Abstract
Phytochrome A (phyA) is the primary photoreceptor for mediating the far-red high irradiance response in Arabidopsis thaliana. FAR-RED ELONGATED HYPOCOTYL1 (FHY1) and its homolog FHY1-LIKE (FHL) define two positive regulators in the phyA signaling pathway. These two proteins have been reported to be essential for light-regulated phyA nuclear accumulation through direct physical interaction with phyA. Here, we report that FHY1 protein is phosphorylated rapidly after exposure to red light. Subsequent exposure to far-red light after the red light pulse reverses FHY1 phosphorylation. Such a phenomenon represents a classical red/far-red reversible low fluence response. The phosphorylation of FHY1 depends on functioning phyA but not on other phytochromes and cryptochromes. Furthermore, we demonstrate that FHY1 and FHL directly interact with phyA by bimolecular fluorescence complementation and that both FHY1 and FHL interact more stably with the Pr form of phyA in Arabidopsis seedlings by coimmunoprecipitation. Finally, in vitro kinase assays confirmed that a recombinant phyA is able to robustly phosphorylate FHY1. Together, our results suggest that phyA may differentially regulate FHY1 and FHL activity through direct physical interaction and red/far-red light reversible phosphorylation to fine-tune their degradation rates and resulting light responses.
INTRODUCTION
Plant growth and development are influenced by complex signaling networks. To perceive light signal, one of the major environmental triggers, plants have evolved several different classes of photoreceptors to monitor light quality and quantity. The red (R)/far-red (FR) light absorbing phytochromes (phy) are among the best characterized. In Arabidopsis thaliana, the five phytochromes (phyA to phyE) are generally categorized into two groups: light labile type I (phyA) and light stable type II (phyB to phyE) (Sharrock and Quail, 1989). In vivo, the phytochromes exist in two distinct but interconvertible forms, the R light–absorbing Pr form and the FR light–absorbing Pfr form. The Pfr form is generally considered to be the biologically active form; nonetheless, there is evidence suggesting that the Pr form also has some biological activity, especially for phyA (Reed, 1999; Shinomura et al., 2000). Three action modes are known for phytochromes. PhyA has been well established to mediate the very low fluence response (VLFR) (Botto et al., 1996; Shinomura et al., 1996; Yanovsky et al., 1997) and the far-red high-irradiance response (HIR) (Whitelam et al., 1993; Yanovsky et al., 1997). VLFRs are activated by extremely low light intensities, whereas HIRs require prolonged exposure to relatively high light intensities and are responsible for the regulation of seedling deetiolation. PhyB is well known to mediate both HIR to red light and the R/FR reversible low-fluence response (LFR) (Shinomura et al., 1996). Recently, there have been several reports that imply phyA might also be involved in LFR (Long and Iino, 2001; Stowe-Evans et al., 2001; Takano et al., 2005). However, direct evidence for a phyA-mediated LFR biochemical process has been lacking.
Genetic studies have led to the identification of a number of mutants that are defective in high irradiance far-red responses, including far-red elongated hypocotyl1 (fhy1), fhy3, and far-red impaired response1 (far1) (Whitelam et al., 1993; Hudson et al., 1999; Yanovsky et al., 2000). The strong etiolated phenotype of the fhy1 mutant in far-red light and the similarity of far-red light-induced genome expression profiles between fhy1 and phyA mutants indicate that FHY1 acts rather close to phyA in the phyA-mediated signal transduction cascade (Desnos et al., 2001; Wang et al., 2002). FHY1-LIKE (FHL), an FHY1 homolog protein, has been reported to share a partially overlapping function with FHY1 in phyA-mediated far-red responses (Zhou et al., 2005). Recent studies revealed that FHY1 and FHL are essential for light-induced phyA nuclear accumulation and subsequent light responses (Hiltbrunner et al., 2005, 2006). FHY3 and FAR1, which encode two proteins related to Mutator-like transposases, work together to activate the transcription of FHY1 and FHL and thus indirectly regulate phyA nuclear accumulation and phyA responses (Lin et al., 2007). FHY1 and FHL have been reported to interact with phyA in vitro (Hiltbrunner et al., 2005, 2006), and FHY1 has been shown to interact with phyA using continuous far-red light (FRc)–grown Arabidopsis seedlings (Saijo et al., 2008). However, direct evidence for interaction of native FHL with phyA in planta is still lacking. Also, the precise nature of the interaction of FHY1 and/or FHL with phyA remains largely unknown.
Protein phosphorylation and dephosphorylation play important roles in regulating phytochrome-mediated signaling pathways. For example, phyA has been reported to be an active kinase (Yeh and Lagarias, 1998). Photoactivated recombinant oat (Avena sativa) phytochromes have been shown by in vitro kinase assays to directly phosphorylate several signaling partners, such as Arabidopsis PHYTOCHROME KINASE SUBSTRATE1 (PKS1) (Fankhauser et al., 1999), NUCLEOSIDE DIPHOSPHATE KINASE2 (NDPK2) (Choi et al., 1999), CRYPTOCHROME1 (cry1) (Ahmad et al., 1998b), and AUXIN/INDOLE-3-ACETIC ACID (AUX/IAA) (Colon-Carmona et al., 2000) in signaling transduction. In addition, two phosphatases, PROTEIN PHOSPHATASE5 (a type 5 phosphatase) and FyPP1 (for PHYTOCHROME-ASSOCIATED SERINE/THREONINE PROTEIN PHOSPHATASE1, a PP6-type Ser/Thr phosphatase), have been reported to interact with and specifically dephosphorylate the Pfr form of phytochromes to modulate light signaling (Kim et al., 2002; Ryu et al., 2005; DeLong, 2006). Phosphorylation also modulates light signaling by influencing protein–protein interactions. For example, the phosphorylation at Ser-598 of oat phyA has been shown to prevent its interaction with putative signal transducers, such as Arabidopsis NDPK2 and PHYTOCHROME INTERACTING FACTOR3 (PIF3) (Kim et al., 2004).
Ubiquitin/proteasome-mediated protein degradation regulates various developmental pathways in phytochrome-mediated signal transduction. The stability of phytochromes, as well as several well-studied photomorphogenesis-related transcription factors, is regulated by phosphorylation. For example, phyA has been reported to be a target of the CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) E3 ubiquitin ligase activity (Seo et al., 2004). Phosphorylated phyA has been shown to preferentially interact with COP1/SPA1 (for SUPPRESSOR OF PHYA-105 1) complex (Saijo et al., 2008), and its stability is regulated by phosphorylation (Trupkin et al., 2007). ELONGATED HYPOCOTYL5 (HY5) is a basic domain/leucine zipper transcription factor that has been shown to be phosphorylated and is targeted for proteasomal degradation in darkness by COP1 (Hardtke et al., 2000; Osterlund et al., 2000; Saijo et al., 2003). Moreover, PIF3, PIF4, PIF5, and LONG HYPOCOTYL IN FAR-RED1 (HFR1) are basic helix-loop-helix transcription factors that undergo phosphorylation in vivo, and they are all degraded through the 26S proteasome pathway (Duek et al., 2004; Al-Sady et al., 2006; Shen et al., 2007). It is believed that rapid degradation of these factors is essential for the regulation of photomorphogenesis (Castillon et al., 2007).
In a previous study (Shen et al., 2005), we reported that FHY1 is most abundant in young seedlings grown in darkness and is quickly downregulated during further seedling development and by light exposure. The light-triggered FHY1 protein reduction is primarily mediated through the 26S proteasome-dependent protein degradation. Furthermore, phyA is directly involved in mediating the light-triggered downregulation of FHY1. Here, we demonstrate that FHY1 protein is rapidly phosphorylated when dark-grown Arabidopsis seedlings are exposed to red light. We show that phyA is responsible for the red light–dependent FHY1 phosphorylation and that this modification is R/FR reversible. We further confirm the occurrence of direct interaction between phyA and FHY1, as well as between phyA and FHL, using bimolecular fluorescence complementation (BiFC) assays. Our coimmunoprecipitation (co-IP) analyses show that FHY1 and FHL bind more stably with the Pr form of phyA, and our kinase assays indicate that phyA can phosphorylate FHY1 in vitro. Therefore, the phyA-dependent phosphorylation and degradation of FHY1 protein likely represent a key regulatory mechanism in phyA signaling.
