Abstract
FSH acts through the Sertoli cell to ensure normal testicular development and function. To identify transcriptional mechanisms through which FSH acts in the testis, we have treated gonadotrophin-deficient hypogonadal (hpg) mice with recombinant FSH and measured changes in testicular transcript levels using microarrays and real-time PCR 12, 24 and 72 h after the start of treatment. Approximately 400 transcripts were significantly altered at each time point by FSH treatment. At 12 h, there was a clear increase in the levels of a number of known Sertoli cell transcripts (e.g. Fabp5, Lgals1, Tesc, Scara5, Aqp5). Additionally, levels of Leydig cell transcripts were also markedly increased (e.g. Ren1, Cyp17a1, Akr1b7, Star, Nr4a1). This was associated with a small but significant rise in testosterone at 24 and 72 h. At 24 h, androgen-dependent Sertoli cell transcripts were up-regulated (e.g. Rhox5, Drd4, Spinlw1, Tubb3 and Tsx) and this trend continued up to 72 h. By contrast with the somatic cells, only five germ cell transcripts (Dkkl1, Hdc, Pou5f1, Zfp541 and 1700021K02Rik) were altered by FSH within the time-course of the experiment. Analysis of canonical pathways showed that FSH induced a general decline in transcripts related to formation and regulation of tight junctions. Results show that FSH acts directly and indirectly to induce rapid changes in Sertoli cell and Leydig cell transcript levels in the hpg mouse but that effects on germ cell development must occur over a longer time-span.
Introduction
Postnatal testicular growth, spermatogenesis and fertility are dependent upon the pituitary gonadotrophins FSH and LH. LH acts directly on Leydig cells to stimulate androgen production, while androgens and FSH stimulate spermatogenesis through direct action on the Sertoli cells (McLachlan et al. 2002). The role of gonadotrophins is clearly seen in the hypogonadal (hpg) mouse that lacks GnRH (Mason et al. 1986) and, consequently, has undetectable circulating levels of LH and FSH (Cattanach et al. 1977). The gonads of the hpg mouse remain in a pre-pubertal state throughout life, with spermatogenesis blocked at early meiosis (Cattanach et al. 1977, Myers et al. 2005) although treatment with exogenous gonadotrophins or androgens will increase testicular growth and restore germ cell development (Charlton et al. 1983, Singh & Handelsman 1996a,b, Haywood et al. 2003). In recent years, generation of mice lacking individual hormones or hormone receptors has allowed us to investigate more clearly the roles played by LH, FSH and androgen in the regulation of testicular function. In particular, study of mice lacking androgen receptors (AR) in the Sertoli cells (SCARKO (De Gendt et al. 2004)) has shown that androgens are essential for spermatocyte progression through meiosis. By contrast, mice lacking FSH (FSHβKO (Kumar et al. 1997)) or the FSH receptor (FSHRKO; Dierich et al. 1998, Abel et al. 2000) are fertile with all stages of spermatogenesis present. Nevertheless, in FSHRKO and FSHβKO mice there is a reduction in sperm number and quality (Krishnamurthy et al. 2001, Wreford et al. 2001) suggesting that FSH action optimises spermatogenesis. In addition, comparison of SCARKO mice with mice lacking both FSHR and AR on the Sertoli cells has shown that FSH acts to increase Sertoli cell number, total germ cell number and the number of germ cells associated with each Sertoli cell (Abel et al. 2008). This is achieved by an increase in the number of spermatogonia and enhanced entry of these cells into meiosis (Abel et al. 2008).
Previous studies have identified a number of Sertoli cell products or mRNA transcripts that are FSH-sensitive including, for example, inhibin, AR, transferrin, doublesex and mab-3 related transcription factor1 (DMRT), androgen-binding protein and inducible cAMP early repressor (Morris et al. 1988, Verhoeven & Cailleau 1988, Skinner et al. 1989, Monaco et al. 1995, Chen & Heckert 2001). In addition, an earlier study has used arrays to examine the short-term effects (up to 24 h) of a single injection of FSH on testicular gene expression in vivo (Sadate-Ngatchou et al. 2004). From these studies, we can now identify a number of transcripts acutely regulated by FSH but we continue to lack a clear understanding of how FSH acts to regulate testicular development and function over the longer term. To address this issue, we have carried out a comprehensive review of the effects of more prolonged FSH treatment (multiple injections up to 72 h) on transcript levels in the testis of the hpg mouse.
Materials and methods
Animals and treatments
hpg mice from the original colony first identified at the MRC Laboratories, Harwell, Oxford (Cattanach et al. 1977) were bred at Oxford. The hpg mutation was identified by PCR analysis of tail DNA as previously reported (Lang 1995). All procedures were carried out in accordance with the UK Animals (Scientific Procedures) Act 1986 and with the approval of a local ethical review committee.
Male hpg mice, 10 weeks of age and in group sizes of 3–4, were injected subcutaneously with 8 IU recombinant human FSH (rhFSH) (Serono Ltd) in 0·2 ml PBS (PBS, pH 7·4, Sigma Aldrich) at the start of the experiment and every 12 h thereafter for 12, 24 or 72 h. This dose of recombinant hormone had previously been shown to induce a significant increase in testis weight in hpg mice when given for 1 week (Abel and Charlton unpublished). Mice were killed 1 h after the last injection, testes removed, snap frozen in liquid nitrogen and stored at −70 °C.
Testicular histology
Three hpg mice treated as above were killed at each time point. The testes were weighed and one testis from each animal was fixed in 1% glutaraldehyde, 4% paraformaldehyde, in phosphate buffer, 0·1 M, pH 7·2 for 24 h at 4 °C, and embedded in araldite. Semi-thin, 1 μm sections were cut and stained with toluidine blue.
DNA microarray
Three or four animals from FSH-treated or control hpg groups were killed at each time point and the RNA from testes of individual animals extracted on RNeasy columns (Qiagen). RNA was quantified using a NanoDrop ND-1000 (NanoDrop, Wilmington, DE, USA) and RNA quality was checked using the Agilent bioanalyzer 2100 (Agilent, Santa Clara, CA, USA). Samples of total RNA (8 μg) from individual animals were reverse transcribed and then in vitro transcribed and hybridised to mouse MOE430A arrays (Affymetrix, Santa Clara, CA, USA) (n=3 or 4 for each group) according to the GeneChip expression technical manual (Affymetrix) as previously reported (Baban & Davies 2008). All the experiments were designed and information compiled in compliance with MIAME guide lines. Gene transcript levels were determined from data image files using algorithms in Gene Chip Operating Software (GCOS1.2, Affymetrix).
The array data were generated in two batches. In the first experiment control, 12 and 72 h FSH groups were extracted and hybridised to the arrays and in a subsequent experiment control and 24 h FSH groups were processed in the same way. Each treatment group was analysed against its own control. Differentially expressed genes were identified using the Welch t-test, variance not assumed equal, P<0·05. Analysis of canonical pathways was carried out using Ingenuity Pathways Analysis (www.ingenuity.com).
The data discussed in this publication have been deposited in NCBIs Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession GSE8924.
Real-time PCR
Total RNA was extracted from individual testes of control or FSH-treated hpg mice using Trizol (Life technologies) and residual genomic DNA was removed by DNAse treatment (DNA-free, Ambion Inc., Austin, TX, USA, supplied by AMS Biotechnology, Abingdon, UK). RNA (1 μg) was reverse transcribed using random hexamers (Ambion) and Moloney murine leukaemia virus reverse transcriptase (Life Technologies) as previously described (Hirst et al. 2004).
Quantitative real-time PCR was used to confirm changes in selected mRNA transcripts identified from the microarray analysis or to examine other transcripts of potential interest. The real-time PCR used either the Taqman (Inha, Inhba, Inhbb and Hdc) or the SYBR green (all other transcripts) method in a 96-well plate format. For Taqman, Universal Taqman master mix, and optimised primer and probe sets were purchased from Applied Biosystems (Warrington, UK) and used according to the manufacturer's recommendations in a 25 μl volume. For SYBR green, each reaction contained 5 μl 2×SYBR mastermix (Stratagene, Amsterdam, Netherlands), primer (100 nM) and template in a total volume of 10 μl. The thermal profile used for amplification was 95 °C for 8 min followed by 40 cycles of 95 °C for 25 s, 63 °C for 25 s and 72 °C for 30 s. At the end of the amplification phase a melting curve analysis was carried out on the products formed. All primers were designed by Primer Express 2.0 (Applied Biosystems) using parameters previously described (Czechowski et al. 2004). No-RT controls for each sample were screened to check for the presence of residual genomic DNA. The primers and probes used for real-time SYBR PCR are shown in Supplementary Table 1, see supplementary data in the online version of the Journal of Molecular Endocrinology at http://jme.endocrinology-journals.org/content/vol42/issue4/. Different animals were used to provide RNA for real-time PCR and microarray studies.
