Abstract
Ubiquitination and proteasome-mediated degradation of proteins are crucial for eukaryotic physiology and development. The largest class of E3 ubiquitin ligases is made up of the cullin-RING ligases (CRLs), which themselves are positively regulated through conjugation of the ubiquitin-like peptide RUB/NEDD8 to cullins. RUB modification is antagonized by the COP9 signalosome (CSN), an evolutionarily conserved eight-subunit complex that is essential in most eukaryotes and cleaves RUB from cullins. The CSN behaves genetically as an activator of CRLs, although it abolishes CRL activity in vitro. This apparent paradox was recently reconciled in different organisms, as the CSN was shown to prevent autocatalytic degradation of several CRL substrate adaptors. We tested for such a mechanism in the model plant Arabidopsis by measuring the impact of a newly identified viable csn2 mutant on the activity and stability of SCFTIR1, a receptor to the phytohormone auxin and probably the best characterized plant CRL. Our analysis reveals that not only the F-box protein TIR1 but also relevant cullins are destabilized in csn2 and other Arabidopsis csn mutants. These results provide an explanation for the auxin resistance of csn mutants. We further observed in vivo a post-translational modification of TIR1 dependent on the proteasome inhibitor MG-132 and provide evidence for proteasome-mediated degradation of TIR1, CUL1, and ASK1 (Arabidopsis SKP1 homolog). These results are consistent with CSN-dependent protection of Arabidopsis CRLs from autocatalytic degradation, as observed in other eukaryotes, and provide evidence for antagonist roles of the CSN and 26S proteasome in modulating accumulation of the plant CRL SCFTIR1.
Post-translational control of protein turnover by ubiquitination and degradation by the 26S proteasome is a highly regulated process essential for all eukaryotes. In plants, it regulates many developmental and physiological responses such as hormone signaling, cell division, floral development, and maintenance of circadian rhythm (1, 2). Ubiquitin is a small peptide that can be covalently transferred to specific target proteins through dedicated enzymatic machinery. Polyubiquitinated substrates are then normally targeted for proteasome-dependent degradation (3). E3 ubiquitin ligases confer substrate specificity to the ubiquitination machinery and are subdivided into different groups based on their mechanistic and structural characteristics (4). Cullin-RING (really interesting new gene) E3 ligases (CRLs)3 are composed of a cullin subunit, a RING protein (RBX1/Hrt1/Roc1), which recruits the E2 ubiquitin-conjugating enzymes, and a substrate receptor that specifically binds proteins to be ubiquitinated. Arabidopsis expresses four functionally relevant cullins giving rise to three distinct classes of CRLs. The functionally redundant cullins CUL3A and CUL3B associate with BTB/POZ (Bric-a-brac, Tramtrack, and broad complex/Pox virus and zinc finger) domain-containing substrate adaptors (5–7). CUL4 associates with a subgroup of WD40 repeat-containing proteins called DWD proteins via the DDB1 adaptor (8, 9). The best characterized SCF (SKP1-cullin-F-box)-type CRLs incorporate CUL1 (10). Substrate specificity of SCF complexes is conferred by F-box proteins, which associate with CUL1 via a SKP1 adaptor protein. Although these different classes of CRLs are important for development in Arabidopsis (6, 11–14), our knowledge of the precise functions and substrates of individual CRLs is limited. A CUL3-ETO1 complex is involved in ethylene biosynthesis (15), and CUL4-based complexes are important for development and photomorphogenesis (9, 13, 14). By contrast, the activities and substrates of several SCFs are known. The best characterized SCFs are the SCFTIR1/AFB complexes, which function as receptors for the plant hormone auxin (16, 17).
Auxins regulate many aspects of plant growth and development (18) and have been shown to directly bind to the F-box protein TIR1 (transport inhibitor response) and the homologous AFB1–3 proteins inside the cell (16, 17, 19). Auxin binding increases the affinity of SCFTIR1/AFB for its substrates, the Aux/IAA proteins, leading to their increased ubiquitination and turnover. As a result, auxin response factors (ARFs), in which activities are normally repressed through dimerization with Aux/IAAs, are liberated and can thus function as transcriptional regulators of auxin responsive genes (18). Suppression of ARF5-dependent transcription by IAA12 was recently shown to depend on the co-repressor TPL (TOPLESS), which binds to IAA12 (20), and the repressor function of other Aux/IAA proteins might also depend on other TPL-related co-factors (21). Mutations affecting SCFTIR1/AFB activity result in the stabilization of Aux/IAA proteins, decreased induction of auxin-regulated genes, and reduced sensitivity to exogenous auxin. The latter phenotype was used extensively for genetic screens leading to the identification of SCF components and regulators. Several auxin-resistant mutants are defective in the conjugation of the ubiquitin-like protein RUB/NEDD8 (related to ubiquitin/neural precursor cell expressed, developmentally down-regulated 8) to cullins. Although the molecular role of RUB modification is not fully understood, it is essential for CRL activity and promotes E3 ligase function both in vivo and in vitro (reviewed in Ref. 22).
The COP9 signalosome (CSN) cleaves RUB/NEDD8 from cullins (23, 24) and is biochemically antagonist to the RUB conjugation machinery. The CSN is conserved in eukaryotes, is structurally related to the 19S lid complex of the 26S proteasome, and is composed of six PCI (proteasome, COP9, eIF3; CSN1–4 and CSN7–8) and two MPN (Mov34, Pad1 N-terminal; CSN5 and CSN6) domain-containing subunits (25). Although the derubylation activity of CSN resides in the JAMM domain of CSN5 (24), Arabidopsis mutants deficient in any subunit share a common derubylation defect, reflected in the accumulation of RUB-modified cullins, and show the severe cop/det/fus phenotype of constitutive photomorphogenesis (cop/det) and accumulation of anthocyanins (fusca), culminating in early seedling lethality (26, 27). This common phenotype is explained by the dissociation of the holocomplex in all csn mutants, accompanied by partial or complete destabilization of all other subunits with the exception of CSN5 (27). Although loss of CSN function is tolerated in some fungi (28–30), CSN is essential for all animal and plant systems studied thus far. Because RUB conjugation promotes E3 ligase activity, RUB cleavage by the CSN is expected to act as a negative regulator of CRL activity. Although this holds true in vitro, genetic studies show that the CSN acts as a promoter of E3 ligase activity in vivo. This apparent paradox was first reconciled in fission yeast, as CSN was shown to protect CRL substrate adaptors from autocatalytic degradation through both its derubylation activity and a CSN-associated deubiquitinating enzyme (31). Similar observations were subsequently made in other systems (29, 32–34). Additionally, destabilization of cullins upon reduction of CSN function has been reported in some but not all model systems (29, 35, 36). Thus, fundamental differences may occur as manifested by the differential impact of CSN on cullin stability in yeast and Drosophila (31, 35). In Arabidopsis, the characterization of csn mutants has been limited by their early seedling lethality. Using a viable/partial csn mutant, we have provided novel insights into CRL assembly and recycling in plant cells.
EXPERIMENTAL PROCEDURES
Plant Material and Growth Conditions—Wild-type Arabidopsis thaliana accessions used were Columbia (Col-0) and Landsberg erecta (Ler). The Ler sgt1b-3 (37), Ler csn2fus12 (line U228 (38)), Col-0 tir1-1 (39), Col-0 csn5a-1 (40), and Col-0 sgt1beta3 (41) mutants are published.
Plants were grown with a light intensity of ∼200 μm photons·m-2·s-1 (16 h light/8 h darkness) at 24 °C/21 °C (light/dark) in soil or sterile conditions: solid 1/10 Murashige and Skoog basal medium (MS medium, Sigma) complemented with Gamborgs vitamins, 0.8% agar, and 0.5% sucrose or liquid MS medium complemented with 1% sucrose and Gamborgs vitamins under shaking. Seeds were stratified for 48 h at 4 °C prior to transfer to growth chambers.
Ethyl Methane Sulfonate Mutagenesis and the eba Mutant Screen—Ler sgt1b-3 seeds were mutagenized as described (42). 3100 individual M2 families were harvested and tested in the root growth inhibition assay on 0.2 μm 2,4-dichlorphenoxyacetic acid (2,4-D; Sigma) as described below. Candidate mutant lines were back-crossed at least three times prior to physiological analyses.
Hormone Treatments and Photomorphogenesis Test—Root growth inhibition assays were performed with methyl jasmonate (Duchefa) or 2,4-D as described (43). For ethylene tests, seeds were grown in darkness for 5 days on horizontal MS plates with 1% sucrose containing 1-aminocyclopropane-1-carboxylic acid as described (44). For photomorphogenesis tests, seeds sown on MS/10 without sucrose were first exposed to light for 6 h and subsequently grown horizontally in darkness or light. All experiments were performed at least in triplicates. Root elongation and hypocotyl length were measured on photographs using NIH ImageJ software.