RESULTS
A Posttranslational Modification of FHY1 Protein Is Rapidly Induced by Red but Not Far-Red Light
We have previously shown that the green fluorescent protein (GFP)-FHY1 protein exhibits rapid proteasome-mediated degradation during dark–light transitions (Shen et al., 2005). To unravel the mechanism underlying the light-triggered FHY1 protein degradation, we examined the molecular weight changes of GFP-FHY1 induced by light exposure using a high-resolution protein gel system. Three-day-old etiolated seedlings (the tissue where FHY1 protein accumulates to the highest level; Shen et al., 2005) were transferred to red light and incubated for time periods ranging from 1 min to 60 min. In this assay, GFP-FHY1 protein shows rapid formation of an upshifted band. The intensity of the upshifted band correlates with increasing length of red light irradiation (Figure 1A). The band is first noticeable after 5-min irradiation with red light and becomes quite intense after 10 min. The ratio of this band to the normal lower molecular weight band peaks at 30 min of red light exposure. After 30 min, the intensity of the high molecular weight band started to decline.
Figure 1.
Detection and Analysis of GFP-FHY1 and FHY1 Phosphorylation.
(A) to (D) Immunoblot analyses of GFP-FHY1 fusion protein or endogenous FHY1 protein extracted from 3-d-old seedlings grown in darkness ([A] to [C]) or in far-red light (D) and then subjected to red light treatment ([A], [B], and [D]) or far-red light treatment (C) for various time periods specified in minutes (m) or hours (h). Wild-type seedlings were used in (B), and 35S:GFP-FHY1 seedlings were used in (A), (C), and (D). Immunoblots were probed with anti-FHY1 antibody. Asterisks indicate modified forms, GFP-FHY1* or FHY1*, that migrate more slowly than the corresponding regular forms of GFP-FHY1 and FHY1. The top panel in (A) shows the ratios of band intensities of GFP-FHY1* to GFP-FHY1.
(E) Three-day-old dark-grown 35S:GFP-FHY1 seedlings were pretreated for 2 h with either DMSO (−, lane 3) or MG132 (+, lane 4) and then exposed to red light for 30 min. In addition, proteins extracted from MG132 pretreated dark-grown seedlings (lane 1) and dark-grown seedlings with 30-min red light treatment (lane 4) were immunoprecipitated (IP) with anti-FHY1 antibody (lanes 2 and 5, respectively). After immunoprecipitation, the samples that had been exposed to red light were divided into three aliquots. One of the aliquots was directly loaded onto lane 5, and the other two aliquots were first treated with either CIP (+) or boiled CIP (+b) for 30 min, and then loaded onto lane 6 and lane 7, respectively. Immunoblots were probed with anti-FHY1 antibody. GFP-FHY1-P and GFP-FHY1 indicate the phosphorylated and nonphosphorylated forms of the protein.
Light conditions are indicated in this and all subsequent figures as D, darkness; R, red light; and FR, far-red light.
To verify this observation, we examined the endogenous FHY1 protein in wild-type seedlings (Figure 1B). We noticed that our FHY1 antibodies recognize two bands in etiolated seedlings, and both bands are larger than the predicted mass (23 kD) of FHY1. Nevertheless, the presence of these two bands in the wild type, phyA-1, phyBDE, cry1 cry2, and other phyA point mutants of Arabidopsis and their absence in the fhy1-1 null mutant indicate that they truly represent the endogenous FHY1 (Figures 3 and 4; see Supplemental Figure 1 online). The larger masses are presumably due to certain unknown protein modification and/or alternative mRNA splicing events. Ten minutes after transferring 3-d etiolated seedlings to red light, a band of higher molecular mass was detected. As was observed with GFP-FHY1, the upshifted band increases in intensity with longer exposures to red light, also reaching maximum intensity at 30 min and declining thereafter. The lower band might undergo the same kind of mobility shift as well; however, the detection of it is obscured by the presence of the upper band.
Figure 3.
Role of phyA in the Phosphorylation of GFP-FHY1 and Endogenous FHY1.
(A) to (D) Immunoblot analysis of proteins extracted from seedlings grown for 3 d in darkness and then transferred to red light for the time periods indicated in minutes (m). Seedlings tested were as follows: 35S:GFP-FHY1 and 35S:GFP-FHY1/phyA-1 (A), wild type and phyA-1 (B), 35S:GFP-FHY1/phyB-1 and 35S:GFP-FHY1/cry1 (C), and phyB-1, phyBDE, and cry1 cry2 (D).
(E) Three-day-old dark-grown wild type, phyA-1, and cry1cry2 seedlings were transferred to blue light (B) for the time periods indicated.
In (A) to (E), immunoblots were probed with anti-FHY1 antibody. Asterisks indicate slower migrating phosphorylated forms of GFP-FHY1 and FHY1.
Figure 4.
Effect of phyA Mutations on FHY1 Phosphorylation.
Immunoblot analysis of proteins extracted from seedlings grown for 3 d in darkness and then transferred to red light for the time periods indicated in minutes (m). Seedlings tested were as follows: wild type (ecotype Columbia), phyA-300D, and phyA-302GFP/phyA-211 (A); wild type (ecotype RLD) and phyA-105 (B); and wild type (ecotype Ler), phyA-201, phyAOX, and phyA S598A (the last two in phyA-201 background) (C). Immunoblots were probed with anti-FHY1 antibody and analyzed for appearance of the slow migrating phosphorylated forms of FHY1 (indicated by asterisks).
It has been shown that GFP-FHY1 is more stable during dark to far-red light transition than during dark to red light transition (Shen et al., 2005). Thus, we investigated whether this difference in stability is correlated with the appearance and subsequent disappearance of the high molecular weight form of GFP-FHY1. As shown in Figure 1C, the upshifted band is not observed when etiolated seedlings were exposed to far-red light, even with up to 24 h of exposure. When seedlings were first grown in far-red light and then transferred to red light (Figure 1D), the high molecular weight form of GFP-FHY1 is clearly detectable after 5 min.
FHY1 Protein Is Rapidly Phosphorylated under Red Light, Forming a Possible Intermediate for 26S Proteasome Degradation
To determine the molecular nature of this light-induced posttranslational modification of GFP-FHY1 and endogenous FHY1, we investigated whether FHY1 is a phosphoprotein. We treated immunoprecipitated GFP-FHY1 protein samples with either active calf-intestinal alkaline phosphatase (CIP) or inactivated (boiled) CIP. The high molecular weight band was abolished only in the sample treated with active CIP (Figure 1E). This result confirms that the upshifted band indeed corresponds to phosphorylated GFP-FHY1. Furthermore, treatment with MG132, which blocks proteasome activity, significantly prevents the loss of the upshifted band of GFP-FHY1 (Figure 1E), indicating that phosphorylated GFP-FHY1 is a preferred substrate for ubiquitin/proteasome-mediated degradation.
Phosphorylation of GFP-FHY1 Is Red/Far-Red Light Reversible
Our data showed that red light could effectively induce FHY1 protein phosphorylation and subsequent degradation, but far-red light could not. To test whether this phosphorylation is R/FR reversible, we performed a series of experiments (Figure 2) using seedlings grown in darkness for 3 d. A 1-min pulse of red light followed by a period of darkness (I) is sufficient to induce phosphorylation of GFP-FHY1 by 20 min, and even more so by 30 min. This is similar to the effect of continuous red light (Figure 1A). As expected, with a 1-min pulse of far-red light (II), phosphorylation of GFP-FHY1 protein is not detected 20 or 30 min later. Intriguingly, with a 1-min pulse of far-red light immediately following the initial red light pulse (III), phosphorylated GFP-FHY1 was not detected. Furthermore, we found that the red light–induced phosphorylation of GFP-FHY1 could be abolished by far-red light only when the far-red light pulse was given within 10 min after the initial pulse of red light (IV, V, and VI). If the seedlings exposed to the pulsed red light were allowed to stay in darkness for >10 min before the far-red light pulse, the far-red light pulse was no longer able to abrogate the induction of phosphorylation (VII and VIII). This R/FR light reversibility suggests that FHY1 phosphorylation follows a typical LFR mode of phytochrome action.
