Abstract
The bacterial helicase-nuclease complex AddAB converts double-stranded DNA breaks into substrates for RecA-dependent recombinational repair. Here we show that the AddB subunit contains a novel class of nuclease domain distinguished by the presence of an iron-sulfur cluster. The cluster is coordinated by an unusual arrangement of cysteine residues that originate from both sides of the AddB nuclease, forming an “iron staple” that is required for the local structural integrity of this domain. Disruption of the iron-sulfur cluster by mutagenesis eliminates the ability of AddAB to bind to duplex DNA ends without affecting the single-stranded DNA-dependent ATPase activity. Sequence analysis suggests that a related iron staple nuclease domain is present in the eukaryotic DNA replication/repair factor Dna2, where it is also associated with a DNA helicase motor.
Double-stranded DNA (dsDNA)2 breaks are caused by a variety of endogenous and exogenous factors including the collapse of impaired DNA replication forks. Unrepaired or misrepaired breaks lead to genomic instability or cell death, and consequently cells have developed mechanisms to repair them (1). In one such mechanism, DNA breaks are salvaged by the recombinational repair machinery, which uses a homologous DNA molecule as a template for the accurate repair of the damage. This process is initiated by the conversion of the free DNA end into a 3′ single-stranded DNA (ssDNA) overhang, which is a substrate for RecA/Rad51 nucleoprotein filament formation (2, 3). In many bacteria, this initiation step is performed by a helicase-nuclease complex, of which there are two distinct classes. The RecBCD-type enzymes are found in Gram (-)ve bacteria, whereas AddAB-type complexes are found in Gram (+)ve bacteria and some proteobacteria (4–8). The prototypical members of each class are the well studied Escherichia coli RecBCD enzyme and the Bacillus subtilis AddAB complex. Eukaryotic cells do not possess obvious structural homologues of either type of complex. In bacteria, the loss of AddAB/RecBCD activity results in defective recombination pathways and sensitivity to DNA damaging agents and, in pathogenic species, to a reduction in the ability to resist oxidative attacks from phagocytic cells (Ref. 9 and references therein).
Both AddAB and RecBCD catalyze the same overall reaction, converting a DNA break into a 3′-ssDNA overhang in a manner that is regulated by a specific DNA sequence called cross-over hotspot instigator (Chi). They bind tightly to the free DNA end and then translocate into and unwind the DNA duplex using energy derived from the hydrolysis of ATP. Prior to Chi recognition, the enzymes degrade both nascent ssDNA strands. However, upon an encounter with the Chi sequence, the nuclease activity on the 3′ → 5′ strand is attenuated, but the enzymes continue to unwind the DNA duplex and degrade the 5′ → 3′ strand. The net result is a dsDNA molecule with a 3′-ssDNA tail terminating with a Chi sequence (10).
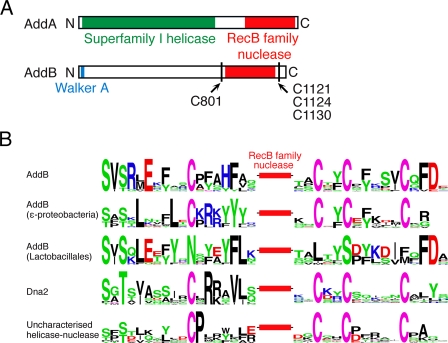
Despite their functional similarity, AddAB- and RecBCD-type complexes are structurally and mechanistically distinct. DNA translocation and unwinding is driven by Superfamily I helicase motors in both cases. However, the RecBCD enzymes employ two motors of opposite polarity to drive translocation along DNA (11, 12), whereas AddAB-type systems rely on a single helicase domain (4). In RecBCD, a single nuclease active site is responsible for all cleavage events on both strands of the duplex (13). This activity resides in a discrete ∼30-kDa domain in the RecB polypeptide (14), which is the prototypical member of the “RecB-like” family of nucleases (15). In contrast, the AddAB-type enzymes employ a dual-nuclease mechanism; two RecB family nuclease domains, found at the C termini of the AddA and AddB subunits, are responsible for cleavage of the 3′ → 5′ and 5′ → 3′ strands, respectively (Fig. 1A) (16, 17). The RecB family of nuclease domains is distinguished by the presence of a family-specific motif containing conserved glutamine and tyrosine residues (15, 18). This motif may transiently stabilize the scissile ssDNA strand at the nuclease active site and is potentially of special importance in the case of enzymes that cleave DNA while moving, as in helicase-nuclease complexes (19). In a subset of RecB family nucleases, three conserved cysteine residues were previously noted that might be responsible for metal cofactor coordination (18). In this report, we demonstrate that the nuclease domain of AddB contains an iron-sulfur cluster and identify four conserved cysteine residues that coordinate this cluster. We have found equivalent conserved residues in other RecB family nucleases, establishing a new group of iron-containing nucleases that includes the essential eukaryotic DNA replication/repair factor Dna2. We further show that, in AddAB, the iron-sulfur cluster stabilizes the local structure of the associated nuclease domain and is essential for the binding and processing of broken DNA.
FIGURE 1.
A, primary structure diagram of the AddA and AddB polypeptides with the helicase and nuclease domains colored green and red, respectively. The positions and residue numbers of the conserved cysteine residues are indicated. B, the conserved cysteine residues that flank RecB family nuclease domains in four different classes of enzyme are shown. Sequence motifs were generated using WebLogo and represent alignments of proteins from a minimum of 10 different organisms. The amino acids are colored according to their chemical properties with green representing polar residues (Gly, Ser, Thr, Tyr, Gln, and Asn), black representing hydrophobic residues (Ala, Val, Leu, Ile, Pro, Trp, Phe, and Met), blue representing basic residues (Lys, Arg, and His), and red representing acidic residues (Asp and Glu). Cysteine residues are shown in purple. The equivalent stretches of amino acids from AddB proteins in the Lactobacillales, which do not contain the conserved cysteine residues, are also shown. The uncharacterized enzyme class is a SF1 helicase-nuclease fusion. See text for discussion.
EXPERIMENTAL PROCEDURES
AddAB Mutagenesis, Expression, and Purification—Mutagenesis, expression, and purification of the AddA and AddAB proteins were performed as described previously with slight modifications (16). We are unable to purify AddB protein separately because it is highly insoluble when over-expressed in the absence of AddA. A detailed description of these procedures is available in the supplemental “Methods.”
Bathophenathroline Iron Chelation Assay—The amount of iron in AddAB preparations was calculated using the bathophenathroline assay (20). AddAB samples ranging from 16 to 33 μm were denatured by incubation with concentrated HCl at 100 °C for 15 min. Following neutralization and the removal of insoluble material, ascorbic acid, followed by disodium bathophenanthrolinedisulfonate salt (Fluka) were added to 0.26 and 0.021%, respectively. The samples were incubated at room temperature for 1 h prior to the absorbance at 535 nm being measured. The concentration of Fe2+ was calculated using the extinction coefficient 22,369 m-1 cm-1 (21).
