Abstract
Ubiquitination is involved in a variety of biological processes, but the exact role of ubiquitination in abiotic responses is not clearly understood in higher plants. Here, we investigated Rma1H1, a hot pepper (Capsicum annuum) homolog of a human RING membrane-anchor 1 E3 ubiquitin (Ub) ligase. Bacterially expressed Rma1H1 displayed E3 Ub ligase activity in vitro. Rma1H1 was rapidly induced by various abiotic stresses, including dehydration, and its overexpression in transgenic Arabidopsis thaliana plants conferred strongly enhanced tolerance to drought stress. Colocalization experiments with marker proteins revealed that Rma1H1 resides in the endoplasmic reticulum (ER) membrane. Overexpression of Rma1H1 in Arabidopsis inhibited trafficking of an aquaporin isoform PIP2;1 from the ER to the plasma membrane and reduced PIP2;1 levels in protoplasts and transgenic plants. This Rma1H1-induced reduction of PIP2;1 was inhibited by MG132, an inhibitor of the 26S proteasome. Furthermore, Rma1H1 interacted with PIP2;1 in vitro and ubiquitinated it in vivo. Similar to Rma1H1, Rma1, an Arabidopsis homolog of Rma1H1, localized to the ER, and its overexpression reduced the PIP2;1 protein level and inhibited trafficking of PIP2;1 from the ER to the plasma membrane in protoplasts. In addition, reduced expression of Rma homologs resulted in the increased level of PIP2;1 in protoplasts. We propose that Rma1H1 and Rma1 play a critical role in the downregulation of plasma membrane aquaporin levels by inhibiting aquaporin trafficking to the plasma membrane and subsequent proteasomal degradation as a response to dehydration in transgenic Arabidopsis plants.
INTRODUCTION
Higher plants are routinely confronted with diverse abiotic stresses throughout their life cycle and have developed unique acclimation mechanisms that increase their tolerance to these stresses. A large and increasing number of stress-inducible genes have been identified by the combination of molecular and genomic methods (Cushman and Bohnert, 2000; Oono et al., 2003; Bohnert et al., 2006; Vij and Tyagi., 2007), but the functional mechanisms of these genes with regard to either stress tolerance or sensitivity in crop plants are largely unknown. Therefore, it is critical to uncover the roles of stress-related genes to develop transgenic crops that have improved tolerance to unfavorable growth conditions. Among these stresses, water deficiency causes a marked reduction of crop yield on as much as half of the world's irrigated land (Boyer, 1982; Cushman and Bohnert, 2000). Diverse genetic and cellular processes that occur under water stress have been widely documented (Bray, 1997; Zhu, 2002; Shinozaki and Yamaguchi-Shinozaki, 2007; Vij and Tyagi, 2007).
Ubiquitination is the posttranslational attachment of ubiquitin (Ub), a highly conserved 8-kD protein, to a wide range of target proteins (Pickart and Eddins, 2004; Mukhopadhyay and Riezman, 2007). This pathway has been found in all eukaryotic cells and plays important roles in the regulation of cellular functions as diverse as cell cycle progression, defense against biotic and abiotic stresses, embryogenesis, endocytosis, hormone responses, protein sorting, and senescence. The best-studied ubiquitination pathway in higher plants is the Ub-26S proteasome system, which leads to rapid degradation of substrate proteins (Vierstra, 2003; Moon et al., 2004; Smalle and Vierstra, 2004; Dreher and Callis, 2007). Ubiquitination can also alter stability, activity, protein–protein interactions, and subcellular localization of target proteins (Pickart and Eddins, 2004; Mukhopadhyay and Riezman, 2007). In the ubiquitination pathway, Ub is attached to substrate proteins in three consecutive steps catalyzed by E1, E2, and E3 enzymes (Kraft et al., 2005; Stone et al., 2005). In higher plants, E3 Ub ligases are encoded by a large gene family comprised of diverse isoforms. Based on the subunit composition, E3s can be classified into two main groups. The HECT and RING/U-box E3 classes consist of a single subunit, whereas the SCF and anaphase-promoting complex E3 ligases consist of multiple polypeptides (Vierstra, 2003; Moon et al., 2004; Smalle and Vierstra, 2004). Regardless of their subunit composition, E3 Ub ligases appear to determine the target specificity, identifying which proteins will be ubiquitinated.
As an initial effort to develop crop plants that are tolerant to drought stress, we previously identified a broad spectrum of cDNAs that were enhanced rapidly in response to dehydration in hot pepper plants (Capsicum annuum) (Park et al., 2003; Hong and Kim, 2005). Among the identified cDNAs, pCa-DI6 encodes a partial protein homologous to a RING domain-containing E3 Ub ligase. In this study, we isolated a full-length Ca-DI6, renamed Rma1H1 for RING membrane-anchor 1 homolog 1. We present results indicating that Rma1H1 is an endoplasmic reticulum (ER) membrane-associated E3 Ub ligase and that overexpression of Rma1H1 confers markedly increased tolerance to severe water deficit in transgenic Arabidopsis plants. To gain further insight into the physiological role of Rma1H1 in relation to stress tolerance, we investigated its possible role in ubiquitination-mediated downregulation of water stress-related proteins and demonstrated that Ram1H1 plays a critical role in lowering the level of PIP2;1, an Arabidopsis plasma membrane–localized water channel protein aquaporin, upon dehydration stress.
RESULTS
Characterization of Rma1H1
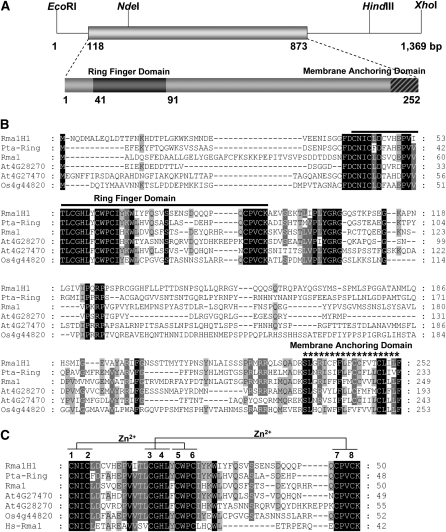
The hot pepper Rma1H1 is predicted to be 28.2 kD with a calculated pI of 7.3 (Figure 1A). A database search revealed that Rma1H1 is 43 and 34% identical to the poplar Pta-Ring and Arabidopsis thaliana RING membrane-anchor 1 (Rma1) (Matsuda and Nakano, 1998) proteins, respectively (Figure 1B). In addition, Rma1H1 shares a significant degree of sequence identity with the Arabidopsis Rma2 (30% identity) and Rma3 (29% identity) and rice (Oryza sativa) RING (29% identity) proteins, whose cellular functions are unknown. Intriguingly, Rma1H1 showed a considerable degree of sequence identity (22% identity) with human RING membrane-anchor 1 protein (Hs-Rma1). Rma1H1 possesses a single RING motif near the N-terminal region, with 57 to 73% identity with the corresponding domain in plant RING proteins (Figure 1C). Rma1H1 also contains a single putative membrane spanning domain in its extreme C terminus, suggesting that it is a membrane-associated protein.
Figure 1.
Sequence Analysis of Hot Pepper Rma1H1.
(A) Restriction enzyme map analysis and schematic structure of the hot pepper Rma1H1 cDNA clone and predicted Rma1H1 protein. Solid bar represents the coding region. Solid lines depict 5′- and 3′-untranslated regions. Dark bar indicates N-terminal RING motif, while hatched bar shows C-terminal membrane anchoring domain.
(B) Comparison of the derived amino acid sequence of hot pepper Rma1H1 with those of the poplar Pta-Ring protein, Arabidopsis RING membrane anchor 1 (Rma1), Rma2 (At4g28270), and Rma3 (At4g27470) proteins, and rice RING (Os4g44820) protein. Amino acid residues that are conserved in at least four of the six sequences are shaded, while amino acids that are identical in all six proteins are shown in black. The solid line denotes the N-terminal RING motif, which is essential for E3 Ub ligase activity. The C-terminal putative membrane anchoring sequence is indicated by an asterisk. Dashes show gaps in the amino acid sequences that were introduced to optimize alignment.