HS::AXR3NT-GUS Experiments—The relevant genotypes were isolated in the F2 progeny of a cross between a Col-0 HS::AXR3NT-GUS transgenic plant (45) and a Ler sgt1b-3 csn2-5 plant. At least two independent lines per genotype were analyzed. Plants were grown for 6–7 days in liquid MS medium in 6-well microtiter plates. To induce expression of AXR3NT-GUS (β-glucuronidase), the plates were placed in a 37 °C water bath for 2 h. GUS staining was performed overnight at 37 °C as described (46).
Map-based Cloning of csn2-5 Mutation—A Ler sgt1b-3 csn2-5 cross to Col sgt1beta3 was used as mapping population. Bulk segregant analysis of ∼100 polymorphic AFLP (amplified fragment length polymorphism) markers was performed on pooled DNAs of 10 resistant or sensitive F2 families selected on 0.2 μm 2,4-D using SacI/TaqI adaptors as described (47). Three linked markers were excised, reamplified, sequenced, and localized to chromosome 2 (see supplemental Fig. 1). Mapping was refined using CAPS, dCAPS, and microsatellite markers (primers available upon request) on ∼1300 DNAs from resistant F2 plants selected from the mapping population. Coding regions of genes from the final 46-kb genetic interval containing csn2-5 were amplified by PCR and treated with SURVEYOR nuclease as described by the manufacturer (Transgenomic).
RT-PCR Analysis—RNA isolated from 6-day-old seedlings grown under sterile conditions was reverse-transcribed using SuperScript reverse transcriptase (Invitrogen) and oligo(dT) primers following the manufacturer's instructions. Relative mRNA accumulation was determined in three independent biological samples under nonsaturating PCR conditions (primer sequences available upon request).
Generation of 35S::CSN2-HAStrep Transgenic Lines—The coding region of CSN2 without stop codon was amplified by PCR from total Col-0 cDNA and cloned into pENTR/D as recommended (Invitrogen) giving pE-CSN2. The csn2-5 mutation was introduced by site-directed mutagenesis resulting in pE-csn2-5. The sequences coding for CSN2 and csn2-5 were recombined into pXCSG-HAStrep giving pXCSG-CSN2-HAStrep and pXCSG-csn2-5-HAStrep. To generate pXCSG-HAStrep, the EcoRV gateway rfB cassette was cloned into pXCS-HAStrep SmaI site (48).
Generation of HA-Strep-tagged TIR1—The genomic Col-0 TIR1 region covering the 1.7-kb promoter, complete 5′-untranslated region, and coding region without stop codon was amplified by PCR and cloned into pExtag-HAStrep, giving pXC-np::TIR1-HAStrep.
Plant Transformations—Stable transformation of Arabidopsis, selection, and genetic analysis was performed as described (43).
MG-132, Cycloheximide, and 2,4-D Treatments—Seedlings were grown in 6-well plates using liquid MS in the presence/absence of 100 μm cycloheximide (Sigma), 50 μm MG-132 (Calbiochem), and 5 μm 2,4-D.
StrepII Affinity Purification—StrepII affinity purifications were performed as described (48). For purification of CSN2-HAStrep, 2 g of tissue from 7-day-old seedlings grown on solid MS/10 medium were used. For TIR1 ubiquitination analysis, seedlings were grown in liquid MS medium with or without a 6-h MG-132 and/or 2,4-D treatment.1g of tissue was used for purification, and bound proteins were eluted in Laemmli buffer.
Gel Filtration Analysis—Soluble protein extracts were prepared from 7-day-old in vitro grown seedlings essentially as described (40). A 100-μl sample was loaded on a Superdex 200 HR 10/30 column (Amersham Biosciences) at 0.2 ml/min flow with extraction buffer. 0.7-ml fractions were sampled, precipitated with 10% trichloroacetic acid, and analyzed by SDS-PAGE. Column calibration was performed as described (43).
SDS-PAGE and Immunoblotting—Total soluble protein extracts were prepared from entire seedlings and separated by SDS-PAGE as described (48). Immunoblots with the Strep-Tactin alkaline phosphatase conjugate (IBA GmbH, Göttingen, Germany) were performed as described (48). The following antibodies were used: mouse anti-LexA (Santa Cruz Biotechnology), rabbit anti-CUL1 (11), rabbit anti-CUL3A (5), rabbit anti-CUL4 (13), rabbit anti-CSN2 (25), rabbit anti-CSN5 (38), rabbit anti-CSN6 (49), rabbit anti-HSC70 (SPA-795, Stressgen) (rat anti-HA, 1867423; Roche Applied Science), mouse antiubiquitin antibody (NB300-130; Novus Biologicals), mouse anti-c-Myc (Santa Cruz Biotechnology). Secondary antibodies were purchased from Sigma (alkaline phosphatase conjugates) and Santa Cruz Biotechnology (horseradish peroxidase conjugates). Alkaline phosphatase and horseradish peroxidase activity was detected with p-nitro blue tetrazolium and enhanced chemiluminescence (SuperSignal West Femto chemiluminescent substrate, Pierce), respectively. To quantify immunoblot results, membranes or films derived from three independent biological samples were scanned and analyzed with ImageJ software. Background values were measured above and below the specific signal and used for corrections. Measurements were repeated at least twice, and measurement errors were <5%. Average values and standard deviations for three biological replicates were calculated.
Schizosaccharomyces pombe Experiments—A Gateway cloning cassette was introduced into pREP3X expression vector giving pREP3XG (details available upon request). The genomic sequence coding for SpCSN2 was amplified by PCR from yeast strain 501 (50) and cloned into pENTR/D giving pE-SpCSN2. The csn2-5 mutation was introduced into pE-SpCSN2 by site-directed mutagenesis, giving pE-Spcsn2-5. Sequences coding for AtCSN2, SpCSN2, and Spcsn2-5 were recombined into pREP3XG. The resulting plasmids were transformed in strain pcu1-MYC csn2-d (28). For UV sensitivity tests, three parallel dilution series were prepared from overnight liquid cultures for each genotype. Dilutions plated on Petri dishes were given a single dose of UV-C (254 nm; 0–100 kJ/m2; without lid) using a Stratalinker 2400 (Stratagene, La Jolla, CA). Colony-forming units were determined in the appropriate dilutions after 4 days at 28 °C. Two independent experiments were performed. Total protein extracts were prepared under denaturing conditions as described (51).
Structure Prediction of CSN2—One PSI-BLAST iteration of the A. thaliana CSN2 query sequence on the NCBI nr Database was performed and only sequences with identity higher than 30% were kept (52). A multiple sequence alignment was built from the 27 retrieved CSN2-like sequences using MUSCLE (53). Secondary structure predictions were performed using PsiPred on this alignment (54). HMM-HMM comparison using the HHpred server on the Protein Data Bank data base was used to search for structural templates suitable for comparative modeling (55). From the results, a structural model of CSN2 A. thaliana could be built from several templates detected with high confidence. Residues 40–321 matched, with highest probabilities, the HEAT repeat from the Danio rerio γ-SNAP (probability = 98.9%, E-value = 2.8e-09, sequence ID = 16%) and the Saccharomyces cerevisiae SEC17 (probability = 98.22%, E-value = 1.1e-05, sequence ID = 11%, Protein Data Bank codes 2ifu and 1qqe, respectively); residues 285–388 matched a region spanning the HAM (HEAT analogous motif) and the WH (winged helix) domain of the Homo sapiens eIF3k subunit (probability = 95.02%, E-value = 0.13, sequence ID = 10%, Protein Data Bank code 1rz4); residues 348 -417 matched the WH C-terminal domain of the Mus musculus CSN4 COP9 subunit (probability = 98.88%, E-value = 1.4e-11, sequence ID = 21%, Protein Data Bank code 1ufm). The four templates were combined into a single alignment together with the A. thaliana CSN2 sequence. The alignment was optimized locally by hand so that secondary structures in the template would be least disrupted. 20 models of CSN2 A. thaliana were generated using Modeler v. 8.1 (56), and the model exhibiting the best consensus between Verify3D (57) and Prosa2003 (58) evaluation functions was selected for analysis. Fig. 2B was done using PyMOL (Delano Scientific).
