Abstract
The essential S-phase kinase Cdc7–Dbf4 acts at eukaryotic origins of replication to trigger a cascade of protein associations that activate the Mcm2–7 replicative helicase. Also known as Dbf4-dependent kinase (DDK), this kinase preferentially targets chromatin-associated Mcm2–7 complexes that are assembled on the DNA during prereplicative complex (pre-RC) formation. Here we address the mechanisms that control the specificity of DDK action. We show that incorporation of Mcm2–7 into the pre-RC increased the level and changes the specificity of DDK phosphorylation of this complex. In the context of the pre-RC, DDK preferentially targets a conformationally distinct subpopulation of Mcm2–7 complexes that is tightly linked to the origin DNA. This targeting requires DDK to tightly associate with Mcm2–7 complexes in a Dbf4-dependent manner. Importantly, we find that DDK association with and phosphorylation of origin-linked Mcm2–7 complexes require prior phosphorylation of the pre-RC. Our findings provide insights into the mechanisms that ensure that DDK action is spatially and temporally restricted to the origin-bound Mcm2–7 complexes that will drive replication fork movement during S phase and suggest new mechanisms to regulate origin activity.
Keywords: Cdc7, DNA replication, Dbf4, Mcm2–7, kinase targeting, prereplication complex
Eukaryotic DNA replication initiates from hundreds of different sites in the genome called origins of replication. These events are tightly controlled during the cell cycle to ensure that no origin initiates more than once in a given cell cycle yet all chromosomal DNA is completely replicated (Arias and Walter 2007). Central to this control is the strict temporal separation of helicase loading (late M and G1 phase) and helicase activation (S phase) during the cell cycle. A cascade of carefully ordered protein–DNA and protein–protein interactions controls these events. The origin recognition complex (ORC) bound to origin DNA initiates helicase loading by recruiting Cdc6 and Cdt1. Together these proteins load the Mcm2–7 helicase onto the origin to form the prereplicative complex (pre-RC) (Stillman 2005). This complex marks and licenses all potential origins of replication. As cells enter S phase, activation of two S-phase-specific protein kinases—Clb5,6/Cdc28 (S-CDK) and the Dbf4-dependent kinase (DDK), Cdc7—triggers initiation of replication at pre-RCs. S-CDK and DDK stimulate a cascade of initiation factor interactions that results in the activation of the Mcm2–7 helicase (for review, see Aparicio et al. 2006; Labib and Gambus 2007). Once activated, Mcm2–7 unwinds origin DNA to provide the ssDNA template required to recruit the remaining DNA synthesis machinery and to assemble a pair of bidirectional replication forks (Takeda and Dutta 2005).
The targets and consequences of S-CDK and DDK during replication initiation are only now being understood. Recent studies have demonstrated that Sld2 and Sld3 are the only replication proteins that must be phosphorylated by S-CDK to allow the initiation of replication in Saccharomyces cerevisiae cells (Tanaka et al. 2007; Zegerman and Diffley 2007). S-CDK phosphorylation of each protein stimulates its association with a third replication factor, Dpb11. Sld3 and Dpb11 are required to recruit Cdc45, GINS, and DNA polymerases to the origin DNA, although exactly how the Sld2/Sld3/Dpb11 complex stimulates replication initiation remains unknown (Labib and Gambus 2007).
The replication proteins targeted by DDK and the consequences of these modifications are less well understood. In vitro assays have shown that DDK can phosphorylate multiple subunits of the Mcm2–7 complex (Weinreich and Stillman 1999; Kihara et al. 2000), Cdc45 (Nougarede et al. 2000), and DNA Pol α (Weinreich and Stillman 1999). Of these proteins, both in vivo and in vitro studies suggest that Mcm2–7 subunits are most likely to be the essential DDK targets (Hardy et al. 1997; Lei et al. 1997; Masai and Arai 2002; Cho et al. 2006; Masai et al. 2006). DDK activity is required for robust association of Cdc45 with chromatin (Walter 2000; Zou and Stillman 2000), and recent studies suggest that Cdc45 and the four-protein GINS complex associate with and activate the helicase activity of the Mcm2–7 complex (Gambus et al. 2006; Moyer et al. 2006; Pacek et al. 2006). This has led to the hypothesis that DDK phosphorylation causes helicase activation by stimulating the formation of this Cdc45/Mcm2–7/GINS (CMG) complex (Moyer et al. 2006).
The localization of DDK to origins of replication is critical to correctly regulate DNA replication. DDK activity is limiting in cells, acting on individual origins as they initiate DNA replication throughout S phase (Bousset and Diffley 1998; Donaldson et al. 1998; Patel et al. 2008). Thus, DDK must phosphorylate its target(s) after association with the origin rather than globally modifying its target(s) prior to their origin association. Consistent with this requirement, in vivo studies suggest that DDK preferentially phosphorylates chromatin-bound Mcm2–7 (Sheu and Stillman 2006), and several lines of evidence indicate that DDK is recruited to origins of replication. One-hybrid assays in S. cerevisiae cells showed that Dbf4 associates with origins in an ORC-dependent manner (Dowell et al. 1994). Studies performed in Xenopus extracts indicate that Cdc7 associates with chromatin in a Dbf4- and pre-RC-dependent manner (Jares and Blow 2000; Edwards et al. 2002; Jares et al. 2004). The pre-RC components Mcm2, Mcm4, Orc2, and Orc3 have each been identified as binding partners for Dbf4 (Duncker et al. 2002; Varrin et al. 2005; Sheu and Stillman 2006), suggesting that Dbf4 recruits Cdc7 to the origin. Despite these observations, the mechanisms of DDK recruitment to the origin and how this event is regulated remain unclear.
We sought to gain a mechanistic understanding of how DDK is targeted to the pre-RC. We found that incorporation into the pre-RC resulted in changes in both the level and specificity of Mcm2–7 phosphorylation by DDK. Using in vitro assembled pre-RCs as a substrate, we found that this activation of Mcm2–7 phosphorylation by DDK required a stable interaction between Mcm2–7 and DDK. In the context of the pre-RC, DDK preferentially phosphorylated Mcm2–7 complexes that were most tightly associated with origin DNA. Intriguingly, DDK binding to and phosphorylation of pre-RC Mcm2–7 required prior phosphorylation of the Mcm2–7 complex, suggesting the existence of a distinct kinase that targets Mcm2–7 proteins only after they are recruited to the origin DNA. Consistent with this hypothesis, we show that pre-RC Mcm2–7 is differentially phosphorylated by a kinase other than DDK. Together, our findings support a model in which changes in Mcm2–7 conformation and phosphorylation result in the selective recruitment of DDK to a subset of origin-bound Mcm2–7 and that this recruitment ensures that DDK acts in the correct spatial and temporal fashion.
Results
DDK preferentially targets pre-RC Mcm2–7 proteins
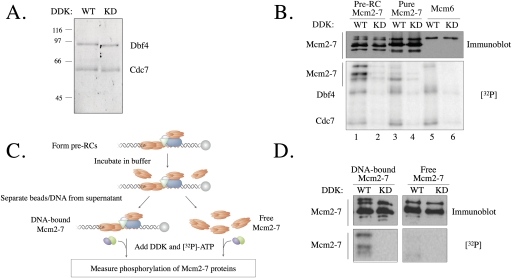
To study the phosphorylation of the pre-RC by DDK, we purified DDK from S. cerevisiae cells that overexpressed epitope-tagged Cdc7 and Dbf4. Both wild-type DDK and a “kinase-deficient” DDK (DDK-KD) that incorporated Cdc7 with a point mutation in the ATP-binding site (Ohtoshi et al. 1997) were purified. The final preparations were composed of Cdc7 and Dbf4 in an equimolar ratio (Fig. 1A). Consistent with previous reports (Kihara et al. 2000), when incubated with [γ-32P]ATP, wild-type DDK exhibited robust autophosphorylation, and this activity was significantly reduced for the DDK-KD preparation (Fig. 1B).
Figure 1.