(C) Sequence alignment of the RING domain of Rma1H1 and other RING proteins. The sequences of RING motifs in hot pepper Rma1H1, Arabidopsis Rma1, Rma2, and Rma3, poplar Pta-Ring protein, rice RING protein, and human Hs-Rma1 are shown. Amino acid residues that are conserved in at least four of the seven sequences are shaded. Amino acids that are identical in all seven proteins are shown in black. Putative Zn2+-interacting amino acid residues are indicated. The numbers on the right indicate the amino acid residues. Dashes show gaps in the amino acid sequences that were introduced to optimize alignment.
Rma1H1 Possesses an E3 Ub Ligase Activity in Vitro
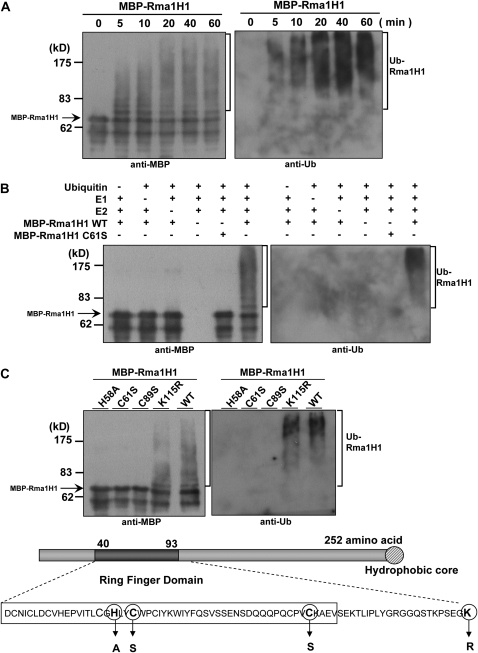
Many RING-containing proteins function as E3 Ub ligases (Kraft et al., 2005; Stone et al., 2005). To test whether Rma1H1 has E3 Ub ligase activity, the full-length Rma1H1 was expressed in Escherichia coli as a fusion protein with maltose binding protein (MBP). The purified MBP-Rma1H1 protein was incubated at 30°C, in the presence or absence of Ub, ATP, E1 (Arabidopsis UBA1), and E2 (Arabidopsis UBC8), for various time points and subjected to immunoblot analysis with anti-MBP or anti-Ub antibody. As shown in Figure 2A, MBP-Rma1H1 gave rise to high molecular mass ubiquitinated smear ladders in a time-dependent manner, while there was no ubiquitinated signal when E1, E2, or Ub was absent (Figure 2B). We next constructed single amino acid substitution mutants of MBP-Rma1H1, in which the His58, Cys61, and Cys89 residues in RING domain were replaced with Ala, Ser, and Ser residues, respectively. When purified, the mutant proteins were almost completely deficient in Ub ligase activity (Figures 2B and 2C). By contrast, mutation of Lys115 to Arg115 did not exert any inhibitory effect on Ub ligase activity, suggesting the specificity of our in vitro ubiquitination enzyme assay. Thus, it appears that Rma1H1 possesses E3 Ub ligase enzyme activity.
Figure 2.
In Vitro Self-Ubiquitination Assay of Rma1H1.
(A) The bacterially expressed MBP-Rma1H1 fusion protein was incubated for the indicated time periods in the presence of E1, E2, ATP, and Ub. Samples were resolved by 8% SDS-PAGE and subjected to immunoblot analysis with anti-MBP antibody (left pane) or anti-Ub antibody (right panel).
(B) MBP-Rma1H1 and MBP-Rma1H1C61S mutant protein were incubated at 30°C for 60 min in the presence or absence of E1, E2, and/or Ub. Samples were analyzed as described above.
(C) Wild-type MBP-Rma1H1 and single amino acid substitution mutants were used in the E3 Ub ligase enzyme assay. Amino acid residues in the RING motif that are used for the substitution mutations are indicated. Arrows indicate nonubiquitinated MBP-Rma1H1.
Expression of the Rma1H1 Gene in Response to Diverse Environmental Factors
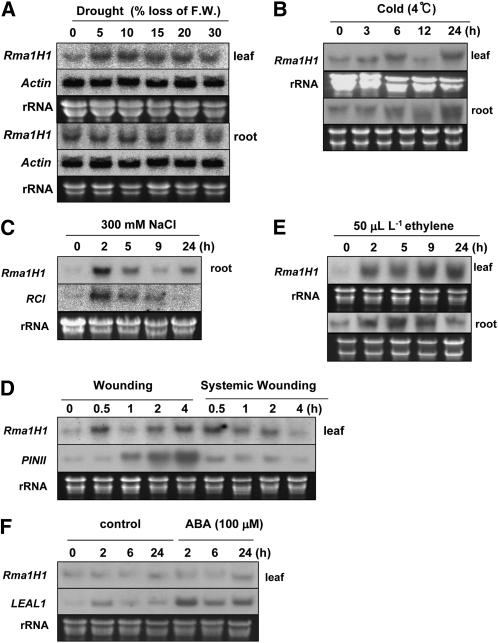
Since Rma1H1 was identified initially as a drought-induced gene (Park et al., 2003), we considered the possibility that the expression of Rma1H1 is regulated by abiotic stresses in hot pepper plants. To test this possibility, its mRNA accumulation profiles were monitored under various stress conditions by RNA gel blot analysis. As shown in Figure 3A, in 2-week-old light-grown seedlings, the Rma1H1 transcript was markedly elevated in response to 5 to 10% water loss in leaves. Subsequently, the expression of Rma1H1 mRNA gradually declined as the leaf tissue was exposed to more severe water loss (15 to 30%). Contrastingly, expression of the gene was unaffected by water deficit in the roots, with the basal transcript level being higher in roots than in leaves. This indicates that leaf tissue is a major site of Rma1H1 induction in hot pepper. Rapid increases of Rma1H transcript levels were also induced by cold (within 3 h at 4°C), high salinity (within 2 h by 300 mM NaCl), mechanical wounding (within 30 min), and ethylene (within 2 h) (Figures 3B to 3E). On the other hand, the expression of Rma1H1 was not induced by abscisic acid (ABA) (Figure 3F). Collectively, these results are consistent with the view that the Rma1H1 gene is subject to regulation by a broad spectrum of abiotic stresses, indicating a role in early events in the abiotic-related defense response in hot pepper plants.
Figure 3.
Induction Kinetics of Rma1H1 in Response to Conditions of Environmental Stress in Hot Pepper Plants.
Light-grown 2-week-old hot pepper seedlings were subjected to drought (A), cold temperature (B), high salinity (C), mechanical wounding (local and systemic) (D), ethylene (E), or ABA (F). Induction profiles of Rma1H1 in leaves and roots (as indicated) were examined by RNA gel blot analysis using 32P-labeled Rma1H1 cDNA as a probe. The ACTIN gene was used as a negative control for drought treatment. The RCI, PINII, and LEAL1 genes were used as positive controls for high salinity, wounding, and ABA, respectively. 18S rRNA was used as a loading control.
Overexpression of Rma1H1 Confers Drought Tolerance in Arabidopsis
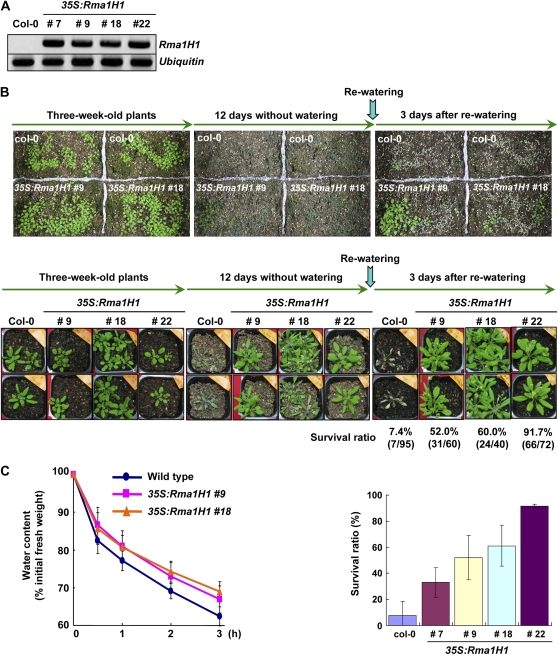
As transgenic work was extremely difficult in hot peppers, in this study, we overexpressed the hot pepper Rma1H1 in Arabidopsis under control of the cauliflower mosaic virus 35S promoter. Previous reports indicate that hot pepper genes are fully functional in heterologous Arabidopsis cells (Cho et al., 2006a, 2006b; Seo et al., 2008). Numerous independent T4 transgenic lines that exhibited markedly enhanced levels of the Rma1H1 transcript under normal growth conditions were chosen for further analysis (Figure 4A).