Electron Paramagnetic Resonance Spectroscopy—AddAB protein (∼10 mg/ml) was oxidized with 0.3 mm K3Fe(CN)6 for 10 min. EPR spectra were measured on a Bruker ESP300 spectrometer with an ER4122 Super-high-Q cavity and an Oxford Instruments ESR900 helium flow cryostat. The conditions were as follows: temperature, 12 K; microwave power, 2 milliwatts; frequency, 9.387 GHz; field modulation amplitude, 0.19 mT; and frequency, 100 kHz.
Analytical Gel Filtration—AddAB samples (concentration, 1 μm; volume, 100 μl) were injected onto a Superdex 200 HR 10/30 gel filtration column (GE Healthcare) equilibrated in 20 mm Tris-Cl, pH 7.5, 0.1 mm EDTA, 150 mm NaCl, 0.1 mm dithiothreitol. Protein from the peak fractions was precipitated with 4 volumes of -20 °C acetone before analysis with 10% SDS-PAGE. The standard curve was generated by running protein standards (Sigma) under identical conditions.
Limited Trypsin Proteolysis—Reactions containing 50 mm Tris acetate, 2 mm magnesium acetate, 1 mm dithiothreitol, and 1 μm AddAB were initiated by addition of trypsin to 1 μg/ml. The reactions were allowed to proceed at room temperature for the indicated times prior to quenching with an equal volume of 63 mm Tris-Cl, pH 6.8, 2% SDS, 25% glycerol, and 715 mm 2-mercaptoethanol. The samples were analyzed on 10% SDS-polyacrylamide gels. Prior to N-terminal sequencing (Proteomics Facility, University of Bristol), the samples were transferred to polyvinylidene difluoride membrane using standard techniques.
Electrophoretic Mobility Shift Assay—DNA substrates consisted of a 15-bp duplex region with four base 3′ overhangs as described previously (16). AddAB (2 nm) and AddAB iron-sulfur mutants (200 nm) were mixed with 1 nm radiolabeled substrate DNA in buffer containing 25 mm Tris acetate, pH 7.5, 2 mm magnesium acetate, 2.5% Ficoll 400, 1 mm dithiothreitol. The samples were incubated at 20 °C for 10 min prior to electrophoresis through 8% polyacrylamide gels in TBE. The gels were dried on DEAE paper, exposed to phosphor screens, and visualized using a Typhoon 9400 with image quant software.
Luminol Metalloprotein Detection—Nondenaturing polyacrylamide gels were stained for the presence of metalloproteins using the chemiluinescent substrate luminol (Sigma) (22, 23). Briefly, 10 μg of proteins were separated by native PAGE through 6% gels using Laemmli buffers without SDS. The gels were washed extensively in Milli-Q water (Millipore) prior to 2 min of incubation in 50 ml of Chelex-100 (Sigma)-treated luminol solution (11 mm luminol, 500 mm Na2CO3, 230 mm H2O2). Luminescence was recorded using a Typhoon 9400 variable mode imager (GE Healthcare). Following imaging, the gels were washed with water and stained for protein with Coomassie Blue (Bio-Rad).
Double-stranded DNA End Processing Reaction—Reactions containing 25 mm Tris acetate pH 7.5, 2 mm magnesium acetate, 2 μm E. coli single-stranded DNA binding protein, 1 nm molecules Cla1 linearized pADG6406, and 1 mm dithiothreitol were initiated by the addition of wild type or mutant AddAB proteins to final concentrations of 1 nm. The reactions were allowed to proceed for 2 min at 37 °C prior to quenching and analysis as described previously (16).
Dye Displacement Helicase Assay—The experiments were performed at 37 °C using an SF61-DX2 stopped flow fluorimeter with a mercury-xenon lamp (TgK Scientific Limited, Bradford-on-Avon, UK). The excitation light was set at 366 nm with a 400-nm cut-off filter. All of the concentrations stated below are final, after mixing in the stopped flow. The experiments were performed by incubating 2 nm AddAB proteins with 0.1 nm EcoR I-linearized pBR322 DNA in a buffer containing 50 mm Tris acetate, pH 7.5, 2 mm magnesium acetate, 1 mm dithiothreitol, 175 nm E. coli SSB, and 200 nm Hoechst 33258 dye for ∼5 min. The reactions were initiated by rapid mixing with an equal volume of 0.5 mm ATP in the same buffer. The fluorescence signals obtained were calibrated using heat-denatured substrate DNA.
ATPase Assays—ATPase activity was measured by coupling the hydrolysis of ATP to the oxidation of NADH. The reactions were carried out in a buffer containing 25 mm Tris acetate, pH 6.5, 2 mm magnesium acetate, 0.5 mm dithiothreitol, 40 units/ml pyruvate dehydrogenase (Sigma), 40 units/ml lactate dehydrogenase (Sigma), 1.5 mm P-enolpyruvate, and 100 μg/ml NADH. For ssDNA-dependent ATPase experiments, the reactions contained 25 μm poly(dT) (in nucleotides), 10 nm AddAB, and varying concentrations of ATP. The initial rates were calculated and fitted to the Michaelis-Menten equation to generate apparent kcat values. Double-strand DNA-dependent ATPase assays were conducted in the above buffer with 0.5 nm molecules λ DNA, 7.5 μm E. coli SSB, 1 mm ATP, and 2.5 nm AddAB. The rates of ATP hydrolysis were calculated over the first 15 s of the reaction for wild type AddAB and 4 min for iron-sulfur mutant enzymes.
RESULTS
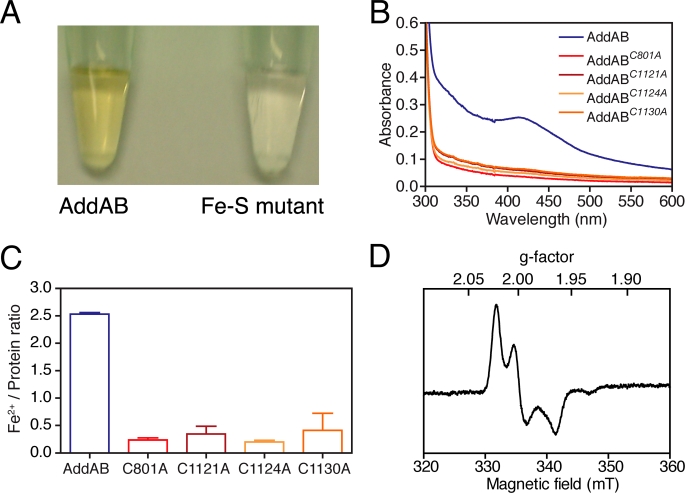
AddAB Contains an Iron-Sulfur Cluster That Is Coordinated by Four Conserved Cysteine Residues Flanking the AddB Nuclease Domain—Purified B. subtilis AddAB complex displays properties that are characteristic of iron-sulfur proteins (Fig. 2). The protein preparation appears yellow-brown at high concentrations (Fig. 2A), and the absorbance spectrum has a broad shoulder of absorbance to ∼450 nm, reminiscent of those displayed by proteins containing [3Fe-4S] and [4Fe-4S] clusters (Fig. 2B) (24). To show directly that purified AddAB contains iron, the sample was denatured and reduced, allowing the chelation of Fe2+ by bathophenathroline and the determination of the iron concentration using a colorimetric assay. Several different preparations of AddAB were found to contain anywhere between 2.5 and 3.5 Fe2+ ions per AddAB heterodimer (Fig. 2C and data not shown).