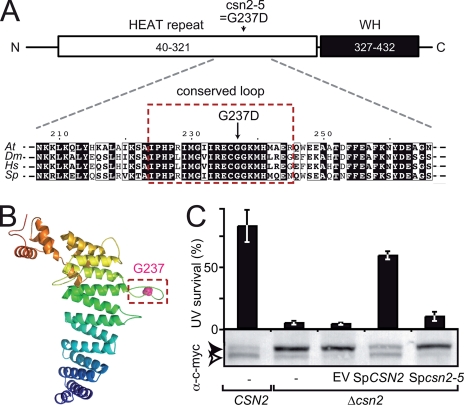
FIGURE 2.
The csn2-5 mutation modifies an evolutionarily conserved loop of CSN2. A, schematic representation of the CSN2 structural domains: N-terminal (N) HEAT repeat (residues 40–321) and C-terminal (C) winged helix (WH) domain (residues 327–432). The position and nature of the amino acid substitution found in csn2-5 mutant protein is shown. Below, an alignment of CSN2 proteins from Arabidopsis (Q8W207; residues 207–266), Drosophila (Q94899; residues 212–271), human (P61201; residues 211–270) and fission yeast (Q9HFR0; residues 213–272) around Arabidopsis Gly237 is shown. Residues boxed in red are highly conserved and belong to a predicted hairpin-like loop structure (framed in B). B, structural model of Arabidopsis CSN2 (residues 40–439) represented as ribbons and colored from the N to the C terminus in blue to red, respectively. Gly237 is highlighted as a pink sphere. C, the csn2-5 mutation abolishes CSN function in S. pombe as assessed by survival after UV-C exposure (50 kJ·m-2·s-1) relative to control. Wild-type CSN2 and mutant Δcsn2 strains expressing wild-type or mutant SpCSN2 were analyzed for complementation of the Δcsn2 UV hypersensitivity. Errors bars represent S.D. between three biological replicates. One representative experiment of two is shown. The level of RUB modification of c-Myc-tagged CUL1 (Pcu1p) expressed in the yeast strains was analyzed by immunoblot of total protein extracts using anti-c-Myc antibody. The open and filled arrowheads correspond to unmodified and RUB-modified cullins, respectively. EV, empty vector.
RESULTS
Isolation of a Novel csn2 Mutant Resistant to Exogenous Auxin—We performed a genetic screen to identify loci contributing to auxin signaling and/or regulators of SCF complexes. The weakly auxin-resistant Ler sgt1b-3 mutant was used as a sensitized genetic background in an auxin resistance screen. SGT1 (suppressor of G2/M allele of skp1) is an essential eukaryotic protein that functions as a co-chaperone of the molecular chaperones HSP70 (heat shock proteins, 70 kDa (43) and HSP90 (59). SGT1 is also needed for optimal activity of several SCFs including SCFTIR1 in Arabidopsis (41), although its exact mode of action on SCFs remains unclear. 3100 individual M2 families originating from ethane methyl sulfonate-mutagenized Ler sgt1b-3 mutant seeds were first germinated on auxin-free medium and then screened for enhanced auxin resistance in a root growth inhibition assay using 0.2 μm synthetic auxin 2,4-D. Under these conditions, root elongation of sgt1b-3 seedlings was fully inhibited (Fig. 1A). The screen identified 13 eba (enhancer of sgt1b-3 on auxin) mutants showing significant and heritable root growth on 0.2 μm 2,4-D. One recessive eba mutation was mapped to a 45-kb region on chromosome II encompassing the CSN2 locus (At2g26990; supplemental Fig. 1). CSN2 is an essential gene coding for CSN subunit 2. Because auxin resistance had been reported previously for partial csn mutants (40, 60), we sequenced CSN2 in our eba mutant and could detect a single base pair exchange (GGT237GAT) causing a predicted G237D amino acid substitution mutation in CSN2. The mutant is hereafter named csn2-5.
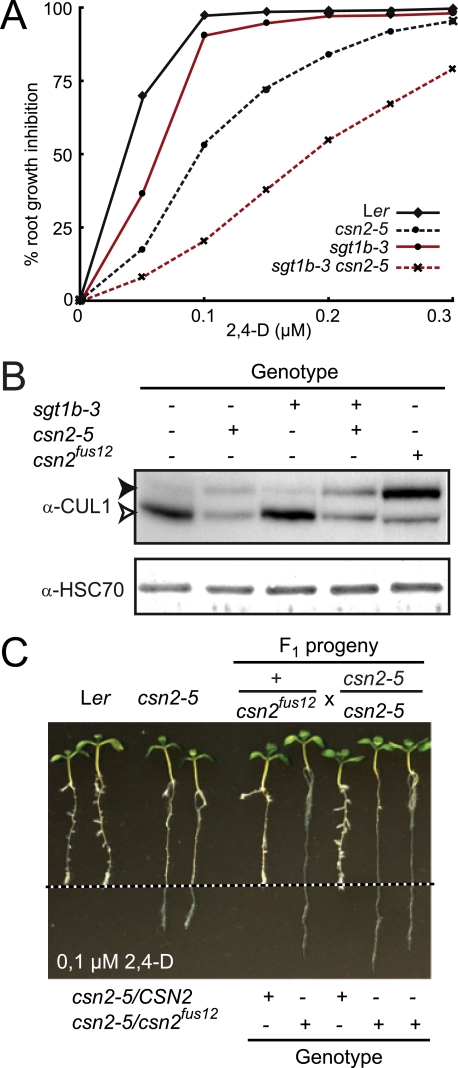
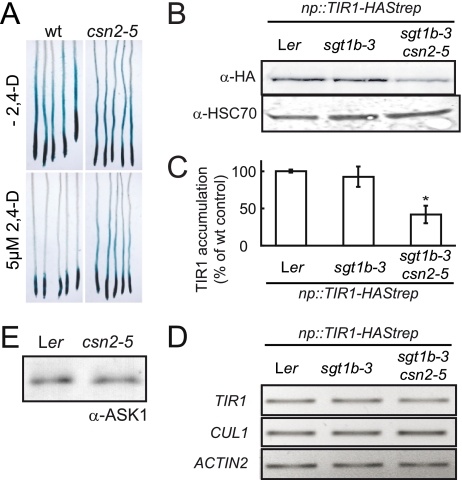
FIGURE 1.
Isolation and characterization of a hypomorphic csn2 mutant. A, inhibition of root elongation by increasing concentrations of synthetic auxin 2,4-D. Data points are averages of at least 10 seedlings, and standard deviations were less than 10%. Measurements of root elongation were performed on 8-day-old seedlings 3 days after transfer on auxin-containing medium. One representative experiment of four repetitions is shown. B, immunoblot analysis of total protein extracts with anti-CUL1 and anti-HSC70 antibodies. Immunodetection of cytosolic/nuclear HSC70 is used as loading control. Open and filled arrowheads correspond to unmodified and RUB-modified CUL1, respectively. + and -, indicate the presence and absence of sgt1b-3 and csn2-5 mutations, respectively. C, allelism test between Ler csn2-5 and Ler csn2fus12 mutants. Plants heterozygous for csn2fus12 were crossed to a homozygous csn2-5 mutant. F1 progeny were analyzed in the root growth inhibition assay on 0.1 μm 2,4-D as described in A. The dashed line indicates the position of the root tip immediately after transfer. F1 seedling genotypes at the CSN2 locus are indicated as determined using allele-specific PCR markers.
To test the dependence of the csn2-5 auxin resistance on the sgt1b-3 background, the double mutant was out-crossed to Ler wild-type plants and the csn2-5 single mutant selected. Although sgt1b-3 clearly enhanced csn2-5 auxin resistance, the csn2-5 mutation alone conferred auxin resistance in root growth inhibition assays (Fig. 1A): Root growth of the csn2-5 single mutant was inhibited by 50% at a concentration of ∼0,1 μm 2,4-D, whereas the originally isolated double mutant was inhibited by 50% at 0,2 μm 2,4-D. These results point toward a genetic interaction between the csn2-5 and sgt1b-3 mutations in auxin response.