Hormone assay
In a separate study adult hpg mice were treated with rhFSH as above and intratesticular levels of testosterone measured by RIA following ethanol extraction as previously described (O'Shaughnessy & Sheffield 1990). The limit of detection of the assay was 25 fmol/testis.
Statistical analysis
With the exception of the array studies described above, the effects of FSH treatment were analysed initially by single-factor ANOVA followed by post hoc analysis using Fisher's test.
Results
Testicular weight and histology after rhFSH treatment
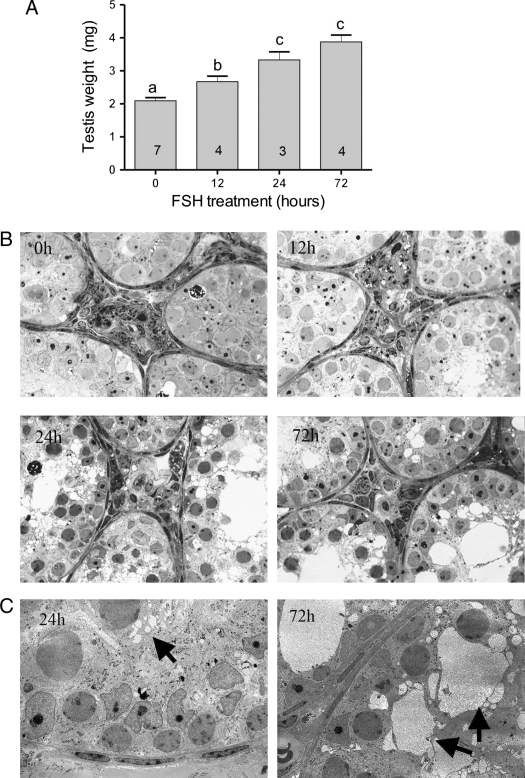
There was a significant increase in testis weight within 12 h of the start of FSH treatment and weight continued to increase up to 24 h (Fig. 1A). This weight increase was accompanied by an apparent increase in tubular diameter with clear formation of a tubular lumen (Fig. 1B). On the semi-thin light micrographs, there was also an apparent increase in vacuolation of the Sertoli cell cytoplasm by 24 h which became more marked by 72 h (Fig. 1B). This was confirmed on electron micrographs with several small vacuoles apparent within the cytoplasm at 24 h and larger vacuoles present at 72 h (Fig. 1C). There was no clear advancement of spermatogenesis within the timescale of the experiment.
Figure 1.
Effect of rhFSH on testis weight and morphology in hpg mice. A) Testis weights of control adult hpg mice and mice treated every 12 h with FSH (n for each group is shown in the histogram). B) Semi-thin sections of testes from control adult hpg mice and mice treated with FSH for 12, 24 and 72 h. Note the appearance of vacuoles within the cytoplasm of the Sertoli cell at 24 and 72 h post-treatment. C) Electron micrographs at 24 and 72 h, arrows indicate the progression from multiple small vacuoles to fewer large vacuoles.
Hormone levels
Intratesticular testosterone levels were undetectable in control hpg mice (<25 fmol/testis (<12 fmol/mg tissue), n=8) and increased to low but consistently detectable levels 24 h after the start of treatment with FSH (65·0±12·4 fmol/testis (19·1±3·6 fmol/mg), n=4) and remained detectable up to 72 h (76·2±45·0 fmol/testis (19·5±11·5 fmol/mg), n=4).
Microarray data
Analysis of the array data showed that there were 182, 164 and 203 transcripts significantly (>2-fold) increased in the hpg testis 12, 24 and 72 h after the start of FSH treatment and 162, 411 and 215 significantly decreased at the same times. Transcripts with the highest fold changes in expression at each time during treatment are listed in Table 1 and the complete list of significantly altered transcripts (>2-fold) is shown in Supplementary Table 2, see supplementary data in the online version of the Journal of Molecular Endocrinology at http://jme.endocrinology-journals.org/content/vol42/issue4/. At 12 h after the start of FSH treatment, there was a clear increase in the levels of a number of transcripts known to be expressed in the Sertoli cells (e.g. Fabp5, Lgals1, Tesc and Scara5; Kingma et al. 1998, Perera et al. 2001, Dettin et al. 2003, Jiang et al. 2006) and, perhaps surprisingly, in the Leydig cells (e.g. Ren1, Cyp17a1, Akr1b7, Star, Ldlr and Nr4a1; Deschepper et al. 1986, Le Goascogne et al. 1991, Song et al. 2001, Baron et al. 2003; Table 1). By 24 h after the start of FSH treatment, androgen-dependent Sertoli cell transcripts appeared in the list of up-regulated transcripts (e.g. Rhox5, Drd4, Spinlw1 and Tubb3; Lindsey & Wilkinson 1996, Cunningham et al. 1998, Denolet et al. 2006, O'Shaughnessy et al. 2007) and this trend became more marked by 72 h. By contrast with the somatic cells, very few germ cell genes appear on the lists of significantly regulated transcripts. Only Hdc (increased 4·0- and 5·9-fold at 24 and 72 h respectively; Safina et al. 2002) and 1700021K02Rik (Spatial) (increased 3·1-fold at 72 h) (Irla et al. 2003) were significantly altered by FSH within the time-span of these studies (Table 1 and Supplementary Table 2).
Table 1.
Effects of FSH treatment on testicular transcript levels – highest-regulated transcripts from microarray studiesa
| Gene symbol | Gene title | Fold change | Gene symbol | Gene title | |
|---|---|---|---|---|---|
| Fold change | |||||
| Transcripts up-regulated 12 h after start of treatment | Transcripts down-regulated 12 h after start of treatment | ||||
| 20·7 | Ren1 | Renin 1 structural | 7·27 | Myh8 | Myosin, heavy polypeptide 8 |
| 18·2 | Dmkn | Dermokine | 6·65 | Rin2 | Ras and Rab interactor 2 |
| 13·3 | Aqp5 | Aquaporin 5 | 5·03 | Pdgfc | Platelet-derived growth factor C |
| 12·1 | Wif1 | Wnt inhibitory factor 1 | 4·55 | Transcribed locusb | |
| 11·0 | Fabp5 | Fatty acid binding protein 5, epidermal | 4·53 | Bcan | Brevican |
| 6·6 | Tubb3 | Tubulin, β 3 | 4·46 | Rgs11 | Regulator of G-protein signalling 11 |
| 6·5 | Cyp17a1 | Cytochrome P450, family 17a 1 | 4·45 | Tmem37 | Transmembrane protein 37 |
| 6·3 | Akr1b7 | Aldo-keto reductase family 1 B7 | 4·43 | Fhod3 | Formin homology 2 domain containing 3 |
| 6·0 | Lgals1 | Lectin, galactose binding, soluble 1 | 4·38 | Derl3 | Der1-like domain family, member 3 |
| 5·9 | Col4a1 | Procollagen, type IV, α1 | 4·01 | Ddit4l | DNA-damage-inducible transcript 4-like |
| 5·7 | Pappa | Pregnancy-associated plasma protein A | 3·98 | BC013672 | cDNA sequence BC013672 |
| 5·2 | Star | Steroidogenic acute regulatory protein | 3·97 | Scin | Scinderin |
| 4·6 | Ldlr | Low density lipoprotein receptor | 3·96 | Ddit4l | DNA-damage-inducible transcript 4-like |
| 4·6 | Tesc | Tescalcin | 3·89 | Cabc1 | Chaperone, ABC1 complex-like |
| 4·6 | Rps6ka2 | Ribosomal protein S6 kinase 2 | 3·86 | Krt20 | Keratin 20 |
| 4·6 | Scara5 | Scavenger receptor class A 5 | 3·74 | Tmem140 | Transmembrane protein 140 |
| 4·4 | Hgsnat | Heparan N-acetyltransferase | 3·71 | Dbp | D site albumin promoter binding protein |
| 4·4 | Hs3st1 | Heparan sulphate 3-O-sulphotransferase 1 | 3·71 | Rnasel | Ribonuclease L |
| 4·2 | Syne1 | Synaptic nuclear envelope 1 | 3·70 | Spsb1 | splA receptor domain and SOCS box 1 |
| 4·1 | Slc38a5 | Solute carrier family 38, member 5 | 3·67 | Tnni3 | Troponin I, cardiac |
| 4·0 | Gpd1 | Glycerol-3-phosphate dehydrogenase 1 | 3·65 | Cdo1 | Cysteine dioxygenase 1, cytosolic |
| 3·9 | Dos | Downstream of Stk11 | 3·57 | Stard8 | START domain containing 8 |
| 3·8 | Svs5 | Seminal vesicle secretory protein 5 | 3·55 | Slc40a1 | Solute carrier family 40, member 1 |
