Abstract
Cattle faeces are considered the most important reservoir for human infection with Escherichia coli O157. We have previously described shedding of E. coli O157 in the faeces of naturally infected cattle cohorts. However, the data require further investigation to quantify the uncertainty and variability in the estimates previously presented. This paper proposes a method for analysing both the presence and the quantity of E. coli O157 in cattle faecal samples, using two isolation procedures, one of which enumerates E. coli O157. The combination of these two measurements, which are fundamentally different in nature and yet measuring a common outcome, has necessitated the development of a novel statistical model for ascertaining the contribution of the various components of variation (both natural and observation induced) and for judging the influence of explanatory variables. Most of the variation within the sampling hierarchy was attributable to multiple samples from the same animal. The contribution of laboratory-level variation was found to be low. After adjusting for fixed and random effects, short periods of increased intensity of shedding were identified in individual animals. We conclude that within-animal variation is greater than between animals over time, and studies aiming to elucidate the dynamics of shedding should focus resources, sampling more within than between animals. These findings have implications for the identification of persistent high shedders and for assessing their role in the epidemiology of E. coli O157 in cattle populations. The development of this non-standard statistical model may have many applications to other microbial count data.
Keywords: Escherichia coli O157, variation, epidemiology, concentration, cattle
1. Introduction
Escherichia coli O157 is a cause of gastrointestinal disease in humans, and direct or indirect contact with cattle faeces has been identified as a risk factor for human infection (Locking et al. 2001). To implement effective control strategies, the dynamics of E. coli O157 shedding in cattle at the individual and group levels are essential. Despite numerous epidemiological studies (Besser et al. 1997, 2001; Hancock et al. 1997; Mechie et al. 1997; Heuvelink et al. 1998; Shere et al. 1998; Sargeant et al. 2000; Khaitsa et al. 2003; Rugbjerg et al. 2003; Synge et al. 2003), the dynamics remain poorly understood. Thus far, shedding profiles of E. coli O157 in naturally infected cattle have been largely described in terms of point estimates with the high levels of variability in prevalence and concentration within and between animals rarely quantified or even discussed (Shere et al. 1998; Omisakin et al. 2003; Robinson et al. 2004a; Widiasih et al. 2004).
Epidemiological studies of cattle within the farm or feedlot environment often involve clustered sampling; for example, taking samples from animals within the same pen or from faecal pats produced by the same animal over time. Thus, there is the potential to quantify within- and between-animal variations. Smith et al. (2001) and Khaitsa et al. (2003) explored the components of variation affecting the shedding of E. coli O157 and concluded that greater variation in shedding occurred within feedlot pens, rather than between pens over time. However, these studies were limited to weekly sampling and presented results as prevalence estimates, with no indication of the concentrations of organisms shed.
Variation and uncertainty are inherent within any observational dataset, especially those containing microbial count data. Although the count data are potentially more informative than the presence/absence data, problems arise when interpreting highly variable data. Methods for measuring and modelling variation and uncertainty within data are desirable as they give rise to more accurate parameter estimates rather than point estimates, which may be subject to many forms of error during sample collection and laboratory processing. Quantifying the sources of variation in count data, including assay variation and natural variation within and between subjects, is complex. However, estimates of both variability and uncertainty associated with each level of the sampling hierarchy can be used to inform food chain quantitative microbial risk assessments (Jordan et al. 1999) and other model-based approaches for comparing control strategies (Turner et al. 2003, 2006; Matthews et al. 2006). Furthermore, apportioning within- and between-animal variations informs sample size calculations and will benefit the design of future studies.
The authors have previously described the shedding profiles of two cohorts of cattle (Robinson et al. 2004a). While the presentation of raw data allowed broad interpretation of the variability within and between animals, it did not reveal how much of the observed variation in counts was attributable to the different levels of the sampling hierarchy. Furthermore, to illustrate infection profiles, we previously averaged four agar plate counts per sample and the concentration for samples positive by the immunomagnetic separation (IMS) assay, which determined the presence/absence, was set at 100 CFU g−1. Here, we use repeated sampling visits to the cohort and replicated samples during the isolation procedures to ascertain the relative importance of several levels of clustering (animal, faecal pat, laboratory). This paper further presents a method analysing both the presence of E. coli O157 in cattle and the intensity of the infection from samples of voided faeces, using two isolation procedure methods. The enumeration method (Robinson et al. 2004b) is based on spiral plating, less sensitive than other presence/absence methods, but provides a measure of the number of colony-forming units in the sample. The IMS method is used as a highly sensitive test for the presence of a pathogen in faecal samples. The combination of these two measurements, which are fundamentally different in nature and yet measuring a common outcome, has necessitated the development of a novel statistical model for ascertaining the contribution of components of variation (both natural and observation induced) and for judging the influence of explanatory variables.