The best characterized biochemical activity of the CSN is in cleavage of RUB/Nedd8 from cullins (23, 61) and mutations in CSN lead to the increased accumulation of rubylated cullins. We therefore determined the consequences of the csn2-5 mutation on CUL1 modification on immunoblots (Fig. 1B). We detected a redistribution of CUL1 toward the higher molecular weight band corresponding to CUL1-RUB in the csn2-5 single mutant, consistent with identification of a novel CSN2 allele. csn2-5 is most likely hypomorphic as its defects in CUL1 derubylation were less severe than for a csn2 null allele, csn2fus12-U228 (25, 62). In agreement with this observation, the csn2-5 mutant is viable and fertile as a homozygote, contrasting with the severe cop/det/fus phenotypes and seedling lethality of null csn mutants (25, 27). To demonstrate unambiguously that the identified mutation in CSN2 is responsible for the increased auxin resistance of the csn2-5 mutant, we conducted an allelism test with the recessive csn2fus12 allele. Because csn2fus12 confers early seedling lethality, heterozygous Ler csn2fus12 was crossed to homozygous Ler csn2-5. A 1:1 segregation of the auxin resistance phenotype was observed among F1 plants (Fig. 1C). Genotyping of individual F1 plants using molecular markers confirmed that heterozygous csn2-5/fus12 seedlings show auxin-resistant root growth. Thus, the csn2-5 mutation causing auxin-resistant root growth is a new csn2 mutant allele.
csn2-5 Is Mutated in an Evolutionarily Conserved Loop of the N-terminal HEAT Domain of CSN2 Important for CSN Accumulation—To address the molecular consequence of the G237D amino acid substitution, we built a structural model of CSN2 exploiting its similarities to several subunits of the 19S proteasome, eukaryotic initiation factor 3 (eIF3) and the COP9 signalosome (63). Sequence analyses revealed that CSN2 is composed of an N-terminal HEAT repeat domain (Huntingtin-EF3-PP2A-TOR1; residues 40–321) followed by a WH domain (winged helix, residues 327–432) at its C terminus (Fig. 2A). The PCI domain (residues 305–409), defined from multiple sequence alignments in the Pfam data base, actually comprises the last helical hairpin of the HEAT repeat and part of the WH domain (63). Gly237 mutated in CSN2-5 is located in a highly conserved part of the HEAT repeat domain that is strictly conserved in CSN2 homologs but absent from the homologous subunit of the proteasome lid complex (Fig. 2A; data not shown). In our structural model of Arabidopsis CSN2 (Fig. 2B; see “Experimental Procedures”), the mutation G237D is located in a loop connecting helices 9 and 10. The predicted secondary structures suggest that the loop may adopt an extended hairpin-like structure. This loop is among the most conserved regions of CSN2, suggesting an important functional role (data not shown). To test this hypothesis, we determined the impact of the csn2-5 mutation in fission yeast. S. pombe Δcsn2 mutants are UV and γ-ray hypersensitive and accumulate RUB-modified cullins (28). We generated a construct coding for SpCSN2 carrying a G243D substitution, equivalent to G237D in AtCSN2. Complementation of UV hypersensitivity and altered RUB modification profile through expression of SpCSN2 or Spcsn2-5 was tested in a Δcsn2 strain expressing Pcu1p-13MYC (28). Although SpCSN2 complemented the UV hypersensitivity and accumulation of SpCUL1-RUB, Spcsn2-5 failed to complement these two phenotypes. Despite a high degree of conservation of plant and yeast CSN2 subunits (44% amino acid identity, 62% similarity), AtCSN2 failed to complement the S. pombe Δcsn2 mutant phenotypes (data not shown). These results show that Gly243/Gly237 is important for CSN function in both S. pombe and Arabidopsis.
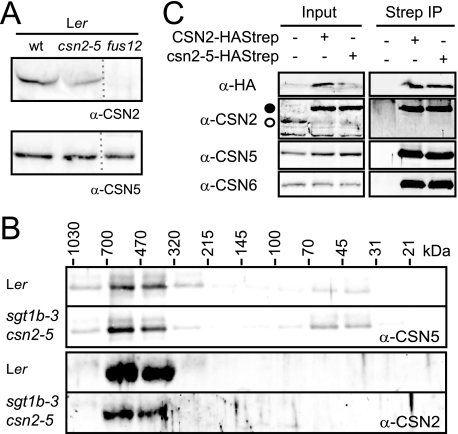
Nonetheless, the normal development of the csn2-5 mutant suggests that csn2-5 protein is still produced and assembled into at least a partially functional CSN complex. In plant extracts, csn2-5 steady-state levels quantified on immunoblots were 50% lower than wild-type CSN2 levels (Fig. 3A). In the same extracts, accumulation of the MPN subunit CSN5 was not altered. To test for CSN2 assembly into the large CSN complex (∼500 kDa), gel filtration experiments were performed. The CSN complex containing the CSN2 and CSN5 proteins eluted in the same molecular mass fractions (320–700 kDa, Fig. 3B; data not shown) from wild type, sgt1b-3, and sgt1b-3 csn2-5 mutant, indicating that the csn2-5 mutant protein becomes incorporated into CSN. In contrast to CSN5, which is also detectable in its monomeric form (31–70-kDa fractions), no free CSN2 or csn2-5 protein was detected. Additionally, the CSN complex was affinity-purified from sgt1b-3 csn2-5 transgenic lines expressing wild-type CSN2-HAStrep or mutant csn2-5-HAStrep under control of the constitutive 35S promoter (Fig. 3C). CSN2-HAStrep and csn2-5-HAStrep accumulated in similar amounts, and CSN5 and CSN6 could be co-purified with equal efficiency. Although it has been reported that AtCSN2 mediates the interaction of CSN with AtCUL1 and AtCUL3 (25, 60), we failed to detect CUL1 and CUL3 co-purification with CSN (data not shown) and therefore could not test whether the csn2-5 mutation affects CSN-cullin interactions. Together, these results show that the csn2-5 mutant protein integrates into the CSN but affects CSN accumulation.
FIGURE 3.
CSN2 accumulation and CSN stability in the csn2-5 mutant. A, steady-state levels of CSN2 and CSN5 proteins were determined in total protein extracts by immunoblot with anti-CSN2 and anti-CSN5 antibodies, respectively. wt, wild type. B, total protein extracts from seedlings were separated by size exclusion chromatography. The collected fractions were analyzed on immunoblots with anti-CSN5 and anti-CSN2 antibodies. Molecular mass range of the fractions is indicated in kDa based on the column calibration. C, total protein extracts (Input) were prepared from untransformed sgt1b-3 csn2-5 plants or plants expressing CSN2-HAStrep or csn2-5-HAStrep and subjected to StrepII affinity purification. Input and elution fractions from the purification (Strep IP) were analyzed on immunoblots with anti-HA, anti-CSN2, anti-CSN5, and anti-CSN6 antibodies. Open and filled circles correspond to CSN2 and CSN2-HAStrep/csn2-5-HAStrep, respectively.
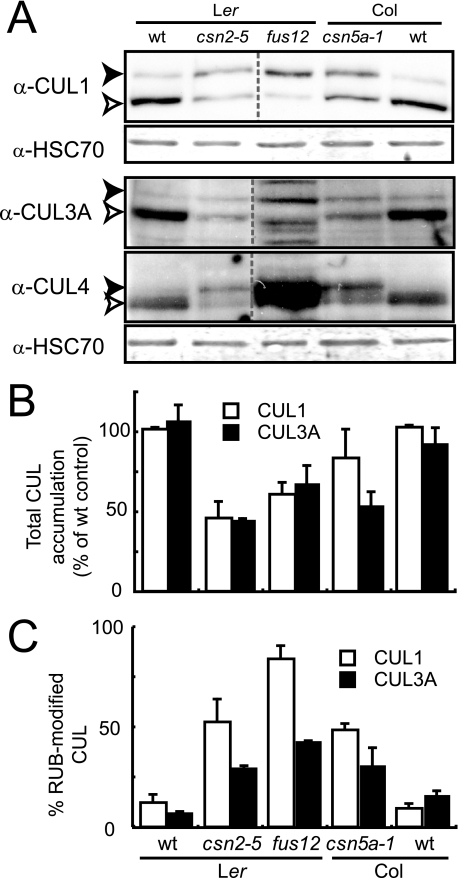
csn Mutants Have Reduced Steady-state Levels of Cullins— We examined cullin modification and accumulation patterns in csn mutants affected in PCI (csn2-5 and csn2fus12) and MPN (csn5a-1) subunits by immunoblotting. Compared with corresponding wild-type extracts, a larger fraction of each cullin could be detected in the RUB-conjugated form in each csn mutant extract, as expected (27, 40). Additionally, we observed a significant decrease in the total amount of at least CUL1 and CUL3 in all tested csn mutants compared with the wild type (∼50% for CUL1 and CUL3A, Fig. 4, A and B). This indicates that CSN is needed for maintenance of cullin steady-state levels, as reported in several other model organisms (27, 29, 33, 35, 36). Because of such a stabilizing function, the CSN would be expected to exert a positive role not only on activities of CUL1-based SCF ubiquitin ligases but also on CUL3- and CUL4-based CRLs.
FIGURE 4.