| 3·8 | Bhmt | Betaine-homocysteine methyltransferase | 3·52 | Hdac5 | Histone deacetylase 5 |
| 3·7 | D9Ertd280e | Chr 9, ERATO Doi 280 | 3·32 | Dbp | D site albumin promoter binding protein |
| 3·7 | Tnfrsf12a | Tumour necrosis factor receptor 12a | 3·23 | Chdh | Choline dehydrogenase |
| 3·7 | 1200016E24Rik | RIKEN cDNA 1200016E24 | 3·23 | 8030411F24Rik | RIKEN cDNA 8030411F24 gene |
| 3·7 | Nr4a1 | Nuclear receptor subfamily 4, group A1 | 3·23 | Per3 | Period homolog 3 (Drosophila) |
| 3·6 | Dkk3 | Dickkopf homolog 3 | 3·22 | Ctnna2 | Catenin, α 2 |
| 3·6 | Cyp51 | Cytochrome P450, family 51 | 3·19 | Trim47 | Tripartite motif protein 47 |
| Transcripts up-regulated 24 h after start of treatment | Transcripts down-regulated 24 h after start of treatment | ||||
| 28·8 | Lin7c | Lin-7 homolog C (C. elegans) | 9·9 | Ddit4l | DNA-damage-inducible transcript 4-like |
| 18·0 | Cyp17a1 | Cytochrome P450, family 17a1 | 9·9 | Rin2 | Ras and Rab interactor 2 |
| 13·6 | Cyp11a1 | Cytochrome P450, family 11a1 | 9·1 | Slc40a1 | Solute carrier family 40, member 1 |
| 11·0 | Fabp5 | Fatty acid binding protein 5, epidermal | 8·0 | Rgs11 | Regulator of G-protein signalling 11 |
| 10·1 | Rhox5 | Reproductive homeobox 5 | 7·6 | Igfbp3 | Insulin-like growth factor binding protein 3 |
| 9·4 | Star | Steroidogenic acute regulatory protein | 7·4 | Myh6 | Myosin, heavy polypeptide 6 α |
| 8·7 | Slc38a5 | Solute carrier family 38, member 5 | 7·2 | Apbb2 | Amyloid precursor protein-binding B2 |
| 7·9 | Aqp5 | Aquaporin 5 | 6·7 | Rassf5 | Ras association domain family 5 |
| 7·4 | Tubb3 | Tubulin, β 3 | 6·3 | Tmem37 | Transmembrane protein 37 |
| 7·0 | Drd4 | Dopamine receptor 4 | 6·3 | Chdh | Choline dehydrogenase |
| 6·3 | Tesc | Tescalcin | 6·2 | Itga9 | Integrin α 9 |
| 5·5 | Lgals1 | Lectin, galactose binding, soluble 1 | 6·2 | Thbd | Thrombomodulin |
| 5·0 | Spinlw1 | Eppin | 5·9 | Fcgr2b | Fc receptor, IgG, low affinity IIb |
| 4·6 | Osr1 | Odd-skipped related 1 | 5·8 | Trim47 | Tripartite motif protein 47 |
| 4·5 | Fads2 | Fatty acid desaturase 2 | 5·7 | Spsb1 | splA receptor domain and SOCS box 1 |
| 4·2 | Pappa | Pregnancy-associated plasma protein A | 5·6 | Ddit4l | DNA-damage-inducible transcript 4-like |
| 4·2 | Scara5 | Scavenger receptor class A5 | 5·5 | Vnn1 | Vanin 1 |
| 4·2 | Pscdbp | Pleckstrin homology binding protein | 5·5 | Fcgr2b | Fc receptor, IgG, low affinity IIb |
| 4·1 | Plac8 | Placenta-specific 8 | 5·4 | Ptprd | Protein tyrosine phosphatase, receptor D |
| 4·0 | Gpt2 | Glutamic pyruvate transaminase 2 | 5·4 | Nkx3-1 | NK-3 transcription factor, locus 1 |
| 4·0 | Rps6ka2 | Ribosomal protein S6 kinase 2 | 5·3 | Fcgr2b | Fc receptor, IgG, low affinity IIb |
| 4·0 | Gpd1 | Glycerol-3-phosphate dehydrogenase 1 | 5·1 | Ctgf | Connective tissue growth factor |
| 4·0 | Hdc | Histidine decarboxylase | 5·0 | H19 | H19 fetal liver mRNA |
| 3·8 | Igf1 | Insulin-like growth factor 1 | 5·0 | Pdgfc | Platelet-derived growth factor C |
| 3·7 | Fah | Fumarylacetoacetate hydrolase | 4·9 | Hsd17b11 | Hydroxysteroid (17β) dehydrogenase 11 |
| 3·7 | Mpzl2 | Myelin protein zero-like 2 | 4·9 | Cabc1 | Chaperone, ABC1 complex-like |
| 3·6 | Insl3 | Insulin-like 3 | 4·9 | 9630031F12Rik | RIKEN cDNA 9630031F12 gene |
| 3·4 | Wif1 | Wnt inhibitory factor 1 | 4·8 | Ptch1 | Patched homolog 1 |
| 3·3 | Inha | Inhibin α | 4·7 | Smoc2 | SPARC related modular calcium binding 2 |
| 3·2 | Col18a1 | Procollagen, type XVIII, α 1 | 4·7 | Fhod3 | Formin homology 2 domain containing 3 |
| Transcripts up-regulated 72 h after start of treatment | Transcripts down-regulated 72 h after start of treatment | ||||
| 35·0 | Drd4 | Dopamine receptor 4 | 13·4 | Igfbp3 | Insulin-like growth factor binding protein 3 |
| 21·1 | Slc38a5 | Solute carrier family 38a5 | 8·2 | Rgs11 | Regulator of G-protein signalling 11 |
| 14·9 | Rhox5 | Reproductive homeobox 5 | 6·6 | Rin2 | Ras and Rab interactor 2 |
| 13·9 | Fabp5 | Fatty acid binding protein 5, epidermal | 6·5 | Clca1 | Chloride channel calcium activated 1 |
| 8·4 | Spinlw1 | Eppin | 6·0 | Slc40a1 | Solute carrier family 40 1 |
| 7·6 | Klk1b24 | Kallikrein 1-related peptidase b24 | 5·6 | Spsb1 | splA receptor domain and SOCS box 1 |
| 6·6 | Tubb3 | Tubulin, β 3 | 4·9 | Ifitm1 | Interferon induced transmembrane 1 |
| 5·9 | Hdc | Histidine decarboxylase | 4·7 | Myh8 | Myosin, heavy polypeptide 8 |
| 5·5 | Fah | Fumarylacetoacetate hydrolase | 4·6 | Fhod3 | Formin homology 2 domain containing 3 |
| 5·5 | Tsx | Testis specific X-linked gene | 4·4 | Bcan | Brevican |
| 5·4 | Zcchc18 | Zinc finger, CCHC domain 18 | 4·4 | Tmem140 | Transmembrane protein 140 |
| 4·6 | Sct | Secretin | 4·4 | BC013672 | cDNA sequence BC013672 |
| 4·6 | Gpd1 | Glycerol-3-phosphate dehydrogenase 1 | 4·3 | Tmem37 | Transmembrane protein 37 |
| 4·5 | Myh1 | Myosin, heavy polypeptide 1 | 4·0 | Erbb3 | v-erb-b2 homolog 3 |
| 4·5 | Fabp4 | Fatty acid binding protein 4, adipocyte | 3·9 | Dst | Dystonin |
| 4·5 | St8sia2 | ST8 sialyltransferase 2 | 3·9 | Xist | Inactive X specific transcripts |
| 4·5 | Tgfb1 | Transforming growth factor, β 1 | 3·8 | Arhgdig | Rho GDP dissociation inhibitor γ |
| 4·3 | Pscdbp | Pleckstrin homology binding protein | 3·7 | Edn1 | Endothelin 1 |
| 4·2 | Igf1 | Insulin-like growth factor 1 | 3·7 | 6330403K07Rik | RIKEN cDNA 6330403K07 gene |
| 4·1 | Scara5 | Scavenger receptor class A, member 5 | 3·7 | Jun | Jun oncogene |
| 4·1 | Sept6 | Septin 6 | 3·6 | Pla2g5 | Phospholipase A2, group V |
| 4·0 | D17H6S56E-5 | Chr 17, human D6S56E 5 | 3·6 | Apbb2 | Amyloid β precursor protein-binding B2 |
| 4·0 | Pappa | Pregnancy-associated plasma protein A | 3·6 | H19 | H19 fetal liver mRNA |
| 4·0 | Klk1 | Kallikrein 1 | 3·6 | Hspb1 | Heat shock protein 1 |
| 3·9 | Dmkn | Dermokine | 3·6 | Fcgr2b | Fc receptor, IgG, low affinity IIb |
| 3·8 | Tpd52l1 | Tumour protein D52-like 1 | 3·5 | Transcribed locusc | |
| 3·8 | Inha | Inhibin α | 3·5 | Adi1 | Acireductone dioxygenase 1 |
| 3·8 | Slc25a5 | Solute carrier family 25, member 5 | 3·4 | H2-T23 | Histocompatibility 2, T region locus 23 |
| 3·7 | Car3 | Carbonic anhydrase 3 | 3·4 | Scin | Scinderin |
| 3·7 | Pde4b | Phosphodiesterase 4B, cAMP specific | 3·3 | Rnasel | Ribonuclease L |
If a transcript is represented more than once on the array only the highest fold change is shown in this table. Supplementary Table 2 shows the complete significant dataset.