Figure 2.
R/FR Reversibility of GFP-FHY1 Phosphorylation.
Immunoblot analysis of proteins extracted from 3-d-old dark-grown (D) 35S:GFP-FHY1 seedlings exposed to 1 min (m) of red light (I), 1 min of far-red light (II), or 1 min of red light followed by 1 min of far-red light (III-VIII), either immediately (III) or after a dark incubation period of 2 (IV), 6 (V), 8 (VI), 10 (VII), or 15 min (VIII). For all treatments, seedlings were analyzed 20 and 30 min after the initial light treatment. Immunoblots were probed with anti-FHY1 antibody. GFP-FHY1-P and GFP-FHY1 indicate the phosphorylated and nonphosphorylated forms of the protein.
Phosphorylation of FHY1 Is Solely Dependent on phyA and Not on Any Other Phytochromes or Cryptochromes
To determine which member of the phytochrome family is responsible for the red light–induced phosphorylation of FHY1 protein, we examined the phosphorylation of GFP-FHY1 and of endogenous FHY1 protein in different mutant backgrounds. Figure 3A shows that phosphorylation of GFP-FHY1 is not observed in a phyA null mutant. This is also true for the endogenous FHY1 protein (Figure 3B). By contrast, GFP-FHY1 in a phyB background is phosphorylated just as in the wild type (Figure 3C). Similarly, red light–induced phosphorylation of endogenous FHY1 is observed in phyB and in phyBDE triple mutants (Figure 3D). To test whether the red light–induced FHY1 phosphorylation is dependent on cryptochromes, the photoreceptors that mainly perceive blue light (Ahmad et al., 1998a), we performed dark-to-red light transition in cryptochrome mutant background. Phosphorylation of GFP-FHY1 and that of FHY1 are observed, clearly not affected by mutations of cry1 and/or cry2 (Figures 3C and 3D). Since phytochromes are known to absorb blue light weakly (Wang and Deng, 2003), we investigated whether blue light could induce phosphorylation of FHY1 and, if so, whether the phosphorylation is phyA and/or cry1/cry2 dependent. It is interesting to note that endogenous FHY1 is phosphorylated upon blue light exposure and that this phosphorylation is also phyA dependent but not affected by mutation in cry1 and cry2 (Figure 3E). Overall, our data suggest that phyA is the major photoreceptor involved in the observed phosphorylation of FHY1.
Phosphorylation of FHY1 Requires phyA to Be Functional
To further define the relationship between FHY1 phosphorylation and phyA, we tested red light–induced phosphorylation of FHY1 in several phyA point mutant backgrounds. We chose to study phyA-300D (Fry et al., 2002), phyA-302GFP/phyA-211 (Yanovsky et al., 2002), and phyA-105 (Xu et al., 1995), mutants that accumulate phyA protein to normal wild-type levels but with phenotypes similar to the phyA null mutant. A marked decrease in the level of red light–induced FHY1 phosphorylation is observed in these mutants (Figures 4A and 4B). Ser-598 (S598) of oat phyA has been shown to be phosphorylated in vivo and in vitro (McMichael and Lagarias, 1990; Lapko et al., 1999). Overexpressing wild-type oat phyA (phyAOX) or S598A point mutant phyA (phyA S598A) complements the Arabidopsis phyA-201 mutant phenotype (Kim et al., 2004). While phyA-201 does not accumulate phosphorylated FHY1 as predicted from its phyA null phenotype, we found evidence of FHY1 phosphorylation in phyA-201 mutants that were transformed with either phyAOX or phyA S598A (Figure 4C). Therefore, S598 is not critical for phyA-mediated FHY1 phosphorylation. Together, these data show the absence of FHY1 phosphorylation in situations where functional phyA is lacking as well as evidence of FHY1 phosphorylation in plants that have complementing phyA.
Analysis of phyA and FHY1 Physical Association
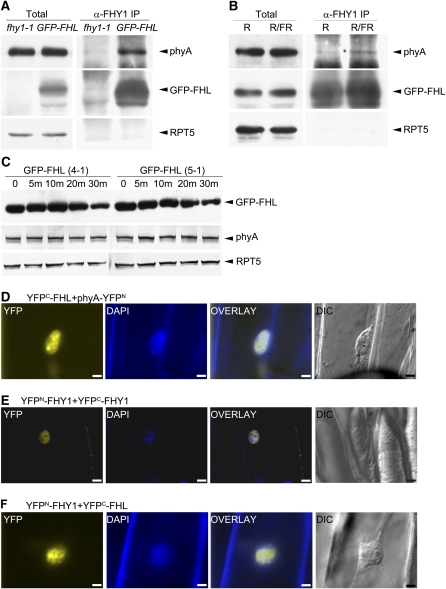
Previous studies have demonstrated the interaction between phyA and FHY1 by yeast two-hybrid assays and in vitro transcription/translation experiments (Hiltbrunner et al., 2005). Recently, Saijo et al. (2008) used co-IP to demonstrate the interaction of these two proteins in FRc-grown Arabidopsis. To rule out the possibility that interaction of proteins found by co-IP could lead to false positives as proteins in different cell compartments are brought together during the homogenization process, we performed BiFC analysis. Plasmids bearing FHY1 fused with the N-terminal half of yellow fluorescent protein (YFP) and phyA fused with the C-terminal half of YFP were introduced into onion epidermal cells by gold particle bombardment. After 24 h of incubation in darkness, followed by 5-min white light, YFP fluorescence was observed in onion cell nuclei (Figure 5A). Since the two fragments of YFP protein must come very close for fluorescence to take place, these results support that phyA and FHY1 can directly associate with each other in living plant cells. Having established this, we performed co-IP experiments using Arabidopsis seedlings. As a positive control of our experimental system, we first tested for interaction of overexpressed FHY1 with endogenous phyA in FRc-grown seedlings of 35S:GFP-FHY1 transgenic plants (in fhy1-1 background). We performed the co-IP analysis using protein A-Sepharose beads coupled with anti-FHY1 antibody. This assay clearly shows that phyA coprecipitated together with GFP-FHY1. When fhy1-1 mutant was used as a negative control, no phyA was brought down by the anti-FHY1 antibody (Figure 5B, top panel). To determine how light regulates the interaction between phyA and FHY1, we tested the level of association between the two proteins in different light conditions. The Pr/Pfr photoconversion of phyA was controlled by 5-min red light exposure (for Pfr form) or 5-min red light exposure immediately followed by 5 min of far-red light pulse (for Pr form). In our co-IP analysis, after a 5-min exposure to red light, the amount of phyA in the Pfr form found associated with GFP-FHY1 was low. However, when that was followed by a 5-min exposure to far-red light, a much larger amount of phyA coprecipitated with GFP-FHY1 (Figure 5B, bottom panel). To confirm that our system could also detect proteins that bind more selectively to phyA in red light, we analyzed the interaction of phyA with PIF3, a protein known to bind selectively to the Pfr form of both phyA and phyB in vitro (Zhu et al., 2000). Our co-IP analysis turned up the expected preferential interaction of PIF3-Myc with the Pfr form of phyA (Figure 5C), further validating our experimental system and strengthening our finding that GFP-FHY1 associates with higher amounts of the Pr form of phyA when a far-red light pulse followed a prior red light exposure.
Figure 5.
Light-Regulated Interaction of phyA and FHY1.
