Abstract
Down-regulation of receptor tyrosine kinases (RTK) through receptor internalization and degradation is critical for appropriate biological responses. The hepatocyte growth factor RTK (also known as Met) regulates epithelial remodeling, dispersal, and invasion and is deregulated in human cancers. Impaired down-regulation of the Met RTK leads to sustained signaling, cell transformation, and tumorigenesis, hence understanding mechanisms that regulate this process is crucial. Here we report that, following Met activation, the endocytic adaptor protein, Eps15, is recruited to the plasma membrane and becomes both tyrosine-phosphorylated and ubiquitinated. Recruitment of Eps15 requires Met receptor kinase activity and involves two distinct Eps15 domains. Unlike previous reports for the EGF RTK, which requires the Eps15 ubiquitin interacting motif, recruitment of Eps15 to Met involves the coiled-coil domain of Eps15 and the signaling adaptor molecule, Grb2, which binds through a proline-rich motif in the third domain of Eps15. Expression of the coiled-coil domain is sufficient to displace the wild-type Eps15 protein complex from Met, resulting in loss of tyrosine phosphorylation of Eps15. Knockdown of Eps15 results in delayed Met degradation, which can be rescued by expression of Eps15 WT but not an Eps15 mutant lacking the coiled-coil domain, identifying a role for this domain in Eps15-mediated Met down-modulation. This study demonstrates a new mechanism of recruitment for Eps15 downstream of the Met receptor, involving the coiled-coil domain of Eps15 as well as interaction of Eps15 with Grb2. This highlights distinct regulation of Eps15 recruitment and the diversity and adaptability of endocytic molecules in promoting RTK trafficking.
Growth factor receptor tyrosine kinases (RTKs)2 regulate multiple key cellular processes, including proliferation, differentiation, migration, and survival. RTK activation must be tightly controlled through multiple levels of regulation to maintain cellular homeostasis. Failure to due so is associated with the development and progression of human disease such as cancer (1–3). Ligand-induced activation of RTKs promotes their rapid removal from the plasma membrane, a key event in their down-regulation, because it is a prerequisite to their lysosomal degradation. The process of RTK internalization modulates levels of RTK at the cell surface and the duration of signals activated in response to growth factors. Ligand-activated RTKs are mainly internalized through clathrin-dependent pathways to be eventually delivered to sorting endosomes (4), although other mechanisms of receptor internalization exist (5). From the sorting endosome, RTKs can recycle back to the plasma membrane or become internalized and accumulate on the limiting and internal membranes of multivesicular bodies. This latter event terminates RTK signaling by sequestering the signaling-competent intracellular domain of RTKs and preventing recycling back to the plasma membrane. Multivesicular bodies subsequently fuse with lysosomes, leading to the degradation of proteins located within intralumenal membranes (6, 7). These internalization and trafficking events are controlled via a complex network of protein-protein and protein-lipid interactions that are evolutionary conserved. Ligand-dependent internalization and trafficking of RTKs is regulated in part through ubiquitination and tyrosine phosphorylation of the receptor (8). In addition, endocytic proteins themselves are modified by RTK-dependent ubiquitin and tyrosine phosphorylation, which serve as signal switches to promote or disassemble protein-protein interactions (8, 9). Many of these interactions have been studied extensively for the epidermal growth factor (EGF) RTK (10), yet the mechanisms regulating internalization and trafficking of other RTKs remain poorly understood.
The hepatocyte growth factor (HGF) RTK (also known as Met) is primarily expressed in epithelial and endothelial cells in the adult. The HGF/Met signaling axis regulates key cellular processes such as scattering of epithelia sheets, as well as epithelial cell proliferation, migration, invasion, and survival (11). HGF/Met signaling is essential for embryonic development, namely the growth and survival of epithelial cells as well as the migration of myogenic precursor cells and the outgrowth of motor neurons (12). Chronic activation of the Met receptor is associated with several human and murine tumors (12, 13) and in the adult, the HGF/Met signaling axis is involved in wound healing and liver regeneration (14, 15).
Activation of the Met receptor by binding to HGF promotes tyrosine phosphorylation of the intracellular domain and recruitment of signaling complexes, including the Cbl E3 ubiquitin ligase (11). Cbl promotes ubiquitination of the Met receptor, an event that is critical for ligand-dependent Met degradation (16–19). Importantly, although deregulation of the Met receptor in human cancers can occur through receptor amplification, point mutations, and chromosomal translocations leading to ligand-independent RTK activation, we and others have recently shown that escape from down-regulation constitutes another mechanism leading to Met RTK deregulation in human cancers (20–22).
Internalization of the Met RTK following HGF stimulation uses canonical pathways and is clathrin-dependent (23, 24), however, little is known of the protein complexes that regulate these events. Multiple proteins have been shown to regulate EGFR internalization. One of these, epidermal growth factor receptor pathway substrate 15 (Eps15), was first identified as a tyrosine-phosphorylated substrate of the EGFR kinase (25). Eps15 has been characterized as a general endocytic adaptor protein, because it is constitutively associated with the adaptor protein complex AP-2, and forms complexes with other endocytic proteins, including Epsin and Hrs (26–29). Studies with the EGFR have shown that Eps15 translocates to the rim of budding clathrin-coated pits upon EGF stimulation and colocalizes with the receptor (30, 31). Eps15 acts as a molecular scaffold by promoting interactions through its many domains. The N terminus contains three Eps15 homology (EH) domains that are required for plasma membrane targeting of Eps15 through engagement with proteins containing the tripeptide motif asparagine-proline-phenylalanine and direct binding to phospholipids (32, 33). Eps15 also contains a central coiled-coil domain, which mediates homo- and heterodimerization (34) and a stretch of aspartate-proline-phenylalanine repeats, which can bind the α-subunit of AP-2. The C terminus harbors two ubiquitin interacting motifs (UIM) of which the second has been shown to participate in ubiquitin binding (35) and is required for recruitment to a ubiquitinated EGFR (36). Mutations within the Eps15 UIM domain that abrogate ubiquitin binding disrupt the localization of Eps15 to the plasma membrane upon EGF stimulation (37). In addition, deletion of the UIM or use of a poorly ubiquitinated EGFR mutant (Y1045F) also abrogates co-immunoprecipitation of the two proteins (36). Hence, Eps15 acts to recognize ubiquitinated EGFR cargo, which is thought to contribute to selection of the EGFR for internalization (36–38).
Although Eps15 has been shown to play a role in the internalization of the EGFR, the extent to which it functions downstream of other RTKs is currently unknown. Where investigated, tyrosine phosphorylation of Eps15 has not been observed downstream of other stimuli such as insulin, platelet-derived growth factor or keratinocyte growth factor (30, 39). Given the biological importance of the more than 50 RTKs encoded in the human genome, elucidation and validation of the molecular mechanisms governing the down-regulation of different RTKs will be important to determine which commonalities and differences exist between these family members. Because deregulation of trafficking of the Met RTK contributes to its oncogenic potential, we have investigated the role of Eps15 downstream of the Met receptor. Here, we report that HGF-induced activation of the Met receptor results in recruitment of Eps15 to the plasma membrane and causes its rapid tyrosine phosphorylation and ubiquitination. In contrast to previous studies with the EGFR, we have mapped the coiled-coiled domain of Eps15 as being sufficient and essential for the interaction of Eps15 with the Met receptor and propose a new mechanism for recruitment and regulation of RTKs by Eps15.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents—Antibody 145 was raised in rabbit against a C-terminal peptide of human Met (40). Phosphotyrosine (4G10) and Met DL-21 antibodies were purchased from Upstate Biotechnology (Lake Placid, NY). Anti-Eps15 (C-20), anti-ubiquitin (P4D1) and anti-GST were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Met antibody AF276 were from R&D Systems (Minneapolis, MN), FLAG and actin antibodies were from Sigma-Aldrich, anti-GFP, Alexa-Fluor 488-, 555-, and 647-conjugated secondary antibodies were obtained from Molecular Probes (Eugene, OR), and phospho-specific Met tyrosine 1234/35 and EGFR were from Cell Signaling Technology (Mississauga, Ontario, Canada). Monoclonal EEA1 antibody and AP-2 (μ2) were obtained from BD Biosciences (Mississauga, Ontario, Canada), HA.11 and Myc 9E10 monoclonal antibodies were from Covance (Berkeley CA) and BabCO (Richmond, CA), respectively. Mannose 6-phosphate receptor, cation-dependent antibody (22d4) was obtained from the developmental studies hybridoma bank (Iowa City, IA). HGF was a kind gift from Genentech (San Francisco, CA). EGF was purchased from Roche Diagnostics (Laval, Quebec, Canada). Cycloheximide purchased from Sigma was reconstituted in water to 10 mg/ml (100×).