DDK preferentially targets pre-RC Mcm2–7 proteins. (A) S. cerevisiae DDK kinase was purified from asynchronous yeast cells overexpressing Cdc7-proA and Dbf4-CBP. Purified DDK protein (625 ng) was analyzed by SDS-PAGE separation and stained by Sypro Orange. Lanes: (WT) wild type; (KD) kinase-deficient. Note that the faster migration of the DDK-KD subunits during SDS-PAGE is due to reduced autophosphorylation. (B) Phosphorylation of different Mcm substrates by DDK. Wild-type or kinase-deficient DDK (350 fmol) was incubated with pre-RC Mcm2–7 (100 fmol), baculovirus-purified Mcm2–7 (100 fmol), or purified Mcm6 (100 fmol) at 25°C for 15 min in the presence of [γ-32P]ATP. Samples were analyzed for the presence of Mcm2–7 by immunoblotting, and phosphorylation was detected by autoradiography. (C) Experimental outline for D. DNA-bound pre-RCs were incubated with H/150 mM KGlut for 30 min at 25°C. Bead-bound DNA was isolated and separated from the buffer supernatant. DDK or DDK-KD and [γ-32P]ATP was added to the proteins remaining bound to the DNA and to the proteins that dissociated from the DNA. (D) DDK preferentially phosphorylates DNA-bound Mcm2–7 complexes. The experiment was performed as described in C.
We tested the kinase activity of the purified DDK using several Mcm substrates: purified Mcm6, purified Mcm2–7 complex, and Mcm2–7 in the context of the pre-RC. The pre-RC substrate was made by in vitro assembly on origin DNA linked to magnetic beads (Seki and Diffley 2000; Bowers et al. 2004). We incubated each of these substrates with wild-type and kinase-deficient DDK in the presence of [γ-32P]ATP and measured their relative levels of phosphorylation. Although the molarity of DDK and Mcm2–7 (or Mcm6) proteins was equal in the three reactions, we observed significantly more phosphorylated protein at the molecular weight of the Mcm2–7 proteins when the pre-RC was used as a substrate (Fig. 1B, cf. lanes 1,3,5). These modifications are DDK-dependent as they are uniformly reduced when DDK-KD is substituted for DDK (Fig. 1B, cf. lanes 1,3,5 and lanes 2,4,6). The two reactions containing Mcm2–7 and wild-type DDK (Fig. 1B, lanes 1,3) showed distinct patterns of phosphorylation, indicating that both the extent and specificity of DDK phosphorylation of Mcm2–7 were altered in the context of the pre-RC. To ask more directly whether association with origin DNA led to enhanced DDK targeting of Mcm2–7, we took advantage of our finding that a subset of Mcm2–7 complexes is released from origin DNA after pre-RC formation (Fig. 1C; Supplemental Fig. 1). Although DDK showed the same robust modification of the proteins that remained associated with origin DNA (which we show are Mcm subunits below), the released Mcm2–7 complexes were phosphorylated at a 10-fold lower level (Fig. 1D).
DDK targets Mcm2, Mcm4, and Mcm6 in the context of the pre-RC
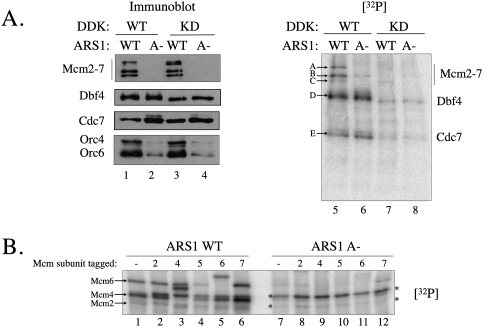
Because other proteins cofractionate with origin DNA after in vitro pre-RC formation (e.g., Abf1) (J.C.W. Randell, unpubl.), it was important to confirm that the proteins modified by DDK were pre-RC components. To this end, we asked whether the DDK-modified proteins required the ORC DNA-binding site to associate with DNA. When pre-RCs were assembled on wild-type origin DNA and treated with DDK, we observed five phosphorylated proteins (Fig. 2A, lane 5, labeled A–E). The two strongly phosphorylated, lower-molecular-weight proteins were the result of DDK autophosphorylation and correspond to Dbf4 and Cdc7 (proteins D and E, respectively). The remaining phosphorylated proteins (A, B, and C) showed the properties of pre-RC components targeted by DDK: They were only present when wild-type origin DNA was used in the pre-RC assembly reaction (Fig. 2A, cf. lanes 5 and 6), and phosphorylation of these proteins was not observed with kinase-deficient DDK (Fig. 2A, cf. lanes 5 and 7). Note that DDK was added after pre-RC formation and purification and is therefore present in all reactions.
Figure 2.
DDK phosphorylates Mcm2, Mcm4, and Mcm6 in the context of the pre-RC. (A) DDK phosphorylation of purified pre-RCs. Pre-RC assembly assays were performed with DNAs including wild-type (WT) or a mutant ARS1 lacking an ORC DNA-binding site (A−). DNA-associated proteins were treated with DDK or DDK-KD in the presence of [γ-32P]ATP for 15 min at 25°C. (Left panel) Samples were separated by SDS-PAGE, and ORC, Cdc7, Dbf4, and Mcm2–7 proteins were detected by immunoblotting. (Right panel) The resulting blot was also analyzed by autoradiography, and the major phosphoproteins are labeled A–E. (B) Mcm2, Mcm4, and Mcm6 are phosphorylated by DDK. Pre-RCs were assembled with wild-type- and A-ARS1-containing DNAs using extracts in which the indicated Mcm subunit was epitope-tagged. An extract with untagged Mcm2–7 subunits was also tested. Pre-RCs were then treated with wild-type DDK kinase in the presence of [γ-32P]ATP. Samples were analyzed by SDS-PAGE followed by immunoblotting and autoradiography. Arrows indicate the bands that were shifted when tagged with BCCP. Asterisks (*) indicate origin-independent phosphoproteins.
To address whether the modified proteins were Mcm2–7 subunits, we assembled pre-RCs using extracts derived from cells that expressed individual epitope-tagged versions of each of the Mcm2–7 proteins (except Mcm3). The use of a large epitope tag (an 80-amino-acid biotin-accepting peptide) ensured that if a Mcm protein were the target of DDK phosphorylation, the corresponding radiolabeled band would have a substantially altered mobility in the epitope-tagged extract. Pre-RC complexes assembled using the Mcm6 and Mcm4 epitope-tagged strains altered the mobility of two radiolabeled proteins (corresponding to A and B in Fig. 2A, respectively), identifying these Mcm proteins as the most prominently phosphorylated subunits (Fig. 2B, lanes 3,5). Likewise, a less prominently phosphorylated protein migrating faster than Mcm4 and Mcm6 was identified as Mcm2 (Fig. 2B, lane 2, corresponding to protein C in A; see Supplemental Fig. 2 for an enlarged view of the relevant lanes). Two remaining labeled proteins were observed both with wild-type and mutant origin templates and, therefore, were not components of the pre-RC and were not analyzed further. Although it was possible that DDK treatment would alter the association of one or more pre-RC components with the origin, we saw no DDK-dependent changes in ORC, Mcm2–7, or Cdc6 origin association (Supplemental Fig. 3; data not shown). Thus, we conclude that Mcm4 and Mcm6, and, to a lesser extent, Mcm2 are targets of DDK in the context of the pre-RC.
DDK preferentially modifies ‘loaded’ Mcm2–7 in an ORC-independent manner
Only a subset of Mcm2–7 complexes in eukaryotic cells participates in DNA replication. Both in vivo and in vitro studies indicate that origin DNA-associated Mcm2–7 complexes can be divided into two classes: (1) associated Mcm2–7 complexes, which require the ongoing presence of other pre-RC components to interact with origin DNA/chromatin; and (2) loaded Mcm2–7 complexes, which maintain their association with origin DNA/chromatin in the absence of other pre-RC components (Donovan et al. 1997; Rowles et al. 1999; Edwards et al. 2002; Bowers et al. 2004). Importantly, studies in Xenopus extracts strongly suggest that the loaded subset of Mcm2–7 complexes is sufficient to direct DNA replication (Rowles et al. 1999). These different forms of Mcm2–7 can be biochemically separated from one another by high-salt extraction of pre-RCs (Donovan et al. 1997; Rowles et al. 1999; Edwards et al. 2002; Bowers et al. 2004). Only loaded Mcm2–7 is retained on the origin DNA after a high-salt wash. ORC, Cdc6, Cdt1, and associated Mcm2–7 are quantitatively removed from the DNA by this treatment.
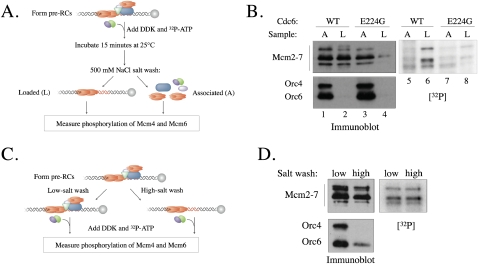
If loaded Mcm2–7 complexes are those that are destined for incorporation into replication forks, targeting the limiting amounts of DDK to this subset of helicases could prevent inappropriate helicase activation and increase the efficiency of replication initiation. To ask if DDK preferentially targets loaded Mcm2–7 complexes, we treated pre-RCs with DDK, allowing equal access to loaded and associated Mcm2–7. Subsequently, we separated loaded from associated Mcm2–7 complexes and determined the level of DDK phosphorylation (Fig. 3A). Interestingly, loaded Mcm2–7 was consistently more heavily phosphorylated than associated Mcm2–7 (Fig. 3B, cf. lanes 5 and 6), and this difference was primarily the result of increased phosphorylation of Mcm4 and Mcm6.