(A) BiFC analysis of YFPN-FHY1 and phyA-YFPC in onion epidermal cells. After bombardment, onion epidermal pieces were incubated in darkness for 24 h followed by a 5-min exposure to white light before observation by fluorescence microscopy. All images were taken at the same magnification. DAPI (4',6-diamidino-2-phenylindole) staining was used to show the positions of nuclei. Overlay: YFP and DAPI images merged. DIC: differential interference contrast image. Bars = 20 μm.
(B) Top panel: protein extract (Total) and protein immunoprecipitated with anti-FHY1 antibody (α-FHY1 IP) from 3-d-old seedlings grown in FRc light. Immunoprecipitates were analyzed by immunoblot analysis using antibodies specific for phyA, FHY1, and RPT5 (the last as a loading control). Seedlings studied were fhy1-1 as a negative control and 35S:GFP-FHY1 in fhy1-1 background. Bottom panel: 3-day-old dark-grown seedlings with 35S:GFP-FHY1 in fhy1-1 background were exposed to 5-min red light (R) or to 5-min red light followed by 5-min far-red light (R/FR) before immunoprecipitation as in the top panel.
(C) Same as the bottom panel of (B) except that 35S:PIF3-Myc seedlings were tested and proteins were immunoprecipitated with antibody to Myc tag. Anti-Myc antibody was used to detect PIF3-Myc.
Similar to FHY1, FHL Physically Associates with phyA in Vivo
FHL is the only homolog of FHY1 in the Arabidopsis genome. Previous studies suggested that FHY1 and FHL share overlapping functions in mediating phyA signaling (Zhou et al., 2005; Hiltbrunner et al., 2006; Shen et al., 2007) (see Supplemental Figure 2A online). To test if FHL can cross-complement the fhy1 mutant phenotype, we introduced a 35S:GFP-FHL transgene into the fhy1-1 mutant background. In the transgenic plants, GFP-FHL fusion protein can be detected by anti-FHY1 antibody, possibly due to the similarity between FHY1 and FHL. The extent that GFP-FHL complements fhy1-1 mutant phenotype in far-red light, as indicated by hypocotyl length, is correlated with the accumulation level of GFP-FHL protein (see Supplemental Figure 2B online). This indicates that FHL protein has the ability to functionally substitute for FHY1 as long as the protein level of FHL in the plant is sufficiently high.
We next performed a co-IP experiment using 35S:GFP-FHL transgenic plants (in fhy1-1 background) to study interaction of GFP-FHL with phyA using anti-FHY1 antibody. We found evidence of interaction of GFP-FHL with phyA in far-red light-grown seedlings (Figure 6A). More importantly, GFP-FHL shows a higher level of association with the Pr form but not the Pfr form of phyA (Figure 6B). These data suggest that GFP-FHL functions similarly to GFP-FHY1. Surprisingly, when 35S:GFP-FHL transgenic seedlings were transferred from darkness to red light, we did not detect a slow migrating band that would suggest phosphorylation of GFP-FHL (Figure 6C). This might be because our gel system does not resolve the phosphorylated and unphosphorylated GFP-FHL, or GFP-FHL is phosphorylated but to a lesser extent compared with GFP-FHY1. Another possibility is that phosphorylated GFP-FHL has a shorter half-life compared with phosphorylated GFP-FHY1. To further confirm the association of FHL and phyA, we performed the BiFC assay with YFPC-FHL and phyA-YFPN. Fluorescence due to reconstituted YFP was observed, further supporting the idea that these two proteins interact directly in living plant cells (Figure 6D). As a previous report suggested that FHY1 forms a homodimer and that FHY1 and FHL form a heterodimer (Zhou et al., 2005), we performed BiFC assays with YFPN-FHY1 and YFPC-FHY1 as well as with YFPN-FHY1 and YFPC-FHL. Observation of YFP fluorescence in onion cell nuclei for both pairs provides further evidence for the formation of homodimers by FHY1 and the heterodimerization of FHY1 and FHL (Figures 6E and 6F).
Figure 6.
Light-Regulated Interaction of phyA and FHL.
(A) Protein extract (Total) and proteins immunoprecipitated with anti-FHY1 antibody (α-FHY1 IP) from 3-d-old seedlings grown in FRc light. Immunoprecipitates were analyzed by immunoblot analysis using antibodies specific for phyA, FHY1 (to detect GFP-FHL), and RPT5 (the last as a loading control). Seedlings studied were fhy1-1 as a negative control and 35S:GFP-FHL in fhy1-1 background.
(B) Three-day-old dark-grown seedlings with 35S:GFP-FHL in fhy1-1 background were exposed to 5-min red light (R) or to 5-min red light followed by 5-min far-red light (R/FR) before immunoprecipitation as in (A).
(C) Immunoblot analysis of proteins extracted from two independent transgenic lines of 35S:GFP-FHL in fhy1-1 background (4-1 and 5-1), which were grown in darkness for 3 d and then transferred to red light for time periods indicated in minutes (m). Immunoblots were probed with anti-FHY1, phyA, and RPT5 antibodies (the latter two as loading controls).
(D) to (F) BiFC analyses of indicated protein pairs in onion epidermal cells. After bombardment, onion epidermal pieces were incubated in darkness for 24 h followed by a 5-min exposure to white light before observation by fluorescence microscopy. All images were taken at the same magnification. DAPI staining was used to show the positions of nuclei. Overlay: YFP and DAPI images merged. DIC: differential interference contrast images. Bars = 20 μm.
FHY1 Protein Is Phosphorylated by phyA in Vitro
Our co-IP and BiFC data show that phyA interacts with FHY1 (Figure 5), and our immunoblot data show that a functional phyA is critically important for FHY1 phosphorylation (Figures 3 and 4). Taken together with the ability of phytochromes to autophosphorylate their Ser/Thr residues (Yeh and Lagarias, 1998), we hypothesized that FHY1 is a direct substrate of phyA kinase activity. To test this, we set up an in vitro kinase assay to check the ability of purified phyA to phosphorylate purified glutathione S-transferase (GST)-FHY1. The phyA used in this assay is recombinant oat phyA expressed in the Pichia expression system (Kim et al., 2004). Wild-type oat phyA has been shown to be physiologically active in the Arabidopsis plant, complementing phyA deficiency and showing slightly shorter hypocotyls than wild-type Ler seedlings (Boylan and Quail, 1991; Kim et al., 2004). After 5 min of exposure of recombinant oat phyA to white light, it was incubated with GST-FHY1 and [γ-32P]ATP in kinase reaction buffer. This led to incorporation of radioactivity in both phyA and GST-FHY1, indicating that both phyA and GST-FHY1 are phosphorylated (Figure 7A). Phosphorylation of GST-FHL was not observed, although autophosphorylation of phyA was observed in the same reaction (Figure 7A). The latter result suggests that phyA does not phosphorylate GST-FHL or requires conditions different from those that worked for GST-FHY1.
Figure 7.
In Vitro Phosphorylation of FHY1 by phyA.
(A) Kinase assays to determine ability of purified recombinant oat phyA to phosphorylate purified recombinant GFP-FHY1 and GFP-FHL proteins. The presence (+) and absence (−) of phyA and potential protein substrates are indicated on the top. The first and fourth lanes are two replicate experiments, as are the second and fifth lanes.
(B) Kinase assays similar to (A) except that phyA was first exposed to 5 min of far-red or red light to obtain predominantly Pr form or Pfr form, respectively.
In (A) and (B), the top panel shows autoradiograms (autorad) of SDS-PAGE, and the bottom panel shows Coomassie blue staining of the protein gel.
[See online article for color version of this figure.]
