N., Kimura, K., Kiyohara, Y., Qu, W. M., Huang, Z. L., Mochizuki, T., Lazarus, M., Kobayashi, T., Kaneko, T., Narumiya, S., Urade, Y., and Hayaishi, O. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 11674–11679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eguchi, N., Minami, T., Shirafuji, N., Kanaoka, Y., Tanaka, T., Nagata, A., Yoshida, N., Urade, Y., Ito, S., and Hayaishi, O. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 726–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsuoka, T., Hirata, M., Tanaka, H., Takahashi, Y., Murata, T., Kabashima, K., Sugimoto, Y., Kobayashi, T., Ushikubi, F., Aze, Y., Eguchi, N., Urade, Y., Yoshida, N., Kimura, K., Mizoguchi, A., Honda, Y., Nagai, H., and Narumiya, S. (2000) Science 287 2013–2017 [DOI] [PubMed] [Google Scholar]
- 9.Hirai, H., Tanaka, K., Yoshie, O., Ogawa, K., Kenmotsu, K., Takamori, Y., Ichimasa, M., Sugamura, K., Nakamura, M., Takano, S., and Nagata, K. (2001) J. Exp. Med. 193 255–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Urade, Y., Fujimoto, N., and Hayaishi, O. (1985) J. Biol. Chem. 260 12410–12415 [PubMed] [Google Scholar]
- 11.Urade, Y., Eguchi, N., and Hayaishi, O. (2006) in Lipocalins (Åkerström, B., Flower, D., and Salier, J.P., eds) Vol. 9, pp. 99–109, Landes Bioscience/Eurekah.com, Georgetown, TX
- 12.Nagata, A., Suzuki, Y., Igarashi, M., Eguchi, N., Toh, H., Urade, Y., and Hayaishi, O. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 4020–4024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Urade, Y., and Hayaishi, O. (2000) Biochim. Biophys. Acta 1482 259–271 [DOI] [PubMed] [Google Scholar]
- 14.Christ-Hazelhof, E., and Nugteren, D. H. (1979) Biochim. Biophys. Acta 572 43–51 [DOI] [PubMed] [Google Scholar]
- 15.Urade, Y., Fujimoto, N., Ujihara, M., and Hayaishi, O. (1987) J. Biol. Chem. 262 3820–3825 [PubMed] [Google Scholar]
- 16.Urade, Y., Mohri, I., Aritake, K., Inoue, T., and Miyano, M. (2006) in Functional and Structural Biology on the Lipo-network (Morikawa, K. T., ed) pp. 135–164, Transworld Research Network, Karala, India
- 17.Ujihara, M., Tsuchida, S., Satoh, K., Sato, K., and Urade, Y. (1988) Arch. Biochem. Biophys. 264 428–437 [DOI] [PubMed] [Google Scholar]
- 18.Beuckmann, C. T., Lazarus, M., Gerashchenko, D., Mizoguchi, A., Nomura, S., Mohri, I., Uesugi, A., Kaneko, T., Mizuno, N., Hayaishi, O., and Urade, Y. (2000) J. Comp. Neurol. 428 62–78 [DOI] [PubMed] [Google Scholar]
- 19.Beuckmann, C. T., Gordon, W. C., Kanaoka, Y., Eguchi, N., Marcheselli, V. L., Gerashchenko, D. Y., Urade, Y., Hayaishi, O., and Bazan, N. G. (1996) J. Neurosci. 16 6119–6124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eguchi, Y., Eguchi, N., Oda, H., Seiki, K., Kijima, Y., Matsu-ura, Y., Urade, Y., and Hayaishi, O. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 14689–14694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerena, R. L., Eguchi, N., Urade, Y., and Killian, G. J. (2000) J. Androl. 21 848–854 [PubMed] [Google Scholar]
- 22.Pinzar, E., Kanaoka, Y., Inui, T., Eguchi, N., Urade, Y., and Hayaishi, O. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 4903–4907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malki, S., Nef, S., Notarnicola, C., Thevenet, L., Gasca, S., Mejean, C., Berta, P., Poulat, F., and Boizet-Bonhoure, B. (2005) EMBO J. 24 1798–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ragolia, L., Palaia, T., Hall, C. E., Maesaka, J. K., Eguchi, N., and Urade, Y. (2005) J. Biol. Chem. 280 29946–29955 [DOI] [PubMed] [Google Scholar]
- 25.Tanaka, R., Miwa, Y., Mou, K., Tomikawa, M., Eguchi, N., Urade, Y., Takahashi-Yanaga, F., Morimoto, S., Wake, N., and Sasaguri, T. (2009) Biochem. Biophys. Res. Commun. 378 851–856 [DOI] [PubMed] [Google Scholar]
- 26.Fujimori, K., Aritake, K., and Urade, Y. (2007) J. Biol. Chem. 282 18458–18466 [DOI] [PubMed] [Google Scholar]
- 27.Hoffmann, A., Conradt, H. S., Gross, G., Nimtz, M., Lottspeich, F., and Wurster, U. (1993) J. Neurochem. 61 451–456 [DOI] [PubMed] [Google Scholar]
- 28.Watanabe, K., Urade, Y., Mader, M., Murphy, C., and Hayaishi, O. (1994) Biochem. Biophys. Res. Commun. 203 1110–1116 [DOI] [PubMed] [Google Scholar]