Cullin modification and accumulation in csn mutants. A, total protein extracts from 7-day-old seedlings were analyzed by immunoblotting with anti-CUL1, anti-CUL3A, anti-CUL4 and anti-HSC70 antibodies. Immunodetection of cytosolic/nuclear HSC70 was used as the loading control. Open and filled arrowheads correspond to unmodified and RUB-modified cullins, respectively. One representative experiment is shown. wt, wild type. B and C, densitometric analysis was performed on anti-CUL1 and anti-CUL3A immunoblots to determine the relative proportion of RUB-modified cullin (B) and total cullin steady-state levels (C) (unmodified plus RUB-modified) for CUL1 or CUL3A protein. Error bars indicate S.D. between three independent biological replicates.
CSN Reduction in the csn2-5 Mutant Has Minor Phenotypic Consequences—Considering the defects in cullin modification and accumulation detected for the csn2-5 mutant together with the phenotypes of previously reported partial or null csn mutants, strong physiological consequences would be anticipated. To our surprise, the csn2-5 mutant did not display major developmental phenotypes other than a mild dwarfism. In contrast, the csn5a-1 mutant, which carries a null mutation in one of two genes encoding the MPN subunit 5 of the CSN complex, is a severe dwarf (40) and displays similar defects in auxin signaling, cullin accumulation, and RUB modification as the csn2-5 mutant (supplemental Fig. 2, A and B). The typical csn mutant phenotypes cop/det (open cotyledons, short hypocotyl and absence of apical hook in darkness) and fusca (accumulation of anthocyanins) were also tested in csn2-5. No obvious fusca phenotype was observed (data not shown). Relative hypocotyl elongation (dark versus light) was similar in wild-type and mutant seedlings (supplemental Fig. 2, C and D), although mutant seedlings were smaller. Puzzled by the absence of a clear cop phenotype, we quantified cotyledon opening after 4 days of darkness (supplemental Fig. 2, C and E). Approximately 50% of csn2-5 seedling had opened cotyledons compared with ∼15% of wild-type and 100% of csn2fus12 null mutant. Thus, the csn2-5 mutant displays a very weak photomorphogenic phenotype. CRL-regulated responses to UV light, γ-rays, and the phytohormones ethylene and jasmonic acid were also not affected in the csn2-5 mutant (data not shown). Such observations highlight the notion that a strict correlation between cullin abundance/RUB modification and development/physiological responses to environmental cues cannot be assumed.
Reduction of Auxin Receptor SCFTIR1 Activity and Accumulation in the csn2-5 Mutant—The observed reduction of CUL1 levels might explain the auxin phenotypes of the csn2-5 and other csn mutants. We therefore tested whether the csn2-5 mutation affects Aux/IAA degradation mediated by SCFTIR1/AFB ubiquitin ligase activity using a nonfunctional heat shock-regulated AXR3/IAA17-GUS reporter fusion (HS:: AXR3NT-GUS (45)). Wild-type and csn2-5 seedlings carrying the HS::AXR3NT-GUS construct were heat shocked for 2 h to induce reporter expression and subsequently stained for GUS activity. No difference in GUS staining between wild type and csn2-5 mutant was observed (Fig. 5A). As auxin stimulates Aux/IAA ubiquitination by SCFTIR1/AFB (for AXR3NT-GUS, see Ref. 45), seedlings were incubated for 1 h with 5 μm 2,4-D prior to GUS staining. AXR3NT-GUS activity almost completely disappeared in the elongation/differentiation zones of wild-type roots but remained visible in csn2-5 roots (Fig. 5A). The csn2-5 mutation also caused a similar stabilization of AXR3NT-GUS in the sgt1b-3 mutant background (data not shown). The impaired degradation of AXR3NT-GUS suggests that a decreased SCFTIR1/AFB ligase activity in csn2-5 leads to stabilization of Aux/IAAs, in line with previous analyses of partial csn mutants (40, 60).
FIGURE 5.
Activity and stability of the SCFTIR1 auxin receptor in the csn2-5 mutant. A, HS::AXR3NT-GUS reporter (Col-0) was crossed into the Ler and Ler csn2-5 background. F3 plants homozygous for HS::AXR3NT-GUS and either the csn2-5 mutation or wild-type CSN2 (wt) were heat-shocked for 2 h to induce reporter gene expression and stained for β-glucuronidase activity following a 60-min incubation at room temperature. 5 μm 2,4-D was added where indicated at the beginning of the room temperature incubation. One representative experiment of three repetitions is shown; three different F3 families were tested for each genotype. B, np::TIR1-HAStrep transgene was initially transformed into Ler and sequentially introgressed into Ler sgt1b-3 and Ler sgt1b-3 csn2-5. TIR1-HAStrep steady-state levels were determined in total soluble protein extracts from seedlings by immunoblot analysis using anti-HA antibody. Immunodetection of cytosolic/nuclear HSC70 with anti-HSC70 antibodies is provided as loading control. C, densitometric analysis of TIR1-HAStrep levels detected by immunoblot analysis in three independent experiments. Error bars indicate S.D. Statistical significance of difference between csn2-5 and controls was evaluated using Student's t test; *, p < 0.001. D, RT-PCR analysis of TIR1 and CUL1 mRNA steady-state levels in wild-type Ler, sgt1b-3, and sgt1b-3 csn2-5 mutants. RT-PCR amplicons were resolved by electrophoresis and visualized by ethidium bromide staining. TIR1 primers amplified both TIR1 endogene and transgene. The ACTIN2 gene was used as constitutive control. E, ASK1 cellular levels were determined on an immunoblot using anti-ASK1 antibody. One representative experiment of three repetitions is shown. The loading control can be found in the first two lanes of the bottom panel of Fig. 4A (anti-HSC70 immunoblot).
Recently, the CSN was shown to stabilize substrate adaptor proteins from cullin-based E3s in S. pombe (31), Neurospora crassa (29), human cells (32, 33), and Caenorhabditis elegans (34). To analyze whether CSN is similarly important for substrate adaptor accumulation in Arabidopsis, we expressed TIR1-HAStrep under the control of its native promoter in transgenic Arabidopsis. TIR1-HAStrep functionality was demonstrated by complementation of the Col tir1-1 mutant (supplemental Fig. 3A). The TIR1-HAStrep transgene derived from a single transformation event in Ler was crossed into the sgt1b-3 and sgt1b-3 csn2-5 backgrounds to evaluate the impact of these mutations on TIR1 stability. sgt1b-3 did not affect TIR1-HAStrep accumulation, as TIR1 steady-state levels remained unchanged compared with the wild-type control (Fig. 5, B and C). In contrast, TIR1-HAStrep signal strength was reduced by 60% in the sgt1b-3 csn2-5 background. This reduced abundance of the auxin receptor TIR1 in the csn2-5 mutant could at least partially explain the auxin resistance phenotype of csn2-5 plants. Consistent with this notion, overexpression of TIR1 (endogenous gene plus transgene) in the TIR1-HAStrep transgenic line partially complemented the csn2-5 auxin resistance phenotype (supplemental Fig. 3B). Finally, accumulation of ASK1 (one Arabidopsis SKP1 homolog) was not affected by the csn2-5 mutation (Fig. 5E). To explain the reduced abundance of TIR1 and CUL1, we analyzed their transcript levels by RT-PCR. TIR1 and CUL1 expression remained unchanged in the different genotypes (Fig. 5D). These results indicate that the CSN directly or indirectly regulates the abundance of SCFTIR1 components (TIR1 and CUL1) by a post-translational mechanism. This activity could at least partially explain the auxin resistance of the csn2-5 mutant.
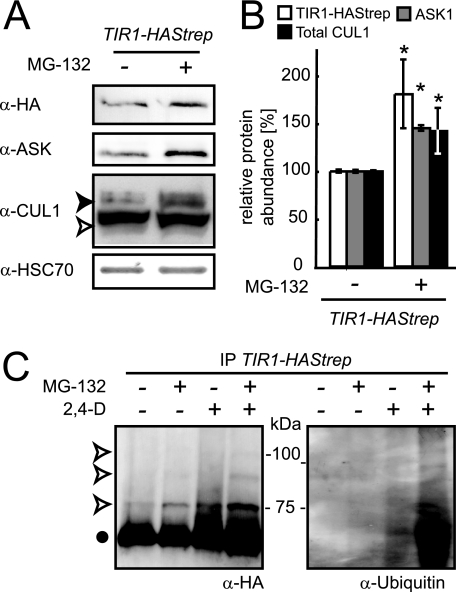
Evidence for SCFTIR1 Ubiquitination and Modulation by the 26S Proteasome—Several recent studies report the degradation of F-box proteins by autoubiquitination and subsequent proteasome-dependent proteolysis (29, 31, 33). To test whether such a mechanism could regulate TIR1, we first measured the impact of the proteasome inhibitor MG-132 on TIR1 protein accumulation. Incubation of wild-type and csn2-5 seedlings expressing TIR1-HAStrep with 50 μm MG-132 for 6 h prior to protein extraction caused an 80% increase in TIR1 accumulation and an ∼40% increase in CUL1 and ASK1 steady-state levels (Fig. 6, A and B; data not shown). These findings show that accumulation of the three SCFTIR1 core components is likely regulated by the 26S proteasome activity in both wild-type and csn2-5 plants.