Cell Culture and Transient Transfections—T47D breast epithelial cells, HeLa cells, and HEK293 cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. T47D stable cell lines expressing wild-type Met was established and described previously (21). K1110A Met stable cells were made through electroporation with pLXSN-Met K1110A cDNA in a similar manor. Transient transfections in HEK293 and HeLa cells were performed using Lipofectamine Plus reagent according to the manufacturer's instructions (Invitrogen).
Immunoprecipitation and Western Blotting—Following stimulation with HGF (0.60 nm unless otherwise indicated), T47D cells, HeLa cells, and HEK293 cells were harvested in TGH lysis buffer (50 mm HEPES, pH 7.5, 150 mm NaCl, 1.5 mm MgCl2, 1 mm EGTA, 1% Triton X-100, 10% glycerol, 1 mm phenylmethylsulfonyl fluoride, 1 mm sodium vanadate, 10 μg/ml aprotinin, and 10 μg/ml leupeptin). HEK293 transfections were harvested 48 h post-transfection. Lysates were incubated with antibody for 1 h at 4 °C with gentle rotation followed by 1-h incubation with protein A- or G-Sepharose beads. For FLAG-immunoprecipitations, lysates were directly incubated with Anti-FLAG M2 beads (Sigma) and incubated for 4 h. Captured proteins were collected by washing three times in their respective lysis buffers, eluted by boiling in SDS sample buffer, resolved by SDS-PAGE, and transferred to a nitrocellulose membrane. Membranes were blocked in 3% bovine serum albumin in TBST (10 mm Tris, pH 8.0, 150 mm NaCl, 2.5 mm EDTA, 0.1% Tween 20) for 1 h and incubated with primary then secondary antibodies in TBST for 1 h. After three washes with TBST, bound proteins were visualized with an ECL detection kit (Amersham Biosciences). To detect Eps15 ubiquitination, cells from a 10-cm dish were lysed in radioimmunoprecipitation assay buffer (0.05% SDS, 50 mm Tris, pH 8.0, 150 nm NaCl, 1% Nonidet P-40, 0.05% sodium deoxycholate) and processed as above. Densitometric analysis of Western blots was performed using NIH ImageJ software.
Confocal Immunofluorescence Microscopy—Cells were seeded at 1 × 104 to 5 × 104 on glass coverslips (Bellco Glass Inc., Vineland, NJ) in 24-well plates (Nalgene Nunc, Rochester, NY) and transfected 24 h later or left untransfected. 24 h post-transfection cells were serum-starved for 2 h prior to HGF treatment in the presence of cycloheximide. Coverslips were washed once with PBS and then fixed with 3% paraformaldehyde (Fisher Scientific) in PBS for 20 min followed by washing four times in PBS. Residual paraformaldehyde was removed with three 5-min washes with 100 mm glycine in PBS. Cells were permeabilized with 0.3% Triton X-100/PBS and blocked for 30 min with blocking buffer (5% bovine serum albumin, 0.2% Triton X-100, 0.05% Tween 20, PBS). Coverslips were incubated with primary and secondary antibodies diluted in blocking buffer for 1 h and 40 min, respectively, at room temperature, and nuclei were counterstained with 4′,6-diamidino-2-phenylindole. Coverslips were mounted with immu-mount (Thermo-Shandon, Pittsburgh, PA). Confocal images were taken using a Zeiss 510 Meta laser scanning confocal microscope (Carl Zeiss, Canada Ltd., Toronto, Ontario, Canada) with 100× objective and 1.5× zoom. Image analysis was carried out using the LSM 5 image browser (Empix Imaging, Mississauga, Ontario, Canada).
Generation of Eps15 Constructs—GFP-Eps15, FLAG-Eps15 WT, delta-I, -II, -III, and -IV, and Y850F constructs were kind gifts from Dr. Edward A. Fon (Montreal, CA) and have been described previously (41). FLAG Eps15-II, -III, and -II/III constructs were made by subcloning fragments amplified by PCR using primers designed with BamHI and XbaI restriction digest sites using the FLAG-Eps15-WT pcDNA3.1zeo as a template. The FLAG-Eps15 P770A point mutation was created using the QuikChange site-directed mutagenesis kit (Stratagene), using FLAG-Eps15-WT pcDNA3.1zeo as a template. Primers were designed to include a BstUI restriction enzyme site (underlined). Forward primer, 5′-GTGAAGATGTGCCCGCGGCACTGCCGCCC-3′; reverse primer, 5′-GGGCGGCAGTGCCGCGGGCACATCTTCAC-3′. GFP-CC construct was made by subcloning a PCR fragment amplified using primers designed with HindIII/BamHI restriction digest site using the FLAG-Eps15-WT cDNA as a template into pEGFP-C2 (Clontech). GFP-Eps15ΔCC was made by subcloning a PCR fragment using primers designed with SalI/BamHI restriction digest sites using FLAG-Eps15ΔII cDNA as a template into pEGFP-C2 (Clontech). All DNA sequences were verified by sequencing.
GST Pulldown Assays—Fusion proteins were produced in DH5, or BL 21 gold Escherichia coli strain, by induction with isopropyl-1-thio-β-d-galactopyranoside. GST and GST-Grb2 constructs have been described previously (42, 43). For pulldown assays, lysates were pre-cleared with glutathione-Sepharose beads for 1 h, whereas 5–10 μg of GST fusion proteins were immobilized to glutathione-Sepharose beads at room temperature for 30 min. Beads were washed three times with lysis buffer containing inhibitors, then pre-cleared lysates were added to the beads. Samples were gently rocked at 4 °C for 1 h, washed three times in TGH with inhibitors, eluted by boiling in SDS sample buffer, and analyzed by Western blotting with the appropriate antibodies.
siRNA—HeLa cells were seeded at 3 × 105 cells/well in a 6-well dish and 24 h later transfected with 10 nm scramble control siRNA or Eps15 siRNA (target sequence: ATG CTG TAG GTT GAA CCA TTA) with HiPerFect as per the manufacturer's instructions (Qiagen). Cells were replated 24 h later at 3 × 105 cells/60-mm dish and re-transfected with siRNA (10 nm) or transfected with siRNA-resistant Eps15 cDNA. The following day cells were stimulated with HGF in the presence of cycloheximide and lysed.
RESULTS
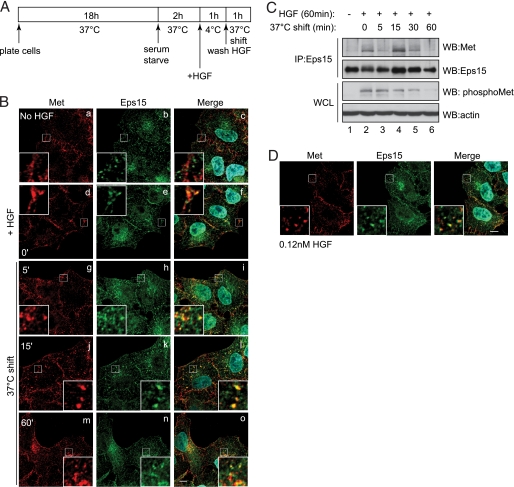
Eps15 Is Recruited to the Met Receptor upon HGF Treatment—To examine the role of Eps15 downstream from the Met RTK, we first determined if Eps15 is recruited to the Met receptor following stimulation with HGF. HeLa cells were stimulated with physiological concentrations of HGF (0.60 nm) for 1 h at 4 °C, fixed, and immunostained for endogenous Eps15 and Met proteins and visualized using confocal microscopy. Under these conditions RTKs do not internalize but bind ligand and accumulate into clathrin-coated pits at 4 °C. Receptors are then synchronously internalized upon temperature release from 4 °C to 37 °C (44–46). This experimental procedure is depicted in Fig. 1A and shall subsequently referred to as a cold load stimulation. Prior to ligand stimulation, Eps15 was localized in punctate structures resembling endosomes as well as being concentrated at the trans-Golgi network, as assessed by co-staining with mannose-6-phosphate receptor (supplemental Fig. S1A), whereas the Met RTK was predominantly localized at the plasma membrane (Fig. 1B, panel a). Following temperature switch to 37 °C post HGF treatment, Met is rapidly internalized and enters the endocytic pathway (21). Eps15 co-localizes with Met following 1 h at 4 °C (Fig. 1B, panels d–f) and as Met internalizes for up to 60 min post temperature switch (Fig. 1B, panels g–o). These structures were confirmed to be endosomes through co-staining for the endosomal marker, EEA-1 (supplemental Fig. S1B).