To further investigate which form of phyA is more active in autophosphorylation and phosphorylation of GST-FHY1, we exposed phyA to either 5 min of red light to obtain the Pfr form or 5 min of far-red light to obtain the Pr form and then incubated each with GST-FHY1 and [γ-32P]ATP in the dark for 30 min. As shown in Figure 7B, phyA autophosphorylation and GST-FHY1 phosphorylation by phyA occurred at about the same levels in these two light conditions. A previous report using the same phyA preparation showed that phyA autophosphorylation activity was similar for both the Pfr form and the Pr form (Kim et al., 2004), while other reports using recombinant oat phyA from different systems showed a range of 1.2- to 2.5-fold higher autophosphorylation activity for the Pfr form of phyA (Yeh and Lagarias, 1998; Fankhauser et al., 1999; Colon-Carmona et al., 2000). These inconsistencies might be due to the different characteristics of recombinant phyA proteins and/or the use of different substrate proteins in the in vitro kinase systems. Nevertheless, it should be noted that, although both Pr and Pfr forms of phyA have the ability to phosphorylate GST-FHY1 in vitro, our in vivo data (Figures 2 and 3) clearly indicate that FHY1 phosphorylation observed in Arabidopsis seedlings is dependent on the active Pfr form of phyA, but not the Pr form.
DISCUSSION
It is well established that in Arabidopsis light signaling, phyA mediates the VLFR (Botto et al., 1996; Shinomura et al., 1996; Yanovsky et al., 1997) and HIR (Whitelam et al., 1993). phyB, on the other hand, mediates the R/FR-reversible LFR (Shinomura et al., 1996). Unlike phyB, phyA's involvement in LFR is less well understood. In this study, we observed that FHY1 undergoes rapid red light–induced phosphorylation (Figure 1) and that the phosphorylation is R/FR reversible (Figure 2), which is a typical LFR-mediated phenomenon. Furthermore, we showed that phyA, but not other phytochromes or cryptochromes, is responsible for the phosphorylation of FHY1 (Figure 3). Although the phytochrome-interacting basic helix-loop-helix transcription factors PIF3 and PIF5 are also phosphorylated in an R/FR-reversible manner, they are regulated by both phyA and phyB and predominantly function in phyB-mediated responses (Al-Sady et al., 2006; Shen et al., 2007). Therefore, the phyA-dependent phosphorylation of FHY1 by an LFR response represents a novel biochemical event that may underlie phyA-mediated physiological responses. Several phyA LFR responses have been previously reported, such as mediation of the swelling response (Long and Iino, 2001), modulation of the magnitude of phototropin-dependent phototropism (Stowe-Evans et al., 2001), and regulation of Lhbc gene expression in rice (Oryza sativa; Takano et al., 2005). Our work complements these previous studies, and more importantly, we provide clear evidence that phyA mediates an R/FR-reversible biochemical event via LFR mode and that this event may play an essential role in regulating phyA signaling.
Our data also suggest that phosphorylation may render FHY1 susceptible to 26S proteasome-mediated degradation under red light (Figure 1). Phosphorylation and targeted degradation of key signaling factors have emerged as a common theme for hormone and light signaling transduction. For example, Arabidopsis BIN2 phosphorylates BZR1 and targets it for degradation, while brassinosteroids inhibit BIN2 activity, leading to BZR1 dephosphorylation and accumulation to promote brassinosteroid signaling (He et al., 2002). In light signaling, several transcription factors, such as PIF3, PIF4, and PIF5, are all rapidly phosphorylated by exposure to light, followed by degradation by the 26S proteasome (Hardtke et al., 2000; Duek et al., 2004; Shen et al., 2005, 2007; Al-Sady et al., 2006; Lorrain et al., 2008). Interestingly, degradation of phyA also appears to be regulated by phosphorylation (Trupkin et al., 2007). Although the physiological significance for the rapid downregulation of phyA and FHY1 level under red light is not apparent at this stage, it is conceivably important, as recent studies documented that phyA has a quantitatively dominant role in red light–induced expression of early responsive genes (Tepperman et al., 2006) and that phyA also serves as an irradiance-dependent red light sensor (Franklin et al., 2007). Furthermore, it has been reported that phyA and FHY1 regulate phyB signaling (Cerdan et al., 1999). Together, these studies suggest that phyA may contribute significantly to the regulation of growth and development in daylight-grown plants. Here, our data imply that phyA-dependent FHY1 phosphorylation followed by 26S proteasome-mediated degradation in red light may be a biochemical mechanism used by Arabidopsis to desensitize FHY1-mediated phyA signaling. Further studies are required to determine the relationship between FHY1 phosphorylation, degradation, and activity regulation in red and far-red light signaling.
FHL, the only homolog of FHY1 in Arabidopsis that shares an overlapping function with FHY1 in mediating far-red light signaling (Zhou et al., 2005), is not visibly phosphorylated in response to red light or by recombinant phyA (at least in our tested conditions). This observation suggests that the stability and activity of FHY1 and FHL could be differentially regulated by phyA. It is conceivable that the regulation of FHY1 is more complex and sophisticated, since it is expressed to a much higher level and plays a more prominent role in light pathways than FHL.
Previous studies have reported that FHY1 and FHL preferentially interact with the Pfr form of phyA in a yeast two-hybrid assay and in an in vitro binding assay. Furthermore, FHY1 and FHL have been reported to colocalize with phyA in mustard seedlings (Hiltbrunner et al., 2005, 2006). In this study, we further confirmed the direct interaction between FHY1 and FHL with phyA using a BiFC assay where constitution of a fluorescent complex requires the fragments of the fluorescent protein to be tethered through a specific interaction between the fusion proteins (Hu and Kerppola, 2003). Thus, the reconstituted YFP fluorescence between YFPC-FHY1 (or FHL) and phyA-YFPN in living plant cells (Figures 5 and 6) provides strong supporting evidence for direct physical interaction between phyA and FHY1 as well as between phyA and FHL. Intriguingly, our co-IP assay using Arabidopsis seedlings revealed that FHY1 and FHL preferably bind the Pr form of phyA (Figures 5 and 6), whereas previously the in vitro binding assays of Hiltbrunner et al. (2005, 2006) showed stronger interaction of these proteins with the Pfr form of phyA. Our co-IP assay is validated by the fact that it shows that PIF3 selectively interacts with the Pfr form of phyA (Figure 5), which is consistent with a previous report (Zhu et al., 2000). These contrasting observations can possibly be explained as follows. First, in our co-IP system, only GFP-FHY1 and GFP-FHL were overexpressed and phyA was at an endogenous level, unlike that in the previous in vitro binding systems. Second, the presence of other proteins (e.g., other phyA interacting proteins) in the plant extracts but not in the in vitro system may modulate the binding of FHY1 and FHL with phyA under our tested conditions. A third possibility is that the interaction of FHY1 and FHL with phyA may be affected by chemical modifications of FHY1/FHL or phyA not present in the in vitro binding system. In fact, a recent study showed that in FRc, unphosphorylated phyA interacts with FHY1/FHY3 more stably, whereas phosphorylated phyA preferentially associates with the COP1/SPA1 complex (Saijo et al., 2008). It is well known that phyA and FHY1/FHL act mainly in far-red light signaling and that the majority (97%) of phyA exists in the Pr form in far-red light. Thus, the more stable binding of FHY1 and FHL with the Pr form of phyA may facilitate the rapid nuclear translocation of phyA upon its exposure to far-red light.
At the same time, it should be noted that the binding of FHY1 with the Pfr form of phyA, although weaker than that with the Pr form (Figure 5), might also be biologically significant, as red light–induced FHY1 phosphorylation might be directly mediated by phyA. This notion is supported by the observations that (1) FHY1 phosphorylation is abrogated in certain phyA point mutant backgrounds where far-red signaling is compromised despite normal accumulation of phyA protein (Figure 4); and (2) recombinant phyA exhibits kinase activity toward recombinant FHY1 (Figure 7). In this regard, it is worth pointing out that previous studies have already identified several potential substrates for phyA kinase activity, such as PKS1, NDPK2, cryptochromes, and the AUX/IAA proteins (Ahmad et al., 1998b; Choi et al., 1999; Fankhauser et al., 1999; Colon-Carmona et al., 2000; Zhu et al., 2000; Seo et al., 2004; Hiltbrunner et al., 2005, 2006). In this study, we demonstrate that phyA might be solely responsible for the observed R/FR-reversible FHY1 phosphorylation.