Net affy number 1454967_at.
Net affy number 1436092_at.
Few of the transcripts down-regulated following FSH treatment have been localised in the testis with the exception of Igfbp3 which has been shown to be of Sertoli cell origin (Smith et al. 1990). In order to identify more of the differentially expressed transcripts on the arrays, which may be of a Sertoli cell origin, up- and down-regulated transcripts were compared with those identified as being of likely Sertoli cell origin by Chalmel et al. (2007) using a cell isolation, GeneChip and clustering approach (Supplementary Table 2A and B). A degree of caution is required as some known Sertoli cell transcripts (e.g. Rhox5) are missing from the list generated by Chalmel et al. (2007) probably due to Chip sensitivity or the subsequent filtering process. Nevertheless, of the transcripts up-regulated at 12 h, 44% matched to the data from Chalmel et al. (2007). Interestingly, this number declined to 35% at 24 h and 27% at 72 h after the start of FSH treatment (Supplementary Table 2A). The number of down-regulated transcripts that matched the Sertoli cell list was 27, 21 and 28% at 12, 24 and 72 h respectively (Supplementary Table 2B).
Real-time PCR
Leydig cell genes
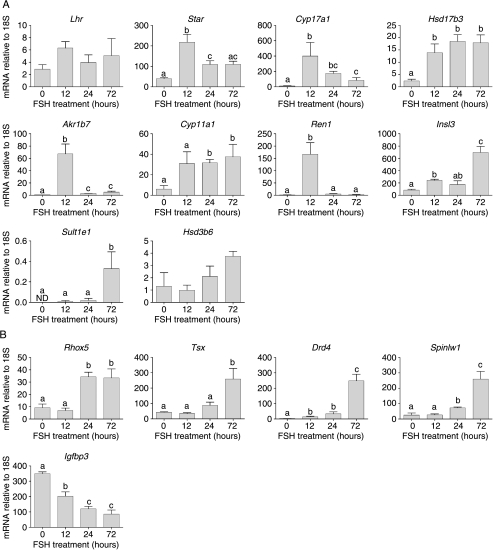
To confirm results from the array studies, real-time PCR was used to measure the effect of FSH treatment on testicular expression of selected transcripts which are known to be expressed exclusively in the Leydig cells (O'Shaughnessy et al. 2002). Eight mRNA species were tested which had shown an increase in transcript levels on the arrays after FSH (Star, Cyp17a1, Hsd17b3, Akr1b7, Lhr, Cyp11a1, Insl3 and Ren1) (Fig. 2A). Results from the real-time PCR studies confirmed that seven of these transcripts are regulated by FSH in the hpg testis, although no change in Lhr was seen. Two other Leydig cell mRNA species (Hsd3b6 and Sult1e1) that had not shown any response to FSH on the arrays were also tested by real-time PCR (Fig. 2A). Levels of Hsd3b6 did not show a response to FSH but there was a significant, if variable, increase in Sult1e1 after 72 h.
Figure 2.
Real-time PCR measurements of mRNA transcript levels in testes from adult hpg mice treated for 0 (control), 12, 24 or 72 h with FSH. Data show results from Leydig cell-specific transcripts (A) and from Sertoli cell-specific, androgen-dependent transcripts (B). The mean±s.e.m. of three or four animals per group is shown. Groups with different letter superscripts are significantly different.
Androgen-dependent genes
The array studies showed clearly that a number of androgen-dependent Sertoli cell transcripts were altered after FSH treatment. Real-time PCR was used to confirm changes in selected transcripts (Rhox5, Tsx, Drd4, Spinlw1 (Eppin) and Igfbp3) shown previously to be androgen-regulated (Lindsey & Wilkinson 1996, Denolet et al. 2006, O'Shaughnessy et al. 2007; Fig. 2B). In agreement with results from the array studies four transcripts (Rhox5, Tsx, Drd4 and Spinlw1) showed increased expression 24–72 h after FSH treatment while one transcript (Igfbp3) showed a significant decrease in expression (Fig. 2B).
Sertoli cell genes
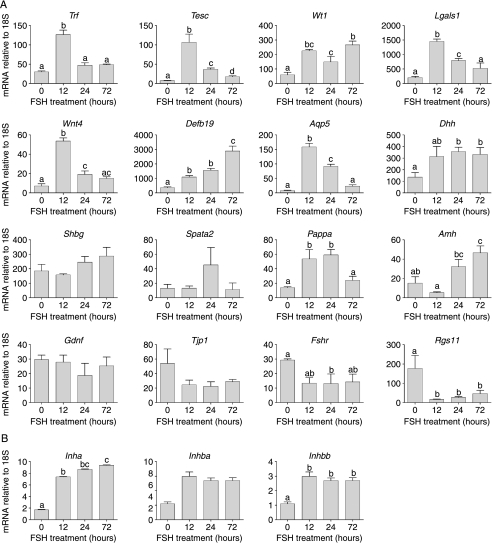
In addition to androgen-dependent Sertoli cell genes described above, 16 other Sertoli cell transcripts were measured by real-time PCR following FSH treatment of adult hpg mice (Fig. 3). Of these transcripts, seven had shown significantly increased expression on the arrays (Tesc, Lgals1, Aqp5, Dhh, Pappa, Wnt4 and Shbg), six had not shown any significant change (Trf, Wt1, Amh, Spata2, Tjp1 and Gdnf), two had shown a significant decrease (Fshr and Rgs11) after FSH treatment and one transcript (Defb19) was not on the array. Results from real-time studies confirmed increased transcript levels for six out of the seven mRNA species identified on the array (the exception was Shbg) and for the one transcript (Defb19) not on the array (Fig. 3A). Both transcripts decreased on the arrays after FSH treatment also showed a significant decrease by real-time PCR (Fig. 3A). Interestingly, however, the real-time PCR data showed there was a significant increase in levels of three out of the six transcripts that were not significantly changed on the arrays (Trf, Wt1 and Amh; Fig. 3A). Two of these transcripts (Trf and Amh) had shown a greater than twofold increase on the arrays but had not reached significance. Differences between results from real-time PCR and arrays may be a matter of sensitivity and variability of the two techniques or may be due to the choice of primers, away from the 3′ region targeted by these arrays.
Figure 3.
Real-time PCR measurements of Sertoli cell-specific mRNA transcript levels in testes from adult hpg mice treated for 0 (control), 12, 24 or 72 h with FSH. Results show the mean±s.e.m. of three or four animals per group. Groups with different letter superscripts are significantly different; where no superscripts are shown there was no difference between groups.
The inhibin subunits are expressed in a number of cell types in the testis (Barakat et al. 2008) although it might be expected that initial responsiveness to FSH would be predominantly localised in the Sertoli cells. On the arrays, both Inha and Inhbb were significantly increased by FSH (Table 1 and Supplementary Table 2) while there was no effect on Inhba. Results from the real-time PCR studies reflected the same pattern of results (Fig. 3B).
Germ cell genes
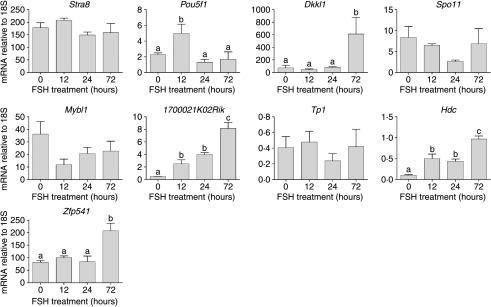
The number of known germ cell transcripts on the array affected by FSH, within the time-span of the study, was not high. This was, therefore, investigated further using real-time PCR. Expression of nine germ cell transcripts known to be expressed predominantly in spermatogonia (Stra8, Pou5f1, Dkkl1 and Spo11), spermatocytes (Mybl1, Zfp541) or spermatids (1700021K02Rik, Hdc and Tp1) was measured following FSH treatment (Fig. 4). Expression levels of Stra8, Spo11, Mybl1 and Tp1 were unaffected by treatment but there was a transient increase in Pou5f1 at 12 h while Dkkl1 and Zfp541 were significantly increased at 72 h. Expression of Hdc and 1700021K02Rik (shown previously to be increased on the array) was increased at all times after treatment.