2. Methods
2.1 Data summary
Data were collected during two cohort studies of E. coli O157 shedding in naturally infected weaned calves. The cohorts for the two studies were kept on separate farms and the studies were conducted at separate times. The first cohort was housed in the same pen that contained 14 replacement heifers aged between six and nine months. Sampling was conducted approximately every 3 hours (between 06.00 and 23.00 hours) for 5 days. The second cohort contained 16 calves of mixed breed and sex all contained in the same pen. Sampling was conducted approximately twice daily for 7 days. A detailed description of the method used for the isolation and enumeration of E. coli O157 can be found elsewhere (Robinson et al. 2004a). For study 1 on each visit, a random selection of faecal samples was processed in duplicate. From these selected samples, after initial mixing, two 2 g samples were processed in parallel. This allowed for laboratory variation to be incorporated in the analysis. Duplicated samples were not used for study 2. The shedding patterns of individual animals in both cohorts are described in Robinson et al. (2004a).
Covariates collected included age (days), sex and breed (cohort 2). Other time-varying covariates such as time since last concentrate feed applied to the whole pen. Faecal pat-level covariates included consistency (subjectively scored 1 (solid)–5 (runny)), colour (cohort 1, scored 3 (pale brown), 4 (dark brown); graded using colour charts) and amount of undigested barley grains (subjectively graded into three categories after examining whole pat (none, less than 10 grains and greater than 10 grains)). The levels of undigested barley grain were recorded only for cohort 2 as the diet of cohort 1 did not contain grain. At the end of study 1, there was found to be very little variation in faecal pat colour and was therefore not recorded in study 2. These variables were measured to gain insight into within-animal faecal variation over time and as a crude measure of gut conditions in each animal. In this instance, faecal pat characteristics are regarded as independent of E. coli O157 levels in an animal but they may, through altering interaction with the rectoanal junction, the proposed site of colonization of E. coli O157 (Naylor et al. 2003), affect the amount of bacteria in a pat, the outcome measured in this study.
2.2 Statistical analysis
The data from the two cohort studies were analysed independently to assess between-farm variation and other between-study variation. The count data from both studies were well described by a Poisson distribution with the addition of a ‘zero-inflated’ component to allow for the preponderance of zeros. We avoided combining the IMS and count data into a single measure as we wished to assess the differences between and commonalities of factors affecting infection status (as detected by IMS) and those affecting the concentration of organisms shed (as indicated by the count data) when an animal is infected.
In our model, animal i was shedding at time t with probability pit, where pit can vary from animal to animal and depend on covariates such as the colour and consistency of the pat sampled. We assume that each IMS measurement Zit is an accurate indication of an animal's infection status, and write the model as
The Bernoulli distribution takes the value 1 with probability pit and 0 otherwise. For study 1, the raw colony count measurement Yitkl from plate l of sample k is assumed to always be zero when the corresponding IMS measurement is negative, and Poisson distributed otherwise, with
The λitk is the unknown true concentration of bacteria in faecal sample k (in CFU g−1), and Sitkl is the quantity of faeces on the plate calculated from the dilution factor of the faecal suspension and the size of the region on the plate from which colonies were counted.
The pit and λitk depend on a vector of covariates Xit with indicator variables for the various possible colour and consistency values, modelled as fixed effects. Furthermore, the IMS and count measurements taken from the same animal are assumed to be more similar than the data from different animals, and correspondingly normally distributed random effects Ai and Bi are used to describe animal i's shedding probability relative to the population average (adjusted for pat colour and consistency, and on the log-odds scale) and average concentration of bacteria in pats voided during shedding episodes. Also, the E. coli O157 concentration is assumed to vary over time, and possibly between laboratory replicates from the same pat, leading us to include pat-level random effects Cit and laboratory-level effects Ditk. The infection probabilities pit and concentrations λitk are modelled as
The parameter vectors β and γ model the effect of colour and consistency on shedding incidence and shedding intensity of E. coli O157, respectively. The use of the multivariate normal for the animal-level random effects Ai and Bi allows for the possibility that an animal's shedding probability and average concentration during shedding episodes are correlated; for instance animals that are frequently colonized shed higher amounts of bacteria than animals that shed only rarely. Finally note that there is no plate-level random effect to allow for extra-Poisson variation in the counts, as this was shown to be unnecessary in a previous analysis (Robinson et al. 2005).
Variation over time is described by the pat-level random effect Cit. If pat t from animal i was produced at time Tit, then Uit=exp(Cit) refers to the concentrations of E. coli O157 in animal i at time Tit relative to what is usual for this animal. A value of Uit greater than 1 indicates that the faecal pat contains higher counts than the average pat produced by animal i. An IMS-negative pat will have zero counts, which corresponds to Uit=0. In this analysis, the Cit is modelled as mutually independent, though a model with the Cit correlated over time would be able to model dependence on shedding over time and, for example, the progression of an infection in an animal. Here, we restrict ourselves to examine plots of Uit against Tit in order to assess the temporal nature of E. coli O157 shedding.