FIGURE 1.
Eps15 is recruited to the Met receptor upon Met activation. A, schematic diagram depicting the experimental protocol for a cold load stimulation, used to assess synchronized internalization and trafficking of RTKs. B, confocal images of endogenous Eps15 and Met upon cold load stimulation. HeLa cells were serum-starved in the presence of cycloheximide to reduce newly synthesized Met staining, then fixed and imaged (a–c) or treated with HGF (0.60 nm) for 1 h at 4 °C (d–f), followed by warming to 37 °C for the indicated times (g–o). Met (red), Eps15 (green), and 4′,6-diamidino-2-phenylindole. Magnification, 100×; zoom, 1.5×; bar = 10 μm. C, HeLa cells were subjected to a cold load stimulation, and endogenous Eps15 was immunoprecipitated (1 mg) and blotted for Met and total Eps15 levels. Whole cell lysates (WCL) were blotted (WB) for phospho-Met (p1234/1235) and actin levels. D, HeLa cells were plated on coverslips. 24 h later, cells were serum-starved in the presence of cycloheximide for 2 h, followed by cold load stimulating with 0.12 nm HGF followed by warming to 37 °C for 15 min. Cells were stained for Met (red), Eps15 (green), and 4′,6-diamidino-2-phenylindole. Bar = 10 μm.
To further validate the association between Eps15 and the Met receptor, endogenous Eps15 was immunoprecipitated from HeLa cells and immunoblotted for the Met receptor (Fig. 1C). Interestingly, a biphasic, HGF-dependent association was observed (lanes 2 and 4), similar to what has been reported for the EGFR (36). Co-immunoprecipitation was also detected in HEK293 cells transiently transfected with constructs expressing these proteins (supplemental Fig. S1C). Thus, taken together, Eps15 can be recruited to a Met receptor complex upon HGF stimulation.
Because ligand concentration has been shown to influence which internalization machinery is utilized by the EGFR (36), we also determined the role of Eps15 in Met trafficking under low concentrations of HGF (0.12 nm) in comparison to the moderate levels (0.60 nm) described above (47). After a cold load stimulation and a 15-min release to 37 °C, Met internalized into early endosomes in response to low HGF concentrations, and no significant differences for the recruitment and co-trafficking of Eps15 and Met were observed (Fig. 1D). This demonstrates that Eps15 is recruited to the Met receptor at multiple and/or various stages of internalization and trafficking irrespective of ligand concentration.
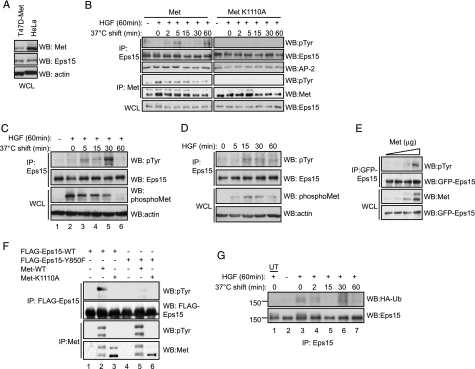
Eps15 Becomes Tyrosine-phosphorylated at Site Tyr-850 and Is Ubiquitinated in Response to Met Activation—Tyrosine phosphorylation of Eps15 has been shown downstream of the EGF receptor but not other growth factor receptors, such as the platelet-derived growth factor (30), insulin (30), or keratinocyte growth factor receptors (39). We therefore established whether HGF could induce tyrosine phosphorylation of Eps15. To test for phosphorylation of Eps15, we utilized T47D breast epithelial cells, which express undetectable levels of the Met receptor, and where we have stably expressed either a wild-type Met or kinase-dead (K1110A) Met receptor mutant (see “Experimental Procedures”) (21). These T47D cells express levels of Met similar to lower than HeLa cells, which contain moderate levels of endogenous Met protein (Fig. 2A) and have previously been used successfully to study the biology of the Met receptor (21). Cells were stimulated under cold load conditions with HGF, and the phosphorylation status of Eps15 was assessed for the indicated times (Fig. 2B). Phosphorylation of Eps15 was induced by 2- to 5-min post temperature release but was attenuated by 15 min (Fig. 2B). A second wave of Eps15 phosphorylation occurred ∼30–60 min after temperature release, consistent with our observation that Eps15 and Met are localized in the same vicinity in a perinuclear compartment at this time point (Fig. 1B). No tyrosine phosphorylation of Eps15 was observed in response to HGF stimulation in the K1110A-expressing T47D cells, showing that phosphorylation of Eps15 is strictly dependent on the kinase activity of the Met receptor. Additionally, no change in Eps15 binding to AP-2 μ-subunit was observed before and after release of HGF treatment, indicating that this complex is still functional under these conditions. To confirm these results using endogenous proteins, Eps15 tyrosine phosphorylation was examined in HeLa cells under both cold load (Fig. 2C) and HGF treatment at 37 °C (Fig. 2D). In both cases, tyrosine phosphorylation of Eps15 was detected, in agreement with our previous results, albeit with distinct but consistent kinetics depending on the condition and cell line used. Additionally, increasing amounts of WT Met receptor, when transiently transfected in HEK293 cells, led to enhanced tyrosine phosphorylation of Eps15 (Fig. 2E), further demonstrating a role for the Met RTK in Eps15 tyrosine phosphorylation.
FIGURE 2.
Eps15 is subject to post-translational modifications in response to HGF stimulation. A, HeLa and T47D stables expressing Met were blotted for protein levels. B, T47D cells stably expressing either Met WT or Met K1110A (kinase-dead) were cold load stimulated with HGF for 1 h at 4 °C then released to 37 °C for the time indicated. Endogenous Eps15 or Met was immunoprecipitated (IP) and blotted for 4G10 anti-phosphotyrosine (pTyr) and total protein levels. AP2, mu2 subunit was also blotted. C, HeLa cells were cold load-stimulated, immunoprecipitated for endogenous Eps15, and blotted for tyrosine phosphorylation. D, HeLa cells were stimulated with HGF at 37 °C for the indicated times, and lysed. Endogenous Eps15 was immunoprecipitated and phosphotyrosine levels were assessed. E, HEK293 cells were co-transfected with GFP-Eps15 and increasing amounts of Met WT cDNA expression vector. Eps15 was immunoprecipitated using anti-GFP antibodies, and phosphotyrosine levels were examined. F, HEK293 cells were transfected with either FLAG-Eps15 WT or Y850F and co-transfected with Met WT or K1110A. Immunoprecipitations were performed using anti-Met or anti-FLAG M2 beads and analyzed through Western blotting as indicated. G, HeLa cells transfected with HA-Ubiquitin (HA-Ub) were cold load-stimulated with HGF and immunoprecipitated for Eps15 then blotted for HA-Ubiquitin, and reprobed for Eps15. UT = untransfected.
Tyrosine residue 850 of Eps15 has been implicated in the internalization of the EGF receptor and has been shown to be phosphorylated downstream of the EGFR (48). To assess whether residue Tyr-850 also plays a role in the tyrosine phosphorylation in Eps15 downstream of Met, the extent of tyrosine phosphorylation of a Y850F Eps15 mutant was compared with the wild-type protein upon Met activation. Although the wild-type Eps15 protein becomes tyrosine-phosphorylated when co-expressed with the activated but not kinase-dead Met receptor (Fig. 2F, lanes 2 and 3), the Y850F mutant was not detectably phosphorylated (Fig. 2F, lanes 5 and 6). These results demonstrate that tyrosine 850 is required for tyrosine phosphorylation of Eps15 downstream of the Met receptor.
Recently, monoubiquitination of Eps15 has been reported to play a regulatory role in the ability of Eps15 to bind to ubiquitinated cargo (49). Monoubiquitination of Eps15 prevents UIM-ubiquitin interactions with other proteins (49). To establish whether Eps15 is ubiquitinated downstream of HGF and hence determine the ability of Eps15 to recognize cargo, HeLa cells were transiently transfected with HA-tagged ubiquitin and subjected to a cold load stimulation (Fig. 2G). Ubiquitination of endogenous Eps15 was detected upon HGF stimulation (lane 3), but not in the absence of HGF (lanes 2) or in non-transfected control cells (lane 1). Ubiquitination of Eps15 was detected with biphasic kinetics at 0–2 min (lanes 3 and 4) and 30 min (lane 6) post-temperature shift.