FIGURE 2.
AddAB contains an iron-sulfur cluster. A, concentrated samples of AddAB have a yellow-brown appearance, whereas the iron-sulfur mutant proteins are colorless. (The C1130A mutant complex is shown as a representative example). B, UV-visible spectra of concentrated AddAB and the four iron-sulfur mutant proteins. C, ratios of Fe2+:AddAB determined using the bathophenathroline assay. D, EPR spectrum of oxidized AddAB protein.
To confirm the presence of an iron-sulfur cluster and to obtain information regarding its structure, we employed electron paramagnetic resonance (EPR) spectroscopy. Neither native AddAB (as prepared) nor the preparation reduced with sodium dithionite produced an EPR signal. However, treatment with the oxidizing agent potassium hexacyanoferrate (III) yielded a strong EPR signal with principal g factors 2.019, 1.997, and 1.962 (Fig. 2D). Signals from iron-sulfur proteins in the oxidized state, with g factors near to 2.0, are characteristic of [3Fe-4S]+ or [4Fe-4S]3+ clusters. The oxidation process was fairly slow, requiring ∼10 min for maximal signal development, conditions under which [4Fe-4S] clusters can become oxidized to produce [3Fe-4S] clusters (25, 26). Hexacyanoferrate (III) facilitates this process by capturing the fourth iron atom as ferric hexacyanoferrate. However, another class of [4Fe-4S] proteins, the high potential iron-sulfur proteins can also give EPR signals in their oxidized state with g factors of ∼2.05, which may resemble that observed for AddAB (25). However, it is more likely that the species observed in the oxidized AddAB is a [3Fe-4S] cluster derived from a [4Fe-4S] cluster, because the high potential iron-sulfur proteins are a rather restricted family of small electron transfer proteins (27).
The type of iron-sulfur cluster present can potentially also be distinguished by considering the sequence around the coordinating amino acids. For example, the most common coordination spheres for [4Fe-4S] centers involve four conserved cysteine ligands, at least three of which are closely spaced in the primary sequence, whereas the fourth can be distant (28). However, there are a few examples in which one cysteine is replaced by either a histidine, aspartate, or serine residue (29). The AddB polypeptide contains exactly four conserved cysteine residues (Fig. 1). The second, third, and fourth candidate ligands are located to the C-terminal side of the AddB nuclease domain in the pattern CX2CX5C. These residues are equivalent to the conserved cysteine triplet previously noted in some RecB family nucleases (18), and their spacing is equivalent to the last three cysteine ligands in certain iron-sulfur containing DNA glycosylases (30, 31). The first candidate cysteine ligand (Cys801) is located some 300 amino acids away from the triplet on the N-terminal side of the nuclease domain (Fig. 1).
To confirm the identity of the putative iron-sulfur cluster ligands, we used site-directed mutagenesis to replace the candidate residues with alanine. Four single cysteine mutant proteins (AddABC801A, AddABC1121A, AddABC1124A, and AddABC1130A) were expressed and purified using the same procedure as for wild type AddAB (supplemental Fig. S1). In all four mutant proteins (hereafter called “iron-sulfur mutants”), the shoulder of absorbance to 450 nm was abolished, and the preparations were colorless at concentrations up to 10 mg/ml (Fig. 2, A and B). Moreover, they contained significantly reduced (∼5-fold) levels of iron compared with wild type AddAB (Fig. 2C). Together with the EPR spectroscopy, the simplest interpretation of these data is that AddAB contains a cubane [4Fe-4S] cluster coordinated by the four conserved cysteine residues Cys801, Cys1121, Cys1124, and Cys1130.
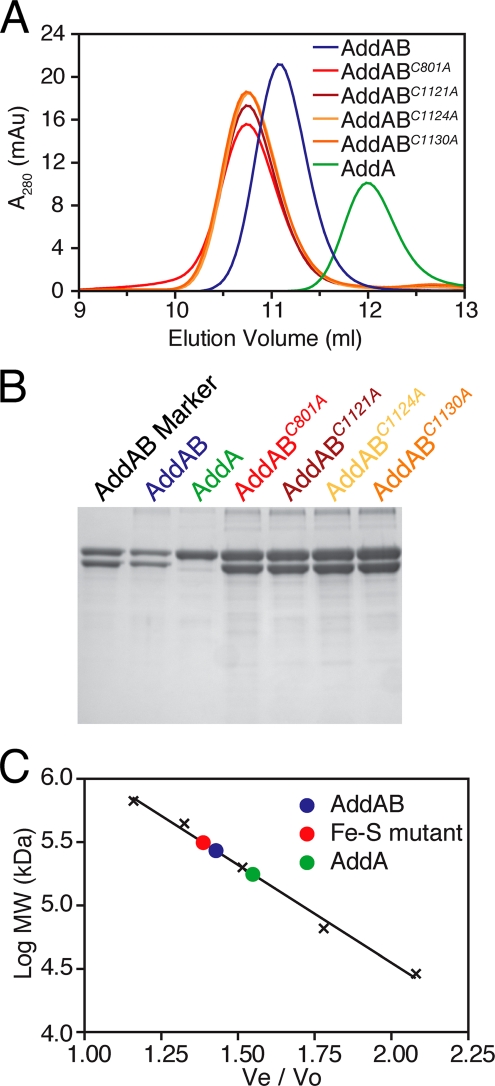
The Iron-Sulfur Cluster in AddB Is Not Required for Interaction with AddA Protein—Our ability to purify the iron-sulfur mutants in the same manner as wild type AddAB suggested that they were not grossly affected in terms of tertiary and quaternary structure. Nevertheless, their structural integrity was investigated further using CD spectroscopy and analytical gel filtration chromatography. The CD spectra for wild type and all four mutant complexes were essentially identical (supplemental Fig. S2) and characteristic of a mixed αβ structure, indicating that most or all of each mutant complex was correctly folded. In gel filtration chromatography experiments (Fig. 3), each iron-sulfur mutant complex eluted as a single symmetrical peak at an apparent molecular mass of 310 kDa. Wild type AddAB eluted at 270 kDa, in excellent agreement with an expected molecular mass for an AddAB heterodimer of 275 kDa. In all cases, the peaks contained the AddA and AddB polypeptides in a 1:1 stoichiometry (Fig. 3B). Purified AddA protein eluted with an apparent molecular mass of 177 kDa compared with an expected value of 143 kDa. There was no evidence for any material running at the position of free AddA in the AddAB preparations. Unfortunately, we were unable to purify and compare the isolated AddB protein in these experiments because of insolubility problems during expression. Together, these data demonstrate that each mutant AddB protein forms a stable heterodimer with AddA under these conditions. The small but reproducible difference in apparent molecular mass between the wild type and mutant complexes may indicate a subtle structural defect in the iron-sulfur mutant AddAB enzymes.