Figure 4.
Increased Tolerance of 35S:Rma1H1 Arabidopsis Transgenic Plants to Water Stress.
(A) RT-PCR analysis of 4-week-old wild-type and four independent 35S:Rma1H1 T4 transgenic plants (lines #7, #9, #18, and #22).
(B) Wild-type and transgenic lines were grown in pots for 3 weeks under normal growth conditions. Thereafter, water was withheld for 12 d, followed by rewatering for 3 d. Dehydration tolerance was assayed as the capability of plants to resume growth when returned to normal conditions following water stress. The survival rate of wild-type and four independent transgenic lines are shown. Error bars are ±sd (n = 5).
(C) Water loss of wild-type and 35S:Rma1H1 leaves before and after drought stress. Water loss is expressed as the percentage of initial fresh weight of detached leaves. Error bars are ±sd (n = 10).
The aforementioned results concerning the RNA expression profile led us to hypothesize that the hot pepper Rma1H1 might function in abiotic stress defense mechanisms. Therefore, we addressed the capacity of wild-type and 35S:Ram1H1 plants to respond to water deficit. Three-week-old Arabidopsis plants were grown in pots. When the soil was allowed to dry by withholding water for 12 d, wild-type plants displayed severe wilting (Figure 4B). After rewatering for 3 d, most control plants (88 out of 95 plants) were unable to recover and eventually died (7.4% survival). By contrast, a majority of the Rma1H1 overexpressing lines appeared to be healthy before and after rewatering, and they survived and continued to grow, unlike wild-type plants, under severe water stress. The survival ratio was 35 to 92% depending on the transgenic lines (#7, #9, #18, and #22). Consistent with these results, detached rosette leaves of 35S:Rma1H1 plants (#9 and #18) lost water more slowly than did those of wild-type plants (Figure 4C). Thus, we concluded that the 35S:Rma1H1 transgenic plants were highly tolerant to severe water deficit. On the other hand, the survival ratio of 35S:Rma1H1 lines did not appear to be correlated with the amount of Rma1H1 transcript (Figures 4A and 4B), suggesting that Rma1H1 protein level might be regulated in transgenic Arabidopsis plants.
Rma1H1 Is Localized to the ER in Arabidopsis
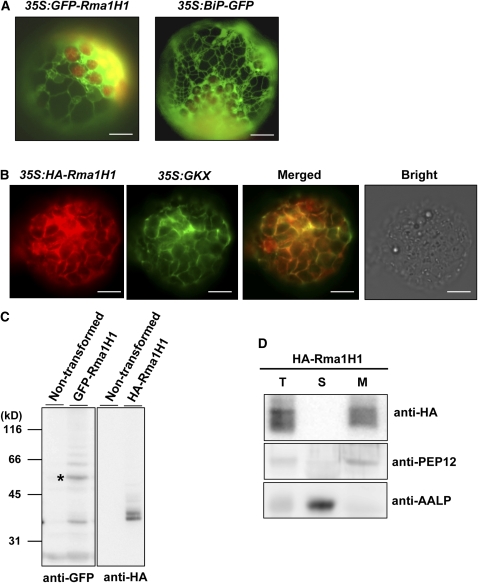
To obtain a clue to the biological role of Rma1H1 with respect to drought tolerance in transgenic Arabidopsis, we generated a green fluorescent protein (GFP)-tagged form of Rma1H1, 35S:GFP-Rma1H1, and introduced it into Arabidopsis protoplasts prepared from leaf tissues. Figure 5A shows that GFP-Rma1H1 displayed a network pattern. It has been well established that ER-localized proteins exhibit a network pattern in protoplasts (Kim et al., 2001). In addition, localization of GFP-Rma1H1 was highly similar to BiP:GFP, a chimeric ER lumenal protein, indicating that Rma1H1 resides in the ER in Arabidopsis. To ensure that its localization was not due to the GFP tag, a hemagglutinin (HA) epitope was added to the N terminus of Rma1H1 and introduced into protoplasts along with GKX, a chimeric ER membrane marker. GKX contains a leader sequence from BiP, the GFP coding region, a transmembrane domain, and an ER membrane retention motif, KKLL (Benghezal et al., 2000). When transformed into protoplasts, green fluorescence signals of GKX exhibited a network pattern and overlapped closely with the red fluorescence signal of BiP-monomeric red fluorescent protein (mRFP) (see Supplemental Figure 1 online). In protoplasts cotransformed with 35S:GKX and 35S:HA-Rma1H1, localization of HA-Rma1H1 was examined by immunostaining using anti-HA antibody, while green fluorescence signals of GKX were observed directly. The result shows that HA-Rma1H1 closely overlapped GKX in the network pattern (Figure 5B), confirming that Rma1H1 localizes to the ER in Arabidopsis cells. The expression of GFP-Rma1H1 and HA-Rma1H1 in protoplasts was examined by immunoblot analysis using anti-GFP and anti-HA antibodies, respectively. GFP-Rma1H1 was detected at the position of 55 kD, an expected size of the fusion protein (indicated by asterisk) and additionally at 35 kD (Figure 5C, left panel). The smaller 35-kD protein may represent the proteolytic product of 55-kD protein. HA-Rma1H1 was detected as two bands at 35 and 36 kD (Figure 5C, right panel). The 36-kD band may be a modified form of HA-Rma1H1. Both GFP-Rma1H1 and HA-Rma1H1 produced several minor bands that migrated more slowly than 55 and 36 kD, respectively. These bands may represent additionally modified forms, such as ubiquitinated products.
Figure 5.
Rma1H1 Localizes to the ER Membrane in Arabidopsis.
(A) Localization of GFP-Rma1H1. Wild-type protoplasts were transformed with 35S:GFP-Rma1H1 or 35S:BiP-GFP, and localization of the green fluorescence signal was examined. Bars = 20 μm.
(B) Colocalization of HA-Rma1H1 with GKX. Protoplasts were cotransformed with 35S:HA-Rma1H1 and 35S:GKX. Localization of HA-Rma1H1 was examined by immunohistochemistry using anti-HA antibody (red signal), while the green fluorescence signal of GKX was observed directly. The green fluorescent signal of 35S:GFP was closely overlapped with the red signal of 35S:HA-Rma1H1. Transformed protoplasts were also viewed under bright-field conditions. Bars = 20 μm.
(C) Immunoblot analyses of GFP-Rma1H1 and HA-Rma1H1. Protein extracts from transformed protoplasts or untransformed control protoplasts were analyzed using anti-GFP or anti-HA antibody. The asterisk indicates 55-kD GFP-Rma1H1 fusion protein.
(D) Subcellular distribution of HA-Rma1H1. Protein extracts from protoplasts transformed with 35S:HA-Rma1H1 were separated into soluble (S) and membrane (M) fractions by ultracentrifugation and analyzed by protein gel blotting using anti-HA antibody. PEP12 and AALP were used as controls for membrane and soluble fractions, respectively. T, total protein extracts.
Rma1H1 contains a single putative transmembrane domain in its C terminus (Figure 1). To determine whether it associates with membranes, protein extracts from protoplasts transformed with 35S:HA-Rma1H1 were separated into soluble and membrane fractions and analyzed by immunoblotting using anti-HA antibody. HA-Rma1H1 was detected in the membrane fraction, indicating that it resides in the membrane (Figure 5D). PEP12 and AALP, used as fractionation controls for membrane and soluble proteins, were found in pellet and soluble fractions, respectively.