The model was fit in a Bayesian framework and the posterior distributions sampled using Markov chain Monte Carlo methods in the software package Winbugs (http://www.mrc-bsu.cam.ac.uk/bugs/winbugs/). Prior distributions were chosen to achieve priors that were flat enough to encompass all parameter values that were felt to be reasonable. Normal priors with zero mean and a standard deviation of 20 were used for β and γ; gamma priors with shape parameter 0.1 and scale parameter 10 were used for the σ parameters; and a Wishart prior with two degrees of freedom and the identity matrix as a scale was applied to the precision matrix Σ−1.
Convergence of the Markov chain was assessed by examining the autocorrelation plots of the simulations, examining sample paths, and by running multiple chains. The posterior means and standard deviations are similar to those obtained by fitting separate univariate models using pseudo-likelihood, which validate the performance of the chain and the choice of prior distributions.
On a number of visits, the IMS test was not performed on the faecal samples for operational reasons. In these instances, the Zit are unobserved latent variables for pats it for which all observed counts Yitkl are zero. In other words, if all spiral plates were free of bacteria, it is not known whether this is due to the pat being uninfected or owing to sampling error resulting in negative samples taken from a positive infected pat. To perform inference on these observations, the likelihood was computed as
being the unconditional probability of all the counts equalling zero.
The data from the second study were modelled in a similar way, but as there were no laboratory replicates the Ditk term was not included.
3. Results
Throughout the findings of the study, the term shedding intensity will be used to refer to the count/concentration of E. coli O157 in faecal pats produced by animals during an incidence of shedding. Shedding incidence refers to a presence/absence of E. coli O157 in the faeces produced by an animal. In this paper, we refer to an animal's shedding probability as the incidence probability, the chance of an animal producing a pat containing E. coli O157.
For the first cohort, a total of 229 faecal samples were collected at 19 visits over the 5-day period. Of these, 113 (49.3%) samples were positive on direct enumeration plates and the remaining 116 samples were subjected to IMS. In total, 134 (58.5%) samples were positive by a combination of IMS and direct plating. The remaining 95 (41.5%) samples were negative by both methods. A total of 321 samples were collected at 22 sampling visits over the 14-day sampling period for the second cohort. Escherichia coli O157 was isolated from 209 (65%) samples with 129 (40.2%) samples positive on direct enumeration plates. The remaining 192 samples were subjected to IMS and 80 (24.8%) samples were positive.
Tables 1 and 2 summarize the posterior distributions of model parameters for cohorts 1 and 2, respectively. The variance estimates from the bivariate multilevel model from both studies are similar. Most of the variation in the data from both studies was attributable to variability in the concentration of E. coli O157 between animals, and between faecal pats produced by the same animal. The degree of variation at the laboratory level was very low in comparison with the variation at other levels. The high value of corr(Ai, Bi) (the correlation between infection probability and intensity) in these models indicates that animals that were more likely to be positive were also more likely to have higher counts. There was more variation in the IMS response between animals in the second study (var(Ai)=3.28) compared with the first (var(Ai)=0.75).
Table 1.
Estimates from the multilevel zero-inflated Poisson model for E. coli O157 incidence and intensity in study 1. (Data are available from IMS tests, indicating the presence/absence of E. coli O157, and bacteria counts. Random effects model the contribution of animal-, pat- and laboratory-level variations to the total variance, and fixed effects relate to the effect of pat colour and consistency on E. coli O157 shedding probabilities and rates.)
| posterior means | posterior s.d. | 95% credible interval | |
|---|---|---|---|
| random effects | |||
| between animal | |||
| incidence: var(Ai) | 0.75 | 0.4 | 0.24, 1.91 |
| intensity: var(Bi) | 3.43 | 2.1 | 1.05, 8.98 |
| corr(Ai,Bi) | 0.73 | 0.2 | 0.23, 0.94 |
| within animal | |||
| 9.53 | 1.6 | 6.83, 13.16 | |
| laboratory | |||
| 0.23 | 0.1 | 0.12, 0.42 | |
| fixed effects | |||
| incidence (IMS) | |||
| baseline (colour=3, consistency=3) | 0.33 | 0.3 | −0.27, 0.99 |
| colour=4 | 0.20 | 0.4 | −0.65, 1.01 |
| consistency=4 | 0.04 | 0.3 | −0.57, 0.65 |
| consistency=5 | 0.45 | 0.6 | −0.63, 1.59 |
| intensity (counts) | |||
| baseline (colour=3, consistency=3) | 5.34 | 0.7 | 3.91, 6.68 |
| colour=4 | −0.78 | 0.8 | −2.32, 0.76 |
| consistency=4 | 0.14 | 0.6 | −1.01, 1.33 |
| consistency=5 | −0.93 | 1.0 | −2.88, 1.02 |
Table 2.