FIGURE 3.
Iron-sulfur mutant AddAB complexes form stable heterodimers. A, gel filtration traces for wild type and iron-sulfur mutant AddAB complexes and the isolated AddA subunit. B, SDS-PAGE analysis of the peak fractions from the gel filtrations runs. C, calibration curve for the gel filtration column generated using protein standards (black crosses). Ve is the elution volume, and Vo is the void volume of the column. The elution positions of the protein samples are shown with filled circles.
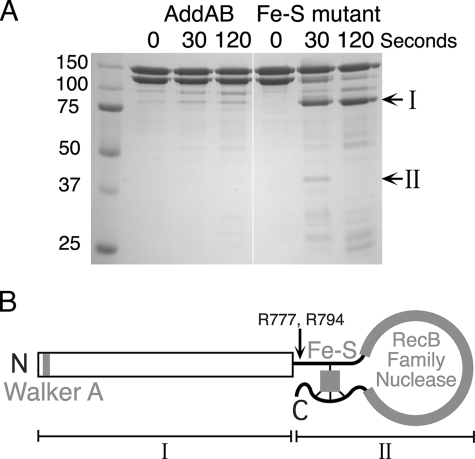
The Iron-Sulfur Cluster Acts as a “Staple” to Stabilize the AddB Nuclease Domain—The intriguing distribution of the four cysteine ligands, immediately flanking either side of the entire RecB family nuclease domain, suggested a potential structural role for the iron-sulfur cluster in stabilizing the local structure of the nuclease domain (Fig. 1). To test for subtle defects in the structure of the AddAB complexes, we employed limited trypsin digestion. Trypsin cleaves exposed peptide bonds to the carboxyl side of arginine and lysine residues. Under the conditions employed in our experiments, wild type AddAB complexes were largely resistant to trypsin digestion, with both the AddA and AddB subunits remaining intact over the time course. In distinct contrast, the AddB subunits of the mutant complexes were sensitive to trypsin proteolysis, rapidly degrading to form a stable fragment of ∼85 kDa (Fig. 4A, band I). Several smaller products were transiently produced, the largest of which was ∼45 kDa (Fig. 4A, band II). The N-terminal sequence of the 85-kDa stable fragment corresponded to the N terminus of AddB. The 45-kDa fragment produced two distinct N-terminal sequences, generated by cleavage after Arg777 and Arg794. We conclude that both of these fragments are produced by cleavage of the entire C-terminal nuclease domain of AddB at a position just upstream of the first iron-sulfur cluster ligand (Cys801). Note that the same region of trypsin sensitivity is formed regardless of which of the four cysteine ligands is mutated, even though the cysteine triplet (Cys1121, Cys1124, and Cys1130) is located ∼300 amino acids away from the site in question (supplemental Fig. S3). This result supports our previous assignment of the iron-sulfur ligands as originating from both flanking regions of the nuclease domain. The cluster acts as an iron staple to pin back the nuclease domain, and its removal results in a localized loss of structural integrity (Fig. 4B). At this stage, we cannot discriminate between the local, but complete unfolding of the AddB nuclease domain or a simple “unhinging” of an otherwise folded domain. This structural defect provides a simple explanation for the apparent molecular mass difference detected by gel filtration analysis (Fig. 3).
FIGURE 4.
AddAB complexes lacking the iron-sulfur cluster are sensitive to trypsin digestion. A, AddAB preparations were treated with trypsin for the indicated time. The reactions were quenched and analyzed by SDS-PAGE. The molecular masses (kDa) of the standards are indicated, as are the positions of two proteolytic products (I and II). The iron-sulfur mutant shown here is C1124A, which is representative for all four mutant AddAB complexes. B, diagram illustrating how the iron-sulfur cluster may act as a staple that is responsible for “pinning back” the AddB nuclease domain. The major proteolytic cleavage sites and products are marked with an arrow and Roman numerals (I and II), respectively.
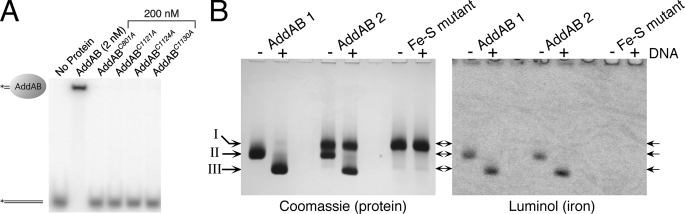
The Iron-Sulfur Cluster Is Essential for dsDNA End Binding— Wild type AddAB binds very tightly to its physiological substrate: a blunt, or nearly blunt, duplex DNA end. Electrophoretic mobility shift assays were used to investigate the DNA end binding properties of the iron-sulfur mutants (Fig. 5A). As reported previously, wild type AddAB displays a subnanomolar affinity for a 15-base pair duplex DNA molecule with short 3′ overhangs, such that 1 nm of the substrate DNA is completely shifted by a small excess of AddAB enzyme. In contrast, no DNA binding was observed by the mutant AddAB complexes even at concentrations of up to 200 nm.
FIGURE 5.
The iron-sulfur cluster is essential for dsDNA end binding. A, electrophoretic mobility shift assays were performed using 1 nm DNA substrate and AddAB at the indicated concentration. B, native polyacrylamide gel showing wild type and iron-sulfur mutant AddAB complexes run in the presence or absence of a 2-fold molar excess of DNA as indicated. The gel was stained for protein and metalloproteins with Coomassie Blue and luminol, respectively. The positions of the protein bands are indicated (I, II, and III). The iron-sulfur mutant shown here is C1124A, which is representative for all four mutant AddAB complexes. AddAB 1 and AddAB 2 are independent wild type AddAB preparations with high and low iron content, respectively.
To directly assess the requirement for the iron-sulfur cluster in dsDNA end binding, we combined reverse electrophoretic mobility shift assays with the method of luminol staining (Fig. 5B) (22, 23). Native polyacrylamide gels were run for wild type and iron-sulfur mutant complexes in the absence and presence of excess DNA-binding substrate and then stained for protein using Coomassie Blue. Each of the iron-sulfur mutant complexes ran as a single band (Fig. 5B, position I, and supplemental Fig. S4), which was shown to contain both the AddA and AddB polypeptides by running SDS-PAGE in a second dimension (supplemental Fig. S4). This confirms our previous conclusion that the mutant AddAB proteins form stable complexes, even at the nanomolar concentrations used in this gel analysis. The mobility of the iron-sulfur mutant complexes was not altered by the presence of DNA, as would be expected based on their inability to shift labeled DNA in the conventional electrophoretic mobility shift assay experiment (Fig. 5A). Wild type AddAB ran as two distinct bands (Fig. 5B, positions I and II), both of which contained the AddA and AddB polypeptides in a ∼1:1 ratio (supplemental Fig. S4) and one of which comigrated with the iron-sulfur mutant complexes (Fig. 5B, position I). The amount of AddAB running in position I was found to vary (5–50% of the total material) in different preparations (compare samples AddAB 1 and AddAB 2 in Fig. 5B). The addition of DNA to wild type AddAB selectively shifted the band in position II but had no effect on the band in position I that comigrates with the iron-sulfur mutants.