Rma1H1 Causes Degradation of PIP2;1 via the 26S Proteasome
In human cells, Hs-Rma1, a homolog of Rma1H1, ubiquitinates misfolded cystic fibrosis transmembrane conductance regulator (CFTR) at the ER membrane for 26S proteasome-mediated degradation (Younger et al., 2006). Likewise, hot pepper Rma1H1 localized to the ER may be involved in ubiquitination of a certain protein followed by proteasomal degradation, with removal of the protein being favorable for transgenic Arabidopsis to survive under dehydration stress. However, in plant cells, not many drought stress-related proteins are known to be ubiquitinated and subjected to proteolytic degradation by the 26S proteasome. As one possible candidate, we selected a plasma membrane aquaporin, PIP2;1, one of the most abundant water channel proteins in Arabidopsis. Plasma membrane aquaporin is generally thought to play a critical role in water relations in plants (Tyerman et al., 1999, 2002; Maurel et al., 2002; Maurel, 2007), and its mRNA levels are downregulated upon dehydration stress (Jang et al., 2004; Alexandersson et al., 2005).
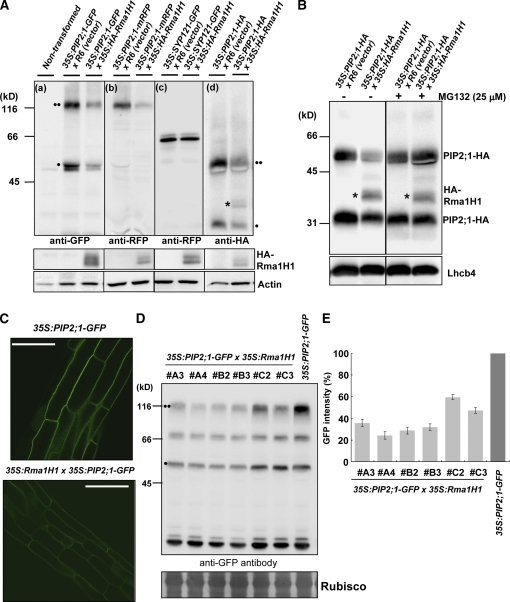
To test this hypothesis, we examined the effect of Rma1H1 on PIP2;1 levels in Arabidopsis protoplasts. PIP2;1 tagged with GFP or mRFP at the C terminus was introduced into protoplasts together with 35S:HA-Rma1H1 or R6, an empty control vector. The protein extracts were then analyzed by immunoblotting using anti-GFP or anti-RFP antibody. Both antibodies detected bands at 50 and 100 kD in control protoplasts (Figures 6A-a and 6A-b). The smaller protein species corresponded to the expected size of PIP2;1-GFP and PIP2;1-mRFP, whereas the upper band appeared to be a dimer. To confirm that the upper band is a dimeric form, we tested various denaturation conditions and found that a prolonged incubation of protein extracts in urea-containing buffer at 50°C solubilized majority of the dimer into monomers (see Supplemental Figure 2 online), which indicates that the upper band represents a dimeric form. Aquaporin that exists as a tetramer in vivo is often observed as a monomer and a dimer in SDS-PAGE (Zelazny et al., 2007). In the presence of HA-Rma1H1, both PIP2;1-GFP and PIP2;1-mRFP levels were markedly reduced compared with the control, suggesting that HA-Rma1H1 causes degradation of both PIP2;1-GFP and PIP2;1-mRFP (Figures 6A-a and 6A-b). The expression of HA-Rma1H1 in the protoplasts was confirmed by immunoblot analysis using anti-HA antibody, while actin was used as a loading control (bottom panel in Figure 6A). SYP121-GFP, a fusion protein between a plasma membrane–localized t-SNARE SYP121 and GFP, was used as a negative control, and its abundance was not affected by HA-Rma1H1 (Figure 6A-c). To demonstrate that the Rma1H1-induced reduction of PIP2;1-GFP and PIP2;1-mRFP protein levels was not due to the C-terminal GFP and mRFP moiety, respectively, 35S:PIP2;1-HA was introduced into protoplasts along with 35S:HA-Rma1H1 or R6, and the respective protein levels were determined. The result revealed that PIP2;1-HA also yielded two bands, at 25 and 50 kD, that corresponded to the monomer and dimer, respectively. PIP2;1-HA levels also were reduced in the presence of coexpressed HA-Rma1H1 (Figure 6A-d), demonstrating that Rma1H1 caused a reduction in PIP2;1 protein levels. One possibility is that Rma1H1 may ubiquitinate PIP2;1 (see below for details).
Figure 6.
HA-Rma1H1 Reduces PIP2;1 Protein Levels in a 26S Proteasome-Dependent Manner.
(A) HA-Rma1H1-induced reduction of PIP2;1 level. Protoplasts were cotransformed with indicated constructs, and protein extracts were analyzed by protein gel blotting using various antibodies as indicated. Actin levels were detected as a loading control with anti-actin antibody. Two circles indicates PIP2;1 in dimeric form, while one circle indicates PIP2;1 as monomer. The asterisk shows HA-Rma1H1, which was detected at the same time as PIP2;1-HA because it is also HA tagged.
(B) Effect of MG132 on PIP2;1-HA levels. Protoplasts cotransformed with 35S:PIP2;1-HA and 35S:HA-Rma1H1 were incubated with MG132, and protein extracts were analyzed using anti-RFP and anti-HA antibodies. Lhcb4 levels were detected as a loading control with anti-Lhcb4 antibody.
(C) to (E) Reduction of PIP2;1-GFP in 35S:Rma1H1/35S:PIP2;1-GFP double transgenic plants. PIP2;1-GFP levels were determined by fluorescent microscopy (C) or by protein gel blot analysis using anti-GFP antibody (D). The degree of PIP2;1-GFP reduction in six independent DT lines was quantified by measuring the intensity of the three bands of PIP2;1-GFP from the protein gel blotting (E). RbcS stained with Coomassie blue was used as a loading control. Bars = 50 μm. Error bars are ±sd (n = 5).
[See online article for color version of this figure.]
To examine whether the Rma1H1-induced reduction of the PIP2;1 protein level was due to proteolytic degradation by the 26S proteasome, protoplasts cotransformed with 35S:HA-RmaH1 and 35S:PIP2;1-HA were treated with MG132, an inhibitor of 26S proteasome, and protein extracts were analyzed using anti-HA antibody. As shown in Figure 6B, MG132 treatment inhibited the Rma1H1-induced reduction in the PIP2;1-HA level, indicating that the 26S proteasome is involved in the Rma1H1-mediated reduction of PIP2;1 levels. As a loading control, endogenous light-harvesting complex b4 (Lhcb4) was detected by anti-Lhcb4 antibody.
We next investigated the functional connection between Rma1H1 and PIP2;1 at the plant level. In this experiment, 35S:Rma1H1 (lines #18 and #22), which are resistant to drought stress (Figures 4B and 4C), and 35S:PIP2;1-GFP plants were crossed, and PIP2;1-GFP levels were analyzed in T3 progenies of 35S:Rma1H1/35S:PIP2;1-GFP double transgenic (DT) plants. In 35S:PIP2;1-GFP transgenic plants, PIP2;1-GFP localized to the plasma membrane in the root epidermal cells (Figure 6C). GFP signals were markedly reduced in the roots of DT plants compared with those of 35S:PIP2;1-GFP plants. This indicates that Rma1H1 reduced the amount of plasma membrane–localized PIP2;1-GFP at the plant level. To further confirm this result at the protein level, protein extracts from roots of both 35S:PIP2;1-GFP and 35S:Rma1H1/35S:PIP2;1-GFP DT plants were analyzed using anti-GFP antibody. As shown in Figure 6D, these transgenic plants produced three protein species. Two species correspond to the monomer and dimer of PIP2;1-GFP, while the third species, a 70-kD band, may represent a partial degradation product. The protein levels in six independent lines of DT plants ranged from 27 to 60% of that of the control (Figures 6D and 6E), consistent with the observations obtained by image analysis of root tissues. These results, together with those from protoplast experiments, strongly argue that Rma1H1 reduces PIP2;1 levels in Arabidopsis.