Figure 4.
Real-time PCR measurements of germ cell-specific mRNA transcript levels in testes from adult hpg mice treated for 0 (control), 12, 24 or 72 h with FSH. Results show the mean±s.e.m. of three or four animals per group. Groups with different letter superscripts are significantly different; where no superscripts are shown there was no difference between groups.
Other transcripts
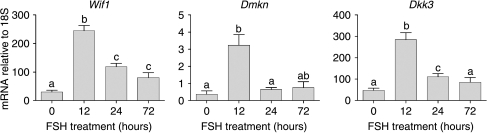
Results from the array studies identified a number of transcripts regulated by FSH but without known function and/or known expression pattern in the testis. Levels of three of these transcripts (Wif1, Dmkn, Dkk3) were measured in hpg testes after FSH treatment (Fig. 5). In all cases, FSH caused a significant increase in transcript levels confirming the results of the array study.
Figure 5.
Real-time PCR measurements of mRNA transcript levels of species with unknown testicular origin. Levels were measured in testes from adult hpg mice treated for 0 (control), 12, 24 or 72 h with FSH. Results show the mean±s.e.m. of three or four animals per group. Groups with different letter superscripts are significantly different.
Canonical pathway analysis
Analysis of canonical pathways showed that components of the cholesterol biosynthetic pathway were significantly increased at 12 h but not at other times (Supplementary Table 3A, see supplementary data in the online version of the Journal of Molecular Endocrinology at http://jme.endocrinology-journals.org/content/vol42/issue4/) and that there was a general decline in transcripts encoding factors involved in formation and regulation of tight junctions (Supplementary Table 3B).
Discussion
FSH is essential for optimum fertility in the adult male but uncertainty remains about how it acts to regulate Sertoli cell activity and spermatogenesis. The hpg mouse is an excellent model system with which to test the effects of FSH since the Sertoli cells have not been exposed to the hormone but express FSHR and are sensitive to FSH action. This study is an extension of earlier work by Sadate-Ngatchou et al. (2004) using a longer treatment period, different array chips with a larger characterised gene set (MOE430A chips (14 000 characterised genes) versus MG U74Av2 chips (6000 characterised genes, 6500 ESTs)), a significantly larger animal cohort and recombinant FSH. In addition, the purpose of this study was to follow changes in testicular transcript levels in the hpg in response to maintained levels of FSH rather than the acute response to a single administration. Together, the two studies complement each other and serve to identify transcripts regulated by FSH over the short and medium term. Interestingly, at 12 h after the start of FSH administration, when both studies can be directly compared with a degree of caution, there were only 44 differentially transcripts common to both studies, 25 up-regulated (e.g. Cyp17, Ren1, Fos, Hdc, Col4a1) and 19 down-regulated (e.g. Rgs11, Cdo1, Erbb3, Ptk2b, Vnn1). This low number of transcripts in common may be due to a combination of the chips used, the age of the animals, the number of animals used and the treatment regime.
In this study, the total number of transcripts altered at each time point did not vary markedly across the treatment period but only 39 transcripts were up-regulated more than twofold at all times indicating that there was a changing pattern of expression as the exposure to FSH was maintained. There were also 63 transcripts down-regulated more than twofold at all times suggesting that the inhibitory effects of FSH are more consistent. Results from the arrays and from real-time PCR showed that FSH treatment caused a general increase in many transcripts encoding known Sertoli cell-specific products such as Tesc, Lgals1, Fabp5 and Aqp5 (Kingma et al. 1998, Perera et al. 2001, Dettin et al. 2003) although some transcripts (e.g. Shbg, Tjp1 (Wang et al. 1989, Byers et al. 1991)) were unaffected while others were decreased (see below) indicating that the effect of FSH was not simply to increase the overall activity of the cells. Comparison of the up-regulated transcripts in this study with the list of Sertoli cell transcripts generated by Chalmel et al. (2007) confirms that, at least initially, a high proportion of the affected transcripts are likely to be of Sertoli cell origin. The declining proportion of Sertoli cell transcripts at later times is likely to be due to increasing activity in other cells such as the Leydig cell. The lower proportion of down-regulated transcripts that match to the Sertoli cell list of Chalmel et al. (2007) may reflect the GeneChip sensitivity involved in generating that list since many of these down-regulated transcripts might be expected to have a low level of expression in the normal animal.
Among the transcripts that showed decreased levels in response to FSH were a number encoding tight junction components. This is consistent with a recent study which reported that gonadotrophins reduce transcript levels of Sertoli cell barrier components but that FSH may act at the level of protein organisation to induce barrier functionality (Tarulli et al. 2008). Other transcripts that showed a significant decrease in levels after FSH treatment included Fshr and Rgs11. It is well established that FSH will cause down-regulation of its receptor by decreasing transcript levels (O'Shaughnessy 1980, Themmen et al. 1991) and a reduction in Fshr is to be expected. RGS11, in contrast, belongs to the regulator of G protein-signalling family which are GTPase-activating proteins that act to inhibit signal transduction and thus play a role in desensitisation (Chasse & Dohlman 2003). The role of RGSs in normal hormonal signalling is not well established but the declining levels of Rgs11 after FSH treatment may act to enhance signal transduction despite a reduction in receptor levels.
In addition to changes in Sertoli cell transcripts induced by FSH, it was clear from the rise in testicular androgen and the array and real-time PCR data that FSH was also acting to induce Leydig cell function. This effect was marked and rapid with a Leydig cell transcript (Ren1) showing the greatest fold change at 12 h (Deschepper et al. 1986). Results from the arrays and real-time PCR show that all components of the androgen biosynthetic pathway were induced at 12 h apart from Hsd3b6. Since Hsd3b1 is already highly expressed in the adult hpg testis (Baker et al. 2003) lack of HSD3B6 is unlikely to affect the steroidogenic potential of the cells. In addition to the steroidogenic enzymes, pathway analysis showed that most components of the cholesterol biosynthetic pathway were induced 12 h after FSH treatment while Ldlr levels are increased. This shows that FSH is acting to increase the capacity of the Leydig cells to produce and sequester cholesterol and to convert cholesterol to androgen. Interestingly, one of the critical components of the cholesterol biosynthetic pathways (Mvk) also acts to inhibit Lhr translation (Nair & Menon 2004) and this may serve to regulate further Leydig cell sensitivity to LH. The hpg mouse testis is likely to contain both adult and fetal-type Leydig cells (Baker et al. 2003) and Hsd3b6 is a marker of adult Leydig cell differentiation (Baker et al. 1999). This might imply that FSH is acting to induce activity in the fetal Leydig cell population but Sult1e1 is a marker of adult Leydig cells (Song et al. 1997) and is increased 72 h after FSH suggesting that the effects of FSH are probably being mediated through the adult Leydig cells.
In the testis, receptors for FSH are only found in the Sertoli cells (Heckert & Griswold 2002) and the effects of FSH must be mediated by a factor or factors released by the Sertoli cells which act on the Leydig cells. In the short-term, the effects of FSH on Leydig cell function in the hpg appear to be more marked than effects of hCG (Baker et al. 2003) and the effects are also very rapid since Sadate-Ngatchou et al. (2004) saw a marked increase in Cyp17a1 after only 4 h of FSH treatment. FSH appears, therefore, to be able to induce a powerful and rapid response in Leydig cells presumably through stimulation of release of potent trophic factors by the Sertoli cells. The presence of such factors has been postulated for a number of years since early studies on perfused testes or hypophysectomised animals treated with FSH (Johnson & Ewing 1971, Chen et al. 1976, Vihko et al. 1991). One report has suggested that the active factors are TIMP1 and Procathepsin L (Boujrad et al. 1995) but this has not been confirmed and we saw no evidence of changes in these factors in our study. Following FSH treatment, our array data showed that there was an increase in Igf1 levels and IGF1 has been suggested to play a role in Leydig cell differentiation (Morera et al. 1987). Interestingly, there was a marked decline in Igfbp3 and an increase in Pappa levels after FSH treatment. Increased Pappa would be expected to increase the bioavailability and activity of IGF1 (Conover et al. 2004) although the effect of altered Igfbp3 may be more complex (Modric et al. 2001). The time-course of changes in expression of Igf1 levels does not appear to fit well with a role in the stimulation of Leydig cell function after FSH treatment although it is possible that early changes in Pappa and Igfbp3 may alter early IGF1 bioavailability. Other secreted molecules showing a marked increase in transcript levels after FSH include Wif1, Dkk3 and Dmkn. Both WIF1 and DKK3 act to regulate WNT signalling and the WNT/CTNNB1 pathway is critical for normal Sertoli cell development (Boyer et al. 2008) although its function in the Leydig cell remains uncertain.