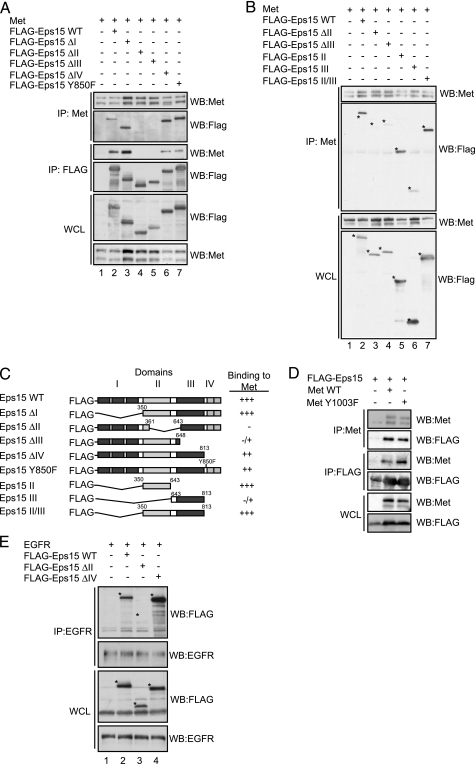
Eps15 Associates with the Met Receptor Primarily through Its Coiled-coil Domain—To determine the structural requirements involved in the Eps15-Met interaction, a panel of Eps15 deletion mutants (41, 50) was tested for their ability to co-immunoprecipitate with the Met receptor (Fig. 3, A and B). Eps15 contains four distinct domains, namely the N terminus domain (domain I), which contains three EH domains, a central coiled-coil (CC) (domain II), a DPF repeats domain (domain III), and domain IV at the C terminus, which contains two UIM domains (Fig. 3C). Recruitment of Eps15 to the membrane and its ability to be tyrosine-phosphorylated upon HGF stimulation occurs with similar kinetics to that reported for EGF (Figs. 1B and 2C) (51). Because the Met RTK is rapidly ubiquitinated in response to HGF (17, 19), we hypothesized that the Met-Eps15 interaction would also follow the proposed model for the EGFR and be dependent on the UIM domains of Eps15 (35, 38, 52, 53). Surprisingly, the ΔUIM Eps15 mutant protein (Eps15ΔIV) co-immunoprecipitated with the Met receptor to the same extent as the WT protein (Fig. 3A, lane 6). The UIM domain of Eps15 has been shown to bind to Lys-48 and Lys-63-liked ubiquitin chains in vitro, with preference for chains over monoubiquitin (54). We verified that Met was ubiquitinated under our experimental conditions (supplemental Fig. S2A). Additionally, removal of the UIMs (Eps15ΔIV mutant) disrupted ubiquitin binding as expected (supplemental Fig. S2B). Furthermore, because Eps15 recruitment to the EGFR is dependent on the integrity of the Eps15 UIM, and the ubiquitination status of the EGFR, we tested the capacity of a poorly ubiquitinated Met receptor (Y1003F) to co-immunoprecipitate with Eps15. We observed no decrease in co-immunoprecipitation with a Met Y1003F mutant, which is poorly ubiquitinated due to impaired Cbl recruitment (17) (Fig. 3D). In contrast, the Eps15ΔII (delta CC domain) mutant failed to co-immunoprecipitate with a WT Met receptor (Fig. 3A, lane 4 and Fig. 3B, lane 3) while the Eps15ΔIII mutant was impaired in its association with Met (Fig. 3A, lane 5, Fig. 3B, lane 4). Consistent with this, expression of the Eps15 CC domain alone was sufficient to interact with the Met receptor, whereas expression of domain III was consistently seen to interact weakly with the receptor (Fig. 3B, lanes 5 and 6, respectively). To test whether the coiled-coil domain also plays a role in the interaction with the EGFR we tested our Eps15-truncated mutants for co-immunoprecipitation with the EGFR. Interestingly, following transient co-transfection the EGFR-Eps15 interaction was disrupted when the coiled-coil domain was removed, whereas, removal of the C-terminal UIMs did not abrogate this association when proteins were overexpressed (Fig. 3E). Hence, unlike previous reports and implications based on the EGFR, Eps15 domain II, namely the coiled-coil domain, is necessary and sufficient for the association of Eps15 with the Met receptor, whereas Eps15 domain III can compensate to a lesser extent to its association.
FIGURE 3.
Eps15 is recruited to a Met complex through its coiled-coil domain. A, HEK293 cells were transiently transfected with Met and truncation mutants of Eps15, followed by immunoprecipitation of Met and FLAG-Eps15 48 h post-transfection and Western blotted (WB) as shown. B, individual domains of Eps15 II, III, or II and III were co-transfected with Met and immunoprecipitated as in A. Eps15 WT, Eps15ΔII and Eps15ΔIII were run out on the same gel for level comparison. C, schematic diagram of Eps15 mutants and a summary of the immunoprecipitation (IP) analysis. Binding to Met was characterized as strong (+++), good (++), weak (–/+) or poorly/not at all (–). D, FLAG-Eps15 and Met WT or Met Y1003F were transiently transfected in HEK293 cells. Lysates were immunoprecipitated for Met and FLAG then Western blotted as indicated. E, HEK293 cells were transiently co-transfected with EGFR and truncation mutants of Eps15. 24 h later cells were serum-starved overnight, then stimulated with 100 ng/ml EGF for 8 min, followed by immunoprecipitation of EGFR, and Western blotted as shown.
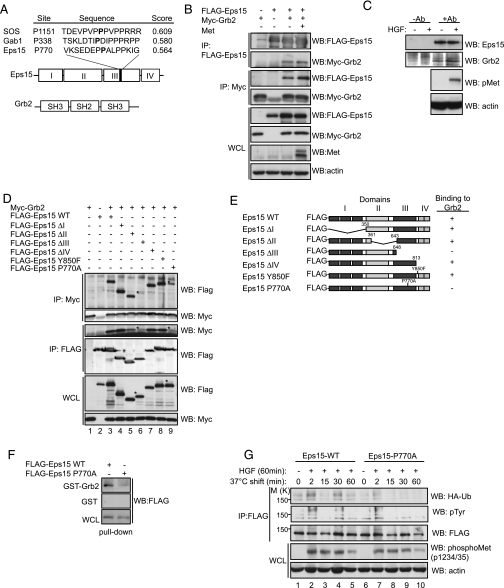
Grb2 Interacts with Eps15 through a Proline-rich Motif—To gain insight into the possible role of Eps15 domain III in complex formation with Met, we used Scansite (55) to identify potential binding protein motifs within domain III of Eps15. Scansite identified a putative Grb2 SH3 binding motif in Eps15, centered on proline residue 770 with a score of 0.564 (Fig. 4A). The known Grb2 binding partner, SOS, by comparison resulted in Scansite scores for three prolines ranging from 0.540 to 0.6345, whereas Gab1, another known Grb2 binding partner, contains a proline-rich domain with three consensus proline motifs with scores ranging from 0.525 to 0.611. Grb2 is an adaptor protein that contains one SH2 domain and two SH3 domains. Upon HGF stimulation, Grb2 is rapidly recruited to the Met receptor through its SH2 domain (56). Grb2 is required for the internalization of the EGFR (57) and recently has been shown to play a role in the clathrin-mediated endocytosis of the Met receptor (58). We therefore investigated whether Grb2 could associate with Eps15. In cells that co-express both FLAG-tagged Eps15 and Myc-tagged Grb2, these proteins co-immunoprecipitate in the presence and absence of an activated Met receptor (Fig. 4B). Parallel co-immunoprecipitations were also carried out between GFP-tagged Eps15 and Gab1 with Myc-tagged Grb2 (supplemental Fig. S3A) for a qualitative comparison between these two Grb2-associating proteins. Additionally, in HeLa cells, endogenous Grb2 was detected in Eps15 immunoprecipitates from both unstimulated and stimulated cells (Fig. 4C) demonstrating a constitutive association indicative of an SH3 domain proline-rich interaction (59). Furthermore, structure function studies demonstrate that neither an Eps15 mutant lacking the putative proline motif (Eps15ΔIII, Fig. 4D, lane 6) nor an Eps15 point mutant (P770A, Fig. 4D, lane 9) fully associates with Grb2, demonstrating the requirement for this proline residue for this interaction (Fig. 4, D and E). We confirmed this interaction using a GST-Grb2 fusion protein and pulldown assays, where we observe a decreased capacity of Grb2 to interact with the P770A mutant as compared with WT Eps15 (Fig. 4F). To assess the functional significance of the Grb2-Eps15 interaction, we tested the ability of the P770A mutant to be tyrosine-phosphorylated and ubiquitinated upon HGF stimulation. The Eps15 P770A mutant showed less ubiquitination and tyrosine phosphorylation when compared with the WT protein upon HGF stimulation (Fig. 4G, compare lanes 1–5 with lanes 6–10). This reduction was not due to reduced recruitment to the Met receptor, because the P770A mutant was able to co-immunoprecipitate with Met to a similar extent as the WT protein (supplemental Fig. S3B). Altogether, these results indicate that Grb2 can bind to Eps15 through an interaction that is dependent of proline 770. This association is maintained upon Met receptor activation and is required for full recruitment and post-translational modification of Eps15.