In summary, our data suggest a dual mechanism of FHY1 regulation: its stable interaction with the Pr form of phyA in far-red light and its rapid phosphorylation by the Pfr form of phyA in red light. This could potentially explain the more rapid degradation of FHY1 in red light conditions (majority of phyA exists in Pfr form in red light compared with far-red light conditions). Whether phosphorylation of FHY1 serves additional regulatory functions in phyA signaling awaits further investigation.
METHODS
Plant Materials and Growth Conditions
The wild-type Arabidopsis thaliana used in this study is of the Ler ecotype, unless otherwise indicated. The fhy1-4 mutant was obtained from the SAIL T-DNA collection (SAIL_291_E01) (Sessions et al., 2002). The phyA-1 (Whitelam et al., 1993), phyA-105 (Xu et al., 1995), phyA-301, phyA-300D (Fry et al., 2002), phyA-201 (Reed et al., 1994), phyB-1 (Reed et al., 1993), phyB phyD phyE (Franklin et al., 2003), cry1-304, cry1-304 cry2-1 (Mockler et al., 1999), fhy1-1 (Desnos et al., 2001), and fhl-1 (Zhou et al., 2005) mutants and the 35S:GFP-FHY1 (Shen et al., 2005), phyA-302GFP/phyA-211 (Yanovsky et al., 2002), phyAOX, phyA S598A (Kim et al., 2004), and 35S:PIF3-MYC (Park et al., 2004) transgenic plants have been described previously. In the text and figures, phyBDE is used to indicate phyB phyD phyE, cry1 is used to indicate cry1-304, and cry2 is used to indicate cry2-1. Growth conditions were as described previously (Shen et al., 2005) with fluence rates of 148 μmol m−2 s−1 for white light, 110 μmol m−2 s−1 for far-red light, 168 μmol m−2 s−1 for red light, and 4 μmol m−2 s−1 for blue light. Light treatment experiments were performed by transferring Arabidopsis seedlings between different light conditions (such as dark, far red, red, and blue) as specified in Results and in figure legends.
Plasmid Construction and Generation of Transgenic Arabidopsis Plants
The full-length cDNA of FHL was amplified by RT-PCR using SuperScript II reverse transcriptase (Invitrogen) and Herculase DNA polymerase (Stratagene), with a forward primer containing a BamHI restriction site (5′-CGCGGATCCGCGATGATAGTTGCTGTGGAATCTCTAGACACA-3′) and a reverse primer containing a NotI restriction site (5′-TTTTCCTTTTGCGGCCGCTTTTTTCCTTCCCATCATGAGTGTAGAAAAGTACTGCTCAAA-3′) and inserted between the BamHI and NotI sites of pGEX-4T1vector (Amersham Biosciences). Then, a BamHI-SpeI fragment of FHL cDNA (amplified by PCR using primers 5′-CGCGGATCCGCGATGATAGTTGCTGTGGAATCTCTAGACACA-3′ and 5′-GGACTAGTCCTTACATCATGAGTGTAGAAAAGTACTGCTCAAA-3′) was subcloned into the BglII and XbaI sites of pRTL2-mGFP (S65T) vector (Torii et al., 1998). For generating the 35S:GFP-FHL construct, a HindIII fragment (containing 35S promoter, transgene, and 3′ end terminator) was subcloned from the above-mentioned pRTL2 construct into pPZP221 (Hajdukiewicz et al., 1994). The 35S:GFP-FHL construct was introduced into the fhy1-1 background via Agrobacterium tumefaciens–mediated transformation (Clough and Bent, 1998). Transgenic plants were selected with Gentamicin (200 μg/mL) (Sigma-Aldrich).
Immunoblot Assays, Antibodies, and Co-IP
Arabidopsis tissues were homogenized in an extraction buffer containing 50 mM Tris-HCl, pH 7.5, 200 mM NaCl, 1 mM EDTA, 10 mM NaF, 2 mM Na3VO4, 25 mM β-glycerolphosphate, 10% glycerol, 0.1% Tween 20, 1 mM DTT, 1mM phenylmethylsulfonyl fluoride, 1× complete protease inhibitor cocktail (Roche), and 40 μM MG132 (EMD Chemicals). For experiments testing the effect of 26S proteasome inhibitors, seedlings were treated with 100 μM MG132 or DMSO only (solvent for MG132) for 2 h followed by different light treatments.
Immunoblotting was performed as previously described (Feng et al., 2004) except that proteins were separated in precast 10% Bis-Tris NuPAGE gels using MOPS buffer as directed by the manufacturer (Invitrogen).
Primary antibodies used in this study include anti-FHY1 (Shen et al., 2005), anti-RPT5 (Kwok et al., 1999), anti-phyA (Xu et al., 1995), and anti-Myc (Cell Signaling) antibodies. The dilutions used were 1:5000 for anti-phyA and 1:1000 for anti-FHY1, anti-RPT5, and anti-Myc antibodies. Anti-mouse IgG antibody conjugated with peroxidase (Sigma-Aldrich) was used as the secondary antibody against anti-phyA and anti-Myc antibodies. Anti-rabbit IgG antibody conjugated with peroxidase (Pierce Scientific) was used as the secondary antibody against anti-FHY1 antibody. Anti-rabbit IgG antibody conjugated with alkaline phosphatase (Sigma-Aldrich) was used as the secondary antibody against anti-RPT5 antibody. The dilution used for secondary antibodies was 1:8000. Immunoblots hybridized with peroxidase-conjugated secondary antibodies were incubated with enhanced chemiluminescence reagents (Pierce Scientific), and signals were detected by BioMax XAR film (Eastman Kodak). Immunoblots hybridized with alkaline phosphatase–conjugated secondary antibodies were visualized by incubating with nitro-blue tetrazolium chloride and 5-bromo-4-chloro-3-indolyphosphate toluidine salt (Sigma-Aldrich).
The co-IP experiments were performed as previously described (Feng et al., 2004) with the following minor modifications. Protein A-Sepharose 4B Fast Flow beads (Sigma-Aldrich) coupled with purified anti-FHY1 antibody (coupling was performed according to a protocol reported in Staub et al., 1996) or anti-Myc antibody conjugated beads (Covance) were used to precipitate the respective proteins. All steps were performed in a darkroom with dim green safe light.
In Vitro Kinase Assays
Recombinant oat (Avena sativa) phytochrome A protein, purified from the Pichia expression system, is a gift from Pill-Soon Song (Gyeongsang National University, Korea) (Kim et al., 2004). The in vitro kinase assays were performed as described previously (Yeh and Lagarias, 1998) with the following minor modifications. Assays were done in a reaction mixture (30 μL) that contains 25 mM Tris-Cl, pH 7.5, 0.2 mM EDTA, 4 mM DTT, 5 mM MgCl2, 100 μM ATP, 2 μg purified GST-FHY1, GST-FHL, or GST alone, and 15 μCi [γ-32P]ATP. The GST fusion proteins were expressed in Escherichia coli and purified as described previously (Shen et al., 2005). The reaction was started by adding 1 μg purified recombinant oat phyA after 5 min of light treatment as specified for each experiment. The reaction was then stopped by adding 6× SDS-PAGE sample loading buffer after incubation for 30 min at 30°C. The samples were subsequently analyzed using 10% SDS-PAGE gels and visualized by Coomassie Brilliant Blue staining followed by autoradiography using BioMax XAR film (Eastman Kodak).