Estimates from the multilevel zero-inflated Poisson model for E. coli O157 incidence and intensity in study 2. (Data are available from IMS tests, indicating the presence/absence of E. coli O157, and bacteria counts. Random effects model the contribution of animal- and pat-level variations to the total variance, and fixed effects relate to the effect of pat consistency and presence of grain in faeces on E. coli O157 shedding probabilities. Numbers reported in italics indicate significant fixed effects (the 95% credible intervals do not include zero).)
| posterior means | posterior s.d. | 95% credible interval | |
|---|---|---|---|
| random effects | |||
| between animal | |||
| incidence: var(Ai) | 3.28 | 1.75 | 1.22, 7.56 |
| intensity: var(Bi) | 3.78 | 2.02 | 1.30, 9.00 |
| corr(Ai,Bi) | 0.81 | 0.12 | 0.50, 0.96 |
| within animal | |||
| 5.96 | 0.91 | 4.40, 7.99 | |
| fixed effects | |||
| incidence (IMS) | |||
| baseline (consistency=3, grain=none) | 1.52 | 0.60 | 0.37, 2.72 |
| consistency=4 or 5 | 0.60 | 0.51 | −0.37, 1.59 |
| consistency=1 or 2 | −0.55 | 0.43 | −1.41, 0.29 |
| grain=1–10 grains | −0.22 | 0.34 | −0.89, 0.48 |
| grain=≤10 grains | −0.51 | 0.23 | −0.99, −0.07 |
| intensity (counts) | |||
| baseline (consistency=3, grain=none) | 4.80 | 0.57 | 3.59, 5.90 |
| consistency=4 or 5 | −0.90 | 0.70 | −2.32, 0.46 |
| consistency=1 or 2 | −0.19 | 0.51 | −1.16, 0.79 |
| grain=1–10 grains | −0.85 | 0.33 | −1.4, 0.20 |
| grain=≤10 grains | 0.93 | 0.39 | 0.07, 1.73 |
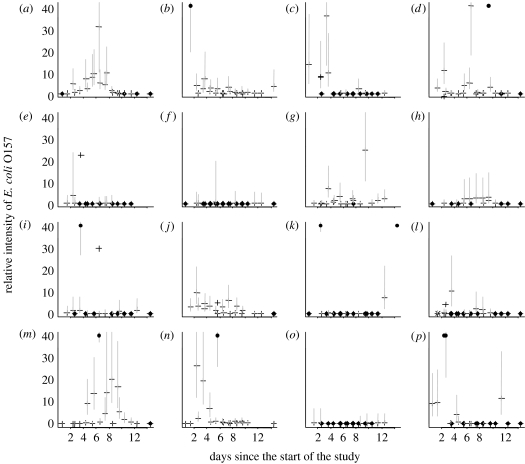
Each individual animal's time trajectory of E. coli O157 intensity was computed by predicting the concentration of bacteria in each pat relative to what would be expected from a pat from the same animal with the same colour and consistency. This was calculated as the posterior mean, the exponentiated pat effects exp(Cit) for pats where E. coli O157 was present and zero for pats in which no E. coli O157 was detected. Plotting these relative intensities over time for the second study illustrated temporal trends in shedding among several cattle (figure 1). No obvious temporal trend in shedding of E. coli O157 was evident in the first study (data not shown), possibly because the animals were observed over a shorter time period. In the second study, there did appear to be several animals in which periods of increased shedding intensity were occurring during the study. This is evident in figure 1a,m,n. Over the 14 days of the study, it was also possible to identify calves in which a shedding episode may have started or finished (figure 1b,j). Other calves within the group may have been between shedding episodes or may have been sporadic shedders. From figure 1a,m,n, it would appear that a shedding episode in this group of cattle might have lasted approximately 7 days.
Figure 1.
(a–p) Temporal changes in the relative shedding intensity of E. coli O157 for each animal in study 2. Each plot refers to an individual animal with each point corresponding to a pat sample. The vertical axis represents the predicted intensity of E. coli O157 in the pat relative to the animal's typical E. coli O157 intensity. For example, a value of 30 on the vertical axis indicates that the pat had 30 times the number of bacteria that would usually be expected from the animal that produced it. Crosses refer to pats with relative intensity greater than 0 but less than 40, with the vertical lines denoting 95% prediction intervals. Circles refer to zeros (pats with no E. coli O157) or relative intensity greater than 40.
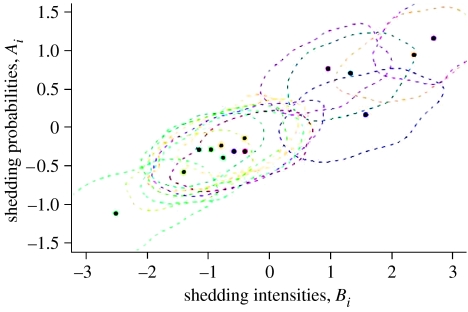
Figure 2 shows the posterior expectation and 50% joint confidence region for the animal-level random effects on E. coli O157 shedding probabilities and intensity for the animals in study 1. The plot confirms the association between presence and intensity, with animals having the highest shedding probabilities also showing the greatest intensity. These animals appear as outliers in figure 2.