FIGURE 6.
Post-translational regulation of SCFTIR1 accumulation in wild-type plants. Ler np::TIR1-HAStrep seedlings were grown in liquid MS for 7 days. A, seedlings were incubated for 6 h with the proteasome inhibitor MG-132 (50 μm (+) or with DMSO (-)). Total protein extracts were analyzed on immunoblots with anti-HA, anti-ASK, anti-CUL1, and anti-HSC70 antibodies. Immunodetection of cytosolic/nuclear HSC70 is used to evaluate relative loading of the samples. The open and filled arrowheads correspond to unmodified and RUB-modified CUL1, respectively. B, densitometric analysis of TIR1-HAStrep, ASK1, and total CUL1 levels (unmodified plus RUB-modified) detected by immunoblots of samples with and without MG-132 treatment was performed on three independent experiments. Error bars indicate S.D. Differences between untreated control and MG-132-treated samples were evaluated for statistical significance using Student's t test; *, p < 0.001. C, TIR1-HAStrep was affinity-purified on StrepTactin-Sepharose 6 h after application of auxin (5 μm 2,4-D) and/or MG-132 (50 μm). Protein extracts were analyzed on immunoblots using the anti-HA and anti-ubiquitin antibodies. The filled dot indicates unmodified TIR1-HAStrep, and the open arrowheads indicate modified TIR1-HAStrep.
Potential ubiquitination of TIR1 was tested in vivo by affinity purification of TIR1-HAStrep from MG-132-treated seedlings, as this treatment would be expected to slow down the proteasome-dependent degradation of polyubiquinated proteins, thereby allowing increased accumulation and detection. Seedlings were treated for 6 h with auxin (5 μm) and/or MG-132 (50 μm). A discrete set of bands (with ∼9–10-kDa increments) of higher molecular mass than TIR1-HAStrep was detected in MG-132-treated samples by HA antibody (Fig. 6C). Consistent with increased persistence of polyubiquitinated TIR1, these signals were markedly weaker in non-MG-132-treated samples. The higher molecular weight bands also cross-reacted with a monoclonal anti-ubiquitin antibody suggesting they may be TIR1-HAStrep-(Ubq)n. These data suggest that a proportion of TIR1 is polyubiquitinated and targeted for degradation by the proteasome under normal physiological conditions in wild-type plants.
The above observations raised the question of TIR1 turnover in plants, as F-box proteins have been shown to possess diverse half-lives ranging from 5 to 30 min in S. cerevisiae to more than 6 h in N. crassa and human cells (29, 33, 64). We wanted to test TIR1-HAStrep turnover by applying cycloheximide (CHX), an inhibitor of de novo protein synthesis. CHX was first added to control seedlings carrying HS::AXR3NT-GUS before induction of transgene expression by heat shock. Although strong GUS activity was detected in untreated samples after heat shock, no GUS staining was observed in the presence of CHX (data not shown), indicating that translation blockage by CHX was effective. Under the same conditions, TIR1-HAStrep remained clearly detectable at 9 h after CHX treatment in wild-type and sgt1b-3 csn2-5 mutant backgrounds, and its levels were not significantly altered in the presence of auxin (supplemental Fig. 3; data not shown). We concluded that TIR1 is a long-lived F-box protein under our experimental conditions.
DISCUSSION
In this study, we investigated the post-translational regulation of CRL ubiquitin ligases in both wild-type plants and csn mutants, focusing on the auxin receptor SCFTIR1. We show that the CSN is needed for the stabilization of cullins and other CRL subunits in Arabidopsis and that SCF components, including the substrate adaptor TIR1, are degraded by the ubiquitin/proteasome machinery even in wild-type plants.
The CSN Is Needed for SCFTIR1 Accumulation: A Molecular Explanation for the CSN Paradox in Arabidopsis—Arabidopsis mutants partially defective in either the RUB/Nedd8 conjugation machinery or the antagonizing CSN are resistant to auxin. Because the CSN acts as a repressor of CRL activity in vitro, these observations were not fully understood formerly. We show here that CSN is important for the stabilization of the substrate receptor TIR1 and CUL1 (Fig. 5). Auxin sensitivity correlates with auxin receptor abundance, as illustrated by the haploinsufficiency of TIR1 (39), the additivity of mutations in TIR1 and AFB1–3 (19), and the increased auxin sensitivity of plants expressing TIR1-HAStrep compared with parental lines (supplemental Fig. 3). Thus destabilization of TIR1 and possibly the related AFB proteins could explain the defects in auxin signaling observed for csn mutants. As CUL1 is similarly haploin-sufficient (65), decreased CUL1 abundance also likely contributes to auxin signaling defects in the csn2-5 mutant and other csn mutants. Although opposing results have been reported for the stability of AtCUL1 (27, 40), both the destabilization of cullins and CRL substrate adaptors in csn mutants are in line with reports from various systems (29, 31, 32, 34–36). In a study analyzing the stability of various F-box proteins in human cells silenced for Csn5/Jab1, the abundance of five of seven tested proteins was significantly reduced upon Csn5 silencing (33). It will therefore be interesting to analyze the dependence of further Arabidopsis substrate receptors on CSN. In this respect, the wild-type phenotypes of the csn2-5 and the csn5a-1 mutants for jasmonic acid- or ethylene-dependent responses (data not shown) suggest that accumulation of the corresponding CRLs might be less dependent on CSN activity than TIR1.
CSN-dependent Protection of SCFTIR1 from Ubiquitination and Degradation—The proteasome inhibitor MG-132 increased accumulation of TIR1, CUL1, and ASK1 (Fig. 6) suggesting that levels of one or more SCFTIR1 components are controlled by ubiquitination and proteasome-dependent degradation in wild-type plants. Although there is, to our knowledge, no evidence for ASK1 (SKP1) post-translational modification, the polyubiquitination and proteasome-dependent degradation of CUL1 has previously been shown in human cells (66). One possible RUB function might thus be the protection of CUL1 from erroneous ubiquitination and degradation by blocking a potential ubiquitination site, although this is unlikely because neddylation of human CUL1 is required for CUL1 in vitro autoubiquitination (66). In Arabidopsis, CUL1 monoubiquitination was reported only recently (67). However, this modification did not induce CUL1 degradation under the tested conditions.
Targeting of the entire SCF to the proteasome through ubiquitination of the single TIR1 subunit seems unlikely, because proteins have to unfold to enter the catalytic core, which implies complex dissociation. Ubiquitination of individual SCF/CRL subunits, possibly including RBX1, through unspecific E3 ligase activity appears more likely, although direct evidence is yet missing. Autoubiquitination and degradation of substrate adaptors has been reported in S. pombe, human cells, Neurospora, and C. elegans (31, 33, 34, 68). In fission yeast, CSN was shown to counteract adaptor degradation through both its derubylating activity and a CSN-associated deubiquitinating activity mediated by Ubp12p, apparently reversing erroneous autoubiquitination of substrate adaptors (31). In csn mutants such as csn2-5, at least one of these activities is diminished, thus enhancing substrate receptor ubiquitination and degradation. Because accumulation of SCF subunits upon MG-132 treatment was also visible in wild-type seedlings, the turnover of SCF complexes is apparently part of their normal post-translational regulation. Despite the presence of a large family of deubiquitinases encoded by the Arabidopsis genome (69), an Arabidopsis Ubp12 orthologue involved in CSN-dependent stabilization of CRL substrate adaptors has yet to be identified.