Rma1H1 Inhibits Trafficking of PIP2;1 from the ER to the Plasma Membrane
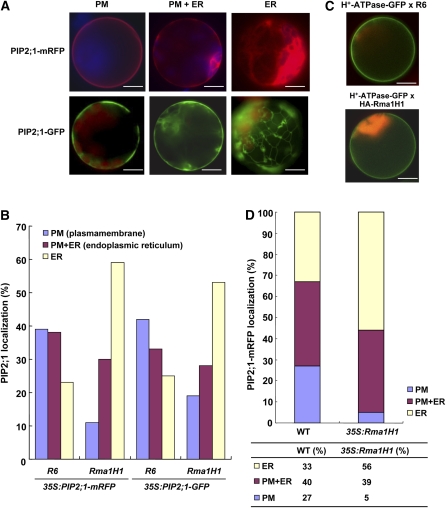
The finding that ER-localized Rma1H1 reduced PIP2;1 levels in a 26S proteasome-dependent manner prompted us to test whether Rma1H1 might also inhibit trafficking of PIP2;1 to the plasma membrane from the ER. We reasoned that, if trafficking of PIP2;1 to the plasma membrane occurred efficiently, ER-localized Rma1H1 may not be able to effectively reduce the PIP2;1 levels. Thus, one possible mechanism would be that Rma1H1 may inhibit either directly or indirectly the plasma membrane trafficking of PIP2;1 at the ER, and subsequently ubiquitinated PIP2;1 is degraded by 26S proteasome. To examine this idea, we first investigated whether PIP2;1 is transported to the plasma membrane in protoplasts. Transient expression of PIP2;1-mRFP or PIP2;1-GFP in wild-type Arabidopsis protoplasts yielded three different localization patterns: the ER network pattern, the plasma membrane pattern, or both plasma membrane and network patterns (Figure 7A). The ER pattern may represent PIP2;1-mRFP/GFP that have not been transported yet to the plasma membrane. We found that 39% of the transformed protoplasts displayed only the plasma membrane pattern, 38% displayed the ER pattern plus the plasma membrane pattern, and 23% displayed only the ER pattern (Figure 7B). When protoplasts were cotransformed with 35:PIP2;1-mRFP and 35S:HA-Rma1H1, the percentage of protoplasts with the plasma membrane pattern was reduced to 11%, and concomitantly the percentage with the ER alone pattern increased to 59%, raising the possibility that coexpressed Rma1H1 inhibits trafficking of PIP2;1-mRFP to the plasma membrane. To confirm that this inhibition was not due to the mRFP domain, we examined trafficking of PIP2;1-GFP to the plasma membrane in the presence of coexpressed HA-Rma1H1 in protoplasts. In the presence of HA-Rma1H1, the plasma membrane pattern of PIP2;1-GFP was reduced to 19% from 42% of the control, and the ER pattern was increased to 53% from 25% of the control protoplasts, indicating that Rma1H1 inhibits trafficking of PIP2;1 to the plasma membrane. As a control, H+-ATPase-GFP, encoding a chimeric protein consisting of the plasma membrane type H+-ATPase and GFP (Kim et al., 2001), was transformed into protoplasts along with HA-Rma1H1 or R6, and the localization of H+-ATPase-GFP was examined. As shown in Figure 7C, HA-Rma1H1 did not affect trafficking of H+-ATPase-GFP to the plasma membrane, indicating that the inhibitory effect of Rma1H1 was specific to PIP2;1 and not due to a general inhibition of trafficking.
Figure 7.
Overexpression of Rma1H1 Inhibits Trafficking of PIP2;1.
(A) Localization of PIP2;1 in protoplasts. Protoplasts were transformed with the indicated constructs, and localization of PIP2;1-mRFP or PIP2;1-GFP was examined. The population of PIP2;1-transformed protoplasts included three different patterns of mRFP or GFP localization: the ER network pattern, plasma membrane pattern, or localization to both the ER network and plasma membrane. Bars = 20 μm.
(B) Quantification of PIP2;1-mRFP and PIP2;1-GFP localization patterns. Protoplasts were cotransformed with the indicated constructs. Protoplasts were counted based on their PIP2;1-mRFP or PIP2-1-GFP localization patterns: the ER-only pattern, plasma membrane only pattern, and the ER plus plasma membrane patterns as shown in (A). More than 150 protoplasts were counted. R6 is the empty vector control.
(C) Effect of HA-Rma1H1 on H+-ATPase-GFP. HA-Rma1H1 was transformed into protoplasts together with H+-ATPase-GFP or R6, and localization of H+-ATPase-GFP was examined. Bars = 20 μm.
(D) Inhibition of PIP2;1-mRFP trafficking by Rma1H1 in transgenic plants. Protoplasts were prepared from wild-type and Rma1H1-overexpressing transgenic plants and then transformed with 35S:PIP2;1-mRFP or R6. Protoplasts were quantified based on the localization pattern of PIP2;1-mRFP. In total, >300 protoplasts were counted in a triplicate experiment.
We then tested whether stable expression of Rma1H1 in transgenic Arabidopsis plants inhibits trafficking of PIP2;1-mRFP. We prepared protoplasts from 35S:Rma1H1 transgenic plants (lines #18 and #22) that displayed strongly enhanced tolerance to dehydration stress (Figures 4B and 4C) and then transformed the 35S:PIP2;1-mRFP construct into the 35S:Rma1H1 protoplasts. Subsequently, the trafficking efficiency was determined based on the localization pattern of PIP2;1-mRFP. In 35S:Rma1H1 protoplasts, PIP2;1-mRFP yielded 5% of the plasma membrane pattern compared with 27% in wild-type protoplasts (Figure 7D). By contrast, the ER pattern was increased to 56% in 35S:Rma1H1 protoplasts compared with 33% of wild-type protoplasts. These results demonstrate that stably expressed Rma1H1 inhibits the PIP2;1-mRFP trafficking from the ER to the plasma membrane, just as was observed with transiently expressed Rma1H1.
Rma1H1 Interacts with PIP2;1
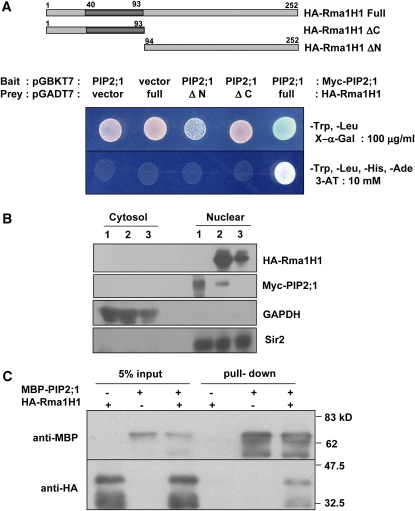
The results in Figures 6 and 7 suggest a functional interaction between Rma1H1 and PIP2;1. To corroborate these results, we first employed a yeast two-hybrid assay. In this experiment, the ability of the yeast strain AH109 to grow in the absence of His was used as a marker for the interaction between Rma1H1 and PIP2;1. We found that His auxotrophy was restored when Rma1H1 was cotransformed with PIP2;1 (Figure 8A). This suggests an interaction occurs between Rma1H1 and PIP2;1 in yeast cells. As a specificity control, we tested the association between PIP2;1 and N- and C-terminal deletion mutants of Rma1H1, respectively. As shown in Figure 8A, both deletion mutants failed to bind PIP2;1, indicating that the full-length Rma1H1 was required for the physical interaction with PIP2;1 in yeast cells.
Figure 8.
Rma1H1 Physically Interacts with PIP2;1.
(A) Yeast two-hybrid assay. PIP2;1 was cloned into pGADT7, and Rma1H1 and its deletion mutant (Rma1H11-93 and Rma1H194-252) were cloned into pGBKT7. Yeast AH109 cells were cotransformed with a combination of the indicated plasmids. To test protein–protein interactions, yeast cells were plated onto SD/-His/-Trp/-Leu medium including 10 mM 3-amino-1,2,4-triazole.
(B) Immunoblot analysis of yeast nuclear and cytosolic proteins. Cytosolic and nuclear protein samples were prepared from yeast cells, in which Myc-PIP2;1 (lane 1), Myc-PIP2;1 + HA-Rma1H1 (lane 2), or HA-Rma1H1 (lane 3) were transformed. Both cytosolic and nuclear extracts of yeast cells were subjected to SDS-PAGE, followed by immunoblot analysis using anti-HA, anti-Myc, anti-GAPDH, or anti-Sir2 antibody. GAPDH and Sir2 were used as fractionation controls for cytosolic and nuclear proteins, respectively.