We hypothesized that the band in position I represented wild type AddAB complex that had lost the iron-sulfur cluster and was consequently unable to bind dsDNA ends. To test this idea, we used the luminol method to stain the same native polyacrylamide gel for transition metals (Fig. 5B and supplemental Fig. S4). As expected, the iron-sulfur mutant complexes were not stained using luminol, indicating that they do not contain detectable levels of iron. In the lanes for wild type AddAB, the material in position II, but not in position I, was shown to contain iron. Because the addition of DNA selectively shifted the band in position II, this gel demonstrates that the iron-sulfur cluster is essential for dsDNA end binding, even in the context of the wild type amino acid sequence. Moreover, because the shifted band (in position III) also stained positive for iron, we can conclude that the iron-sulfur cluster remains intact in the AddAB-DNA ternary complex.
This method revealed that wild type AddAB preparations contain variable proportions of the complex that lacks the iron-sulfur cluster. The amount of iron-free protein detected by native gel analysis correlates with the amount of iron found in different AddAB preparations using the bathophenanthroline method (see above). The wild type AddAB 1 and AddAB 2 samples shown in Fig. 5B are the highest and lowest iron-content preparations (respectively) that we have obtained to date. We have also observed that the amount of iron-free protein in each sample affects the exact position of the elution peak in gel filtration experiments (data not shown) and that the iron-free material in a wild type preparation is sensitive to proteolysis in the same manner as an iron-sulfur mutant complex (supplemental Fig. S3). Furthermore, iron can be removed from the wild type protein simply by incubating at 37 °C for long periods with a concomitant loss of DNA binding activity (data not shown).
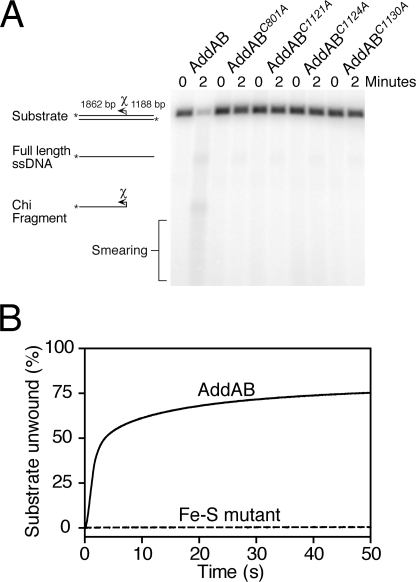
The Iron-Sulfur Cluster Is Essential for DNA Break Processing— Based on their inability to bind free dsDNA ends, one would expect the mutant AddAB complexes to be severely defective for any activity dependent on loading of the complex at its physiological substrate, a DNA break site. To test this, we compared the DNA helicase and DNA break processing activities of wild type and mutant AddAB complexes using established assays. We first monitored the processing of linearized plasmid DNA containing a single B. subtilis Chi sequence (Fig. 6A). As expected based on previous work (32), wild type AddAB rapidly unwinds and degrades ∼80% of the substrate DNA, generating a smear of ssDNA products and a Chi-specific ssDNA fragment. Under identical conditions, the iron-sulfur mutants display severely reduced substrate utilization, consistent with an inability to initiate DNA duplex unwinding and degradation from DNA ends. To confirm this observation, we employed a real-time helicase assay (33) (Fig. 6B). Wild type AddAB unwinds a linearized plasmid substrate with a maximum observed rate of 550 bp s-1 (per DNA end). The iron-sulfur mutants displayed virtually no helicase activity, the traces being comparable with a control experiment with no enzyme. Furthermore, the iron-sulfur mutants were unable to compete with wild type AddAB for the free ends of the DNA, because preincubation of excess iron-sulfur mutant with the substrate had no detectable effect on DNA unwinding by wild type AddAB (data not shown).
FIGURE 6.
The iron-sulfur cluster is essential for dsDNA end processing. A, reactions containing 1 nm linearized plasmid DNA, containing a single Chi sequence, were initiated by the addition of AddAB to a final concentration of 1 nm. The substrate and products of the end processing reactions are indicated. The position of the Chi sequence and the direction in which it is recognized by AddAB is illustrated by an arrow. B, dye displacement helicase assays. Saturating AddAB protein (2 nm) was preincubated with linearized plasmid DNA (0.1 nm) and then rapidly mixed with ATP to initiate unwinding. DNA unwinding is monitored as a decrease in fluorescence from a dsDNA binding dye and is calibrated using heat-denatured substrate DNA. The iron-sulfur mutant shown here is C1124A, which is representative for all four mutant AddAB complexes.
The Iron-Sulfur Mutants Retain ssDNA-dependent ATPase and ssDNA Translocation Activities—Given that our site-directed mutations had targeted the iron-sulfur cluster associated with the AddB subunit, we were interested in determining to what extent biochemical activities associated with the AddA subunit were retained in mutant AddAB complexes. The AddA subunit is the location of the single set of helicase motifs that are found in the AddAB complex. These motifs are a blueprint for a Superfamily I DNA helicase motor that is closely related to the well studied UvrD/Rep/PcrA family of helicases. These enzymes couple ATP hydrolysis to directional translocation along a single strand of DNA, and their ATPase activity is strongly stimulated by single-stranded but not double-stranded DNA (Refs. 34 and 35 and this work). The AddAB- and RecBCD-type helicase-nucleases contain SF1 helicase motors of this family that are, unusually, activated by both single- and double-stranded DNA (36). However, the crystal structure of RecBCD shows how the dsDNA-dependent ATPase activity results from the ability of the complex to load at free dsDNA ends and melt several base pairs, thereby presenting single-stranded DNA to the SF1 helicase motor in the canonical fashion (37). Presumably, single-stranded DNA activates the ATPase by direct binding to the helicase domain, as is the case in other UvrD/Rep/PcrA family enzymes.