Figure 2.
Animal-level random effects for intensities and shedding probabilities of E. coli O157. Circles refer to posterior expectations, with dashed lines being the posterior 50% credible region. The horizontal and vertical axes represent relative log-transformed intensities and logit-transformed probabilities, respectively.
The time since the last concentrate feed did not appear to be related to shedding probability or intensity, and was therefore not included in the final model. In study 1, the colour and consistency of faeces varied with scores of 3 (pale) or 4 (dark) and 3 (solid), 4 or 5 (runny), respectively. In study 2, there were no changes in the colour of faeces but consistency ranged from 1 to 5. Of the covariates included in the models, neither colour nor consistency was found to markedly affect the concentration or the presence of E. coli O157 in faeces (tables 1 and 2). In study 2, the presence of undigested barley grains in the faeces of the calves was associated with a marked reduction (credible interval does not include 0) in the probability of detecting E. coli O157 in faeces. However, if E. coli O157 was present in the faecal pat, then the presence of less than 10 pieces of undigested barley grains was associated with a marked increase in the intensity of E. coli O157.
Values in table 1 represent the posterior distributions of model parameters from a sample of size 951, which resulted from a run of 10 000 iterations with the first 500 discarded as burn-in and subsequently retaining only every 10th realization to minimize autocorrelation.
4. Discussion
Several studies combining direct enumeration methods for E. coli O157 in cattle faeces with more sensitive isolation methods such as IMS have reported data in a bivariate format (Brown et al. 1997; Cray et al. 1998; Cobbold & Desmarchelier 2002). The data from this paper were first considered by Robinson et al. (2004a), where the two datasets were examined separately. This paper presents a more rigorous and comprehensive analysis of the data with an integrated model for IMS and count data through the development of a statistical model that accommodates longitudinal and animal-level variation in both the shedding incidence and the intensity of E. coli O157 in faecal samples. To the authors' knowledge, this is the first comprehensive analysis of longitudinal variation in count data for E. coli O157.
A significant finding was that within the sampling hierarchy, most of the variation in the concentration with which cattle shed E. coli O157 was attributable to variation within animals over time, with average intensities varying little between animals. This was not evident from a simple graphical presentation of the raw data (Robinson et al. 2004a). Although there was variability in concentrations shed between animals, this was relatively low after accounting for variation between faecal pats from the same animal. This is a surprising finding; it would be expected that repeated samples from the same animals would be highly correlated and such temporal autocorrelation is likely to produce relatively similar values within, compared to between, animals. The high between-pat variation existed despite the inclusion of pat-specific explanatory variables such as consistency and undigested grain content of pats. Furthermore, the observed variation could not be attributed to between-strain variation; all of the E. coli O157 isolated from these cattle had indistinguishable pulsed-field gel electrophoresis (PFGE) patterns (Robinson et al. 2004a). Hence, natural fluctuations within an animal over time may be caused by more intrinsic factors such as gut flora, gut health, behaviour or other anatomical, physiological or physical stresses.
The amount of variation at the laboratory level in this study was relatively low suggesting that the method for enumerating E. coli O157 used in this study was reasonably consistent at both the plating and mixing stages. When comparing different studies, it is helpful to consider the contribution of laboratory variation to estimates of bacterial counts. Variation within faecal pats may also contribute to the observed within-animal variation; however, the pooling of samples taken from multiple sites within faecal pats would reduce this source of variation (Robinson et al. 2005).
Estimation of pat-level intensities, allowing for explanatory variables and sources of variation, appears to show short periods of high shedding in a number of calves within the cohort. The majority of other calves that appear to be largely non-shedders indicate that within the same group of calves different carriage states may be present. A visual inspection of the longitudinal variation in E. coli O157 concentrations shows shedding episodes clustering together in periods of one or two weeks, with some animals showing peaks in shedding. The differences in average shedding intensities observed between animals could be caused by the particular sampling period in this study corresponding to a high-shedding state in some animals and a low-shedding state in others. It may be that sampling the group of animals at other times may have led to different conclusions. Definite conclusions regarding the shape and duration of shedding episodes are not possible with this study, as the data collection period was not sufficiently long and the statistical model used did not explicitly model dependence on time. However, the periods of high shedding occurring in the cattle during this study provides evidence that the intensity and duration of shedding are co-dependent. This will have implications for the rate of transmission of E. coli O157 between animals occupying the same pen.