Figure 3.
DDK preferentially phosphorylates loaded Mcm2–7 complexes. (A) Experimental outline for B. DNA-bound pre-RCs were first incubated with either DDK or DDK-KD and [γ-32P]ATP at 25°C. After 15 min, the reactions were incubated with H/500 mM NaCl (which also inactivates DDK) (Kihara et al. 2000). The abundance and levels of DDK phosphorylation of associated (A) and loaded (L) Mcm2–7 complexes were analyzed by immunoblotting and autoradiography. (B) DDK preferentially phosphorylates loaded Mcm2–7 complexes. The experiment was performed as described in A. Pre-RCs were assembled using either wild-type Cdc6 (WT) or the ATPase-defective Cdc6-E224G (E224G) as indicated. (C) Experimental outline for D. Pre-RCs were assembled and either washed with H buffer containing 300 mM KGlut (“low-salt wash”) or 500 mM NaCl (“high-salt wash”). The DNA beads were exchanged into kinase buffer and incubated with DDK and [γ-32P]ATP. DDK phosphorylation and Mcm2–7 and ORC association with origin DNA were measured as in Figure 2A. (D) ORC is not required for DDK phosphorylation of Mcm2–7. The experiment was performed as described in C.
To address the preferential targeting of loaded Mcm2–7 in a different manner, we assembled pre-RCs using an ATPase-defective Cdc6 (Cdc6-E224G) that inhibits the formation of loaded Mcm2–7 (Fig. 3B, lane 3; Perkins and Diffley 1998; Randell et al. 2006). Consistent with loaded Mcm2–7 complexes being a preferred substrate, DDK showed significantly reduced levels of Mcm2–7 phosphorylation when Cdc6-E224G was used during pre-RC formation (Fig. 3B, lanes 7,8). Due to residual ATPase activity, Cdc6-E224G supports a low level of Mcm2–7 loading (Fig. 3B, lane 4; Randell et al. 2006). It is noteworthy that these loaded Mcm2–7 complexes are also preferentially modified (Fig. 3B, cf. Mcm2–7 levels in lanes 3,4 and Mcm4 and Mcm6 phosphorylation in lanes 7,8). We conclude that DDK preferentially phosphorylates loaded Mcm2–7.
Previous two-hybrid studies indicate that Dbf4 binds to multiple ORC subunits (Duncker et al. 2002), suggesting that such an interaction could facilitate preferential DDK targeting of pre-RC Mcm2–7. In support of this hypothesis, in both situations in which we observe differential DDK phosphorylation of Mcm2–7 (Figs. 1D, 3B), the preferred targets were associated with ORC-bound DNA. To address whether ORC facilitates DDK targeting, we compared DDK phosphorylation of pre-RC Mcm2–7 before and after high-salt extraction (Fig. 3C). Because high-salt extraction removes ORC from origin DNA, only the reactions that are not salt-extracted will retain ORC. Although there was less Mcm2–7 complex in the high-salt-extracted reactions (due to removal of associated Mcm2–7), autoradiography showed similar levels of DDK phosphorylation for both populations of Mcm2–7 complexes (Fig. 3D). Thus, the presence of ORC in the pre-RC was not required for the preferential phosphorylation of loaded Mcm2–7 by DDK.
Loaded and associated Mcm2–7 complexes are conformationally distinct
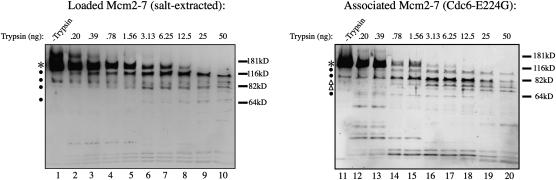
Because DDK retains its preferential targeting of loaded Mcm2–7 in the absence of other pre-RC components, we asked if there was a conformational difference between loaded and associated Mcm2–7. To test this possibility, we compared their protease accessibility. Associated Mcm2–7 showed significantly more susceptibility to proteolysis compared with loaded Mcm2–7 (Fig. 4). This difference was revealed by the relative stability of full-length Mcm2–7 protein (Fig. 4, cf. lanes 5 and 15), the appearance of degradation intermediates (cf. the 110-kDa and 82-kDa intermediates), and the distinct pattern of degradation intermediates. In each case, these changes required significantly more protease in the case of the loaded Mcm2–7 compared with the associated Mcm2–7. The relative protease resistance of the loaded complexes is not due to other pre-RC components since the high-salt wash used to isolate loaded Mcm2–7 removes all other pre-RC proteins from the DNA (Randell et al. 2006). It is more likely that the altered accessibility of loaded Mcm2–7 is due to the mechanism by which these complexes maintain tight association with the origin DNA (e.g., by encircling the DNA). These findings strongly suggest that the dissimilar conformations of loaded and associated Mcm2–7 present a distinctly different target for the enzymes acting on them whether these enzymes are proteases or kinases.
Figure 4.
Loaded and associated Mcm2–7 complexes are conformationally distinct. Limited trypsin digestion of loaded and associated Mcm2–7. To isolate loaded Mcm2–7, assembled pre-RCs were washed with high-salt buffer. To enrich for associated Mcm2–7, pre-RCs were assembled using Cdc6-E224G. The resulting Mcm2–7 complexes were washed with buffer compatible with trypsin and treated with the indicated amounts of TPCK-treated trypsin for 25 min at 25°C. Samples were analyzed by SDS-PAGE followed by immunoblotting for the Mcm2–7 complexes. (*) Full-length Mcm2–7 proteins; (●) degradation intermediates present in both proteolysis profiles; (△) degradation intermediates present in the proteolysis profiles of the associated Mcm2–7 complexes but not in that of the loaded Mcm2–7 complexes. Note that the use of a 4%–20% polyacrylamide gel resulted in the intact Mcm2–7 proteins migrating as a single broad band.
DDK binds to the pre-RC
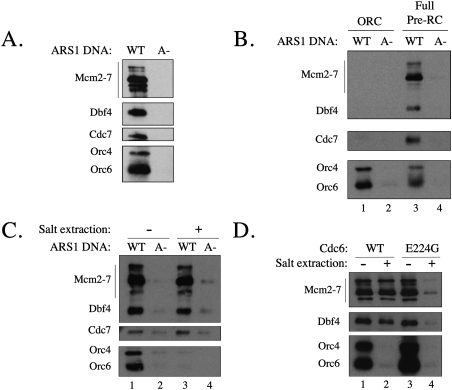
Previous studies established that Dbf4 is recruited to an origin in vivo (Dowell et al. 1994) and that Dbf4 binds to multiple ORC subunits, Mcm2 and Mcm4 (Duncker et al. 2002; Varrin et al. 2005; Sheu and Stillman 2006). Thus, DDK binding to the pre-RC might be a factor in targeting of pre-RC Mcm2–7 complexes. To test this possibility, we asked if purified DDK specifically bound pre-RCs. We repeated the pre-RC phosphorylation experiment described above; however, after DDK addition, we reisolated the origin DNA and any associated DDK. Importantly, this analysis revealed that in addition to the pre-RC components, both Cdc7 and Dbf4 copurified with the wild-type origin DNA but not DNA containing a mutant origin (Fig. 5A).
Figure 5.
DDK associates with the pre-RC in an Mcm2–7-dependent manner. (A) DDK associates with the pre-RC. Pre-RC assembly assays were performed using DNA containing wild-type (WT) or a mutant ARS1 lacking an ORC DNA-binding site (A−). DNA-associated proteins were then incubated with DDK and ATP. After DDK incubation, the DNA-associated proteins were isolated and analyzed by SDS-PAGE and immunoblotting. (B) ORC is not sufficient to recruit DDK. DDK association with the pre-RC was measured as in A except Cdc6 was not included in the pre-RC reactions labeled “ORC” (shown in lanes 1,2). Without Cdc6, ORC binds to the DNA but cannot recruit Cdt1 or the Mcm2–7 complex (Randell et al. 2006). (C) The Mcm2–7 complex is sufficient for recruitment of DDK. DDK association with the pre-RC was measured as in A except that the pre-RC complexes in the reactions shown in lanes 3 and 4 were washed with high-salt buffer before DDK addition. (D) DDK binds associated and loaded Mcm2–7 with similar affinity. DDK association with the pre-RC was measured as in A except that only wild-type ARS1 DNA was used. Pre-RC complexes were formed with wild-type Cdc6 (lanes 1,2) and with the ATPase-defective Cdc6E224G (lanes 3,4). After assembly, the pre-RC complexes shown in lanes 1 and 3 were washed with low-salt buffer and in lanes 2 and 4, with high-salt buffer. Note: To normalize the levels of Mcm2–7, more DNA was used in lane 3 as compared with lane 1, resulting in increased ORC levels.