BiFC Assay
All vectors used in the BiFC assays were derived from pSY728, pSY735, pSY736, and pSY738 plasmids that were described previously (Bracha-Drori et al., 2004). The full-length open reading frames of FHY1, FHL, and PHYA were amplified by PCR, and the PCR products were cloned into the pSY vectors containing either N-terminal (1 to 155 amino acids) or C-terminal (156 to 239 amino acid) fragments of the YFP fluorescent protein (YFPN and YFPC) (see Supplemental Table 1 online). Possible pairwise combinations (see Supplemental Table 2 online) of these constructs (1 μg of each plasmid coated onto 1-μm gold particles) (Bio-Rad) were cobombarded into onion epidermal pieces on agar plates containing 1× Murashige and Skoog salts (Sigma-Aldrich) using a Biolistic PDS-1000/He system (Bio-Rad). YFP fluorescence was visualized using a Diaphot 200 inverted fluorescence microscope (Nikon) with appropriate filter sets for detection of YFP (exciter, HQ 500/20; emitter, HQ 535/30). Images were recorded with a Coolsnap HQ CCD digital camera (Roper Scientific) controlled by MetaMorph software (Molecular Devices).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL database under accession numbers NM_001084549 (FHY1), NM_120298 (FHL), and NM_100828 (PHYA).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. FHY1 Protein in Various Genetic Backgrounds.
Supplemental Figure 2. Characterization of fhl Mutants with Respect to fhy1 and phyA Mutants.
Supplemental Table 1. Constructs Used for Bimolecular Fluorescence Complementation.
Supplemental Table 2. Pairwise Combinations of Constructs Used for Bimolecular Fluorescence Complementation.
Supplementary Material
Acknowledgments
We thank Peter Quail for anti-phyA antibody and phyA-105 seeds, Jorge J. Casal for phyA-302GFP/phyA-211 seeds, Pill-Soon Song for phyAOX, phyA S598A seeds and recombinant oat phyA protein, Giltsu Choi for 35S:PIF3-MYC seeds, and Nir Ohad for pSY728, pSY735, pSY736, and pSY738 plasmids. This work was supported by a National Institutes of Health grant (GM047850) to X.W.D.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Xing Wang Deng (xingwang.deng@yale.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Ahmad, M., Jarillo, J.A., and Cashmore, A.R. (1998. a). Chimeric proteins between cry1 and cry2 Arabidopsis blue light photoreceptors indicate overlapping functions and varying protein stability. Plant Cell 10 197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad, M., Jarillo, J.A., Smirnova, O., and Cashmore, A.R. (1998. b). The CRY1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Mol. Cell 1 939–948. [DOI] [PubMed] [Google Scholar]
- Al-Sady, B., Ni, W., Kircher, S., Schafer, E., and Quail, P.H. (2006). Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23 439–446. [DOI] [PubMed] [Google Scholar]
- Botto, J.F., Sanchez, R.A., Whitelam, G.C., and Casal, J.J. (1996). Phytochrome A mediates the promotion of seed germination by very low fluences of light and canopy shade light in Arabidopsis. Plant Physiol. 110 439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan, M.T., and Quail, P.H. (1991). Phytochrome a overexpression inhibits hypocotyl elongation in transgenic Arabidopsis. Proc. Natl. Acad. Sci. USA 88 10806–10810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracha-Drori, K., Shichrur, K., Katz, A., Oliva, M., Angelovici, R., Yalovsky, S., and Ohad, N. (2004). Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J. 40 419–427. [DOI] [PubMed] [Google Scholar]
- Castillon, A., Shen, H., and Huq, E. (2007). Phytochrome interacting factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 12 514–521. [DOI] [PubMed] [Google Scholar]
- Cerdan, P.D., Yanovsky, M.J., Reymundo, F.C., Nagatani, A., Staneloni, R.J., Whitelam, G.C., and Casal, J.J. (1999). Regulation of phytochrome B signaling by phytochrome A and FHY1 in Arabidopsis thaliana. Plant J. 18 499–507. [DOI] [PubMed] [Google Scholar]
- Choi, G., Yi, H., Lee, J., Kwon, Y.K., Soh, M.S., Shin, B., Luka, Z., Hahn, T.R., and Song, P.S. (1999). Phytochrome signalling is mediated through nucleoside diphosphate kinase 2. Nature 401 610–613. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Colon-Carmona, A., Chen, D.L., Yeh, K.C., and Abel, S. (2000). Aux/IAA proteins are phosphorylated by phytochrome in vitro. Plant Physiol. 124 1728–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong, A. (2006). Switching the flip: protein phosphatase roles in signaling pathways. Curr. Opin. Plant Biol. 9 470–477. [DOI] [PubMed] [Google Scholar]
- Desnos, T., Puente, P., Whitelam, G.C., and Harberd, N.P. (2001). FHY1: A phytochrome A-specific signal transducer. Genes Dev. 15 2980–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duek, P.D., Elmer, M.V., van Oosten, V.R., and Fankhauser, C. (2004). The degradation of HFR1, a putative bHLH class transcription factor involved in light signaling, is regulated by phosphorylation and requires COP1. Curr. Biol. 14 2296–2301. [DOI] [PubMed] [Google Scholar]
- Fankhauser, C., Yeh, K.C., Lagarias, J.C., Zhang, H., Elich, T.D., and Chory, J. (1999). PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 284 1539–1541. [DOI] [PubMed] [Google Scholar]
- Feng, S., Shen, Y., Sullivan, J.A., Rubio, V., Xiong, Y., Sun, T.P., and Deng, X.W. (2004). Arabidopsis CAND1, an unmodified CUL1-interacting protein, is involved in multiple developmental pathways controlled by ubiquitin/proteasome-mediated protein degradation. Plant Cell 16 1870–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin, K.A., Allen, T., and Whitelam, G.C. (2007). Phytochrome A is an irradiance-dependent red light sensor. Plant J. 50 108–117. [DOI] [PubMed] [Google Scholar]
- Franklin, K.A., Praekelt, U., Stoddart, W.M., Billingham, O.E., Halliday, K.J., and Whitelam, G.C. (2003). Phytochromes B, D, and E act redundantly to control multiple physiological responses in Arabidopsis. Plant Physiol. 131 1340–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry, R.C., Habashi, J., Okamoto, H., and Deng, X.W. (2002). Characterization of a strong dominant phytochrome A mutation unique to phytochrome A signal propagation. Plant Physiol. 130 457–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25 989–994. [DOI] [PubMed] [Google Scholar]
- Hardtke, C.S., Gohda, K., Osterlund, M.T., Oyama, T., Okada, K., and Deng, X.W. (2000). HY5 stability and activity in arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J. 19 4997–5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, J.X., Gendron, J.M., Yang, Y., Li, J., and Wang, Z.Y. (2002). The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 99 10185–10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltbrunner, A., Tscheuschler, A., Viczian, A., Kunkel, T., Kircher, S., and Schafer, E. (2006). FHY1 and FHL act together to mediate nuclear accumulation of the phytochrome A photoreceptor. Plant Cell Physiol. 47 1023–1034. [DOI] [PubMed] [Google Scholar]
- Hiltbrunner, A., Viczian, A., Bury, E., Tscheuschler, A., Kircher, S., Toth, R., Honsberger, A., Nagy, F., Fankhauser, C., and Schafer, E. (2005). Nuclear accumulation of the phytochrome A photoreceptor requires FHY1. Curr. Biol. 15 2125–2130. [DOI] [PubMed] [Google Scholar]
- Hu, C.D., and Kerppola, T.K. (2003). Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 21 539–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, M., Ringli, C., Boylan, M.T., and Quail, P.H. (1999). The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes Dev. 13 2017–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D.H., Kang, J.G., Yang, S.S., Chung, K.S., Song, P.S., and Park, C.M. (2002). A phytochrome-associated protein phosphatase 2A modulates light signals in flowering time control in Arabidopsis. Plant Cell 14 3043–3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.I., Shen, Y., Han, Y.J., Park, J.E., Kirchenbauer, D., Soh, M.S., Nagy, F., Schafer, E., and Song, P.S. (2004). Phytochrome phosphorylation modulates light signaling by influencing the protein-protein interaction. Plant Cell 16 2629–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok, S.F., Staub, J.M., and Deng, X.W. (1999). Characterization of two subunits of Arabidopsis 19S proteasome regulatory complex and its possible interaction with the COP9 complex. J. Mol. Biol. 285 85–95. [DOI] [PubMed] [Google Scholar]