Theoretical studies suggest that between-host heterogeneity is epidemiologically important for persistence within the farm and that 80 per cent of transmission is due to 20 per cent of infections (Matthews et al. 2006). Matthews et al. (2006) developed a model that includes within- and between-animal variations, though they were unable to estimate the proportion of within-animal variation from the cross-sectional data available. Regardless of this, they estimate between-animal variation at 21 per cent, although the data are not given. Our findings substantiate the estimate of 21 per cent for between-animal variation arrived at in their study. The longitudinal data available in our study have enabled the separation of within- and between-animal variations, and the model used by Matthews et al. (2006) would likely be able to identify the model parameters for infection and recovery rate were it applied to this longitudinal dataset. We have presented data that could support future theoretical approaches by providing accurate estimates of within- and between-animal variability and the associated credible intervals. These kinds of data can be further incorporated into food chain quantitative microbial risk assessments, and could guide future studies in how to best allocate resources to minimize uncertainty in their shedding incidence and intensity estimates. For example, the results reported here suggest that studies wishing to accurately estimate prevalence, incidence or concentration of shedding may be more efficient in sampling relatively fewer animals over a longer time period.
The substantial correlation between the two animal-level latent variables in the model suggests that an animal with a higher probability of infection (as measured by IMS tests on faecal pats) will have higher bacterial counts in its faeces. Although this may due in part to an increased probability of detection using IMS at higher concentrations, this finding is in agreement with reports in the literature of persistent high shedders (greater than 104 CFU g−1; Liu et al. 2005).
It has been suggested that removing ‘super-shedders’ would be the most effective control strategy for reducing on-farm prevalence of E. coli O157 (Matthews et al. 2006). A recent study used mixture distribution analysis of retrospective data to classify super-shedders within groups of animals shedding E. coli O157 (Chase-Topping et al. 2007). They suggest that cattle shedding over 103 or 104 CFU g−1 should be considered as super-shedders. Our findings, while supporting the idea of the presence of super-shedders within groups of cattle, show the difficulty in defining animals as super-shedders based on count data due to its highly variable nature. However, incorporating the estimates of variability and uncertainty presented in this study could lead to the development of a formal field definition of a ‘super-shedder’.
To control E. coli O157 at the farm level, it may be crucial to understand the risk factors for the emergence of high-shedding animals. In this study, in addition to assessing variance estimates, covariates at the pat level were also considered. As the groups of cattle and their environment remained relatively constant during the respective studies (no change in diet, disruption of the group), pat-level covariates changing over time, such as consistency, colour and barley grain content, were included in the model. In the first cohort, all cattle were of the same sex and breed, and in the second cohort initial inspection of the data revealed no evidence of age, breed or sex effects. In this study, the presence of undigested pieces of barley grain in the faeces was found to be negatively associated with the presence, but positively associated with the concentration of E. coli O157 in faeces. Although these appear to be contradictory findings, they could be the result of complex dynamics that are likely to affect populations of E. coli O157 in the bovine gut. Furthermore, they may also explain inconsistencies between the previous studies that have assessed the effect of inclusion or exclusion of grain in the diet (Kudva et al. 1995; Dargatz et al. 1997; Buchko et al. 2000; Tkalcic et al. 2000). However, the association between the presence of undigested barley grain in faeces may be confounded with several other factors including increased growth or cell proliferation rate in cattle ingesting more grain, or increased friction between faecal material and the rectoanal junction. When evaluating the effects of different diets on the shedding of E. coli O157 in cattle, it may be important in future to identify the effect of risk factors on both the duration and the concentration with which cattle shed E. coli O157. Consideration of potentially correlated risk factors, and clustering are essential for making correct inferences from the studies of dietary variables and E. coli O157 carriage. Collection of more detailed data at the animal level and within-animal level over time may reveal further risk factors that influence shedding of E. coli O157 in cattle.
This study assessed the variability in the shedding of E. coli O157, and by attributing it to the various different levels of the sampling hierarchy, we conclude that the concentration of E. coli O157 shed by cattle varies markedly between different faecal pats. However, the sampling time frame proved insufficient to fully explore the durations of and time between shedding episodes and so the temporal component of the model was deliberately simplistic to be more easily identifiable. Longer follow-up periods combined with models incorporating a biologically informed mechanistic model for the evolution of shedding episodes should be considered for future studies in this area. An additional contribution of this study was to separate residual temporal variation in shedding intensity from variability attributable to changes in diet, housing, water source, pen dynamics or personnel, and from variations in shedding incidence. A better understanding of the biological basis for the large amount of residual within-animal variation in shedding intensity may be critical to the development of on-farm control measures for this important food-borne zoonotic agent.
Acknowledgments
The authors wish to thank the farmers for compliance and agreement to be involved in the study and the Department for Environment, Food and Rural Affairs for funding the study as part of the DEFRA Epidemiology Fellowship Unit at the University of Liverpool. We also thank Prof. P. Diggle for assistance in the study design.