- 29.Gerena, R. L., Irikura, D., Urade, Y., Eguchi, N., Chapman, D. A., and Killian, G. J. (1998) Biol. Reprod. 58 826–833 [DOI] [PubMed] [Google Scholar]
- 30.Tokugawa, Y., Kunishige, I., Kubota, Y., Shimoya, K., Nobunaga, T., Kimura, T., Saji, F., Murata, Y., Eguchi, N., Oda, H., Urade, Y., and Hayaishi, O. (1998) Biol. Reprod. 58 600–607 [DOI] [PubMed] [Google Scholar]
- 31.Clausen, E., and Pedersen, J. (1961) Ugeskr. Laeger. 123 620–622 [PubMed] [Google Scholar]
- 32.Pervaiz, S., and Brew, K. (1987) FASEB J. 1 209–214 [DOI] [PubMed] [Google Scholar]
- 33.Tanaka, T., Urade, Y., Kimura, H., Eguchi, N., Nishikawa, A., and Hayaishi, O. (1997) J. Biol. Chem. 272 15789–15795 [DOI] [PubMed] [Google Scholar]
- 34.Beuckmann, C. T., Aoyagi, M., Okazaki, I., Hiroike, T., Toh, H., Hayaishi, O., and Urade, Y. (1999) Biochemistry 38 8006–8013 [DOI] [PubMed] [Google Scholar]
- 35.Mohri, I., Taniike, M., Okazaki, I., Kagitani-Shimono, K., Aritake, K., Kanekiyo, T., Yagi, T., Takikita, S., Kim, H. S., Urade, Y., and Suzuki, K. (2006) J. Neurochem. 97 641–651 [DOI] [PubMed] [Google Scholar]
- 36.Kanekiyo, T., Ban, T., Aritake, K., Huang, Z. L., Qu, W. M., Okazaki, I., Mohri, I., Murayama, S., Ozono, K., Taniike, M., Goto, Y., and Urade, Y. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 6412–6417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanaoka, Y., Ago, H., Inagaki, E., Nanayama, T., Miyano, M., Kikuno, R., Fujii, Y., Eguchi, N., Toh, H., Urade, Y., and Hayaishi, O. (1997) Cell 90 1085–1095 [DOI] [PubMed] [Google Scholar]
- 38.Urade, Y., and Hayaishi, O. (2000) Vitam. Horm. 58 89–120 [DOI] [PubMed] [Google Scholar]
- 39.Urade, Y., and Eguchi, N. (2002) Prostaglandins Other Lipid Mediat. 68 375–382 [DOI] [PubMed] [Google Scholar]
- 40.Islam, F., Watanabe, Y., Morii, H., and Hayaishi, O. (1991) Arch. Biochem. Biophys. 289 161–166 [DOI] [PubMed] [Google Scholar]
- 41.Matsumura, H., Takahata, R., and Hayaishi, O. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 9046–9050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aritake, K., Kado, Y., Inoue, T., Miyano, M., and Urade, Y. (2006) J. Biol. Chem. 281 15277–15286 [DOI] [PubMed] [Google Scholar]
- 43.Fujimori, K., Aritake, K., and Urade, Y. (2008) Gene (Amst.) 426 72–80 [DOI] [PubMed] [Google Scholar]
- 44.Inui, T., Ohkubo, T., Emi, M., Irikura, D., Hayaishi, O., and Urade, Y. (2003) J. Biol. Chem. 278 2845–2852 [DOI] [PubMed] [Google Scholar]
- 45.Irikura, D., Kumasaka, T., Yamamoto, M., Ago, H., Miyano, M., Kubata, K. B., Sakai, H., Hayaishi, O., and Urade, Y. (2003) J. Biochem. (Tokyo) 133 29–32 [DOI] [PubMed] [Google Scholar]
- 46.Inoue, T., Irikura, D., Okazaki, N., Kinugasa, S., Matsumura, H., Uodome, N., Yamamoto, M., Kumasaka, T., Miyano, M., Kai, Y., and Urade, Y. (2003) Nat. Struct. Biol. 10 291–296 [DOI] [PubMed] [Google Scholar]
- 47.Lazarus, M., Kubata, B. K., Eguchi, N., Fujitani, Y., Urade, Y., and Hayaishi, O. (2002) Arch. Biochem. Biophys 397 336–341 [DOI] [PubMed] [Google Scholar]
- 48.Yamamoto, S. (1982) Methods Enzymol. 86 55–60 [DOI] [PubMed] [Google Scholar]
- 49.Shimamoto, S., Yoshida, T., Inui, T., Gohda, K., Kobayashi, Y., Fujimori, K., Tsurumura, T., Aritake, K., Urade, Y., and Ohkubo, T. (2007) J. Biol. Chem. 282 31373–31379 [DOI] [PubMed] [Google Scholar]
- 50.Mahmud, I., Ueda, N., Yamaguchi, H., Yamashita, R., Yamamoto, S., Kanaoka, Y., Urade, Y., and Hayaishi, O. (1997) J. Biol. Chem. 272 28263–28266 [DOI] [PubMed] [Google Scholar]
- 51.Trivedi, S. G., Newson, J., Rajakariar, R., Jacques, T. S., Hannon, R., Kanaoka, Y., Eguchi, N., Colville-Nash, P., and Gilroy, D. W. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 5179–5184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fujitani, Y., Kanaoka, Y., Aritake, K., Uodome, N., Okazaki-Hatake, K., and Urade, Y. (2002) J. Immunol. 168 443–449 [DOI] [PubMed] [Google Scholar]
- 53.Craik, D. J., and Wilce, J. A. (1997) Methods Mol. Biol. 60 195–232 [DOI] [PubMed] [Google Scholar]
- 54.White, D. M., Takeda, T., DeGroot, L. J., Stefansson, K., and Arnason, B. G. (1997) J. Biol. Chem. 272 14387–14393 [DOI] [PubMed] [Google Scholar]