Which pre-RC protein factors are required to recruit DDK to origin DNA? We first addressed whether ORC could recruit DDK independently of other pre-RC components. Using extracts lacking Cdc6, we prepared DNA templates associated with ORC but lacking other pre-RC components (Fig. 5B, lane 1). Although we observed robust association of ORC with the DNA, no DDK association was detected (Fig. 5B, lanes 1,2). Thus, ORC was not sufficient to recruit DDK.
To address the role of Mcm2–7 in recruiting DDK to origin DNA, we assembled pre-RCs and treated them with a high-salt wash. After normalizing the amount of Mcm2–7 in the high-salt- and low-salt-washed pre-RCs, we determined DDK association with each fraction. DDK associated with both complexes to a similar extent (Fig. 5C, cf. lanes 1 and 3). We conclude that DDK is recruited to the origin DNA through interactions with the Mcm2–7 complex. Quantification of the DNA-associated ORC, Mcm2–7 and DDK support this conclusion. At saturating DDK levels, DDK and MCM complexes are present on origin DNA in equimolar amounts (∼100 fmol per pmol of origin DNA), whereas ORC is present at approximately fivefold higher levels. This is consistent with our previous findings that only a fraction of ORC-bound origins successfully recruit the Mcm2–7 complex (Bowers et al. 2004).
The findings presented thus far suggest that the differential ability of DDK to phosphorylate different Mcm2–7 populations is mediated by distinct DDK binding or phosphorylation efficiency. To determine if loaded Mcm2–7 is preferentially bound by DDK, we prepared equal amounts of loaded (salt-resistant), associated (prepared using Cdc6-E224G), or a mixed population (standard pre-RC assembly reaction) of Mcm2–7 complexes and measured DDK binding to each preparation. DDK associated with all three populations of the Mcm2–7 complex to a similar extent (Fig. 5D, cf. lanes 1 [mixed], 2 [loaded], 3 [associated]). Salt extraction of the reaction assembled with Cdc6-E224G removed all but a small amount of the Mcm2–7 protein (Fig. 5D, lane 4), demonstrating that the majority of the Mcm2–7 complexes shown in Figure 5D, lane 3, were associated with the DNA, rather than loaded. Moreover, only a small amount of DDK binding was observed in this reaction, demonstrating that DDK was specifically binding to Mcm2–7 protein (Fig. 5D, lane 4). These findings indicate that DDK binds equally well to loaded and associated Mcm2–7. Thus, DDK binding to the Mcm2–7 complex is not responsible for the differential phosphorylation of these two populations by DDK.
Previous studies have found that Mcm10 stimulates DDK phosphorylation of Mcm2–7 (Lee et al. 2003). To address the role of Mcm10 in the context of our pre-RC/DDK assay, we immunodepleted Mcm10 or mock-depleted a G1-arrested yeast extract and then assembled pre-RCs (Supplemental Fig. 4A). Consistent with in vivo studies (Ricke and Bielinsky 2004), depletion of Mcm10 did not reduce pre-RC formation, nor did it reduce DDK binding to or phosphorylation of pre-RC Mcm2–7 (Supplemental Fig. 4B). Therefore, Mcm10 is not involved in the observed preferential phosphorylation of pre-RC Mcm2–7.
The N terminus of Dbf4 mediates Mcm2–7 binding and phosphorylation
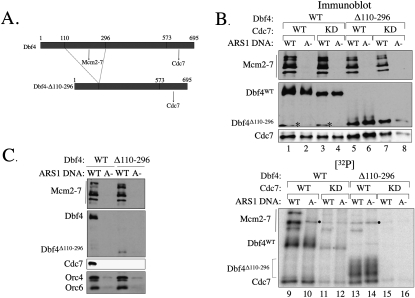
We next addressed what part of DDK is required for association with the pre-RC and what the consequences of disrupting this association are. Previous studies showed that the N-terminal half of Dbf4 is critical for recruitment of DDK to the origin and that the C-terminal half is required to bind the Cdc7 kinase (Fig. 6A; Dowell et al. 1994). For this reason, we asked if DDK with amino acids 110–296 deleted from the N-terminal region of Dbf4 was able to bind and phosphorylate pre-RCs. This Dbf4 mutant is unable to complement a DBF4 deletion and is defective in two-hybrid interactions with Mcm2 and Orc2, but retains the ability to interact with Cdc7 (Duncker et al. 2002; Varrin et al. 2005). We found that the variant DDK (DDK–Dbf4Δ110–296) retained similar levels of autophosphorylation, indicating that DDK kinase activity was not defective in the mutant (Fig. 6B, cf. lanes 9,10 and lanes 13,14). In contrast, DDK–Dbf4Δ110–296 was strongly defective in the association with the pre-RC (Fig. 6C) and phosphorylation of Mcm4 and Mcm6 (Fig. 6B, cf. lanes 9 and 13). Interestingly, the DDK–Dbf4Δ110–296 kinase retained substantial Mcm2 phosphorylation, suggesting that DDK uses a distinct mechanism to target this subunit. These findings indicate that the N-terminal region of Dbf4 mediates the association of DDK with pre-RC Mcm2–7 and that this association is required for DDK to phosphorylate Mcm4 and Mcm6.
Figure 6.
DDK–Dbf4Δ110-296 is an active kinase but cannot bind the Mcm2–7 complex or phosphorylate Mcm4 or Mcm6. (A) Dbf4Δ110–296 mutant. The Dbf4 mutant used in this figure is depicted. Regions required for Mcm2–7 and Cdc7 interaction are indicated (Varrin et al. 2005). (B) DDK–Dbf4Δ110–296 has autophosphorylation activity but does not phosphorylate Mcm4 and Mcm6. The experiment was performed as described in Figure 2A. (*) Detection of nonspecific proteins in lanes 1 and 3 of Western; (●) origin-independent phosphoprotein. (C) DDK–Dbf4Δ110–296 is defective for pre-RC association. DDK and DDK–Dbf4Δ110–296 association with pre-RCs was measured as described in Figure 5A.
Prior phosphorylation of the pre-RC is required for DDK to target Mcm2–7
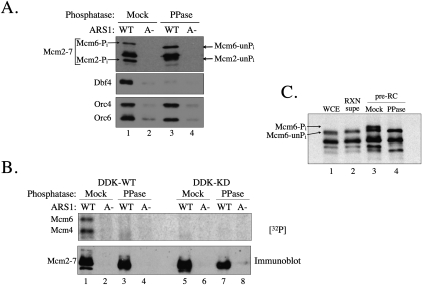
Previous studies have shown that DDK phosphorylation of Mcm2 is dramatically increased when it has been previously phosphorylated (Kihara et al. 2000; Cho et al. 2006; Montagnoli et al. 2006). Furthermore, several Mcm2–7 subunits have been shown to be phosphoproteins in G1 cells in vivo (Young and Tye 1997; Li et al. 2007). Therefore, we asked whether prior phosphorylation of pre-RC Mcm2–7 complexes influenced their ability to be bound and/or phosphorylated by DDK. To test this hypothesis, pre-RCs were prepared and treated with λ-phosphatase. After washing away the phosphatase, the ability of the dephosphorylated Mcm2–7 to bind to and act as a substrate for DDK was determined.
Dephosphorylation of the pre-RC had two consequences. First, dephosphorylated Mcm2–7 lost the ability to bind DDK (Fig. 7A, cf. lanes 1 and 3). This loss of binding is not due to the displacement of Mcm2–7 proteins from the DNA, as immunoblotting showed no change in ORC or Mcm2–7 origin association after phosphatase treatment (Fig. 7A). Second, prior phosphatase treatment dramatically reduced DDK phosphorylation of Mcm4 and Mcm6 (Fig. 7B). Importantly, we continued to see robust autophosphorylation of Cdc7 and Dbf4 after phosphatase treatment (Supplemental Fig. 5), indicating that the defect in Mcm4 and Mcm6 phosphorylation was not due to incomplete phosphatase removal. Overall, these data provide strong evidence that targeting of pre-RC Mcm2–7 by DDK is regulated by prior phosphorylation.