References
- Besser T.E., Hancock D.D., Pritchett L.C., McRae E.M., Rice D.H., Tarr P.I. Duration of detection of faecal excretion of Escherichia coli O157:H7 in cattle. J. Infect. Dis. 1997;175:726–729. doi: 10.1093/infdis/175.3.726. [DOI] [PubMed] [Google Scholar]
- Besser T.E., Richards B.L., Rice D.H., Hancock D.D. Escherichia coli O157:H7 infection of calves: infectious dose and direct contact transmission. Epidemiol. Infect. 2001;127:555–560. doi: 10.1017/S095026880100615X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.A., Harmon B.G., Zhao T., Doyle M.P. Experimental Escherichia coli O157:H7 carriage in calves. Appl. Environ. Microbiol. 1997;63:27–32. doi: 10.1128/aem.63.1.27-32.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchko S.J., Holley R.A., Olson W.O., Gannon V.P., Veira D.M. The effect of different grain diets on faecal shedding of Escherichia coli O157:H7 by steers. J. Food Protect. 2000;63:1467–1474. doi: 10.4315/0362-028x-63.11.1467. [DOI] [PubMed] [Google Scholar]
- Chase-Topping M.E., et al. Risk factors for the presence of high-level shedders of Escherichia coli O157 on Scottish farms. J. Clin. Microbiol. 2007;45:1594–1603. doi: 10.1128/JCM.01690-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbold R., Desmarchelier P. Horizontal transmission of Shiga toxin-producing Escherichia coli within groups of dairy calves. Appl. Environ. Microbiol. 2002;68:4148–4152. doi: 10.1128/AEM.68.8.4148-4152.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cray W.C., Jr, Casey T.A., Bosworth B.T., Rasmussen M.A. Effect of dietary stress on faecal shedding of Escherichia coli O157:H7 in calves. Appl. Environ. Microbiol. 1998;64:1975–1979. doi: 10.1128/aem.64.5.1975-1979.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dargatz D.A., Wells S.J., Thomas L.A., Hancock D.D., Garber L.P. Factors associated with the presence of Escherichia coli O157 in the faeces of feedlot cattle. J. Food Protect. 1997;60:466–470. doi: 10.4315/0362-028X-60.5.466. [DOI] [PubMed] [Google Scholar]
- Hancock D.D., Besser T.E., Rice D.H., Herriott D.E., Tarr P.I. A longitudinal study of Escherichia coli O157 in fourteen cattle herds. Epidemiol. Infect. 1997;118:193–195. doi: 10.1017/S0950268896007212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuvelink A.E., van den Biggelaar F.L.A.M., Zwartkruis-Nahuis J.T.M., Herbes R.G., Huyben R., Nagelkerke N., Melchers W.J.G., Monnens L.A.H., de Boer E. Occurrence of verocytotoxin-producing Escherichia coli O157 on Dutch dairy farms. J. Clin. Microbiol. 1998;36:3480–3487. doi: 10.1128/jcm.36.12.3480-3487.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan D., McEwen S.A., Lammerding A.M., McNab W.B., Wilson J.B. Pre-slaughter control of Escherichia coli O157 in beef cattle: a simulation study. Prev. Vet. Med. 1999;41:55–74. doi: 10.1016/S0167-5877(99)00032-X. [DOI] [PubMed] [Google Scholar]
- Khaitsa M.L., Smith D.R., Stoner J.A., Parkhurst A.M., Hinkley S., Klopfenstein T.J., Moxley R.A. Incidence, duration and prevalence of Escherichia coli O157:H7 faecal shedding by feedlot cattle during the finishing period. J. Food Protect. 2003;66:1972–1977. doi: 10.4315/0362-028x-66.11.1972. [DOI] [PubMed] [Google Scholar]
- Kudva I.T., Hatfield P.G., Hovde C.J. Effect of diet on the shedding of Escherichia coli O157:H7 in a sheep model. Appl. Environ. Microbiol. 1995;61:1363–1370. doi: 10.1128/aem.61.4.1363-1370.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W.C., Jenkins C.J., Shaw D.J., Matthews L., Pearce M.C., Low J.C., Gunn G.J., Smith H.R., Frankel G., Woolhouse M.E.J. Modelling the epidemiology of verocytotoxin-producing Escherichia coli serogroups in young calves. Epidemiol. Infect. 2005;133:449–458. doi: 10.1017/S0950268804003644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locking M.E., O'Brien S.J., Reilly W.J., Wright E.M., Campbell D.M., Coia J.E., Browning L.M., Ramsay C.N. Risk factors for sporadic cases of Escherichia coli O157 infection: the importance of contact with animal excreta. Epidemiol. Infect. 2001;127:215–220. doi: 10.1017/s0950268801006045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews L., et al. Heterogeneous shedding of Escherichia coli O157 in cattle and its implications for control. Proc. Natl Acad. Sci. USA. 2006;103:547–552. doi: 10.1073/pnas.0503776103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechie S.C., Chapman P.A., Siddons C.A. A fifteen month study of Escherichia coli O157:H7 in a dairy herd. Epidemiol. Infect. 