(C) In vitro pull-down assay. MBP-PIP2;1 was incubated with HA-Rma1H1 and amylose affinity resin. The bound protein was eluted from resin and probed with anti-HA or anti-MBP antibody.
[See online article for color version of this figure.]
Rma1H1 contains a C-terminal membrane anchoring domain (Figure 1). In addition, PIP2;1 is a highly hydrophobic protein and contains six membrane-spanning domains (Maurel, 2007). To examine whether Rma1H1 and PIP2;1 proteins were effectively targeted to nuclei in our yeast two-hybrid experiment, we performed immunoblot analysis. Yeast cells were transformed with Myc-PIP2;1 alone, Myc-PIP2;1 along with HA-Rma1H1, or HA-Rma1H1 alone. They were then separated into nuclear and cytoplasmic fractions and proteins were extracted. These fractions were relatively free of cross-contamination, as judged by the absence of cross-reaction with antibodies to cytosolic and nuclear proteins, GAPDH and Sir2, respectively (Figure 8B). Immunoblotting using anti-HA or anti-Myc antibody revealed that both Rma1H1 and PIP2;1 proteins were predominantly localized in the nuclear fraction in our yeast two-hybrid conditions, indicating the association of Rma1H1 and PIP2;1 in yeast nuclei (Figure 8B).
We next performed an in vitro pull-down assay. Rma1H1 was expressed in E. coli and efficiently purified as an HA fusion protein. By contrast, it was extremely difficult to obtain purified PIP2;1 protein as it was highly insoluble and precipitated in E. coli cells. Consequently, we were able to obtain only a minimal amount of soluble MBP-PIP2;1 for pull down. The two fusion proteins were coincubated with an amylose affinity matrix, followed by extensive washing. The bound protein was then eluted from resin by 10 mM maltose and immunoblotted with anti-MBP and anti-HA antibodies. Figure 8C shows that HA-Rma1H1 protein was pulled down from the amylose affinity resin only in the presence of MBP-PIP2;1, but not by HA-Rma1H1 alone. These data are consistent with a direct physical interaction between Rma1H1 and PIP2;1, in agreement with the results of the yeast two-hybrid assay.
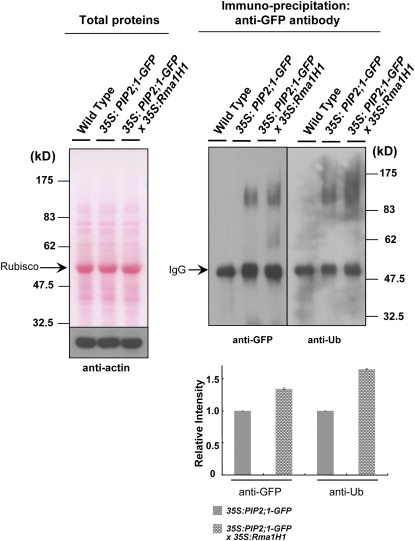
Ubiquitination of PIP2;1 by Rma1H1 in Vivo
We were unable to conduct in vitro ubiquitination assay, because the insolubility of PIP2;1 in E. coli cells made it difficult to purify sufficient protein. To circumvent this problem and to enhance our understanding of the interaction between Rma1H1 and PIP2;1, we performed an in vivo ubiquitination experiment using 35S:PIP2;1-GFP and 35S:Rma1H1/35S:PIP2;1-GFP DT plants. Light-grown 2-week-old wild-type and T3 transgenic whole seedlings were incubated with 10 μM MG132 for 4 h. The crude extracts were then prepared from treated tissues and subjected to immunoprecipitation using anti-GFP antibody and protein A-sepharose. The immunoprecipitated proteins were subsequently analyzed by immunoblotting with anti-GFP antibody. Figure 9 reveals the production of high molecular mass smear ladders when PIP2;1-GFP was overexpressed in Arabidopsis. More importantly, both the amount and the intensity of these smear bands became significantly stronger in 35S:Rma1H1/35S:PIP2;1-GFP DT plants compared with those in 35S:PIP2;1-GFP plants. Subsequent immunoblot analysis using anti-Ub antibody indicated that the high molecular mass smear ladders were indeed ubiquitinated PIP2;1-GFP (Figure 9). These argue that PIP2;1 is ubiquitinated in a Rma1H1-dependent manner in Arabidopsis.
Figure 9.
In Vivo Ubiquitination of PIP2;1.
Intact whole seedlings of wild-type and transgenic T3 seedlings (35S:PIP2;1-GFP and 35S: PIP2;1-GFP/35S:Rma1H1) were incubated with MG132. The whole cell free extracts containing 100 μg proteins were prepared, separated on an 8% SDS-PAGE, and visualized by staining with the Ponceau S solution (left panel). Proteins (500 μg) were then incubated with anti-GFP antibody along with 40 μL protein A-sepharose. Immunoprecipitated proteins were eluted and detected by anti-GFP or anti-Ub-antibody (right panel). Actin was detected as a loading control. Ribulose-1,5-bisphosphate carboxylase/oxygenase (left panel) and IgG (right panel) are indicated by arrows. A PIP2;1-GFP protein band is masked by the IgG signal. The magnitude of relative ubiquitination of PIP2;1-GFP in 35S:PIP2;1-GFP and 35S:PIP2;1-GFP/35S:Rma1H1 transgenic plants was quantified and normalized to 1.00 for the ubiquitinated bands of PIP2;1-GFP in 35S:PIP2;1-GFP plants. Error bars are ±sd (n = 3). [See online article for color version of this figure.]
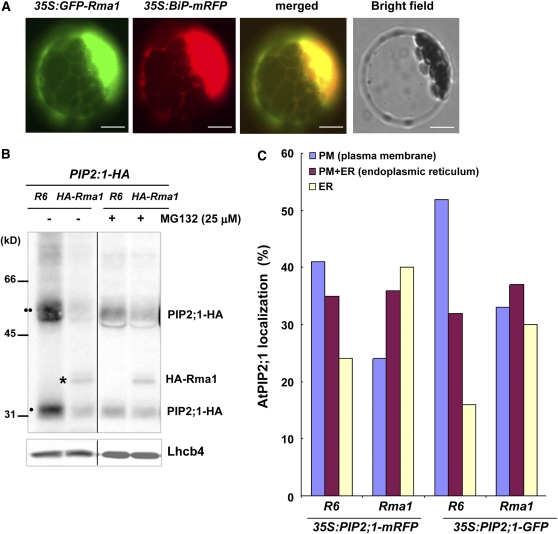
Arabidopsis Rma1 Reduced the PIP2;1 Protein Level and Inhibited Trafficking of PIP2;1 from the ER to the Plasma Membrane
Aforementioned results described in Figures 4 to 9 suggest that constitutive expression of heterologous hot pepper Rma1H1 in Arabidopsis confers drought tolerance via downregulation of PIP2;1 through the ubiquitination pathway. Although Rma1H1 appears to function in Arabidopsis, it might be still questionable whether an Arabidopsis Rma1H1-related protein plays a similar function. To answer this important question, we examined the localization of Rma1, an Arabidopsis homolog of Rma1H1 (Figure 1), as well as its overexpression effect on PIP2;1. The 35S:GFP-Rma1 and 35S:BiP-mRFP constructs were transformed into protoplasts and their localizations were determined. Rma1 produced a network pattern and overlapped closely with the red fluorescence signal of BiP-mRFP, indicating that it localized to the ER as observed with RmaH1 (Figure 10A). This result raised the possibility that Rma1 may have similar functions to RmaH1. To examine further the biological role of Rma1, 35S:PIP2;1-HA construct was transformed into protoplasts along with 35S:HA-Rma1 or R6, a control vector. The protein extracts were subsequently assayed by immunoblotting using anti-HA antibody. Figure 10B shows that, in the presence of HA-Rma1, the abundance of both mono- and dimeric forms of PIP2;1-HA were significantly reduced, indicating that HA-Rma1 caused degradation of PIP2;1-HA in protoplasts. The Rma1-induced reduction in the PIP2;1-HA level was inhibited by MG132 treatment, suggesting that the 26S proteasome participates in the Rma1-mediated reduction of PIP2;1 levels. As a loading control, Lhcb4 protein was detected by anti-Lhcb4 antibody.