- Lapko, V.N., Jiang, X.Y., Smith, D.L., and Song, P.S. (1999). Mass spectrometric characterization of oat phytochrome A: Isoforms and posttranslational modifications. Protein Sci. 8 1032–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, R., Ding, L., Casola, C., Ripoll, D.R., Feschotte, C., and Wang, H. (2007). Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318 1302–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, C., and Iino, M. (2001). Light-dependent osmoregulation in pea stem protoplasts. photoreceptors, tissue specificity, ion relationships, and physiological implications. Plant Physiol. 125 1854–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain, S., Allen, T., Duek, P.D., Whitelam, G.C., and Fankhauser, C. (2008). Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53 312–323. [DOI] [PubMed] [Google Scholar]
- McMichael, R.W., Jr., and Lagarias, J.C. (1990). Phosphopeptide mapping of Avena phytochrome phosphorylated by protein kinases in vitro. Biochemistry 29 3872–3878. [DOI] [PubMed] [Google Scholar]
- Mockler, T.C., Guo, H., Yang, H., Duong, H., and Lin, C. (1999). Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126 2073–2082. [DOI] [PubMed] [Google Scholar]
- Osterlund, M.T., Wei, N., and Deng, X.W. (2000). The roles of photoreceptor systems and the COP1-targeted destabilization of HY5 in light control of Arabidopsis seedling development. Plant Physiol. 124 1520–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, E., Kim, J., Lee, Y., Shin, J., Oh, E., Chung, W.I., Liu, J.R., and Choi, G. (2004). Degradation of phytochrome interacting factor 3 in phytochrome-mediated light signaling. Plant Cell Physiol. 45 968–975. [DOI] [PubMed] [Google Scholar]
- Reed, J.W. (1999). Phytochromes are Pr-ipatetic kinases. Curr. Opin. Plant Biol. 2 393–397. [DOI] [PubMed] [Google Scholar]
- Reed, J.W., Nagatani, A., Elich, T.D., Fagan, M., and Chory, J. (1994). Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 104 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, J.W., Nagpal, P., Poole, D.S., Furuya, M., and Chory, J. (1993). Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu, J.S., et al. (2005). Phytochrome-specific type 5 phosphatase controls light signal flux by enhancing phytochrome stability and affinity for a signal transducer. Cell 120 395–406. [DOI] [PubMed] [Google Scholar]
- Saijo, Y., Sullivan, J.A., Wang, H., Yang, J., Shen, Y., Rubio, V., Ma, L., Hoecker, U., and Deng, X.W. (2003). The COP1–SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 17 2642–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo, Y., Zhu, D., Li, J., Rubio, V., Zhou, Z., Shen, Y., Hoecker, U., Wang, H., and Deng, X.W. (2008). Arabidopsis COP1/SPA1 complex and FHY1/FHY3 associate with distinct phosphorylated forms of phytochrome A in balancing light signaling. Mol. Cell 31 607–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, H.S., Watanabe, E., Tokutomi, S., Nagatani, A., and Chua, N.H. (2004). Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev. 18 617–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions, A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock, R.A., and Quail, P.H. (1989). Novel phytochrome sequences in Arabidopsis thaliana: Structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev. 3 1745–1757. [DOI] [PubMed] [Google Scholar]
- Shen, Y., Feng, S., Ma, L., Lin, R., Qu, L.J., Chen, Z., Wang, H., and Deng, X.W. (2005). Arabidopsis FHY1 protein stability is regulated by light via phytochrome A and 26S proteasome. Plant Physiol. 139 1234–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Y., Khanna, R., Carle, C.M., and Quail, P.H. (2007). Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol. 145 1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura, T., Nagatani, A., Hanzawa, H., Kubota, M., Watanabe, M., and Furuya, M. (1996). Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 93 8129–8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura, T., Uchida, K., and Furuya, M. (2000). Elementary processes of photoperception by phytochrome A for high-irradiance response of hypocotyl elongation in Arabidopsis. Plant Physiol. 122 147–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub, J.M., Wei, N., and Deng, X.W. (1996). Evidence for FUS6 as a component of the nuclear-localized COP9 complex in Arabidopsis. Plant Cell 8 2047–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowe-Evans, E.L., Luesse, D.R., and Liscum, E. (2001). The enhancement of phototropin-induced phototropic curvature in Arabidopsis occurs via a photoreversible phytochrome A-dependent modulation of auxin responsiveness. Plant Physiol. 126 826–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano, M., Inagaki, N., Xie, X., Yuzurihara, N., Hihara, F., Ishizuka, T., Yano, M., Nishimura, M., Miyao, A., Hirochika, H., and Shinomura, T. (2005). Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice. Plant Cell 17 3311–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperman, J.M., Hwang, Y.S., and Quail, P.H. (2006). phyA dominates in transduction of red-light signals to rapidly responding genes at the initiation of Arabidopsis seedling de-etiolation. Plant J. 48 728–742. [DOI] [PubMed] [Google Scholar]
- Torii, K.U., McNellis, T.W., and Deng, X.W. (1998). Functional dissection of Arabidopsis COP1 reveals specific roles of its three structural modules in light control of seedling development. EMBO J. 17 5577–5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trupkin, S.A., Debrieux, D., Hiltbrunner, A., Fankhauser, C., and Casal, J.J. (2007). The serine-rich N-terminal region of Arabidopsis phytochrome A is required for protein stability. Plant Mol. Biol. 63 669–678. [DOI] [PubMed] [Google Scholar]
- Wang, H., and Deng, X.W. (2003). Dissecting the phytochrome A-dependent signaling network in higher plants. Trends Plant Sci. 8 172–178. [DOI] [PubMed] [Google Scholar]
- Wang, H., Ma, L., Habashi, J., Li, J., Zhao, H., and Deng, X.W. (2002). Analysis of far-red light-regulated genome expression profiles of phytochrome A pathway mutants in Arabidopsis. Plant J. 32 723–733. [DOI] [PubMed] [Google Scholar]
- Whitelam, G.C., Johnson, E., Peng, J., Carol, P., Anderson, M.L., Cowl, J.S., and Harberd, N.P. (1993). Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5 757–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y., Parks, B.M., Short, T.W., and Quail, P.H. (1995). Missense mutations define a restricted segment in the C-terminal domain of phytochrome A critical to its regulatory activity. Plant Cell 7 1433–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky, M.J., Casal, J.J., and Luppi, J.P. (1997). The VLF loci, polymorphic between ecotypes Landsberg erecta and Columbia, dissect two branches of phytochrome A signal transduction that correspond to very-low-fluence and high-irradiance responses. Plant J. 12 659–667. [DOI] [PubMed] [Google Scholar]
- Yanovsky, M.J., Luppi, J.P., Kirchbauer, D., Ogorodnikova, O.B., Sineshchekov, V.A., Adam, E., Kircher, S., Staneloni, R.J., Schafer, E., Nagy, F., and Casal, J.J. (2002). Missense mutation in the PAS2 domain of phytochrome A impairs subnuclear localization and a subset of responses. Plant Cell 14 1591–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky, M.J., Whitelam, G.C., and Casal, J.J. (2000). fhy3-1 retains inductive responses of phytochrome A. Plant Physiol. 123 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh, K.C., and Lagarias, J.C. (1998). Eukaryotic phytochromes: Light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc. Natl. Acad. Sci. USA 95 13976–13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Q., Hare, P.D., Yang, S.W., Zeidler, M., Huang, L.F., and Chua, N.H. (2005). FHL is required for full phytochrome A signaling and shares overlapping functions with FHY1. Plant J. 43 356–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y., Tepperman, J.M., Fairchild, C.D., and Quail, P.H. (2000). Phytochrome B binds with greater apparent affinity than phytochrome A to the basic helix-loop-helix factor PIF3 in a reaction requiring the PAS domain of PIF3. Proc. Natl. Acad. Sci. USA 97 13419–13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.