Figure 7.
Prior phosphorylation of the pre-RC Mcm2–7 is required for DDK binding to and phosphorylation of the Mcm2–7 complex. (A) DDK does not bind to dephosphorylated pre-RCs. Pre-RCs were assembled, then either treated or mock-treated with λ-phosphatase for 5 min at 25°C. After washing away the phosphatase, the pre-RCs were washed and incubated with DDK or DDK-KD and ATP. The DNA-bound proteins were isolated following the DDK incubation and analyzed by immunoblotting. (B) DDK requires prior phosphorylation of the Mcm2–7 complex to phosphorylate Mcm4 and Mcm6. Pre-RCs were assembled and washed in H/500 mM NaCl, then either treated or mock-treated with λ-phosphatase for 5 min at 25°C. Treated Mcm2–7 complexes were then washed and incubated with wild-type and mutant DDK and [γ-32P]ATP. The DNA-bound proteins were isolated following the DDK incubation and analyzed by SDS-PAGE followed by autoradiography. (C) Mcm6 is phosphorylated in the context of the pre-RC even in the absence of DDK. Whole-cell extract (WCE; lane 1) was analyzed by SDS-PAGE followed by immunoblotting, along with the supernatant and DNA-bound fractions from a pre-RC assembly reaction performed without subsequent DDK treatment (lanes 2,3), and phosphatase-treated pre-RCs (lane 4).
These observations raise the interesting question of whether the priming phosphorylation event is itself specific to DNA-bound Mcm2–7. We found that Mcm6 often migrates as a doublet even in experiments with kinase-deficient DDK (e.g., Fig. 6B). We asked whether this doublet is due to DDK-independent but pre-RC-specific phosphorylation of Mcm6. We compared the migration pattern of Mcm2–7 from whole-cell extract (Fig. 7C, lane 1) with Mcm2–7 found in the supernatant (Fig. 7C, lane 2) and DNA fractions (Fig. 7C, lane 3) of the pre-RC assembly reaction. We found that only Mcm6 that had been incorporated into the pre-RC showed a clear doublet. Importantly, this doublet was eliminated by phosphatase treatment (Fig. 7C, cf. lanes 3 and 4). Because these reactions were performed in the absence of DDK and the G1 extracts used for the assembly reaction lack Dbf4, this phosphorylation must be due to either the preferential incorporation of a small subset of phosphorylated Mcm2–7 or Mcm6 is phosphorylated by an unknown kinase during or after pre-RC assembly. Such an activity could be a key contributor to the targeting of DDK to Mcm2–7 complexes at the origin.
Discussion
DDK activity is required to recruit Cdc45 and GINS to origins (Zou and Stillman 2000; Kanemaki and Labib 2006), both of which are likely activators of the Mcm2–7 helicase in vivo (Moyer et al. 2006). Only a fraction of Mcm2–7 is recruited to origins (Donovan et al. 1997; Liang and Stillman 1997; Mendez and Stillman 2000), and only a subset of these chromatin-associated Mcm2–7 complexes participate in replication forks (Edwards et al. 2002; Gambus et al. 2006; Ge et al. 2007). Thus, it is likely that Mcm2–7 helicase activation is restricted in the cell. One mechanism to achieve this restriction would be to target DDK to a subset of Mcm2–7 complexes. Consistent with this hypothesis, studies of DDK action suggest that it is either limiting or restricted in its function during S phase (Bousset and Diffley 1998; Donaldson et al. 1998; Patel et al. 2008), and a separate study suggests that the chromatin-associated subset of Mcm2–7 is preferentially targeted by DDK (Sheu and Stillman 2006).
Here we used a biochemical approach to investigate the mechanisms that direct DDK to phosphorylate Mcm2–7 in the context of the pre-RC. Our findings indicate that assembly into the pre-RC changes the Mcm2–7 subunits phosphorylated by DDK and increases the overall level of phosphorylation. DDK stably associates with origin-bound Mcm2–7 complexes, and this interaction is required for DDK phosphorylation of origin-bound Mcm4 and Mcm6. Interestingly, we find that DDK preferentially phosphorylates the loaded subset of Mcm2–7 complexes that are most tightly bound to the origin. Finally, we provide evidence that prior phosphorylation of Mcm2–7 is required for DDK to bind to and preferentially modify Mcm2–7 in the context of the pre-RC. Taken together, our studies support a model in which the loading of Mcm2–7 onto origin DNA facilitates their subsequent activation by enhancing their recruitment of and further phosphorylation by DDK.
Incorporation into the pre-RC activates Mcm2–7 for DDK phosphorylation
Incorporation into the pre-RC alters DDK phosphorylation of Mcm2–7 in several ways, each of which serves to focus DDK activity on the subset of Mcm2–7 complexes that are most likely to participate in chromosome replication. First, the absolute level of Mcm2–7 phosphorylation by DDK is increased in the context of the pre-RC. This observation can explain the previously observed preference of DDK to modify chromatin-associated Mcm2–7 (Sheu and Stillman 2006). Second, the pattern of Mcm subunit phosphorylation is altered in the context of the pre-RC. Mcm4 and Mcm6 are the preferred substrates, whereas Mcm2 is preferred when the complex is away from the origin. Consistent with this preference, analysis of DDK function in S. cerevisiae and Schizosaccharomyces pombe cells suggests that Mcm4 is a critical target of DDK (Masai et al. 2006; Sheu and Stillman 2006). Finally, we see that the loaded subset of Mcm2–7 complexes is the preferred substrate for DDK. Studies in Xenopus extracts indicate that these tightly DNA-linked Mcm2–7 complexes are sufficient for full replication activity, suggesting that this subset is normally selected for replication fork assembly (Rowles et al. 1999). Overall, these findings can explain how the limiting amount of DDK in cells is targeted to a critical fraction of Mcm2–7 at the origin.
The changes in Mcm2–7 phosphorylation upon pre-RC formation require the ability of DDK to stably interact with Mcm2–7. We observe that DDK binds tightly to the Mcm2–7 complex. Interfering with this Mcm2–7–DDK docking event by deleting a N-terminal segment of the Dbf4 protein (Dbf4Δ110–296) also eliminates preferential targeting of pre-RC Mcm2–7 (Fig. 6). This mutation does not alter DDK autophosphorylation, suggesting that the primary defect of this mutant is in kinase targeting. This view is supported by studies that indicate that this region of Dbf4 is required for a one-hybrid interaction with origin DNA and interacts with at least one Mcm2–7 subunit (Dowell et al. 1994; Varrin et al. 2005). This Dbf4 mutant is unable to complement a DBF4 deletion (Varrin et al. 2005), indicating that the function mediated by this region is critical for DNA replication, which is the only essential function for Dbf4 in mitotic cells (Jackson et al. 1993; Hardy et al. 1997). Dephosphorylation of pre-RC Mcm2–7 also inhibits DDK–Mcm2–7 docking. Importantly, phosphatase treatment results in the same reduction in Mcm4 and Mcm6 phosphorylation as seen for the Dbf4Δ110–296 mutation, further supporting the conclusion that DDK docking to Mcm2–7 is required for enhanced Mcm2–7 phosphorylation upon pre-RC formation.
Our finding that the Mcm2–7 complex is central to recruit DDK to the pre-RC is consistent with previous observations. Studies in Xenopus egg extracts showed that Cdc7 chromatin association is dependent on Dbf4 and pre-RC formation, but does not require the continued association of ORC or Cdc6 with chromatin (Jares and Blow 2000; Edwards et al. 2002; Jares et al. 2004). Studies of the S. cerevisiae proteins also support an interaction between Dbf4 and Mcm2–7 subunits (Varrin et al. 2005; Sheu and Stillman 2006). Most notably, a direct interaction between Dbf4 and the N-terminal 333 amino acids of Mcm4 has been detected (Sheu and Stillman 2006). In this study, the C-terminal region of this fragment binds Dbf4 and is important for processive hyperphosphorylation of Mcm4 by DDK. Other studies in S. cerevisiae support an interaction between ORC and Dbf4 (Duncker et al. 2002; Varrin et al. 2005) that we did not observe. There are several reasons that we might not have detected such an interaction including different modification states of ORC or that the interaction is too transient to be detected in our assay. Importantly, studies in both S. cerevisiae cells and Xenopus extracts indicate that an interaction with ORC is not required for DDK to perform its essential function. In both species, elimination of ORC prior to DDK action does not inhibit the ability of DDK to activate previously assembled pre-RCs to replicate the genome (Jares and Blow 2000; Shimada et al. 2002).