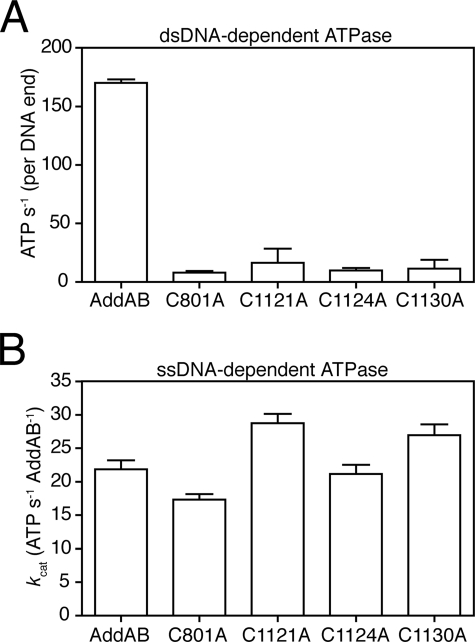
Using a coupled ATPase assay, we measured the initial rate of dsDNA-dependent ATP turnover in wild type AddAB as 170 s-1 (per DNA end) (Fig. 7A). The iron-sulfur mutant complexes displayed reduced ATP hydrolysis (>10-fold) under identical conditions. In stark contrast to these results with duplex DNA as the cofactor, all four iron-sulfur mutant complexes displayed wild type ssDNA-stimulated ATPase activity (Fig. 7B). This activity of the AddAB complexes is entirely dependent on the AddA helicase domain because it is abolished by mutation of the AddA helicase motif I3 but is distinct from the equivalent activity of the free AddA protein, which possesses a more potent ssDNA-dependent ATPase (data not shown). These results suggest that ssDNA motor activity may be retained in the iron-sulfur mutants but that it cannot be targeted to free DNA ends to promote DNA break processing. We tested this idea further by monitoring ATP-dependent translocation on single-stranded DNA using a streptavidin displacement assay (38). All four mutant proteins were indeed capable of displacing streptavidin from the 5′ end of biotinylated oligonucleotides, albeit with greatly reduced efficacy compared with wild type AddAB (supplemental Fig. S5).
FIGURE 7.
A, the dsDNA-dependent ATPase activity of wild type and iron-sulfur mutant AddAB enzymes was measured in reactions containing 0. 5 nm molecules λ DNA and saturating ATP (1 mm) and AddAB enzyme (2.5 nm). The initial rates of ATP hydrolysis, calculated per DNA end binding site, are shown and represent the average and standard error for at least two repeats. B, ssDNA-dependent ATPase activity. Rates of ATP hydrolysis were calculated in reactions containing 25 μm nucleotides poly(dT), 10 nm AddAB, and varying concentrations of ATP. The rates shown are apparent kcat values, generated by least squares fitting of those data to the Michaelis-Menten equation, with the error bars representing the errors on the fits.
DISCUSSION
We have shown that the prototypical AddAB-type helicase-nuclease from B. subtilis contains an iron-sulfur cluster. The cluster is associated with the C-terminal nuclease domain of AddB, which is responsible for the 5′ → 3′ nuclease activity of AddAB (16). Using site-directed mutagenesis, we identified four conserved cysteine residues that are required for cluster coordination. Three are closely spaced in a group to the C-terminal side of the AddB nuclease domain, whereas the final cysteine is located over 300 amino acids away on the N-terminal side of the domain (Fig. 1). Therefore, formation of the cluster pins back the nuclease domain, linking the extreme C-terminal region of the AddB polypeptide to a region at the start of the nuclease domain, by way of an iron-sulfur cluster staple. Loss of the cluster by mutagenesis of any of the four cysteine ligands results in a localized loss of structural integrity at the nuclease domain but does not detectably affect the global folding or stability of the AddAB complex. The cluster is required, directly or indirectly, for binding of AddAB to dsDNA ends and for all associated activities. In contrast, the ssDNA-dependent ATPase activity of AddAB is unaffected by the loss of the iron-sulfur cluster, and iron-sulfur mutant complexes retain the ability to translocate directionally on ssDNA, at least to a limited extent. We conclude that disruption of the iron-sulfur cluster eliminates the ability of the AddAB complex to correctly target its helicase motor domain to DNA break sites.
The AddAB complex contains a second nuclease domain associated with the AddA subunit. This does not contain an iron-sulfur cluster, despite being otherwise homologous to the AddB nuclease domain. Likewise, the single nuclease domain of the functionally related RecBCD complex does not contain an iron-sulfur cluster and is not involved in DNA end binding. Interestingly, the C-terminal region of the RecC subunit shows partial structural similarity with a RecB family nuclease domain, although the critical catalytic residues are no longer present (39, 40). This region of the protein forms a channel at the front of the RecBCD complex that accommodates the 5′-terminated DNA strand at the junction with duplex DNA (37). It is tempting to speculate that the AddB nuclease domain resides in an equivalent position to the RecC nuclease-like domain with respect to the DNA substrate. In that case, the iron staple nuclease would be positioned at the front of the enzyme in a position where it could be involved with dsDNA end binding as we have demonstrated. Moreover, the nuclease domain would be appropriately positioned to degrade the 5′ → 3′ DNA strand during DNA translocation as observed (16).
The iron-sulfur cluster is conserved in all AddB sequences with a few interesting exceptions (Fig. 1B). Despite possessing a high degree of overall sequence similarity to other AddB proteins, those found in lactic acid bacteria do not possess any of the four conserved cysteine residues. Not only are the cysteine residues missing, they have actually been conservatively replaced with alternative amino acids. Therefore, it seems plausible that AddAB complexes in lactic acid bacteria employ an alternative structural element to maintain the nuclease domain staple, but why should they do this? Unusually, lactic acid bacteria are relatively content to grow in medium lacking iron and have been found to contain extremely low intracellular iron levels, possibly as part of a defense mechanism against the oxidative damage caused by hydroxyl radicals (41–44). Consequently, it is common for lactic acid bacteria to substitute iron-metalloproteins with proteins that lack iron or contain alternative metal cofactors (45, 46). Therefore, although somewhat counter-intuitive, the conserved replacement of all four cysteine ligands in this niche of AddB sequences actually re-enforces our conclusion that they are involved in iron-sulfur cluster coordination in “standard” AddB proteins. The AddB sequences of some pathogenic Campylobacter species, which face intense competition for iron during colonization of the human intestine (47), also lack the conserved cysteines.
The B. subtilis AddAB helicase-nuclease is the latest addition to a rapidly growing list of nucleic acid enzymes that contain iron-sulfur clusters. This list includes enzymes involved in a variety of DNA transactions including DNA restriction, RNA primer synthesis, base excision repair, nucleotide excision repair, and mRNA synthesis (20, 31, 48–51), but this is the first example of an iron-sulfur cluster protein associated with a double-stranded DNA break repair pathway. Interestingly, most of these nucleic acid enzymes contain cubane-type [4Fe-4S] clusters, as we strongly suspect to be the case in AddAB. Each of these clusters is coordinated by four conserved cysteine residues, but the spacing and arrangement of the cysteine residues varies from enzyme to enzyme. For example, in both EndoIII and MutY all four cysteine residues are found in close proximity (with the spacing CX6CX2CX5C), and the clusters form discrete domains that were shown to be involved in DNA binding (52). The residues between the first and second cysteine ligands form an iron-sulfur cluster loop that is involved in making DNA backbone contacts (52–54). In AddAB, the entire AddB nuclease domain resides in the region equivalent to the iron-sulfur cluster loop, and so it seems unlikely that AddAB contacts the DNA backbone in exactly the same manner as the DNA glycosylases. The XPD-like DNA repair enzymes were the first helicases to be shown to contain iron-sulfur clusters (20). Loss of the cluster results in the uncoupling of ssDNA-stimulated ATPase activity from DNA translocation and unwinding, and it was suggested that the cluster may target the helicase to the junction with dsDNA (55). Our results are consistent with a similar role for the iron-sulfur cluster in AddAB.