Following the increase in Leydig cell activity after FSH treatment, there was a significant change in the levels of known androgen-dependent Sertoli cell-specific transcripts. It is possible that changes in these transcripts are due to direct effects of FSH treatment but the known androgen-dependence of the transcripts makes it more likely that changes are related to increased Leydig cell androgen production induced by FSH. The rise in intratesticular androgen after FSH treatment was significant but levels remained very low, probably because FSH stimulates synthesis of the components of the steroidogenic pathway without being able to stimulate the pathway itself. The apparent effect of these low levels of androgen on Sertoli cell transcript levels suggests that the Sertoli cells are extremely sensitive to androgen stimulation.
Treatment of hpg mice with FSH increased vacuolation in the Sertoli cells and induced formation of a lumen within the seminiferous tubules but had little apparent effect on germ cell morphology or progression up to 72 h. Changes in the Sertoli cell and tubule diameter correlate with a marked rise in Aqp5 at 12 h suggesting that increased water movement across the Sertoli cell membrane may contribute to increased tubular diameter and testis weight. The absence of spermatogenic progression over the time-course studied is likely to reflect the inactive state of the Sertoli cell in the adult hpg testis and the time required for the Sertoli cell to become active enough to support germ cell maturation. Real-time PCR studies of a small number of known germ cell genes showed that there was a variable response of germ cell transcripts to FSH stimulation. POU5F1 has been shown to be necessary for primordial germ cell survival (Kehler et al. 2004) and the increase in response to FSH, albeit small, may facilitate an increase in spermatogonial number within the testis. DKKL1 and ZFP541 are expressed in both spermatocytes and spermatids (Kohn et al. 2005, Choi et al. 2008) while HDC and SPATIAL are associated with round spermatids and the later stages of spermatogenesis (Safina et al. 2002, Irla et al. 2003). Lack of a general increase in germ cell transcripts (data from both arrays and real-time PCR) would indicate that the effects seen are not due to an overall increase in germ cell number but are more likely to be part of an early specific response to FSH stimulation. It has been shown previously that more prolonged treatment of hpg mice with FSH will stimulate an increase in germ cell number and development (O'Shaughnessy et al. 1992, Singh & Handelsman 1996b, Baines et al. 2008) but the stimulatory effect of FSH on the Leydig cells makes interpretation of the FSH effects on the germ cells difficult because of the known stimulatory effect of testosterone on germ cell development in the hpg mouse (O'Shaughnessy & Sheffield 1990, Singh et al. 1995).
In this study, FSH treatment of hpg mice for up to 72 h induced significant changes in Sertoli cell transcript levels and led to indirect stimulation of Leydig cell function. The changes in Leydig cell activity probably induced further changes in androgen-dependent Sertoli cell transcripts. While FSH is known to be required for optimal germ cell development (Abel et al. 2000, 2008), treatment of hpg mice for 72 h did not have a marked effect on germ cell differentiation suggesting that longer-term action of FSH is required to induce germ cell proliferation and progression.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Funding
This study was supported by the Wellcome Trust (grant number 078137).
Acknowledgements
We would like to thank Ana Monteiro for technical assistance, Mohan Masih for histology and animal care staff for assistance with this project. Sheena Lee is supported by the Wellcome Trust funded OXION initiative.
Footnotes
(D Baban is now at Genomics Group, The Wellcome Trust Centre for Human Genetics, University of Oxford, Roosevelt Drive, Oxford OX3 7BN, UK)
References
- Abel MH, Wootton AN, Wilkins V, Huhtaniemi I, Knight PG, Charlton HM. The effect of a null mutation in the follicle-stimulating hormone receptor gene on mouse reproduction. Endocrinology. 2000;141:1795–1803. doi: 10.1210/endo.141.5.7456. [DOI] [PubMed] [Google Scholar]
- Abel MH, Baker PJ, Charlton HM, Monteiro A, Verhoeven G, De Gendt K, Guillou F, O'Shaughnessy PJ. Spermatogenesis and Sertoli cell activity in mice lacking Sertoli cell receptors for follicle stimulating hormone and androgen. Endocrinology. 2008;149:3279–3285. doi: 10.1210/en.2008-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baban D, Davies KE. Microarray analysis of mdx mice expressing high levels of utrophin: therapeutic implications for dystrophin deficiency. Neuromuscular Disorders. 2008;18:239–247. doi: 10.1016/j.nmd.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Baines H, Nwagwu MO, Hastie GR, Wiles RA, Mayhew TM, Ebling FJ. Effects of estradiol and FSH on maturation of the testis in the hypogonadal (hpg) mouse. Reproductive Biology and Endocrinology. 2008;6:4. doi: 10.1186/1477-7827-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker PJ, Sha JA, McBride MW, Peng L, Payne AH, O'Shaughnessy PJ. Expression of 3beta-hydroxysteroid dehydrogenase type I and VI isoforms in the mouse testis during development. European Journal of Biochemistry. 1999;260:911–916. doi: 10.1046/j.1432-1327.1999.00245.x. [DOI] [PubMed] [Google Scholar]
- Baker PJ, Johnston H, Abel MH, Charlton HM, O'Shaughnessy PJ. Differentiation of adult-type Leydig cells occurs in gonadotrophin-deficient mice. Reproductive Biology and Endocrinology. 2003;1:4. doi: 10.1186/1477-7827-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat B, O'Connor A, Gold E, de Kretser D, Loveland K. Inhibin, activin, follistatin and follicle stimulating hormone serum levels and testicular production are highly modulated during the first spermatogenic wave in mice. Reproduction. 2008;136:345–359. doi: 10.1530/REP-08-0140. [DOI] [PubMed] [Google Scholar]
- Baron S, Manin M, Aigueperse C, Berger M, Jean C, Veyssiere G, Morel L. Hormonal and developmental regulation of the mouse aldose reductase-like gene akr1b7 expression in Leydig cells. Journal of Molecular Endocrinology. 2003;31:71–81. doi: 10.1677/jme.0.0310071. [DOI] [PubMed] [Google Scholar]
- Boujrad N, Ogwuegbu SO, Garnier M, Lee CH, Martin BM, Papadopoulos V. Identification of a stimulator of steroid-hormone synthesis isolated from testis. Science. 1995;268:1609–1612. doi: 10.1126/science.7777858. [DOI] [PubMed] [Google Scholar]
- Boyer A, Hermo L, Paquet M, Robaire B, Boerboom D. Seminiferous tubule degeneration and infertility in mice with sustained activation of WNT/CTNNB1 signaling in Sertoli cells. Biology of Reproduction. 2008;79:475–485. doi: 10.1095/biolreprod.108.068627. [DOI] [PubMed] [Google Scholar]
- Byers S, Graham R, Dai HN, Hoxter B. Development of Sertoli cell junctional specializations and the distribution of the tight-junction-associated protein ZO-1 in the mouse testis. American Journal of Anatomy. 1991;191:35–47. doi: 10.1002/aja.1001910104. [DOI] [PubMed] [Google Scholar]
- Cattanach BM, Iddon CA, Charlton HM, Chiappa SA, Fink G. Gonadtrophin releasing hormone deficiency in a mutant mouse with hypogonadism. Nature. 1977;269:338–340. doi: 10.1038/269338a0. [DOI] [PubMed] [Google Scholar]