FIGURE 4.
Grb2 associates with Eps15 through a proline-rich motif. A, schematic of Eps15 and Grb2 domains. Proline 770 of Eps15 within the PXXP motif with surrounding amino acid identified using Scansite are indicated along with the score. The scores for one of the proline residues of SOS and Gab1 as identified by Scansite are also given for comparison. B, FLAG-Eps15 and Myc-Grb2 expression constructs were transfected with or without Met and immunoprecipitated (IP) as indicated followed by blotting for Eps15 (FLAG), Grb2 (myc), or Met levels. C, HeLa cells left unstimulated or stimulated with HGF for 15 min were lysed and immunoprecipitated using Eps15 antibodies (+Ab) and blotted for Grb2. Parallel control immunoprecipitates were carried out without antibody addition (–Ab). D, a panel of FLAG-Eps15 truncation mutants were co-transfected with myc-Grb2 in HEK293 cells and immunoprecipitated and blotted as indicated. E, summary of co-immunoprecipitations of Eps15 and Grb2, (+) = co-immunoprecipitation observed, (–) = no co-immunoprecipitation observed. F, GST pulldown assays using GST-Grb2 using lysates transfected with Eps15 WT or Eps15 P770A expression constructs. GST-alone was used as a control. G, HeLa cells transfected with Eps15 WT or P770A and HA-Ub were cold load-stimulated with 0.60 nm HGF and immunoprecipitated for Eps15 then blotted for HA and phosphotyrosine. Met activation was assessed through Western blotting using phosphospecific Met tyrosine 1234/35 antibodies.
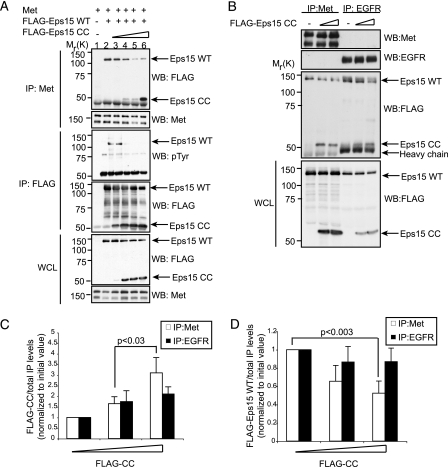
The Coiled-coil Domain Is Sufficient to Displace Eps15-Met Interactions—To further elucidate the role of the coiled-coil domain of Eps15 upon Met activation, we used increasing levels of Eps15 CC domain to determine whether this region of Eps15 could directly compete for binding to Met (Fig. 5A). Titrating increasing amounts of the Eps15 coiled-coil domain resulted in a displacement of the WT Eps15 from its association with the Met receptor and coincident loss of tyrosine phosphorylation of Eps15 (Fig. 5A, first and third panels, respectively). Binding of both FLAG-Eps15 and FLAG-CC was specific, because no binding was observed in parallel immunoprecipitations where no antibody was used (data not shown). Thus, the coiled-coil domain of Eps15 is sufficient to displace the wild-type Eps15 protein from a Met complex, and results in a loss of tyrosine phosphorylation of Eps15 following its uncoupling from the Met RTK. Because the mechanisms for tyrosine phosphorylation of Eps15 downstream of HGF are the same as downstream of EGF (Fig. 2F), we next determined the ability of the coiled-coil domain to compete in the presence of EGF stimulation. The coiled-coil domain of Eps15 was able to co-immunoprecipitate with EGFR, however, in contrast to the Met receptor, the coiled-coil domain of Eps15 associated poorly with the EGFR and was less efficient at displacing the association of the full-length Eps15 protein with the EGFR (Fig. 5, B–D). This demonstrates that, unlike the Met RTK, the coiled-coil domain of Eps15 does not play a significant role in the recruitment of Eps15 to the EGFR.
FIGURE 5.
Coiled-coil domain is sufficient for displacing a WT-Eps15-Met complex but not an EGFR-Eps15 complex. A, HEK293 cells were transfected with Met and Eps15 WT expression constructs in the presence of increasing amounts of the coiled-coil domain (Eps15 CC). Immunoprecipitations (IP) were performed on the lysates as indicated. Arrows show migration of wild-type (Eps15 WT) and Eps15 CC proteins. Molecular weight protein marker migration is indicated to the left of the blots. B, HEK293 cells transfected with either Met or EGFR and FLAG-Eps15 expression constructs with increasing amounts of FLAG-CC. EGFR cells were stimulated with 100 ng/ml EGF for 8 min prior to lysing. Lysates were immunoprecipitated with anti-Met and anti-EGFR antibodies and blotted for FLAG. Arrows show migration of wild-type (Eps15 WT) and Eps15 CC proteins and heavy chain. Molecular weight protein marker migration is indicated to the left of the blots. C, quantification of coiled-coil protein levels co-immunoprecipitated with anti-Met or anti-EGFR antibodies with increasing CC domain protein levels. Results were quantified using densitometric analysis from at least three independent experiments. For Met immunoprecipitations; p < 0.03, Student's t test, mean ± S.E. D, quantification of FLAG-Eps15 WT protein levels co-immunoprecipitated in the presence of increasing amounts of coiled-coil domain using anti-Met and anti-EGFR antibodies. Results are quantified using densitometric analysis from at least three independent experiments. Shown is the mean ± S.E. For Met co-immunoprecipitation; p < 0.003, Student's t test.
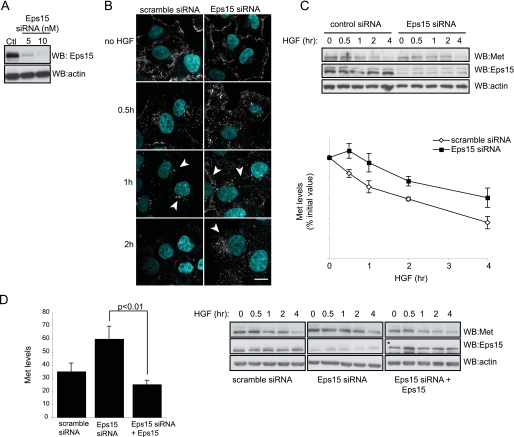
Eps15 Knockdown Delays Met Degradation—Knockdown of Eps15 has been reported to cause a modest reduction in the internalization of the EGFR (60); however, infection of the bacterium Listeria monocytogenes, which can occur via a Met-dependent mechanism, was drastically reduced upon Eps15 silencing (61). To directly assess the role of Eps15 in Met down-regulation, siRNA was used to reduce Eps15 protein expression levels in HeLa cells. A dose-dependent knockdown of Eps15 was observed with increasing concentrations of siRNA (Fig. 6A), with optimal knockdown consistently observed to be >70% at a concentration of 10 nm (Fig. 6A). Knockdown was specific, as a control scramble siRNA did not affect levels of endogenous Eps15. Cells with knockdown of Eps15 showed a delay in Met trafficking to a perinuclear compartment in comparison to scramble negative control cells (Fig. 6B). By 60 min, control cells were observed to be in a perinuclear compartment, whereas Eps15 siRNA-treated cells showed a less defined cellular localization in the cell (Fig. 6B). By 2 h of HGF treatment, Met is normally degraded and poorly detected by immunofluorescence, however in Eps15 siRNA-treated cells, Met was still detected in a perinuclear compartment (Fig. 6B). This delay in trafficking was found to correlate with enhanced stability of protein levels as observed by Western blot analysis (Fig. 6C). Densitometric analysis of Met protein levels was also used to demonstrate a delay in Met degradation (Fig. 6C). We verified the specificity of the siRNA knockdown by re-expressing siRNA-resistant full-length Eps15. Low levels of Eps15 restored Met degradation dynamics to that of scramble siRNA-treated cells, confirming that the delayed degradation is due to a loss of Eps15 protein levels and not due to off-target effects (Fig. 6D).
FIGURE 6.