Iron-sulfur clusters fulfill a multitude of different functions including electron transport, iron storage, substrate binding, regulation of gene expression, and structural roles (29). At this stage, the available evidence minimally suggests that the iron-sulfur cluster in AddAB acts as a structural element that is important for DNA end binding, but the exact details of this interaction will await high resolution structural information. We do not know whether the iron-sulfur cluster in AddAB is redox active, but its apparent absence from the Lactobacillus niche seems inconsistent with any mechanism invoking redox sensing or electron transfer as an essential feature. This question will be the subject of further investigation.
This study defines AddB as the prototypical member of a new family of iron-sulfur cluster associated nuclease domains. The data presented here show that the iron-sulfur cluster is intimately involved in the stability of the region of AddB that contains the nuclease domain, although the precise mechanism by which this stabilization is afforded remains to be determined. We have found putative iron staple nuclease domains in several other proteins by searching for the motif CX2CX5C in close proximity to RecB family nuclease motifs (Fig. 1B and supplemental Fig. S6). This cysteine triplet was originally observed to the C-terminal side of RecB family nucleases by Koonin and co-workers (18). Crucially, we always find an additional conserved cysteine ligand (equivalent to Cys801 in AddB) distantly located on the N-terminal side of each of these domains. In AddB proteins, the iron staple nuclease domain is coupled to an active SF1 helicase by virtue of the protein-protein interaction with AddA (Fig. 1). Indeed, it may be that iron staple nucleases always function in conjunction with a DNA motor, because we also found them as direct fusions to both termini of SF1 helicase domains. Of particular interest is the Dna2 protein, which contains a putative iron staple nuclease linked to a C-terminal SF1 helicase domain. Dna2 is a conserved and essential eukaryotic protein that is thought to be involved in Okazaki fragment processing, DNA break repair, and telomere maintenance (56–60). Intriguingly, very recent reports implicate Dna2 as one of the major nucleases responsible for the processing of DNA breaks for recombinational repair (61, 62). Together with the unexpected structural link between Dna2 and AddB, these data suggest that the functional and/or mechanistic parallels between bacterial helicase-nucleases and Dna2 may be stronger than have been previously appreciated.
Supplementary Material
Acknowledgments
We thank Dr. Chris Kay for providing access to EPR facilities and Drs. Beth Bromley and Derek Woolfson for providing access to and assistance with CD measurements. We are grateful to Drs. John Eccleston, Emma Longman, Nigel Savery, and Mark Szczelkun for helpful discussions and/or for providing their comments on the manuscript.
This work was supported by a Biotechnology and Biological Sciences Research Council doctoral training grant (to J. T. P. Y.) and a Royal Society University Research Fellowship (to M. S. D.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental text and Figs. S1–S6.
Footnotes
The abbreviations used are: dsDNA, double-stranded DNA; ssDNA, single-stranded DNA; Chi, cross-over hot spot instigator; EPR, electron paramagnetic resonance.
J. T. P. Yeeles and M. S. Dillingham, unpublished data.
References
- 1.Wyman, C., and Kanaar, R. (2006) Annu. Rev. Genet. 40 363-383 [DOI] [PubMed] [Google Scholar]
- 2.Kowalczykowski, S. C. (2000) Trends Biochem. Sci. 25 156-165 [DOI] [PubMed] [Google Scholar]
- 3.San Filippo, J., Sung, P., and Klein, H. (2008) Annu. Rev. Biochem. 77 229-257 [DOI] [PubMed] [Google Scholar]
- 4.Chedin, F., and Kowalczykowski, S. C. (2002) Mol. Microbiol. 43 823-834 [DOI] [PubMed] [Google Scholar]
- 5.Rocha, E. P., Cornet, E., and Michel, B. (2005) PLoS Genet. 1 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zuniga-Castillo, J., Romero, D., and Martinez-Salazar, J. M. (2004) J. Bacteriol. 186 7905-7913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amundsen, S. K., Fero, J., Hansen, L. M., Cromie, G. A., Solnick, J. V., Smith, G. R., and Salama, N. R. (2008) Mol. Microbiol. 69 994-1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mertens, K., Lantsheer, L., Ennis, D. G., and Samuel, J. E. (2008) Mol. Microbiol. 69 1411-1426 [DOI] [PubMed] [Google Scholar]
- 9.Dillingham, M. S., and Kowalczykowski, S. C. (2008) Microbiol. Mol. Biol. Rev. 72 642-671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson, D. G., and Kowalczykowski, S. C. (1997) Genes Dev. 11 571-581 [DOI] [PubMed] [Google Scholar]
- 11.Dillingham, M. S., Spies, M., and Kowalczykowski, S. C. (2003) Nature 423 893-897 [DOI] [PubMed] [Google Scholar]
- 12.Taylor, A. F., and Smith, G. R. (2003) Nature 423 889-893 [DOI] [PubMed] [Google Scholar]
- 13.Wang, J., Chen, R., and Julin, D. A. (2000) J. Biol. Chem. 275 507-513 [DOI] [PubMed] [Google Scholar]
- 14.Yu, M., Souaya, J., and Julin, D. A. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 981-986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aravind, L., Makarova, K. S., and Koonin, E. V. (2000) Nucleic Acids Res. 28 3417-3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeeles, J. T., and Dillingham, M. S. (2007) J. Mol. Biol. 371 66-78 [DOI] [PubMed] [Google Scholar]
- 17.Quiberoni, A., Biswas, I., El Karoui, M., Rezaiki, L., Tailliez, P., and Gruss, A. (2001) J. Bacteriol. 183 4071-4078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aravind, L., Walker, D. R., and Koonin, E. V. (1999) Nucleic Acids Res. 27 1223-1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sisakova, E., Stanley, L. K., Weiserova, M., and Szczelkun, M. D. (2008) Nucleic Acids Res. 36 3939-3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rudolf, J., Makrantoni, V., Ingledew, W. J., Stark, M. J., and White, M. F. (2006) Mol. Cell 23 801-808 [DOI] [PubMed] [Google Scholar]