- Chalmel F, Rolland AD, Niederhauser-Wiederkehr C, Chung SS, Demougin P, Gattiker A, Moore J, Patard JJ, Wolgemuth DJ, Jégou B, et al. The conserved transcriptome in human and rodent male gametogenesis. PNAS. 2007;104:8346–8351. doi: 10.1073/pnas.0701883104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton HM, Halpin DMG, Iddon CA, Rosie R, Levy G, McDowell IFW, Megson A, Morris JF, Bramwell A, Speight A, et al. The effects of daily administration of single and multiple injections of gonadotrophin-releasing hormone on pituitary and gonadal function in the hypogonadal (hpg) mouse. Endocrinology. 1983;113:535–544. doi: 10.1210/endo-113-2-535. [DOI] [PubMed] [Google Scholar]
- Chasse SA, Dohlman HG. RGS proteins: G protein-coupled receptors meet their match. Assay and Drug Development Technologies. 2003;1:357–364. doi: 10.1089/154065803764958649. [DOI] [PubMed] [Google Scholar]
- Chen JK, Heckert LL. Dmrt1 expression is regulated by follicle-stimulating hormone and phorbol esters in postnatal Sertoli cells. Endocrinology. 2001;142:1167–1178. doi: 10.1210/endo.142.3.8021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YI, Payne AH, Kelch RP. FSH stimulation of Leydig cell function in the hypophysectomized immature rat. Proceedings of the Society for Experimental Biology and Medicine. 1976;153:473–475. doi: 10.3181/00379727-153-39571. [DOI] [PubMed] [Google Scholar]
- Choi E, Han C, Park I, Lee B, Jin S, Choi H, Kim DH, Park ZY, Eddy EM, Cho C. A novel germ cell-specific protein, SHIP1, forms a complex with chromatin remodeling activity during spermatogenesis. Journal of Biological Chemistry. 2008;283:35283–35294. doi: 10.1074/jbc.M805590200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conover CA, Bale LK, Overgaard MT, Johnstone EW, Laursen UH, Fuchtbauer EM, Oxvig C, van Deursen J. Metalloproteinase pregnancy-associated plasma protein A is a critical growth regulatory factor during fetal development. Development. 2004;131:1187–1194. doi: 10.1242/dev.00997. [DOI] [PubMed] [Google Scholar]
- Cunningham DB, Segretain D, Arnaud D, Rogner UC, Avner P. The mouse Tsx gene is expressed in Sertoli cells of the adult testis and transiently in premeiotic germ cells during puberty. Developmental Biology. 1998;204:345–360. doi: 10.1006/dbio.1998.9004. [DOI] [PubMed] [Google Scholar]
- Czechowski T, Bari RP, Stitt M, Scheible WR, Udvardi MK. Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant Journal. 2004;38:366–379. doi: 10.1111/j.1365-313X.2004.02051.x. [DOI] [PubMed] [Google Scholar]
- Denolet E, De Gendt K, Allemeersch J, Engelen K, Marchal K, Van Hummelen P, Tan KA, Sharpe RM, Saunders PT, Swinnen JV, et al. The effect of a Sertoli cell-selective knockout of the androgen receptor on testicular gene expression in prepubertal mice. Molecular Endocrinology. 2006;20:321–334. doi: 10.1210/me.2005-0113. [DOI] [PubMed] [Google Scholar]
- Deschepper CF, Mellon SH, Cumin F, Baxter JD, Ganong WF. Analysis by immunocytochemistry and in situ hybridization of renin and its mRNA in kidney, testis, adrenal, and pituitary of the rat. PNAS. 1986;83:7552–7556. doi: 10.1073/pnas.83.19.7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettin L, Rubinstein N, Aoki A, Rabinovich GA, Maldonado CA. Regulated expression and ultrastructural localization of galectin-1, a proapoptotic beta-galactoside-binding lectin, during spermatogenesis in rat testis. Biology of Reproduction. 2003;68:51–59. doi: 10.1095/biolreprod.102.006361. [DOI] [PubMed] [Google Scholar]
- Dierich A, Sairam MR, Monaco L, Fimia GM, Gansmuller A, LeMeur M, Sassone-Corsi P. Impairing follicle-stimulating hormone (FSH) signaling in vivo: targeted disruption of the FSH receptor leads to aberrant gametogenesis and hormonal imbalance. PNAS. 1998;95:13612–13617. doi: 10.1073/pnas.95.23.13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gendt K, Swinnen JV, Saunders PT, Schoonjans L, Dewerchin M, Devos A, Tan K, Atanassova N, Claessens F, Lecureuil C, et al. A Sertoli cell-selective knockout of the androgen receptor causes spermatogenic arrest in meiosis. PNAS. 2004;101:1327–1332. doi: 10.1073/pnas.0308114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Goascogne C, Sananes N, Gouezou M, Takemori S, Kominami S, Baulieu EE, Robel P. Immunoreactive cytochrome P450 (17alpha) in rat and guinea-pig gonads, adrenal-glands and brain. Journal of Reproduction and Fertility. 1991;93:609–622. doi: 10.1530/jrf.0.0930609. [DOI] [PubMed] [Google Scholar]
- Haywood M, Spaliviero J, Jimemez M, King NJ, Handelsman DJ, Allan CM. Sertoli and germ cell development in hypogonadal (hpg) mice expressing transgenic follicle-stimulating hormone alone or in combination with testosterone. Endocrinology. 2003;144:509–517. doi: 10.1210/en.2002-220710. [DOI] [PubMed] [Google Scholar]
- Heckert LL, Griswold MD. The expression of the follicle-stimulating hormone receptor in spermatogenesis. Recent Progress in Hormone Research. 2002;57:129–148. doi: 10.1210/rp.57.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst RC, Abel MH, Wilkins V, Simpson C, Knight PG, Zhang FP, Huhtaniemi I, Kumar TR, Charlton HM. Influence of mutations affecting gonadotropin production or responsiveness on expression of inhibin subunit mRNA and protein in the mouse ovary. Reproduction. 2004;128:43–52. doi: 10.1530/rep.1.00176. [DOI] [PubMed] [Google Scholar]
- Irla M, Puthier D, Le Goffic R, Victorero G, Freeman T, Naquet P, Samson M, Nguyen C. Spatial, a new nuclear factor tightly regulated during mouse spermatogenesis. Gene Expression Patterns. 2003;3:135–138. doi: 10.1016/s1567-133x(03)00024-3. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Oliver P, Davies KE, Platt N. Identification and characterization of murine SCARA5, a novel class A scavenger receptor that is expressed by populations of epithelial cells. Journal of Biological Chemistry. 2006;281:11834–11845. doi: 10.1074/jbc.M507599200. [DOI] [PubMed] [Google Scholar]
- Johnson BH, Ewing LL. Follicle-stimulating hormone and the regulation of testosterone secretion in rabbit testes. Science. 1971;173:635–637. doi: 10.1126/science.173.3997.635. [DOI] [PubMed] [Google Scholar]
- Kehler J, Tolkunova E, Koschorz B, Pesce M, Gentile L, Boiani M, Lomeli H, Nagy A, McLaughlin KJ, Scholer HR, et al. Oct4 is required for primordial germ cell survival. EMBO Reports. 2004;5:1078–1083. doi: 10.1038/sj.embor.7400279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingma PB, Bok D, Ong DE. Bovine epidermal fatty acid-binding protein: determination of ligand specificity and cellular localization in retina and testis. Biochemistry. 1998;37:3250–3257. doi: 10.1021/bi972520l. [DOI] [PubMed] [Google Scholar]
- Kohn MJ, Kaneko KJ, DePamphilis ML. DkkL1 (Soggy), a Dickkopf family member, localizes to the acrosome during mammalian spermatogenesis. Molecular Reproduction and Development. 2005;71:516–522. doi: 10.1002/mrd.20314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy H, Babu PS, Morales CR, Sairam MR. Delay in sexual maturity of the follicle-stimulating hormone receptor knockout male mouse. Biology of Reproduction. 2001;65:522–531. doi: 10.1095/biolreprod65.2.522. [DOI] [PubMed] [Google Scholar]
- Kumar TR, Wang Y, Lu N, Matzuk MM. Follicle stimulating hormone is required for ovarian follicle maturation but not male fertility. Nature Genetics. 1997;15:201–204. doi: 10.1038/ng0297-201. [DOI] [PubMed] [Google Scholar]
- Lang J. Assay for deletion in GnRH (hpg) locus using PCR. Mouse Genome. 1995;89:857. [Google Scholar]
- Lindsey JS, Wilkinson MF. Pem: a testosterone-regulated and LH-regulated homeobox gene expressed in mouse sertoli cells and epididymis. Developmental Biology. 1996;179:471–484. doi: 10.1006/dbio.1996.0276. [DOI] [PubMed] [Google Scholar]