Eps15 knockdown delays Met degradation and is rescued by WT Eps15. A, HeLa cells transfected with control scramble siRNA (10 nm) or Eps15 siRNA at 5 nm or 10 nm concentrations were harvested 72 h post-transfection and assessed for protein levels. B, HeLa cells transfected with scramble or Eps15 siRNA were stimulated with HGF in the presence of cycloheximide then stained for Met and 4′,6-diamidino-2-phenylindole and imaged using confocal microscopy for the indicated times. Arrows point to Met-positive staining. Bar = 20 μm. C, Western blots of HeLa cells transfected with scramble and Eps15 siRNA. Cells were stimulated with HGF in the presence of cycloheximide for the indicated amount of time. Lysates were blotted for Met, Eps15, and actin (top panel). Mean Met protein levels were quantified through densitometric analysis and graphed as percentage of initial protein levels. Error bars represent S.E. Results shown are from four independent experiments (bottom panel). D (left panel): HeLa cells transfected with Eps15 siRNA were transiently transfected with siRNA-resistant Eps15 cDNA and compared with scramble control or Eps15 siRNA-treated cells. Met protein levels were quantified as in C, after HGF stimulation in the presence of cycloheximide. Shown is the mean value of Met levels at the 4-h time point. Results are from four independent experiments. The difference between Eps15-depleted cells versus rescued cells is statistically significant; p < 0.01, Student's t test. Right panel: Western blots of siRNA-treated cells; *, GFP-Eps15 protein levels.
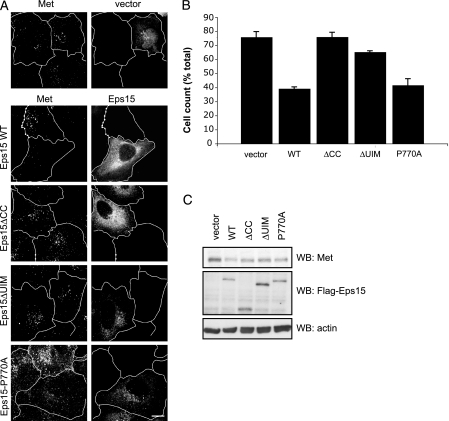
Eps15 Knockdown Is Rescued by Full-length and P770A Mutant but Not Eps15ΔCC Protein or Eps15ΔUIM Mutant—We have shown that the coiled-coil domain of Eps15 is responsible for mediating Eps15-Met interactions. To assess the importance of the coiled-coil domain-mediated Met interaction for Met degradation, we sought to re-introduce siRNA-resistant Eps15ΔCC protein and compare this with the full-length Eps15 protein for its ability to restore Met degradation. After HGF stimulation for 2 h, HeLa cells were fixed and assessed through indirect immunofluorescence for Met staining in the perinuclear compartment. Cells transfected with siRNA to Eps15 and transfected with Eps15ΔCC displayed persistent Met perinuclear localization 2 h post-HGF treatment in a similar manner to control knockdown cells transfected with vector (Fig. 7, A and B). In contrast, knockdown cells transfected with WT Eps15 exhibited less Met protein as observed through immunofluorescence (Fig. 7A) and Western blot (Fig. 7C). We next determined whether the Eps15 P770A and Eps15ΔIV (Eps15ΔUIM) proteins could also rescue the delay in Met degradation. Expression of Eps15 P770A rescued Met levels similar to Eps15 WT-expressing cells (Fig. 7). Interestingly, the Eps15ΔUIM was unable to fully rescue Met degradation as detected by immunofluorescence but was not as effective as the Eps15ΔCC in preventing Met degradation (Fig. 7B). Hence these results support that the coiled-coil domain is critical for Eps15-mediated Met trafficking and degradation and that ubiquitin-Eps15 UIM domain interactions are also critical for promoting Met trafficking and degradation.
FIGURE 7.
Eps15 knockdown is rescued by WT and P770A mutant but not rescued by Eps15ΔCC or Eps15ΔUIM mutants. A, HeLa cells treated with siRNA as described under “Experimental Procedures,” were transfected with either vector, siRNA-resistant Eps15 WT, Eps15ΔCC, Eps15 P770A, or Eps15ΔUIM. 24 h later, cells were treated with HGF in the presence of cycloheximide for 2 h, fixed, stained for Met and Eps15 (or vector), and viewed using confocal microscopy. Magnification, 100×; zoom, 1.5; bar = 10 μm. B, Eps15 knockdown cells expressing Eps15 constructs from A were counted for anti-Met staining. Results shown are from three independent experiments with at least 30 cells counted per condition. C, Western blot of knockdown cells expressing Eps15 constructs. Cells were stimulated with HGF for 2 h in the presence of cycloheximide, and protein levels were blotted as indicated.
DISCUSSION
Here we report induced recruitment and post-translational modification of Eps15 downstream from the Met receptor tyrosine kinase. Few studies have directly addressed the role of Eps15 for ligand-dependent endocytosis of RTKs other than for the EGFR. In fact, where investigated, tyrosine phosphorylation of Eps15 has not been reported for other stimuli (30, 39). Although Eps15 is commonly used as a marker for clathrin-dependent endocytosis, many details beyond its interaction with other endocytic proteins are lacking. In this study we have performed the first systematic analysis of Eps15 downstream from the Met RTK. Similar to reports carried out with the EGFR (25), Eps15 is a downstream target for the Met receptor. In this study, we show that Eps15 is recruited to the plasma membrane and undergoes rapid but biphasic tyrosine phosphorylation and ubiquitination in response to HGF (Figs. 1B, 2B, 2C, and 2G).
Using structure-function analysis, we have mapped the region of Eps15 responsible for the interaction with Met to the coiled-coil domain and a proline-rich domain. This is in contrast to published data based on the EGFR supporting a model whereby Eps15 is recruited to the EGF receptor through the UIM domain of Eps15 (62). Our data demonstrate that an Eps15 construct that lacks its UIM domains associates with the Met receptor to a similar level as the wild-type Eps15 protein (Fig. 3A) and argue that the mechanism for Eps15 recruitment to the Met receptor does not follow this model. In further support of this, Eps15 co-immunoprecipitated equally well with a mutant Met receptor (Y1003F) that is poorly ubiquitinated due to impaired Cbl recruitment (Fig. 3D). Instead, we demonstrate a novel mode of recruitment of Eps15 to the Met receptor, and we show that removal of the coiled-coil domain is sufficient to disrupt the recruitment of Eps15 to an activated Met complex (Fig. 3, A and B). Moreover an siRNA-resistant Eps15 mutant lacking the coiled-coil domain was unable to promote Met receptor degradation as observed upon expression of an siRNA-resistant WT Eps15, further confirming a role for the coiled-coil domain in the recruitment of Eps15 to Met (Fig. 7). Interestingly, removal of the coiled-coil domain also disrupts the recruitment of Eps15 to the EGFR (Fig. 3E). Whereas previous data have mapped the UIM domain of Eps15 for recruitment to the EGFR, a role for the coiled-coil domain was not addressed (36). Although the UIM domain of Eps15 is not essential for recruitment to the Met receptor, an Eps15 mutant lacking this domain failed to promote degradation of the Met receptor (Fig. 7). While this mutant was not as effective as the Eps15ΔCC mutant (Fig. 7B), it nonetheless highlights the importance of ubiquitin-UIM interaction in the overall function of Eps15 downstream of the Met receptor, which is in agreement with previous studies carried out with the EGFR (36, 37).
The coiled-coil domain of Eps15 has been shown to be involved in forming homo- and hetero-dimers and tetramers (34). The formation of these higher order molecular complexes may be required to stabilize low affinity associations of Eps15 with the Met receptor. Yet, because the coiled-coil domain alone is sufficient for binding to the Met receptor, and can compete with the wild-type protein for binding, an alternative (but not exclusive) hypothesis is that this domain is directly involved in binding to the receptor. As endogenous Eps15 protein is present, we also cannot exclude the possibility that endogenous Eps15 may dimerize with the coiled-coil domain to contribute to its recruitment to a Met complex. This interaction, however, appears to be minimal as an Eps15 mutant (ΔIII) with an intact coiled-coil domain was also impaired in its ability to associate with the Met receptor (Fig. 3A). Additionally, the coiled-coil domain of Eps15 has been reported to mediate interactions with another coiled-coil-containing protein, Hrs (29, 63). We therefore cannot rule out indirect protein interactions, although recently, for the case of Hrs, it has been shown that Hrs preferentially interacts with Eps15b rather than Eps15 in vivo (64).