Function of prior Mcm2–7 phosphorylation
Our studies indicate that dephosphorylation of the origin-bound Mcm2–7 complex eliminates both DDK binding to and phosphorylation of Mcm4 and Mcm6. This observation strongly suggests that prior phosphorylation of the Mcm2–7 complexes is required for these downstream events. We can envision two possible roles for this “priming” phosphorylation: (1) creating a docking site for DDK, or (2) creating a target peptide for DDK phosphorylation. These two explanations are not mutually exclusive, and both could contribute to DDK specificity.
Our data strongly support the presence of a phosphorylation-dependent docking site for DDK on Mcm2–7 (Fig. 7), although the nature of this binding site remains elusive. It is noteworthy that the deleted region of Dbf4Δ110–296 includes a motif distantly related to the BRCA1 C-terminal (BRCT) motif (Gabrielse et al. 2006), and pairs of BRCT motifs are known to act as phosphopeptide-binding domains (Manke et al. 2003). Although there is only one distantly related BRCT motif in Dbf4, it is possible that this motif participates in the phosphorylation-dependent docking of DDK with origin-bound Mcm2–7 (Fig. 7). We attempted to make smaller Dbf4 deletions to test this possibility directly, but all the resulting DDK mutants lacked kinase activity. The phosphorylation-dependent DDK docking site(s) on the Mcm2–7 complex has also not been identified. Previous studies have identified a DDK docking domain in the N-terminal tail of Mcm4 (Sheu and Stillman 2006). These studies were performed with Escherichia coli produced Mcm4 so it is unlikely that phosphorylation was involved in this interaction, but it is possible that the observed interaction is stimulated by phosphorylation. Given that DDK docking enhances phosphorylation of Mcm4 and Mcm6, it is also likely that there are multiple docking sites on Mcm2–7. Further analysis of Mcm2–7 phosphorylation and DDK binding will be required to map this interaction more precisely.
Does the priming phosphorylation play a role in DDK target peptide recognition? This possibility is supported by several studies suggesting that DDK can target serines or threonines adjacent to phosphoserine or phosphothreonine (Cho et al. 2006; Masai et al. 2006; Montagnoli et al. 2006; Wan et al. 2008). Indeed, a perusal of the sequences of the N terminus of Mcm4 and Mcm6 shows a large number of clusters of Ser/Thr. Phosphorylation of a C-terminal Ser/Thr in such a cluster could create a DDK target site.
The kinase responsible for the priming phosphorylation is unknown. One interesting possibility would be a cyclin-dependent kinase (CDK). Consistent with this possibility, a previous study in S. cerevisiae cells suggests that S-CDK activity is required for DDK to perform its essential function (Nougarede et al. 2000). It is clear that Mcm2–7 proteins are modified by CDKs (Montagnoli et al. 2006; Devault et al. 2008), and it has been observed that CDK modification can create DDK target sites (Masai et al. 2006; Wan et al. 2008). Indeed, there are numerous SerSerPro motifs in the N termini of Mcm4 and Mcm6 that could represent DDK target sequences after being phosphorylated by S-CDK (CDK typically targets Ser/Thr–Pro sites). It is noteworthy, however, that the extracts used to assemble pre-RCs are derived from G1-arrested cells, and no S-CDK and little G1-CDK activity is expected to be present. Other kinases also could mediate the priming phosphorylation. For example, previous studies of human Mcm2 phosphorylation have identified phosphorylation at sites consistent with casein kinase II or ATM/ATR-related kinases (Montagnoli et al. 2006). Identification of the sites of Mcm2–7 protein phosphorylation and further fractionation of the pre-RC assembly extracts will be required to identify the responsible kinase.
Analysis of Mcm2–7 before and after in vitro pre-RC assembly in the absence of DDK shows that Mcm6 is phosphorylated only when in the context of the pre-RC. This finding indicates that there is a kinase in the pre-RC assembly extracts derived from G1-arrested cells that targets Mcm2–7 specifically in the context of the pre-RC. Such an activity would be well suited to act as the priming kinase. It will be interesting to use this system to isolate the responsible kinase and to directly address whether this modification is linked to DDK function in the cell.
Distinguishing loaded from associated Mcm2–7
Our studies demonstrate that DDK prefers to modify Mcm2–7 that is loaded onto origin DNA. Our data eliminate several possible explanations for this specificity, including proximity to origin DNA (Fig. 3B), a role for ORC (Fig. 3D), and differential DDK binding to associated and loaded Mcm2–7 (Fig. 5D). The distinct conformation of loaded and associated Mcm2–7 complexes (Fig. 4) could facilitate DDK phosphorylation by exposing additional target peptides. Alternatively, differential priming phosphorylation could mediate the preferential targeting of loaded Mcm2–7 complexes. If so, then the kinase responsible for priming phosphorylation must distinguish between loaded and associated/free Mcm2–7. Of course, differential conformation and phosphorylation could work together to make the loaded form of the Mcm2–7 complex a preferred DDK substrate.
The importance of DDK targeting
Cell cycle progression requires the precise temporal and spatial coordination of the events of DNA replication. The mechanisms of DDK targeting described here are likely to make important contributions to this coordination. First and foremost, this targeting would ensure that DDK phosphorylates and activates only the subset of Mcm2–7 that is most tightly associated with the origin. Such targeting would spatially restrict the activation of the replicative helicase and prevent Mcm2–7 that is free in solution or readily released from the origin from being inappropriately activated. A requirement for pre-RC formation and phosphorylation to direct DDK to a subset of Mcm2–7 could also provide mechanisms to order the events of replication initiation such that helicase activation does not precede helicase loading. Indeed, if the priming kinase is an S-phase CDK, such a mechanism could ensure that no helicase activation occurs until after S-phase entry. The use of priming phosphorylation to direct DDK to certain Mcm2–7 complexes could also play a regulatory role. If the extent or timing of such phosphorylation was not uniform, priming phosphorylation of the Mcm2–7 complex could regulate origin efficiency or the timing of origin activation within S phase. As sites targeted by this priming phosphorylation are identified, it will be intriguing to determine how the level of modification varies across the genome and during the cell cycle.
Materials and methods
Yeast strains and plasmids
Yeast strains expressing wild-type (yLF52) and kinase-deficient DDK (yLF53) were constructed by transforming W303 with plasmids pLF8 (WT Cdc7) and pLF9 (Cdc7D163N), respectively. pLF8 and pLF9 were made using the pESC Split-Tap plasmid (D'Souza and Walker 2006). Cdc7 and Dbf4 were integrated into the plasmid using FseI/AsiSI and NotI/AscI, respectively, resulting in the fusion of a fragment of Protein A with the C terminus of Cdc7 and the fusion of calmodulin-binding peptide (CBP) with the C terminus of Dbf4. Yeast strains expressing biotin carboxyl carrier protein (BCCP)-tagged Mcm subunits (VTy173, VTy174, VTy175, VTy176, VTy177) were constructed by integrating the BCCP-coding sequence (Cronan 1990) at the 3′-end of the indicated MCM gene using the pVT104 integrating plasmid. The yeast strains used in this study are listed in Supplemental Table S1.
Protein purification
Asynchronous yeast cells were grown to mid-log phase in 1% raffinose, and expression of Dbf4-CBP/Cdc7-proA was induced with addition of 2% galactose for 3 h. Cells were resuspended in 1 volume of calmodulin elution buffer (Puig et al. 2001) and frozen in liquid nitrogen. Extracts were made by grinding frozen cell pellets in a motorized mortar/pestle (Retsch RM100) followed by centrifugation at 53,000g for 1 h. Forty milliliters of extract (20 mg/mL) were incubated with 400 μL of calmodulin resin for 1 h at 4°C in calmodulin-binding buffer as described (Puig et al. 2001). After binding, the resin was washed with binding buffer then eluted with calmodulin elution buffer for 45 min at 4°C. The eluate from the calmodulin column was incubated with 400 μL of heparin resin for 1 h at 4°C. The proA tag was cleaved from Cdc7 (while bound to the heparin column) by incubation with TEV cleavage buffer and 20 units of TEV (Puig et al. 2001). The resin was washed twice with buffer H/150 mM KCl (50 mM HEPES at pH 7.6, 5 mM Mg-acetate, 1 mM EDTA, 1 mM EGTA, 10% glycerol, 0.02% NP-40, 150 mM KCl), and bound proteins were eluted using H/600 mM KCl. Peak fractions were pooled and dialyzed with H/300 mM potassium glutamate (KGlut). Mcm6-TAP was purified as described (Puig et al. 2001). Mcm2–7 was expressed and purified from baculovirus-infected insect cells as described previously (Schwacha and Bell 2001).