Figure 10.
Arabidopsis Rma1 Reduces PIP2;1 Protein Levels and Inhibits Trafficking of PIP2;1 from ER to the Plasma Membrane in the Protoplasts.
(A) Localization of GFP-Rma1. Wild-type protoplasts were transformed with 35S:GFP-Rma1 and 35S:BiP-mRFP, and localization of the proteins was examined. Bars = 20 μm.
(B) HA-Rma1-induced reduction of PIP2;1-HA level. Protoplasts were cotransformed with indicated constructs, incubated with or without MG132, and protein extracts were analyzed by protein gel blotting using anti-HA antibody. R6 is the empty vector control. Lhcb4 levels were detected as a loading control with anti-Lhcb4 antibody. Two circles indicates PIP2;1-HA in dimeric form, while one circle indicates PIP2;1-HA as monomer. The asterisk shows HA-Rma1, which was detected at the same time as PIP2;1-HA because it is also HA tagged.
(C) Quantification of PIP2;1-mRFP and PIP2;1-GFP localization patterns in the presence or absence of Arabidopsis Rma1. Protoplasts were transformed with the indicated constructs, and localization of PIP2;1-mRFP or PIP2;1-GFP was examined. Protoplasts were counted based on their PIP2;1-mRFP or PIP2-1-GFP localization patterns: the ER only pattern, plasma membrane only pattern, and the ER plus plasma membrane patterns. More than 150 protoplasts were counted.
We also observed that Rma1 inhibits trafficking of PIP2;1 from the ER to the plasma membrane. When protoplasts were cotransformed with 35S:PIP2;1-mRFP and 35S:HA-Rma1, the percentage of protoplasts with the plasma membrane pattern of PIP2;1-mRFP decreased from 41 to 24%, while the percentage of ER alone pattern increased from 24 to 40% (Figure 10C). We obtained similar results with PIP2;1-GFP. In the presence of HA-Rma1, the plasma membrane pattern of PIP2;1-GFP was reduced from 53 to 33%, and the ER alone pattern was increased from 16 to 30% (Figure 10C). Thus, these results show that Rma1 reduced the PIP2;1 protein level and inhibited trafficking of PIP2;1 from the ER to the plasma membrane.
We next constructed 35S:HA-Rma1 transgenic plants and examined their phenotype. Under our experimental conditions, the HA-Rma1-overexpressing lines were indistinguishable from wild-type plants in terms of drought tolerance and development. We found that, although HA-Rma1 mRNA was expressed in transgenic lines, the level of corresponding protein was extremely low, probably due to the rapid degradation (see Supplemental Figure 3 online). Immunoblotting using anti-HA antibody shows that the HA-Rma1H1 protein was barely detectable only after transgenic plants were treated with MG132 and UCH-L3, an inhibitor of deubiquitinase, for a long time (24 h) (see Supplemental Figure 3 online), indicating that endogenous level of HA-Rma1 protein was tightly regulated via an Ub-26S proteasome pathway.
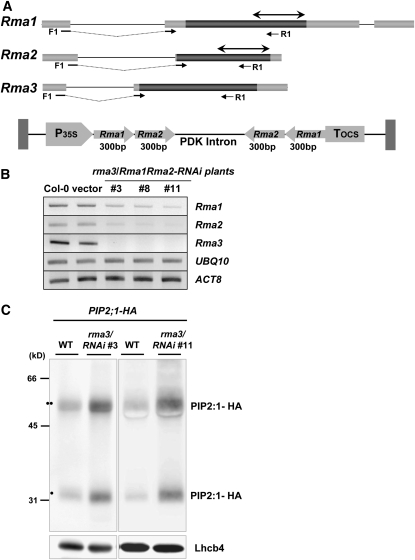
Reduced Expression of Arabidopsis Rma Homologs Results in the Increased Level of PIP2;1
In Arabidopsis, there are three Rma1H1 homologs, referred to as Rma1 (34% identity) (Matsuda and Nakano, 1998), Rma2 (At4g28270, 30% identity), and Rma3 (At4g27470, 29% identity) (Figure 1). To explore the in vivo functions of Arabidopsis Rma homologs, we analyzed mutants carrying T-DNA insertions in the Rma1, Rma2, and Rma3 genes. The rma1, rma2, and rma3 mutants have a T-DNA insertion after nucleotide 365 (line GT_5_68771), after nucleotide 1045 (line SALK_136700), and after nucleotide 619 (line Sail_218_G01), respectively, on chromosome 4 (see Supplemental Figure 4 online). The rma mutant plants did not differ in appearance from wild-type plants. In addition, these single mutant lines did not display detectable phenotypes in response to drought stress compared with the wild-type plants. These results might be due to the complementation of three homologous genes. Also, we were not able to construct double or triple mutants for these genes because they reside in the same chromosome. Alternatively, RNA interference (RNAi) constructs for Rma1 and Rma2 genes were introduced into the rma3 knockout mutant line (Figure 11A). RT-PCR analysis revealed that transcripts for both Rma1 and Rma2 were significantly reduced with a negligible amount of Rma3 mRNA in several independent rma3-knockout/Rma1Rma2-RNAi-knockdown plants (Figure 11B). As a next step, the 35S:PIP2;1-HA construct was transformed into protoplasts prepared from these rma3/Rma1Rma2-RNAi plants (lines #3 and #11) and from wild-type Arabidopsis. Finally, the amount of PIP2;1-HA protein was compared in the transformed protoplasts by immunoblotting using anti-HA antibody. The results in Figure 11C demonstrate that the level of PIP2;1-HA protein is significantly higher in 35S:PIP2;1-HA/rma3/Rma1Rma2-RNAi protoplasts relative to that in 35S:PIP2;1-HA protoplasts. As a loading control, Lhcb4 was detected with anti-Lhcb4 antibody. We interpret these results as the evidence that reduced expression of Rma homologs results in the increased level of PIP2;1 protein in Arabidopsis.
Figure 11.
Reduced Expression of Rma Homologs Results in the Increased Level of PIP2;1 in the Protoplasts.
(A) Schematic representation of the RNAi construct for Rma1 and Rma2. Dark bars indicate coding regions, while gray bars show the 5′- and 3′-untranslated regions. Solid lines reveal introns. Gene-specific forward and reverse primers (F1 and R1) used in the genotyping and RT-PCR are shown with arrows. Double arrows in the 3′-ends of Rma1 and Rma2 indicate the 300-bp region used for RNAi constructs.
(B) RT-PCR analysis of Rma1, Rma2, Rma3, Actin8, and Ubiquitin10 mRNAs in wild-type, vector-control transgenic plants, and rma3-knockout/Rma1Rma2-RNAi-knockdown plants (#3, #8, and #11) using gene-specific primers.
(C) Quantification of PIP2;1-HA protein levels in 35S:PIP2;1-HA and 35S:PIP2;1-HA/ rma3/Rma1Rma2-RNAi protoplasts. Protoplasts were prepared from wild-type and rma3/Rma1Rma2-RNAi plants (lines #3 and #11), transformed with 35S:PIP2;1-HA, and protein extracts were analyzed by protein gel blotting using anti-HA antibody. Lhcb4 levels were detected as a loading control with anti-Lhcb4 antibody. Two circles indicates PIP2;1-HA in dimeric form, while one circle indicates PIP2;1-HA as monomer.
DISCUSSION
Plant RING E3 Ub ligases have recently attracted much interest due to growing evidence that they play important roles in the mediation of cellular responses to environmental stresses. For example, they are known to participate in cold stress signal transduction (Lee et al., 2001; Dong et al., 2006), in the responses to salt and osmotic stress through increased ABA biosynthesis (Ko et al., 2006), and in the ABA-mediated drought signaling pathway (Zhang et al., 2007). In addition, DRIP-RING E3 ligases function as negative regulators in drought response by ubiquitinating the DREB2A transcription factor (Qin et al., 2008). These findings prompted us to construct transgenic Arabidopsis plants that constitutively expressed Rma1H1, which encodes a hot pepper abiotic stress-induced RING membrane-anchor E3 Ub ligase. Our search for phenotypic properties indicated that 35S:Rma1H1 transgenic plants were highly tolerant to severe drought stress (Figure 4). Thus, it seems most likely that Rma1H1 is functionally relevant in the heterologous Arabidopsis cells, thereby effectively altering a subset of the response to water deficit in transgenic lines. With this in mind, we speculated that the E3 Ub ligase Rma1H1 was involved in the ubiquitination of as yet unidentified proteins, which might function in the process of drought response in transgenic Arabidopsis plants.