Recruitment of Eps15 to Met depends on the kinase activity of the Met receptor (Fig. 2A). Here we have shown that a pool of Grb2 is constitutively bound to Eps15 though a proline-rich motif and that this interaction is maintained upon Met activation (Fig. 4, B and C). An Eps15 mutant uncoupled from Grb2 (P770A) shows decreased ubiquitination and tyrosine phosphorylation when compared with the WT Eps15 protein, in particular at the later (30 min) time point (Fig. 4G), supporting a role for Grb2 in the recruitment of Eps15 in vivo.
Interestingly, this proline motif within Eps15 has been shown to interact with the SH3 domain of the adaptor protein Crk (65). Crk and Grb2 would therefore be predicted to compete for binding to Eps15. Although, whether this competition occurs in vivo and the significance of this competition remain to be tested.
Several studies have demonstrated a requirement for Grb2 in mediating RTK internalization (57). Knockdown of both Eps15 and Grb2 has been shown to inhibit Met-dependent bacterial entry of L. monocytogenes (61). Studies depleting or, conversely, overexpressing Grb2 have also been used to show a requirement for Grb2 in the clathrin-dependent internalization of the Met receptor (58). Grb2 is rapidly recruited to the Met receptor following phosphorylation of Tyr-1356, a consensus Grb2 SH2 domain binding site in the C terminus of Met (66). Grb2 recruitment to Met is essential for the full morphogenic response elicited by HGF (56) and required for the tyrosine phosphorylation of Cbl and Gab1 signaling adaptor proteins (67). By analogy, Grb2 is likely involved as an adaptor bridging endocytic proteins such as Eps15 to the activated Met receptor, serving as an additional factor in the recruitment of Eps15 to the vicinity of the Met receptor.
Recently, a second isoform of Eps15, Eps15b, has been reported to localize to endosomes (64). This isoform lacks the EH domains and differs in the first 32 N terminus residues from Eps15. Knockdown of both Eps15 and Eps15b resulted in delayed EGFR degradation, whereas siRNA to Eps15 alone had no effect (64). Disparity between these results and ours may be due to receptor-specific differences between the Met receptor and EGFR, because the coiled-coil domain, as opposed to the UIM domain of Eps15, is required for Met receptor-Eps15 interactions (Fig. 3, A and B). Additionally, we observed that high levels of Eps15 were not as effective as low levels in rescuing our Eps15 knockdown. As a biphasic tyrosine phosphorylation and ubiquitination of Eps15 was observed (Fig. 2, B and G), it is possible that a pool of Eps15 may dimerize with Eps15b to cause this secondary wave.
In conclusion, we demonstrate a novel mode of recruitment of Eps15 to the Met RTK. As Eps15 is a key scaffold protein involved in the internalization of RTKs, this study reinforces the need to gain a better understanding of the molecular mechanisms of the interactions of Eps15 and its interacting partners downstream of growth factors.
Supplementary Material
Acknowledgments
We thank members of the Park laboratory for helpful comments on the manuscript. We thank Genetech Inc. for HGF as well as Dr. E. Fon and Dr. P. M. P. van Bergen en Henegouwen for Eps15 reagents.
This work was supported in part by a fellowship (to C. P.) from the Canadian Institutes of Health Research (CIHR) Canada Graduate Scholarship Doctoral Award and by an operating grant (to M. P.) from CIHR. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
Footnotes
The abbreviations used are: RTK, receptor tyrosine kinase; EGF, epidermal growth factor; HGF, hepatocyte growth factor; E3, ubiquitin-protein isopeptide ligase; Eps15, epidermal growth factor receptor pathway substrate 15; EGFR, EGF receptor; UIM, ubiquitin interacting motif; GST, glutathione S-transferase; PBS, phosphate-buffered saline; siRNA, small interference RNA; CC, coiled-coil; WT, wild type; HA, hemagglutinin.
References
- 1.Lee, J.-H., Han, S.-U., Cho, H., Jennings, B., Gerrard, B., Dean, M., Schmidt, L., Zbar, B., and Woude, G. F. V. (2000) Oncogene 19 4947–4953 [DOI] [PubMed] [Google Scholar]
- 2.Ma, P. C., Kijima, T., Maulik, G., Fox, E. A., Sattler, M., Griffin, J. D., Johnson, B. E., and Salgia, R. (2003) Cancer Res. 63 6272–6281 [PubMed] [Google Scholar]
- 3.Schmidt, L., Duh, F.-M., Chen, F., Kishida, T., Glenn, G., Choyke, P., Scherer, S. W., Zhuang, Z., Lubensky, I., Dean, M., Allikmets, R., Chidambaram, A., Bergerheim, U. R., Feltis, J. T., Casadevall, C., Zamarron, A., Bernues, M., Richard, S., Lips, C. J. M., Walther, M. M., Tsui, L.-C., Geil, L., Orcutt, M. L., Stackhouse, T., Lipan, J., Slife, L., Brauch, H., Decker, J., Niehans, G., Hughson, M. D., Moch, H., Storkel, S., Lerman, M. I., Linehan, W. M., and Zbar, B. (1997) Nat. Genet. 16 68–73 [DOI] [PubMed] [Google Scholar]
- 4.Marmor, M. D., and Yarden, Y. (2004) Oncogene 23 2057–2070 [DOI] [PubMed] [Google Scholar]
- 5.Mayor, S., and Pagano, R. E. (2007) Nat. Rev. Mol. Cell. Biol. 8 603–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hicke, L., and Dunn, R. (2003) Annu. Rev. Cell Dev. Biol. 19 141–172 [DOI] [PubMed] [Google Scholar]
- 7.Raiborg, C., Rusten, T. E., and Stenmark, H. (2003) Curr. Opin. Cell Biol. 15 446–455 [DOI] [PubMed] [Google Scholar]
- 8.Sorkin, A., and von Zastrow, M. (2002) Nat. Rev. Mol. Cell. Biol. 3 600–614 [DOI] [PubMed] [Google Scholar]
- 9.Hicke, L. (2001) Cell 106 527–530 [DOI] [PubMed] [Google Scholar]
- 10.Le Roy, C., and Wrana, J. L. (2005) Nat. Rev. Mol. Cell. Biol. 6 112–126 [DOI] [PubMed] [Google Scholar]
- 11.Peschard, P., and Park, M. Oncogene 26 1276–1285 [DOI] [PubMed]
- 12.Birchmeier, C., Birchmeier, W., Gherardi, E., and Vande Woude, G. F. (2003) Nat. Rev. Mol. Cell. Biol. 4 915–925 [DOI] [PubMed] [Google Scholar]
- 13.Jeffers, M., Rong, S., and Woude, G. F. (1996) J. Mol. Med. 74 505–513 [DOI] [PubMed] [Google Scholar]
- 14.Huh, C.-G., Factor, V. M., Sanchez, A., Uchida, K., Conner, E. A., and Thorgeirsson, S. S. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 4477–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borowiak, M., Garratt, A. N., Wustefeld, T., Strehle, M., Trautwein, C., and Birchmeier, C. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 10608–10613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammond, D. E., Urbe, S., Vande Woude, G. F., and Clague, M. J. (2001) Oncogene 20 2761–2770 [DOI] [PubMed] [Google Scholar]
- 17.Peschard, P., Fournier, T. M., Lamorte, L., Naujokas, M. A., Band, H., Langdon, W. Y., and Park, M. (2001) Mol. Cell 8 995–1004 [DOI] [PubMed] [Google Scholar]
- 18.Peschard, P., Ishiyama, N., Lin, T., Lipkowitz, S., and Park, M. (2004) J. Biol. Chem. 279 29565–29571 [DOI] [PubMed] [Google Scholar]
- 19.Jeffers, M., Taylor, G. A., Weidner, K. M., Omura, S., and Vande Woude, G. F. (1997) Mol. Cell. Biol. 17 799–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peschard, P., and Park, M. (2003) Cancer Cell 3 519–523 [DOI] [PubMed] [Google Scholar]