- Mason AJ, Hayflick JS, Zoeller RT, Young WS, Phillips HS, Nikolics K, Seeburg TA. A deletion truncating the GnRH gene is responsible for hypogonadism in the hpg mouse. Science. 1986;234:1366–1371. doi: 10.1126/science.3024317. [DOI] [PubMed] [Google Scholar]
- McLachlan RI, O'Donnell L, Meachem SJ, Stanton PG, de Kretser DM, Pratis K, Robertson DM. Identification of specific sites of hormonal regulation in spermatogenesis in rats, monkeys, and man. Recent Progress in Hormone Research. 2002;57:149–179. doi: 10.1210/rp.57.1.149. [DOI] [PubMed] [Google Scholar]
- Modric T, Silha JV, Shi Z, Gui Y, Suwanichkul A, Durham SK, Powell DR, Murphy LJ. Phenotypic manifestations of insulin-like growth factor-binding protein-3 overexpression in transgenic mice. Endocrinology. 2001;142:1958–1967. doi: 10.1210/endo.142.5.8165. [DOI] [PubMed] [Google Scholar]
- Monaco L, Foulkes NS, Sassone-Corsi P. Pituitary follicle-stimulating hormone (FSH) induces CREM gene expression in Sertoli cells: involvement in long-term desensitization of the FSH receptor. PNAS. 1995;92:10673–10677. doi: 10.1073/pnas.92.23.10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morera AM, Chauvin MA, De Peretti E, Binoux M, Benahmed M. Somatomedin C/insulin-like growth factor 1: an intratesticular differentiative factor of Leydig cells? Hormone Research. 1987;28:50–57. doi: 10.1159/000180925. [DOI] [PubMed] [Google Scholar]
- Morris PL, Vale WW, Cappel S, Bardin CW. Inhibin production by primary Sertoli cell-enriched cultures: regulation by follicle-stimulating hormone, androgens, and epidermal growth factor. Endocrinology. 1988;122:717–725. doi: 10.1210/endo-122-2-717. [DOI] [PubMed] [Google Scholar]
- Myers M, Ebling FJ, Nwagwu M, Boulton R, Wadhwa K, Stewart J, Kerr JB. Atypical development of Sertoli cells and impairment of spermatogenesis in the hypogonadal (hpg) mouse. Journal of Anatomy. 2005;207:797–811. doi: 10.1111/j.1469-7580.2005.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair AK, Menon KMJ. Isolation and characterization of a novel trans-factor for luteinizing hormone receptor mRNA from ovary. Journal of Biological Chemistry. 2004;279:14937–14944. doi: 10.1074/jbc.M309484200. [DOI] [PubMed] [Google Scholar]
- O'Shaughnessy PJ. FSH receptor autoregulation and cyclic AMP production in the immature rat testis. Biology of Reproduction. 1980;23:810–814. doi: 10.1095/biolreprod23.4.810. [DOI] [PubMed] [Google Scholar]
- O'Shaughnessy PJ, Sheffield JW. Effect of testosterone on testicular steroidogenesis in the hypogonadal (hpg) mouse. Journal of Steroid Biochemistry. 1990;35:729–734. doi: 10.1016/0022-4731(90)90315-j. [DOI] [PubMed] [Google Scholar]
- O'Shaughnessy PJ, Bennett MK, Scott IS, Charlton HM. Effects of FSH on Leydig cell morphology and function in the hypogonadal mouse. Journal of Endocrinology. 1992;135:517–525. doi: 10.1677/joe.0.1350517. [DOI] [PubMed] [Google Scholar]
- O'Shaughnessy PJ, Willerton L, Baker PJ. Changes in Leydig cell gene expression during development in the mouse. Biology of Reproduction. 2002;66:966–975. doi: 10.1095/biolreprod66.4.966. [DOI] [PubMed] [Google Scholar]
- O'Shaughnessy PJ, Abel M, Charlton HM, Hu B, Johnston H, Baker PJ. Altered expression of genes involved in regulation of vitamin A metabolism, solute transportation, and cytoskeletal function in the androgen-insensitive tfm mouse testis. Endocrinology. 2007;148:2914–2924. doi: 10.1210/en.2006-1412. [DOI] [PubMed] [Google Scholar]
- Perera EM, Martin H, Seeherunvong T, Kos L, Hughes IA, Hawkins JR, Berkovitz GD. Tescalcin, a novel gene encoding a putative EF-hand Ca(2+)-binding protein, Col9a3, and renin are expressed in the mouse testis during the early stages of gonadal differentiation. Endocrinology. 2001;142:455–463. doi: 10.1210/endo.142.1.7882. [DOI] [PubMed] [Google Scholar]
- Sadate-Ngatchou PI, Pouchnik DJ, Griswold MD. Follicle-stimulating hormone induced changes in gene expression of murine testis. Molecular Endocrinology. 2004;18:2805–2816. doi: 10.1210/me.2003-0203. [DOI] [PubMed] [Google Scholar]
- Safina F, Tanaka S, Inagaki M, Tsuboi K, Sugimoto Y, Ichikawa A. Expression of l-histidine decarboxylase in mouse male germ cells. Journal of Biological Chemistry. 2002;277:14211–14215. doi: 10.1074/jbc.M200702200. [DOI] [PubMed] [Google Scholar]
- Singh J, Handelsman DJ. Neonatal administration of FSH increases Sertoli cell numbers and spermatogenesis in gonadotropin-deficient (hpg) mice. Journal of Endocrinology. 1996a;151:37–48. doi: 10.1677/joe.0.1510037. [DOI] [PubMed] [Google Scholar]
- Singh J, Handelsman DJ. The effects of recombinant FSH on testosterone-induced spermatogenesis in gonadotropin-deficient (Hpg) mice. Journal of Andrology. 1996b;17:382–393. [PubMed] [Google Scholar]
- Singh J, O'Neill C, Handelsman DJ. Induction of spermatogenesis by androgens in gonadotropin-deficient (Hpg) mice. Endocrinology. 1995;136:5311–5321. doi: 10.1210/endo.136.12.7588276. [DOI] [PubMed] [Google Scholar]
- Skinner MK, Schlitz SM, Anthony CT. Regulation of Sertoli cell differentiated function: testicular transferrin and androgen-binding protein expression. Endocrinology. 1989;124:3015–3024. doi: 10.1210/endo-124-6-3015. [DOI] [PubMed] [Google Scholar]
- Smith EP, Dickson BA, Chernausek SD. Insulin-like growth factor binding protein-3 secretion from cultured rat sertoli cells: dual regulation by follicle stimulating hormone and insulin-like growth factor-I. Endocrinology. 1990;127:2744–2751. doi: 10.1210/endo-127-6-2744. [DOI] [PubMed] [Google Scholar]
- Song WC, Qian Y, Sun X, Negishi M. Cellular localization and regulation of expression of testicular estrogen sulfotransferase. Endocrinology. 1997;138:5006–5012. doi: 10.1210/endo.138.11.5512. [DOI] [PubMed] [Google Scholar]
- Song KH, Park JI, Lee MO, Soh J, Lee K, Choi HS. LH induces orphan nuclear receptor Nur77 gene expression in testicular Leydig cells. Endocrinology. 2001;142:5116–5123. doi: 10.1210/endo.142.12.8525. [DOI] [PubMed] [Google Scholar]
- Tarulli GA, Meachem SJ, Schlatt S, Stanton PG. Regulation of testicular tight junctions by gonadotrophins in the adult Djungarian hamster in vivo. Reproduction. 2008;135:867–877. doi: 10.1530/REP-07-0572. [DOI] [PubMed] [Google Scholar]
- Themmen AP, Blok LJ, Post M, Baarends WM, Hoogerbrugge JW, Parmentier M, Vassart G, Grootegoed JA. Follitropin receptor down-regulation involves a cAMP-dependent post-transcriptional decrease of receptor mRNA expression. Molecular and Cellular Endocrinology. 1991;78:R7–R13. doi: 10.1016/0303-7207(91)90130-k. [DOI] [PubMed] [Google Scholar]
- Verhoeven G, Cailleau J. Follicle-stimulating hormone and androgens increase the concentration of the androgen receptor in Sertoli cells. Endocrinology. 1988;122:1541–1550. doi: 10.1210/endo-122-4-1541. [DOI] [PubMed] [Google Scholar]
- Vihko KK, Lapolt PS, Nishimori K, Hsueh AJ. Stimulatory effects of recombinant follicle-stimulating hormone on Leydig cell function and spermatogenesis in immature hypophysectomized rats. Endocrinology. 1991;129:1926–1932. doi: 10.1210/endo-129-4-1926. [DOI] [PubMed] [Google Scholar]
- Wang Y-M, Sullivan PM, Petrusz P, Yarbrough W, Joseph DR. The androgen-binding protein gene is expressed in CD1 mouse testis. Molecular and Cellular Endocrinology. 1989;63:85–92. doi: 10.1016/0303-7207(89)90084-1. [DOI] [PubMed] [Google Scholar]
- Wreford NG, Rajendra Kumar T, Matzuk MM, de Kretser DM. Analysis of the testicular phenotype of the follicle-stimulating hormone beta-subunit knockout and the activin type II receptor knockout mice by stereological analysis. Endocrinology. 2001;142:2916–2920. doi: 10.1210/endo.142.7.8230. [DOI] [PubMed] [Google Scholar]