Preparation of whole-cell extracts and pre-RC assembly
The WCEs used in the pre-RC assembly assays were made as described previously (Bowers et al. 2004). Pre-RC assembly assays were performed as described previously (Randell et al. 2006). To assemble non-high-salt-washed pre-RCs, extracts were incubated with origin DNA, and then the DNA was washed three times in H/300 mM KGlut. To assemble high-salt-washed pre-RCs, extracts were incubated with origin DNA, then the DNA was washed once with H/300 mM KGlut, incubated for 2 min with H/500 mM NaCl (high-salt wash), then washed again with H/300 mM KGlut.
DDK phosphorylation and binding assays
Pre-RCs assembled from a 40-μL pre-RC assembly reaction were incubated in a 30-μL kinase reaction with 125 ng of either DDK or DDK-KD, 0.1 mM ATP, 3 μCi of [γ-32P]ATP (3000 Ci/mmol), and kinase buffer (50 mM HEPES at pH 7.6, 5 mM Mg-acetate, 1 mM EDTA, 1 mM EGTA, 10% glycerol, 0.01% NP-40) containing 150 mM KGlut for 15 min at 25°C. Kinase assays were stopped by the addition of 12 μL of 5× SDS-PAGE sample buffer and analyzed by immunoblotting and autoradiography. For pre-RC/DDK-binding assays, after the 15-min DDK kinase reaction, the beads/origin DNA and any associated proteins were separated from the supernatant. Thirty microliters of 1× SDS-PAGE sample buffer were added to the beads/origin DNA, 12 μL of 5× SDS sample buffer were added to the supernatant, and the samples were analyzed by SDS-PAGE, immunoblotting, and autoradiography. To isolate free Mcm2–7 complexes, pre-RCs were assembled, washed, and resuspended in H/150 mM KGlut for 30 min. The magnetic bead/origin DNA-bound Mcm2–7 was separated from the released/free Mcm2–7 using a magnet. DDK kinase assays with purified Mcm6 were performed as for the pre-RC/DDK activity assay, except that 15 ng of purified Mcm6-CBP was used in the reaction.
Trypsin sensitivity assays
Loaded Mcm2–7 complexes were obtained by incubation of assembled pre-RCs with H/500 mM NaCl for 15 min at 25°C, then washed with H/300 mM KGlut. Associated MCM complexes were obtained by assembling pre-RCs with Cdc6E224G (Randell et al. 2006). MCM complexes (25 ng) were then treated with the indicated quantity of TPCK-treated trypsin in H+/300 mM KGlut for 25 min at 25°C. Tryptic digestions were quenched by addition of SDS-PAGE sample buffer followed immediately by boiling for 4 min. Samples were analyzed by SDS-PAGE using a 4%–20% gel followed by immunoblotting for the Mcm2–7.
Acknowledgments
We thank Tania Baker, Frank Solomon, Ryan Heller, and Adam Matthews for critical reading and helpful comments on the manuscript. We thank Vasiliki Tsakraklides for providing yeast strains and extracts containing BCCP-tagged Mcm subunits. The work described here was supported by a grant from the National Institutes of Health to S.P.B. (GM52339) and the Howard Hughes Medical Institute. S.P.B. is an employee of the Howard Hughes Medical Institute. J.C.W.R. was a Special Fellow of the Leukemia and Lymphoma Society. T.J.T. was supported by a National Institutes of Health Training grant (GM007287). L.U. was supported by an Undergraduate Biological Science Education Program Award from the Howard Hughes Medical Institute.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1759609.
Supplemental material is available at http://www.genesdev.org.
References
- Aparicio T., Ibarra A., Mendez J. Cdc45–MCM–GINS, a new power player for DNA replication. Cell Div. 2006;1:18. doi: 10.1186/1747-1028-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias E.E., Walter J.C. Strength in numbers: Preventing rereplication via multiple mechanisms in eukaryotic cells. Genes & Dev. 2007;21:497–518. doi: 10.1101/gad.1508907. [DOI] [PubMed] [Google Scholar]
- Bousset K., Diffley J.F. The Cdc7 protein kinase is required for origin firing during S phase. Genes & Dev. 1998;12:480–490. doi: 10.1101/gad.12.4.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers J.L., Randell J.C., Chen S., Bell S.P. ATP hydrolysis by ORC catalyzes reiterative Mcm2–7 assembly at a defined origin of replication. Mol. Cell. 2004;16:967–978. doi: 10.1016/j.molcel.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Cho W.H., Lee Y.J., Kong S.I., Hurwitz J., Lee J.K. CDC7 kinase phosphorylates serine residues adjacent to acidic amino acids in the minichromosome maintenance 2 protein. Proc. Natl. Acad. Sci. 2006;103:11521–11526. doi: 10.1073/pnas.0604990103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan J.E., Jr Biotination of proteins in vivo. A post-translational modification to label, purify, and study proteins. J. Biol. Chem. 1990;265:10327–10333. [PubMed] [Google Scholar]
- Devault A., Gueydon E., Schwob E. Interplay between S-cyclin-dependent kinase and Dbf4-dependent kinase in controlling DNA replication through phosphorylation of yeast Mcm4 N-terminal domain. Mol. Biol. Cell. 2008;19:2267–2277. doi: 10.1091/mbc.E07-06-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson A.D., Fangman W.L., Brewer B.J. Cdc7 is required throughout the yeast S phase to activate replication origins. Genes & Dev. 1998;12:491–501. doi: 10.1101/gad.12.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan S., Harwood J., Drury L.S., Diffley J.F. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc. Natl. Acad. Sci. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell S.J., Romanowski P., Diffley J.F. Interaction of Dbf4, the Cdc7 protein kinase regulatory subunit, with yeast replication origins in vivo. Science. 1994;265:1243–1246. doi: 10.1126/science.8066465. [DOI] [PubMed] [Google Scholar]
- D'Souza S., Walker G.C. Novel role for the C terminus of Saccharomyces cerevisiae Rev1 in mediating protein–protein interactions. Mol. Cell. Biol. 2006;26:8173–8182. doi: 10.1128/MCB.00202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncker B.P., Shimada K., Tsai-Pflugfelder M., Pasero P., Gasser S.M. An N-terminal domain of Dbf4p mediates interaction with both origin recognition complex (ORC) and Rad53p and can deregulate late origin firing. Proc. Natl. Acad. Sci. 2002;99:16087–16092. doi: 10.1073/pnas.252093999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards M.C., Tutter A.V., Cvetic C., Gilbert C.H., Prokhorova T.A., Walter J.C. MCM2–7 complexes bind chromatin in a distributed pattern surrounding the origin recognition complex in Xenopus egg extracts. J. Biol. Chem. 2002;277:33049–33057. doi: 10.1074/jbc.M204438200. [DOI] [PubMed] [Google Scholar]
- Gabrielse C., Miller C.T., McConnell K.H., DeWard A., Fox C.A., Weinreich M. A Dbf4p BRCA1 C-terminal-like domain required for the response to replication fork arrest in budding yeast. Genetics. 2006;173:541–555. doi: 10.1534/genetics.106.057521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambus A., Jones R.C., Sanchez-Diaz A., Kanemaki M., van Deursen F., Edmondson R.D., Labib K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- Ge X.Q., Jackson D.A., Blow J.J. Dormant origins licensed by excess Mcm2–7 are required for human cells to survive replicative stress. Genes & Dev. 2007;21:3331–3341. doi: 10.1101/gad.457807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy C.F., Dryga O., Seematter S., Pahl P.M., Sclafani R.A. mcm5/cdc46-bob1 bypasses the requirement for the S phase activator Cdc7p. Proc. Natl. Acad. Sci. 1997;94:3151–3155. doi: 10.1073/pnas.94.7.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A.L., Pahl P.M., Harrison K., Rosamond J., Sclafani R.A. Cell cycle regulation of the yeast Cdc7 protein kinase by association with the Dbf4 protein. Mol. Cell. Biol. 1993;13:2899–2908. doi: 10.1128/mcb.13.5.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jares P., Blow J.J. Xenopus cdc7 function is dependent on licensing but not on XORC, XCdc6, or CDK activity and is required for XCdc45 loading. Genes & Dev. 2000;14:1528–1540. [PMC free article] [PubMed] [Google Scholar]