1997;118:17–25. doi: 10.1017/S0950268896007194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor S.W., Low J.C., Besser T.E., Mahajan A., Gunn G.J., Pearce M.C., McKendrick I.J., Smith D.G.E., Gally D.L. Lymphoid follicle-dense mucosa at the terminal rectum is the principal site of colonisation of enterohaemorrhagic Escherichia coli O157:H7 in the bovine host. Infect. Immun. 2003;71:1505–1512. doi: 10.1128/IAI.71.3.1505-1512.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omisakin F., MacRae M., Ogden I.D.J., Strachan N.J. Concentration and prevalence of Escherichia coli O157 in cattle faeces at slaughter. Appl. Environ. Microbiol. 2003;69:2444–2447. doi: 10.1128/AEM.69.5.2444-2447.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson S.E., Wright E.J., Hart C.A., Bennett M., French N.P. Intermittent and persistent shedding of Escherichia coli O157 in cohorts of naturally infected calves. J. Appl. Microbiol. 2004a;97:1045–1053. doi: 10.1111/j.1365-2672.2004.02390.x. [DOI] [PubMed] [Google Scholar]
- Robinson S.E., Wright E.J., Williams N.J., Hart C.A., French N.P. Development and application of a spiral plating method for the enumeration of Escherichia coli O157 in bovine faeces. J. Appl. Microbiol. 2004b;97:581–589. doi: 10.1111/j.1365-2672.2004.02339.x. [DOI] [PubMed] [Google Scholar]
- Robinson S.E., Brown P.E., Wright E.J., Bennett M., Hart C.A., French N.P. Heterogeneous distributions of Escherichia coli O157 within naturally infected bovine faecal pats. FEMS Microbiol. Lett. 2005;244:291–296. doi: 10.1016/j.femsle.2005.01.056. [DOI] [PubMed] [Google Scholar]
- Rugbjerg H., Nielsen E.M., Andersen J.S. Risk factors associated with faecal shedding of verocytotoxin-producing Escherichia coli O157 in eight known-infected Danish dairy herds. Prev. Vet. Med. 2003;58:101–113. doi: 10.1016/S0167-5877(03)00023-0. [DOI] [PubMed] [Google Scholar]
- Sargeant J.M., Gillespie J.R., Oberst R.D., Phebus R.K., Hyatt D.R., Bohra L.K., Galland J.C. Results of a longitudinal study of the prevalence of Escherichia coli O157:H7 on cow-calf farms. Am. J. Vet. Res. 2000;61:1375–1379. doi: 10.2460/ajvr.2000.61.1375. [DOI] [PubMed] [Google Scholar]
- Shere J.A., Bartlett K.J., Kaspar C.W. Longitudinal study of Escherichia coli O157:H7 dissemination on four dairy farms in Wisconsin. Appl. Environ. Microbiol. 1998;64:1390–1399. doi: 10.1128/aem.64.4.1390-1399.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D., Blackford M., Younts S., Moxley R., Gray J., Hungerford L., Milton T., Klopfenstein T. Ecological relationships between the prevalence of cattle shedding Escherichia coli O157:H7 and characteristics of the cattle or conditions of the feedlot pen. J. Food Protect. 2001;64:1899–1903. doi: 10.4315/0362-028x-64.12.1899. [DOI] [PubMed] [Google Scholar]
- Synge B.A., et al. Factors influencing the shedding of verocytotoxin-producing Escherichia coli O157 by beef suckler cows. Epidemiol. Infect. 2003;130:301–312. doi: 10.1017/S0950268802008208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkalcic S., et al. Effects of diet on rumen proliferation and faecal shedding of Escherichia coli O157:H7 in calves. J. Food Protect. 2000;63:1630–1636. doi: 10.4315/0362-028x-63.12.1630. [DOI] [PubMed] [Google Scholar]
- Turner J., Begon M., Bowers R.G., French N.P. A model appropriate to the transmission of a human food-borne pathogen in a multigroup managed herd. Prev. Vet. Med. 2003;57:175–198. doi: 10.1016/S0167-5877(03)00006-0. [DOI] [PubMed] [Google Scholar]
- Turner J., Bowers R.G., Begon M., Robinson S.E., French N.P. A semi-stochastic model of the transmission of Escherichia coli O157 in a typical UK dairy herd: dynamics, sensitivity analysis and intervention/prevention strategies. J. Theor. Biol. 2006;241:806–822. doi: 10.1016/j.jtbi.2006.01.020. [DOI] [PubMed] [Google Scholar]
- Widiasih D.A., Ido N., Omoe K., Sugii S., Shinagawa K. Duration and magnitude of faecal shedding of Shiga toxin-producing Escherichia coli from naturally infected cattle. Epidemiol. Infect. 2004;132:67–75. doi: 10.1017/S0950268803001468. [DOI] [PMC free article] [PubMed] [Google Scholar]