An Updated Model for CRL Regulation in Plants—The conflicting in vitro and genetic data for CSN impact on CRL activity led to the formulation of a cycling model with cycles of cullin modification and RUB cleavage necessary for CRL activity (70, 71). Although all experimental results strongly support dynamic CRL assembly and activity, more recent analyses suggest that CSN is needed rather for the maintenance of substrate adaptor or CRL stability (e.g. see Refs. 29 and 31), whereas cycling is not needed for CRL activity per se (reviewed in Ref. 71). We therefore display our current view of the post-translational regulation of CRLs exemplified by SCFTIR1 as a sequence of reversible reactions (supplemental Fig. 4). This illustrates that each step forms an unstable equilibrium that can be perturbed using pharmacological and/or genetic tools. Initially, unmodified CUL1 is associated with CAND1 and RBX1. The TIR1-ASK1 substrate receptor module, possibly with bound auxin and substrate-loaded, can dissociate CAND1 from CUL1. Competition of the substrate receptor module and CAND1 for CUL1 binding has been described in vitro (72, 73) and is indicated by the phenotypical complementation of mutations in CUL1 affecting ASK1 binding (axr6-1/-2) through a mutation in CAND1 (74). Substrate receptor binding and CAND1 dissociation exposes the RUB modification site allowing rubylation (75, 76). Irrespective of the precise function of rubylation at this point, active SCFTIR1 can ubiquitinate its Aux/IAA substrate proteins, triggering their proteasome-dependent degradation. Eventually, SCFTIR1 is inactivated through the cleavage of RUB from CUL1 by CSN, allowing recycling of the SCF subunits. At a low frequency, at least TIR1 might itself become ubiquitinated under normal physiological conditions. Ultimately, SCFTIR1 is degraded by the proteasome. Although metabolically costly, this may represent a natural process for SCF inactivation/recycling in vivo, as excessive amounts of CUL1 are apparently also deleterious for plants (11). In the case of a partial or complete loss of CSN activity, assembled SCFs are neither sufficiently protected from degradation by a putative CSN-associated deubiquinating activity nor recycled by RUB cleavage. Consequently, ubiquitinated SCF complexes accumulate and their proteasome-dependent degradation is unimpeded. In this case, de novo protein synthesis would not be sufficient to compensate for degradation, resulting in reduced SCFTIR1 abundance and CRL activity. This model predicts that CUL1-substrate adaptor module interaction should trigger the destabilization of all SCF subunits. Consistent with this notion, mutant CUL1 proteins affected in interactions to ASK1, as inferred from crystal structure analyses (77), were found to accumulate in cells (65, 78).4 When tested, the respective CUL1 mutations also caused an increase in RBX1 (65) and ASK1 (78) steady-state levels. Therefore, destabilization of the SCF subunits appears to depend on the assembly of a functional SCF complex. Future experiments should tell us whether such a working model holds true for further SCFs and other cullin-based ubiquitin ligases in Arabidopsis.
Supplementary Material
Acknowledgments
We are grateful to Pascal De Guio for expert technical assistance and to Thierry Desnos for valuable discussions. We thank Thomas Lahaye and Doreen Gürlebeck for contributing SacI/TaqI AFLP primers. Xing Wang Deng and Claus Schwechheimer are acknowledged for contributing various published antibodies and csn mutant lines. We also thank the groupe de recherches appliquées en Phytotechnologie members for maintenance of the plant growth facility.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
Footnotes
The abbreviations used are: CRL, cullin-RING ligase; SCF, SKP1-cullin-F-box; Aux, auxin; CSN, COP9 signalosome; GUS, β-glucuronidase; RT, reverse transcription; CHX, cycloheximide; HA, hemagglutinin; 2,4-D, 2,4-dichlorphenoxyacetic acid; IAA, indoleacetic acid.
J. Stuttmann and L. D. Noël, unpublished data.
References
- 1.Smalle, J., and Vierstra, R. D. (2004) Annu. Rev. Plant Biol. 55 555-590 [DOI] [PubMed] [Google Scholar]
- 2.Moon, J., Parry, G., and Estelle, M. (2004) Plant Cell 16 3181-3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hershko, A., and Ciechanover, A. (1998) Annu. Rev. Biochem. 67 425-479 [DOI] [PubMed] [Google Scholar]
- 4.Pickart, C. M. (2001) Annu. Rev. Biochem. 70 503-533 [DOI] [PubMed] [Google Scholar]
- 5.Dieterle, M., Thomann, A., Renou, J. P., Parmentier, Y., Cognat, V., Lemonnier, G., Muller, R., Shen, W. H., Kretsch, T., and Genschik, P. (2005) Plant J. 41 386-399 [DOI] [PubMed] [Google Scholar]
- 6.Figueroa, P., Gusmaroli, G., Serino, G., Habashi, J., Ma, L., Shen, Y., Feng, S., Bostick, M., Callis, J., Hellmann, H., and Deng, X. W. (2005) Plant Cell 17 1180-1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gingerich, D. J., Gagne, J. M., Salter, D. W., Hellmann, H., Estelle, M., Ma, L., and Vierstra, R. D. (2005) J. Biol. Chem. 280 18810-18821 [DOI] [PubMed] [Google Scholar]
- 8.Lee, J., and Zhou, P. (2007) Mol. Cell 26 775-780 [DOI] [PubMed] [Google Scholar]
- 9.Lee, J. H., Terzaghi, W., Gusmaroli, G., Charron, J. B., Yoon, H. J., Chen, H., He, Y. J., Xiong, Y., and Deng, X. W. (2008) Plant Cell 20 152-167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lechner, E., Achard, P., Vansiri, A., Potuschak, T., and Genschik, P. (2006) Curr. Opin. Plant Biol. 9 631-638 [DOI] [PubMed] [Google Scholar]
- 11.Shen, W. H., Parmentier, Y., Hellmann, H., Lechner, E., Dong, A., Masson, J., Granier, F., Lepiniec, L., Estelle, M., and Genschik, P. (2002) Mol. Biol. Cell 13 1916-1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomann, A., Brukhin, V., Dieterle, M., Gheyeselinck, J., Vantard, M., Grossniklaus, U., and Genschik, P. (2005) Plant J. 43 437-448 [DOI] [PubMed] [Google Scholar]
- 13.Bernhardt, A., Lechner, E., Hano, P., Schade, V., Dieterle, M., Anders, M., Dubin, M. J., Benvenuto, G., Bowler, C., Genschik, P., and Hellmann, H. (2006) Plant J. 47 591-603 [DOI] [PubMed] [Google Scholar]
- 14.Chen, H., Shen, Y., Tang, X., Yu, L., Wang, J., Guo, L., Zhang, Y., Zhang, H., Feng, S., Strickland, E., Zheng, N., and Deng, X. W. (2006) Plant Cell 18 1991-2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang, K. L., Yoshida, H., Lurin, C., and Ecker, J. R. (2004) Nature 428 945-950 [DOI] [PubMed] [Google Scholar]
- 16.Kepinski, S., and Leyser, O. (2005) Nature 435 446-451 [DOI] [PubMed] [Google Scholar]
- 17.Dharmasiri, N., Dharmasiri, S., and Estelle, M. (2005) Nature 435 441-445 [DOI] [PubMed] [Google Scholar]
- 18.Teale, W. D., Paponov, I. A., and Palme, K. (2006) Nat. Rev. Mol. Cell Biol. 7 847-859 [DOI] [PubMed] [Google Scholar]
- 19.Dharmasiri, N., Dharmasiri, S., Weijers, D., Lechner, E., Yamada, M., Hobbie, L., Ehrismann, J. S., Jurgens, G., and Estelle, M. (2005) Dev. Cell 9 109-119 [DOI] [PubMed] [Google Scholar]
- 20.Szemenyei, H., Hannon, M., and Long, J. A. (2008) Science 319 1384-1386 [DOI] [PubMed] [Google Scholar]
- 21.Lau, S., Jurgens, G., and De Smet, I. (2008) Plant Cell 20 1738-1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parry, G., and Estelle, M. (2004) Semin. Cell Dev. Biol. 15 221-229 [DOI] [PubMed] [Google Scholar]
- 23.Lyapina, S., Cope, G., Shevchenko, A., Serino, G., Tsuge, T., Zhou, C. S., Wolf, D. A., Wei, N., and Deshaies, R. J. (2001) Science 292 1382-1385 [DOI] [PubMed] [Google Scholar]
- 24.Cope, G. A., Suh, G. S., Aravind, L., Schwarz, S. E., Zipursky, S. L., Koonin, E. V., and Deshaies, R. J. (2002) Science 298 608-611 [DOI] [PubMed] [Google Scholar]