- Jares P., Luciani M.G., Blow J.J. A Xenopus Dbf4 homolog is required for Cdc7 chromatin binding and DNA replication. BMC Mol. Biol. 2004;5:5–19. doi: 10.1186/1471-2199-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanemaki M., Labib K. Distinct roles for Sld3 and GINS during establishment and progression of eukaryotic DNA replication forks. EMBO J. 2006;25:1753–1763. doi: 10.1038/sj.emboj.7601063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara M., Nakai W., Asano S., Suzuki A., Kitada K., Kawasaki Y., Johnston L.H., Sugino A. Characterization of the yeast Cdc7p/Dbf4p complex purified from insect cells. Its protein kinase activity is regulated by Rad53p. J. Biol. Chem. 2000;275:35051–35062. doi: 10.1074/jbc.M003491200. [DOI] [PubMed] [Google Scholar]
- Labib K., Gambus A. A key role for the GINS complex at DNA replication forks. Trends Cell Biol. 2007;17:271–278. doi: 10.1016/j.tcb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Lee J.K., Seo Y.S., Hurwitz J. The Cdc23 (Mcm10) protein is required for the phosphorylation of minichromosome maintenance complex by the Dfp1–Hsk1 kinase. Proc. Natl. Acad. Sci. 2003;100:2334–2339. doi: 10.1073/pnas.0237384100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M., Kawasaki Y., Young M.R., Kihara M., Sugino A., Tye B.K. Mcm2 is a target of regulation by Cdc7–Dbf4 during the initiation of DNA synthesis. Genes & Dev. 1997;11:3365–3374. doi: 10.1101/gad.11.24.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Gerber S.A., Rudner A.D., Beausoleil S.A., Haas W., Villen J., Elias J.E., Gygi S.P. Large-scale phosphorylation analysis of α-factor-arrested Saccharomyces cerevisiae. J. Proteome Res. 2007;6:1190–1197. doi: 10.1021/pr060559j. [DOI] [PubMed] [Google Scholar]
- Liang C., Stillman B. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes & Dev. 1997;11:3375–3386. doi: 10.1101/gad.11.24.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manke I.A., Lowery D.M., Nguyen A., Yaffe M.B. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- Masai H., Arai K. Cdc7 kinase complex: A key regulator in the initiation of DNA replication. J. Cell. Physiol. 2002;190:287–296. doi: 10.1002/jcp.10070. [DOI] [PubMed] [Google Scholar]
- Masai H., Taniyama C., Ogino K., Matsui E., Kakusho N., Matsumoto S., Kim J.M., Ishii A., Tanaka T., Kobayashi T., et al. Phosphorylation of MCM4 by Cdc7 kinase facilitates its interaction with Cdc45 on the chromatin. J. Biol. Chem. 2006;281:39249–39261. doi: 10.1074/jbc.M608935200. [DOI] [PubMed] [Google Scholar]
- Mendez J., Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: Assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnoli A., Valsasina B., Brotherton D., Troiani S., Rainoldi S., Tenca P., Molinari A., Santocanale C. Identification of Mcm2 phosphorylation sites by S-phase-regulating kinases. J. Biol. Chem. 2006;281:10281–10290. doi: 10.1074/jbc.M512921200. [DOI] [PubMed] [Google Scholar]
- Moyer S.E., Lewis P.W., Botchan M.R. Isolation of the Cdc45/Mcm2–7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl. Acad. Sci. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nougarede R., Della Seta F., Zarzov P., Schwob E. Hierarchy of S-phase-promoting factors: Yeast Dbf4–Cdc7 kinase requires prior S-phase cyclin-dependent kinase activation. Mol. Cell. Biol. 2000;20:3795–3806. doi: 10.1128/mcb.20.11.3795-3806.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtoshi A., Miyake T., Arai K., Masai H. Analyses of Saccharomyces cerevisiae Cdc7 kinase point mutants: Dominant-negative inhibition of DNA replication on overexpression of kinase-negative Cdc7 proteins. Mol. Gen. Genet. 1997;254:562–570. doi: 10.1007/s004380050452. [DOI] [PubMed] [Google Scholar]
- Pacek M., Tutter A.V., Kubota Y., Takisawa H., Walter J.C. Localization of MCM2–7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol. Cell. 2006;21:581–587. doi: 10.1016/j.molcel.2006.01.030. [DOI] [PubMed] [Google Scholar]
- Patel P.K., Kommajosyula N., Rosebrock A., Bensimon A., Leatherwood J., Bechhoefer J., Rhind N. The Hsk1(Cdc7) replication kinase regulates origin efficiency. Mol. Biol. Cell. 2008;19:5550–5558. doi: 10.1091/mbc.E08-06-0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins G., Diffley J.F. Nucleotide-dependent prereplicative complex assembly by Cdc6p, a homolog of eukaryotic and prokaryotic clamp-loaders. Mol. Cell. 1998;2:23–32. doi: 10.1016/s1097-2765(00)80110-0. [DOI] [PubMed] [Google Scholar]
- Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Seraphin B. The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- Randell J.C., Bowers J.L., Rodriguez H.K., Bell S.P. Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2–7 helicase. Mol. Cell. 2006;21:29–39. doi: 10.1016/j.molcel.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Ricke R.M., Bielinsky A.K. Mcm10 regulates the stability and chromatin association of DNA polymerase-α. Mol. Cell. 2004;16:173–185. doi: 10.1016/j.molcel.2004.09.017. [DOI] [PubMed] [Google Scholar]
- Rowles A., Tada S., Blow J.J. Changes in association of the Xenopus origin recognition complex with chromatin on licensing of replication origins. J. Cell Sci. 1999;112:2011–2018. doi: 10.1242/jcs.112.12.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha A., Bell S.P. Interactions between two catalytically distinct MCM subgroups are essential for coordinated ATP hydrolysis and DNA replication. Mol. Cell. 2001;8:1093–1104. doi: 10.1016/s1097-2765(01)00389-6. [DOI] [PubMed] [Google Scholar]
- Seki T., Diffley J.F. Stepwise assembly of initiation proteins at budding yeast replication origins in vitro. Proc. Natl. Acad. Sci. 2000;97:14115–14120. doi: 10.1073/pnas.97.26.14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu Y.J., Stillman B. Cdc7–Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol. Cell. 2006;24:101–113. doi: 10.1016/j.molcel.2006.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada K., Pasero P., Gasser S.M. ORC and the intra-S-phase checkpoint: A threshold regulates Rad53p activation in S phase. Genes & Dev. 2002;16:3236–3252. doi: 10.1101/gad.239802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B. Origin recognition and the chromosome cycle. FEBS Lett. 2005;579:877–884. doi: 10.1016/j.febslet.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Takeda D.Y., Dutta A. DNA replication and progression through S phase. Oncogene. 2005;24:2827–2843. doi: 10.1038/sj.onc.1208616. [DOI] [PubMed] [Google Scholar]
- Tanaka S., Umemori T., Hirai K., Muramatsu S., Kamimura Y., Araki H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- Varrin A.E., Prasad A.A., Scholz R.P., Ramer M.D., Duncker B.P. A mutation in Dbf4 motif M impairs interactions with DNA replication factors and confers increased resistance to genotoxic agents. Mol. Cell. Biol. 2005;25:7494–7504. doi: 10.1128/MCB.25.17.7494-7504.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J.C. Evidence for sequential action of cdc7 and cdk2 protein kinases during initiation of DNA replication in Xenopus egg extracts. J. Biol. Chem. 2000;275:39773–39778. doi: 10.1074/jbc.M008107200. [DOI] [PubMed] [Google Scholar]
- Wan L., Niu H., Futcher B., Zhang C., Shokat K.M., Boulton S.J., Hollingsworth N.M. Cdc28–Clb5 (CDK-S) and Cdc7–Dbf4 (DDK) collaborate to initiate meiotic recombination in yeast. Genes & Dev. 2008;22:386–397. doi: 10.1101/gad.1626408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich M., Stillman B. Cdc7p–Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53 checkpoint pathway. EMBO J. 1999;18:5334–5346. doi: 10.1093/emboj/18.19.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M.R., Tye B.K. Mcm2 and Mcm3 are constitutive nuclear proteins that exhibit distinct isoforms and bind chromatin during specific cell cycle stages of Saccharomyces cerevisiae. Mol. Biol. Cell. 1997;8:1587–1601. doi: 10.1091/mbc.8.8.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P., Diffley J.F. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–285. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- Zou L., Stillman B. Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p–Dbf4p kinase. Mol. Cell. Biol. 2000;20:3086–3096. doi: 10.1128/mcb.20.9.3086-3096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]