- 21.Pieroni, L., Khalil, L., Charlotte, F., Poynard, T., Piton, A., Hainque, B., and Imbert-Bismut, F. (2001) Clin. Chem. 47 2059-2061 [PubMed] [Google Scholar]
- 22.Hogbom, M., Ericsson, U. B., Lam, R., Bakali, H. M., Kuznetsova, E., Nordlund, P., and Zamble, D. B. (2005) Mol. Cell. Proteomics 4 827-834 [DOI] [PubMed] [Google Scholar]
- 23.Ferrer, M., Golyshina, O. V., Beloqui, A., Golyshin, P. N., and Timmis, K. N. (2007) Nature 445 91-94 [DOI] [PubMed] [Google Scholar]
- 24.Bruschi, M., Hatchikian, C., Le Gall, J., Moura, J. J., and Xavier, A. V. (1976) Biochim. Biophys. Acta 449 275-284 [DOI] [PubMed] [Google Scholar]
- 25.Beinert, H., and Thomson, A. J. (1983) Arch. Biochem. Biophys. 222 333-361 [DOI] [PubMed] [Google Scholar]
- 26.Thomson, A. J., Robinson, A. E., Johnson, M. K., Cammack, R., Rao, K. K., and Hall, D. O. (1981) Biochim. Biophys. Acta 637 423-432 [Google Scholar]
- 27.Meyer, J. (2008) J. Biol. Inorg. Chem. 13 157-170 [DOI] [PubMed] [Google Scholar]
- 28.Meyer, J. (1988) Trends Ecol. Evol. 3 222-226 [DOI] [PubMed] [Google Scholar]
- 29.Johnson, D. C., Dean, D. R., Smith, A. D., and Johnson, M. K. (2005) Annu. Rev. Biochem. 74 247-281 [DOI] [PubMed] [Google Scholar]
- 30.Michaels, M. L., Pham, L., Nghiem, Y., Cruz, C., and Miller, J. H. (1990) Nucleic Acids Res. 18 3841-3845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cunningham, R. P., Asahara, H., Bank, J. F., Scholes, C. P., Salerno, J. C., Surerus, K., Munck, E., McCracken, J., Peisach, J., and Emptage, M. H. (1989) Biochemistry 28 4450-4455 [DOI] [PubMed] [Google Scholar]
- 32.Chedin, F., Ehrlich, S. D., and Kowalczykowski, S. C. (2000) J. Mol. Biol. 298 7-20 [DOI] [PubMed] [Google Scholar]
- 33.Eggleston, A. K., Rahim, N. A., and Kowalczykowski, S. C. (1996) Nucleic Acids Res. 24 1179-1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lohman, T. M., Tomko, E. J., and Wu, C. G. (2008) Nat. Rev. Mol. Cell. Biol. 9 391-401 [DOI] [PubMed] [Google Scholar]
- 35.Singleton, M. R., Dillingham, M. S., and Wigley, D. B. (2007) Annu. Rev. Biochem. 76 23-50 [DOI] [PubMed] [Google Scholar]
- 36.Roman, L. J., and Kowalczykowski, S. C. (1989) Biochemistry 28 2873-2881 [DOI] [PubMed] [Google Scholar]
- 37.Singleton, M. R., Dillingham, M. S., Gaudier, M., Kowalczykowski, S. C., and Wigley, D. B. (2004) Nature 432 187-193 [DOI] [PubMed] [Google Scholar]
- 38.Morris, P. D., and Raney, K. D. (1999) Biochemistry 38 5164-5171 [DOI] [PubMed] [Google Scholar]
- 39.Kinch, L. N., Ginalski, K., Rychlewski, L., and Grishin, N. V. (2005) Nucleic Acids Res. 33 3598-3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rigden, D. J. (2005) BMC Struct. Biol. 5 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daly, M. J., Gaidamakova, E. K., Matrosova, V. Y., Vasilenko, A., Zhai, M., Venkateswaran, A., Hess, M., Omelchenko, M. V., Kostandarithes, H. M., Makarova, K. S., Wackett, L. P., Fredrickson, J. K., and Ghosal, D. (2004) Science 306 1025-1028 [DOI] [PubMed] [Google Scholar]
- 42.Posey, J. E., and Gherardini, F. C. (2000) Science 288 1651-1653 [DOI] [PubMed] [Google Scholar]
- 43.Pandey, A., Bringel, F., and Meyer, J. M. (1994) Appl. Microbiol. Biotechnol. 40 735-739 [Google Scholar]
- 44.Archibald, F. (1983) FEMS Microbiol. Lett. 19 29-32 [Google Scholar]
- 45.Gostick, D. O., Green, J., Irvine, A. S., Gasson, M. J., and Guest, J. R. (1998) Microbiology 144, 705-717 [DOI] [PubMed] [Google Scholar]
- 46.Jang, S., and Imlay, J. A. (2007) J. Biol. Chem. 282 929-937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holmes, K., Mulholland, F., Pearson, B. M., Pin, C., McNicholl-Kennedy, J., Ketley, J. M., and Wells, J. M. (2005) Microbiology 151 243-257 [DOI] [PubMed] [Google Scholar]
- 48.Klinge, S., Hirst, J., Maman, J. D., Krude, T., and Pellegrini, L. (2007) Nat. Struct. Mol. Biol. 14 875-877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lambert, A. R., Sussman, D., Shen, B., Maunus, R., Nix, J., Samuelson, J., Xu, S. Y., and Stoddard, B. L. (2008) Structure 16 558-569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weiner, B. E., Huang, H., Dattilo, B. M., Nilges, M. J., Fanning, E., and Chazin, W. J. (2007) J. Biol. Chem. 282 33444-33451 [DOI] [PubMed] [Google Scholar]
- 51.Hirata, A., Klein, B. J., and Murakami, K. S. (2008) Nature 451 851-854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thayer, M. M., Ahern, H., Xing, D., Cunningham, R. P., and Tainer, J. A. (1995) EMBO J. 14 4108-4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guan, Y., Manuel, R. C., Arvai, A. S., Parikh, S. S., Mol, C. D., Miller, J. H., Lloyd, S., and Tainer, J. A. (1998) Nat. Struct. Biol. 5 1058-1064 [DOI] [PubMed] [Google Scholar]
- 54.Chepanoske, C. L., Golinelli, M. P., Williams, S. D., and David, S. S. (2000) Arch. Biochem. Biophys. 380 11-19 [DOI] [PubMed] [Google Scholar]
- 55.Pugh, R. A., Honda, M., Leesley, H., Thomas, A., Lin, Y., Nilges, M. J., Cann, I. K., and Spies, M. (2008) J. Biol. Chem. 283 1732-1743 [DOI] [PubMed] [Google Scholar]
- 56.Bae, S. H., Bae, K. H., Kim, J. A., and Seo, Y. S. (2001) Nature 412 456-461 [DOI] [PubMed] [Google Scholar]
- 57.Budd, M. E., and Campbell, J. L. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 7642-7646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Budd, M. E., and Campbell, J. L. (2000) Mutat. Res. 459 173-186 [DOI] [PubMed] [Google Scholar]
- 59.Fiorentino, D. F., and Crabtree, G. R. (1997) Mol. Biol. Cell 8 2519-2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tomita, K., Kibe, T., Kang, H. Y., Seo, Y. S., Uritani, M., Ushimaru, T., and Ueno, M. (2004) Mol. Cell. Biol. 24 9557-9567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liao, S., Toczylowski, T., and Yan, H. (2008) Nucleic Acids Res. 36 6091-6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu, Z., Chung, W. H., Shim, E. Y., Lee, S. E., and Ira, G. (2008) Cell 134 981-994 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.