- 25.Serino, G., Su, H., Peng, Z., Tsuge, T., Wei, N., Gu, H., and Deng, X. W. (2003) Plant Cell 15 719-731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Serino, G., and Deng, X. W. (2003) Annu. Rev. Plant Biol. 54 165-182 [DOI] [PubMed] [Google Scholar]
- 27.Gusmaroli, G., Figueroa, P., Serino, G., and Deng, X. W. (2007) Plant Cell 19 564-581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mundt, K. E., Liu, C., and Carr, A. M. (2002) Mol. Biol. Cell 13 493-502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He, Q., Cheng, P., He, Q., and Liu, Y. (2005) Genes Dev. 19 1518-1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Busch, S., Schwier, E. U., Nahlik, K., Bayram, O., Helmstaedt, K., Draht, O. W., Krappmann, S., Valerius, O., Lipscomb, W. N., and Braus, G. H. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 8089-8094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wee, S., Geyer, R. K., Toda, T., and Wolf, D. A. (2005) Nat. Cell Biol. 7 387-391 [DOI] [PubMed] [Google Scholar]
- 32.Denti, S., Fernandez-Sanchez, M. E., Rogge, L., and Bianchi, E. (2006) J. Biol. Chem. 281 32188-32196 [DOI] [PubMed] [Google Scholar]
- 33.Cope, G. A., and Deshaies, R. J. (2006) BMC Biochem. 7 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luke-Glaser, S., Roy, M., Larsen, B., Le Bihan, T., Metalnikov, P., Tyers, M., Peter, M., and Pintard, L. (2007) Mol. Cell. Biol. 27 4526-4540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu, J. T., Lin, H. C., Hu, Y. C., and Chien, C. T. (2005) Nat. Cell Biol. 7 1014-1020 [DOI] [PubMed] [Google Scholar]
- 36.Schweitzer, K., Bozko, P. M., Dubiel, W., and Naumann, M. (2007) EMBO J. 26 1532-1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Austin, M. J., Muskett, P., Kahn, K., Feys, B. J., Jones, J. D., and Parker, J. E. (2002) Science 295 2077-2080 [DOI] [PubMed] [Google Scholar]
- 38.Kwok, S. F., Solano, R., Tsuge, T., Chamovitz, D. A., Ecker, J. R., Matsui, M., and Deng, X. W. (1998) Plant Cell 10 1779-1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruegger, M., Dewey, E., Gray, W. M., Hobbie, L., Turner, J., and Estelle, M. (1998) Genes Dev. 12 198-207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dohmann, E. M., Kuhnle, C., and Schwechheimer, C. (2005) Plant Cell 17 1967-1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gray, W. M., Muskett, P. R., Chuang, H. W., and Parker, J. E. (2003) Plant Cell 15 1310-1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weigel, D., and Glazebrook, J. (2002) Arabidopsis: A Laboratory Manual, pp. 24-25, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 43.Noël, L. D., Cagna, G., Stuttmann, J., Wirthmuller, L., Betsuyaku, S., Witte, C. P., Bhat, R., Pochon, N., Colby, T., and Parker, J. E. (2007) Plant Cell 19 4061-4076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alonso, J. M., Stepanova, A. N., Solano, R., Wisman, E., Ferrari, S., Ausubel, F. M., and Ecker, J. R. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 2992-2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gray, W. M., Kepinski, S., Rouse, D., Leyser, O., and Estelle, M. (2001) Nature 414 271-276 [DOI] [PubMed] [Google Scholar]
- 46.Jefferson, R. A. (1987) Plant Mol. Biol. Rep. 5 387-405 [Google Scholar]
- 47.Vos, P., Hogers, R., Bleeker, M., Reijans, M., van de Lee, T., Hornes, M., Frijters, A., Pot, J., Peleman, J., Kuiper, M., and Zabeau, M. (1995) Nucleic Acids Res. 23 4407-4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Witte, C. P., Noël, L. D., Gielbert, J., Parker, J. E., and Romeis, T. (2004) Plant Mol. Biol. 55 135-147 [DOI] [PubMed] [Google Scholar]
- 49.Peng, Z., Serino, G., and Deng, X. W. (2001) Development (Camb.) 128 4277-4288 [DOI] [PubMed] [Google Scholar]
- 50.Murray, J. M., Doe, C. L., Schenk, P., Carr, A. M., Lehmann, A. R., and Watts, F. Z. (1992) Nucleic Acids Res. 20 2673-2678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caspari, T., Dahlen, M., Kanter-Smoler, G., Lindsay, H. D., Hofmann, K., Papadimitriou, K., Sunnerhagen, P., and Carr, A. M. (2000) Mol. Cell. Biol. 20 1254-1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D. J. (1997) Nucleic Acids Res. 25 3389-3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Edgar, R. C. (2004) Nucleic Acids Res. 32 1792-1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jones, D. T. (1999) J. Mol. Biol. 292 195-202 [DOI] [PubMed] [Google Scholar]
- 55.Soding, J. (2005) Bioinformatics (Oxf.) 21 951-960 [DOI] [PubMed] [Google Scholar]
- 56.Sali, A., and Blundell, T. L. (1993) J. Mol. Biol. 234 779-815 [DOI] [PubMed] [Google Scholar]
- 57.Luthy, R., Bowie, J. U., and Eisenberg, D. (1992) Nature 356 83-85 [DOI] [PubMed] [Google Scholar]
- 58.Sippl, M. J. (1993) Proteins 17 355-362 [DOI] [PubMed] [Google Scholar]
- 59.Takahashi, A., Casais, C., Ichimura, K., and Shirasu, K. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 11777-11782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schwechheimer, C., Serino, G., Callis, J., Crosby, W. L., Lyapina, S., Deshaies, R. J., Gray, W. M., Estelle, M., and Deng, X. W. (2001) Science 292 1379-1382 [DOI] [PubMed] [Google Scholar]
- 61.Zhou, C., Seibert, V., Geyer, R., Rhee, E., Lyapina, S., Cope, G., Deshaies, R. J., and Wolf, D. A. (2001) BMC Biochem. 2 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Misera, S., Muller, A. J., Weiland-Heidecker, U., and Jurgens, G. (1994) Mol. Gen. Genet. 244 242-252 [DOI] [PubMed] [Google Scholar]
- 63.Scheel, H., and Hofmann, K. (2005) BMC Bioinformatics 6 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Galan, J. M., and Peter, M. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 9124-9129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hellmann, H., Hobbie, L., Chapman, A., Dharmasiri, S., Dharmasiri, N., del Pozo, C., Reinhardt, D., and Estelle, M. (2003) EMBO J. 22 3314-3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morimoto, M., Nishida, T., Nagayama, Y., and Yasuda, H. (2003) Biochem. Biophys. Res. Commun. 301 392-398 [DOI] [PubMed] [Google Scholar]
- 67.Manzano, C., Abraham, Z., Lopez-Torrejon, G., and Del Pozo, J. C. (2008) Plant Mol. Biol. 68 145-158 [DOI] [PubMed] [Google Scholar]
- 68.He, Q., and Liu, Y. (2005) Biochem. Soc. Trans. 33 953-956 [DOI] [PubMed] [Google Scholar]
- 69.Yan, N., Doelling, J. H., Falbel, T. G., Durski, A. M., and Vierstra, R. D. (2000) Plant Physiol. 124 1828-1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wolf, D. A., Zhou, C., and Wee, S. (2003) Nat. Cell Biol. 5 1029-1033 [DOI] [PubMed] [Google Scholar]
- 71.Cope, G. A., and Deshaies, R. J. (2003) Cell 114 663-671 [DOI] [PubMed] [Google Scholar]
- 72.Liu, J., Furukawa, M., Matsumoto, T., and Xiong, Y. (2002) Mol. Cell 10 1511-1518 [DOI] [PubMed] [Google Scholar]
- 73.Oshikawa, K., Matsumoto, M., Yada, M., Kamura, T., Hatakeyama, S., and Nakayama, K. I. (2003) Biochem. Biophys. Res. Commun. 303 1209-1216 [DOI] [PubMed] [Google Scholar]
- 74.Zhang, W., Ito, H., Quint, M., Huang, H., Noël, L. D., and Gray, W. M. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 8470-8475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Goldenberg, S. J., Cascio, T. C., Shumway, S. D., Garbutt, K. C., Liu, J., Xiong, Y., and Zheng, N. (2004) Cell 119 517-528 [DOI] [PubMed] [Google Scholar]
- 76.Bornstein, G., Ganoth, D., and Hershko, A. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 11515-11520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zheng, N., Schulman, B. A., Song, L., Miller, J. J., Jeffrey, P. D., Wang, P., Chu, C., Koepp, D. M., Elledge, S. J., Pagano, M., Conaway, R. C., Conaway, J. W., Harper, J. W., and Pavletich, N. P. (2002) Nature 416 703-709 [DOI] [PubMed] [Google Scholar]
- 78.Moon, J., Zhao, Y., Dai, X., Zhang, W., Gray, W. M., Huq, E., and Estelle, M. (2007) Plant Physiol. 143 684-696 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.