- 21.Abella, J. V., Peschard, P., Naujokas, M. A., Lin, T., Saucier, C., Urbe, S., and Park, M. (2005) Mol. Cell. Biol. 25 9632–9645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong-Beltran, M., Seshagiri, S., Zha, J., Zhu, W., Bhawe, K., Mendoza, N., Holcomb, T., Pujara, K., Stinson, J., Fu, L., Severin, C., Rangell, L., Schwall, R., Amler, L., Wickramasinghe, D., and Yauch, R. (2006) Cancer Res. 66 283–289 [DOI] [PubMed] [Google Scholar]
- 23.Hammond, D. E., Carter, S., McCullough, J., Urbe, S., Vande Woude, G., and Clague, M. J. (2003) Mol. Biol. Cell 14 1346–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, N., Xiang, G.-S., Dokainish, H., Ireton, K., and Elferink, L. A. (2005) Traffic 6 459–473 [DOI] [PubMed] [Google Scholar]
- 25.Fazioli, F., Minichiello, L., Matoskova, B., Wong, W. T., and Fiore, P. P. D. (1993) Mol. Biol. Cell 13 5814–5828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benmerah, A., Gagnon, J., Begue, B., Megarbane, B., Dautry-Varsat, A., and Cerf-Bensussan, N. (1995) J. Cell Biol. 131 1831–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benmerah, A., Bègue, B., Dautry-Varsat, A., and Cerf-Bensussan, N. (1996) J. Biol. Chem. 271 12111–12116 [DOI] [PubMed] [Google Scholar]
- 28.Chen, H., Fre, S., Slepnev, V. I., Capua, M. R., Takei, K., Butler, M. H., Di Fiore, P. P., and De Camilli, P. (1998) Nature 394 793–797 [DOI] [PubMed] [Google Scholar]
- 29.Bean, A. J., Davanger, S., Chou, M. F., Gerhardt, B., Tsujimoto, S., and Chang, Y. (2000) J. Biol. Chem. 275 15271–15278 [DOI] [PubMed] [Google Scholar]
- 30.Delft, S. v., Schumacher, C., Hage, W., Verkleij, A. J., and van Bergen en Henegouwen, P. M. (1997) J. Cell Biol. 136 811–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tebar, F., Sorkina, T., Sorkin, A., Ericsson, M., and Kirchhausen, T. (1996) J. Biol. Chem. 271 28727–28730 [DOI] [PubMed] [Google Scholar]
- 32.Naslavsky, N., Rahajeng, J., Chenavas, S., Sorgen, P. L., and Caplan, S. (2007) J. Biol. Chem. 282 16612–16622 [DOI] [PubMed] [Google Scholar]
- 33.Benmerah, A., Bayrou, M., Cerf-Bensussan, N., and Dautry-Varsat, A. (1999) J. Cell Sci. 112 1303–1311 [DOI] [PubMed] [Google Scholar]
- 34.Cupers, P., ter Haar, E., Boll, W., and Kirchhausen, T. (1997) J. Biol. Chem. 272 33430–33434 [DOI] [PubMed] [Google Scholar]
- 35.Polo, S., Sigismund, S., Faretta, M., Guidi, M., Capua, M. R., Bossi, G., Chen, H., De Camilli, P., and Di Fiore, P. P. (2002) Nature 416 451–455 [DOI] [PubMed] [Google Scholar]
- 36.Sigismund, S., Woelk, T., Puri, C., Maspero, E., Tacchetti, C., Transidico, P., Di Fiore, P. P., and Polo, S. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 2760–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Melker, A. A., van der Horst, G., and Borst, J. (2004) J. Cell Sci. 117 5001–5012 [DOI] [PubMed] [Google Scholar]
- 38.Riezman, H. (2002) Nature 416 381–383 [DOI] [PubMed] [Google Scholar]
- 39.Belleudi, F., Visco, V., Ceridono, M., Leone, L., Muraro, R., Frati, L., and Torrisi, M. R. (2003) FEBS Lett. 553 262–270 [DOI] [PubMed] [Google Scholar]
- 40.Rodrigues, G. A., Naujokas, M. A., and Park, M. (1991) Mol. Cell. Biol. 11 2962–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klapisz, E., Sorokina, I., Lemeer, S., Pijnenburg, M., Verkleij, A. J., and van Bergen en Henegouwen, P. M. (2002) J. Biol. Chem. 277 30746–30753 [DOI] [PubMed] [Google Scholar]
- 42.Lock, L. S., Maroun, C. R., Naujokas, M. A., and Park, M. (2002) Mol. Biol. Cell 13 2132–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peschard, P., Kozlov, G., Lin, T., Mirza, I. A., Berghuis, A. M., Lipkowitz, S., Park, M., and Gehring, K. (2007) Mol. Cell 27 474–485 [DOI] [PubMed] [Google Scholar]
- 44.Gorden, P., Carpentier, J. L., Cohen, S., and Orci, L. (1978) Proc. Natl. Acad. Sci. U. S. A. 75 5025–5029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sorkina, T., Huang, F., Beguinot, L., and Sorkin, A. (2002) J. Biol. Chem. 277 27433–27441 [DOI] [PubMed] [Google Scholar]
- 46.Hillman, G. M., and Schlessinger, J. (1982) Biochemistry 21 1667–1672 [DOI] [PubMed] [Google Scholar]
- 47.Funakoshi, H., and Nakamura, T. (2003) Clin. Chim. Acta 327 1–23 [DOI] [PubMed] [Google Scholar]
- 48.Confalonieri, S., Salcini, A. E., Puri, C., Tacchetti, C., and Di Fiore, P. P. (2000) J. Cell Biol. 150 905–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoeller, D., Crosetto, N., Blagoev, B., Raiborg, C., Tikkanen, R., Wagner, S., Kowanetz, K., Breitling, R., Mann, M., Stenmark, H., and Dikic, I. (2006) Nat. Cell Biol. 8 163–169 [DOI] [PubMed] [Google Scholar]
- 50.Fallon, L., Belanger, C. M. L., Corera, A. T., Kontogiannea, M., Regan-Klapisz, E., Moreau, F., Voortman, J., Haber, M., Rouleau, G., Thorarinsdottir, T., Brice, A., van Bergen en Henegouwen, P. M., and Fon, E. A. (2006) Nat Cell Biol. 8 834–842 [DOI] [PubMed] [Google Scholar]
- 51.Torrisi, M. R., Lotti, L. V., Belleudi, F., Gradini, R., Salcini, A. E., Confalonieri, S., Pelicci, P. G., and Di Fiore, P. P. (1999) Mol. Biol. Cell 10 417–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Melker, A. A., van der Horst, G., and Borst, J. (2004) J. Biol. Chem. 279 55465–55473 [DOI] [PubMed] [Google Scholar]
- 53.Stang, E., Blystad, F. D., Kazazic, M., Bertelsen, V., Brodahl, T., Raiborg, C., Stenmark, H., and Madshus, I. H. (2004) Mol. Biol. Cell 15 3591–3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barriere, H., Nemes, C., Lechardeur, D., Khan-Mohammad, M., Fruh, K., and Lukacs, G. L. (2006) Traffic 7 282–297 [DOI] [PubMed] [Google Scholar]
- 55.Obenauer, J. C., Cantley, L. C., and Yaffe, M. B. (2003) Nucleic Acids Res. 31 3635–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fournier, T. M., Kamikura, D., Teng, K., and Park, M. (1996) J. Biol. Chem. 271 22211–22217 [DOI] [PubMed] [Google Scholar]
- 57.Jiang, X., Huang, F., Marusyk, A., and Sorkin, A. (2003) Mol. Biol. Cell 14 858–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li, N., Lorinczi, M., Ireton, K., and Elferink, L. A. (2007) J. Biol. Chem. 282 16764–16775 [DOI] [PubMed] [Google Scholar]
- 59.Pawson, T., and Scott, J. D. (1997) Science 278 2075–2080 [DOI] [PubMed] [Google Scholar]
- 60.Huang, F., Khvorova, A., Marshall, W., and Sorkin, A. (2004) J. Biol. Chem. 279 16657–16661 [DOI] [PubMed] [Google Scholar]
- 61.Veiga, E., and Cossart, P. (2005) Nat. Cell Biol. 7 894–900 [DOI] [PubMed] [Google Scholar]
- 62.Aguilar, R. C., and Wendland, B. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 2679–2680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bache, K. G., Raiborg, C., Mehlum, A., and Stenmark, H. (2003) J. Biol. Chem. 278 12513–12521 [DOI] [PubMed] [Google Scholar]
- 64.Roxrud, I., Raiborg, C., Pedersen, N. M., Stang, E., and Stenmark, H. (2008) J. Cell Biol. 180 1205–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schumacher, C., Knudsen, B. S., Ohuchi, T., Fiore, P. P. D., Glassman, R. H., and Hanafusa, H. (1995) J. Biol. Chem. 270 15341–15347 [DOI] [PubMed] [Google Scholar]
- 66.Ponzetto, C., Bardello, A., Zhen, Z., Maina, F., Zonca, P. d., Giordano, S., Graziani, A., Panayotou, G., and Comoglio, P. M. (1994) Cell 77 261–271 [DOI] [PubMed] [Google Scholar]
- 67.Fixman, E. D., Holgado-Madruga, M., Nguyen, L., Kamikura, D. M., Fournier, T. M., Wong, A. J., and Park, M. (1997) J. Biol. Chem. 272 20167–20172 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.