Since the water channel protein aquaporin has been initially identified in red blood cells (Preston et al., 1992), several lines of evidence indicate that aquaporins contribute to diverse cellular processes in higher plants, including water transport, nutrient and solute transport, and stress responses (Tyerman et al., 1999, 2002; Johanson et al., 2001; Maurel et al., 2002; Maurel, 2007). Among different families of aquaporins, plasma membrane–localized PIPs appear to function in intercellular water transport. Antisense transgenic tobacco (Nicotiana tabacum) plants, in which the expression of aquaporin gene Nt-AQP1 was downregulated, displayed reduced root hydraulic conductivity and lower water stress resistance, suggesting the importance of aquaporin-mediated symplastic water transport (Siefritz et al., 2002). Likewise, the Arabidopsis knockout mutant of PIP2;2, which is predominantly expressed in roots, exhibited marked decrease in the hydraulic conductivity of root cortex cells (Javot et al., 2003). This suggested that the PIP2;2 aquaporin was specialized to function in osmotic fluid transport in roots. On the other hand, Aharon et al. (2003) reported that overexpression of the plasma membrane aquaporin PIP1b in tobacco caused hypersensitivity to drought stress. Based on this result, it was proposed that enhanced symplastic water transport via plasma membrane aquaporins may have a deleterious effect on the plants during water stress. Moreover, expression of aquaporin gene families is generally downregulated by abiotic stresses, including dehydration, in Arabidopsis (Jang et al., 2004; Alexandersson et al., 2005; Boursiac et al., 2005). After rewatering, the expression levels of all PIP genes were restored to the same level as in the control plants (Alexandersson et al., 2005). It was suggested that aquaporins might function as osmosensors and turgor sensors more than as water transporters (Hill et al., 2004). Thus, there are some contradictory results concerning the role of aquaporins in dehydration tolerance.
Our results indicate a functional relationship between Rma1H1 and PIP2;1, a plasma membrane–specific aquaporin, in the process of drought tolerance in transgenic Arabidopsis plants, as evidenced by the following observations. Ectopic expression of Rma1H1 effectively inhibits the trafficking of PIP2;1 from the ER to the plasma membrane (Figure 7) and leads to reduced levels of PIP2;1 in both protoplasts and 35S:Rma1H1/35S:AtPIP2:1-GFP DT roots compared with their respective control samples (Figure 6). PIP2;1 physically interacts with Rma1H1 (Figure 8) and, more importantly, is ubiquitinated in DT cells in a Rma1H1-dependent fashion (Figure 9). Thus, it is attractive to propose that Rma1H1 ubiquitinates PIP2;1 at the ER. However, it is not clear whether Rma1H1 acts directly to inhibit trafficking of PIP2;1 to the plasma membrane. One possibility is that ubiquitination of PIP2;1 by Rma1H1 may act as a signal to inhibit trafficking to the plasma membrane, as observed for lipoprotein receptor-related proteins 5/6 (LRP5/6) in HeLa cells, whose ubiquitinated form is retained in the ER (Abrami et al., 2008). In human cells, Derlin-1, which forms a complex with Rma1, appears to be responsible for retaining cystic fibrosis transmembrane conductance regulator in the ER membrane (Younger et al., 2006). Thus, another possibility is that a putative Rma1H1-interacting protein, functionally similar to Derlin-1, is responsible for inhibiting PIP2;1 trafficking from the ER to the plasma membrane. The ER-retained PIP2;1 may be ubiquitinated by Rma1H1 for proteasomal degradation. Accordingly, the amount of plasma membrane–localized PIP2;1 is reduced to lower levels, which in turn results in the increased tolerance to water deficit. In this regard, it should be noted that transgenic tobacco plants constitutively expressing the plasma membrane aquaporin PIP1b gene showed faster wilting in response to dehydration stress (Aharon et al., 2003). In addition, the level of PIP2;1 mRNA declined upon water stress and returned to the normal level after rehydration (Alexandersson et al., 2005). Taken together, these results may indicate that expression of PIP2;1 in the plasma membrane is not beneficial to plants under water stress conditions.
Although our results indicate that constitutive expression of Rma1H1 in heterologous Arabidopsis results in drought tolerance through ubiquitination of PIP2;1, one might wonder whether the Arabidopsis Rma1 homolog plays similar roles. To address this critical issue, we employed gain- and loss-of-function studies of Arabidopsis Rma homologs. After a series of careful experiments, we learned that gain- and loss-of-function studies of the Rma genes were extremely difficult due to the following reasons: (1) Although HA-Rma1 mRNA was effectively expressed, the level of corresponding HA-Rma1 protein was very low due to the rapid degradation in transgenic plants. The HA-Rma1 protein was detectable only after transgenic plants were incubated with inhibitors for 26S proteasome (MG132) and deubiquitinase (UCH-L3) for a long time (24 h) (see Supplemental Figure 3 online); (2) loss-of-function single knockout mutant plants for Rma1, Rma2, and Rma3 did not show detectable phenotypes because of complementation of these three homologous genes; and (3) all three homologs reside in the chromosome 4 so that we could not generate a triple mutant (see Supplemental Figure 4 online). To overcome these problems, we employed protoplast transfection assays, and the results revealed that, as was the case for Rma1H1, Arabidopsis Rma1, which localized to the ER, also reduced the PIP2;1 protein levels and inhibited trafficking of PIP2;1 from the ER to the plasma membrane (Figure 10). Furthermore, we constructed rma3-knockout/Rma1Rma2-RNAi-knockdown plants and obtained the results that the PIP2:1 protein levels were upregulated in protoplasts from rma3/Rma1Rma2-RNAi plants relative to those in wild-type protoplasts (Figure 11). Taken together, these results provide additional evidence that Rma homologs participate in the regulation of PIP2;1 protein levels in Arabidopsis. This is consistent with the notion that PIP2;1-GFP is significantly ubiquitinated without Rma1H1 in wild-type plants (see lane 35S:PIP2;1-GFP in right panel of Figure 9). However, there was an intriguing difference between hot pepper RmaH1 and Arabidopsis Rma1. RmaH1 was relatively more stable than Rma1 in Arabidopsis when overexpressed transiently in protoplasts and stably in transgenic plants. The difference in the protein stability, which is under the regulation of ubiquitin/26S proteasome, appears to be the underlying cause of the phenotypic difference between RmaH1- and Rma1-overexpressing transgenic plants under dehydration stresses.
Rma1H1 overexpressors did not display enhanced tolerance to high salinity (see Supplemental Figure 5 online), suggesting that PIP2;1 may play a specific role in drought response. This hypothesis is in line with the suggestion that, although there are multiple homologous isoforms, individual aquaporins function nonredundantly in plant cells (Javot et al., 2003). It is also worth noting that mannitol-induced water imbalance resulted in increased Mc-TIP1;2 amounts in the tonoplast and a shift in Mc-TIP1;2 distribution to other membrane fractions, indicating aquaporin relocalization during osmotic stress in ice plant (Mesembryanthemum crystallinum) (Vera-Estrella et al., 2004). Subcellular redistribution of plasma membrane aquaporin was also detected in Arabidopsis roots in response to salt stress. Boursiac et al. (2005) observed intracellular structures containing not only the TIP2;1 isoform, but also PIP1 and PIP2 homologs after salt stress. Most recently, it was reported that environmental stimuli induced an H2O2-mediated internalization of PIPs to downregulate root water transport (Boursiac et al., 2008). Therefore, we cannot rule out the possibility that Rma1H1 may also play a role in internalization of PIP2;1 from the plasma membrane. In fact, ubiquitin acts as a signal for endocytosis of plasma membrane proteins (Kölling and Hollenberg, 1994), and PIP2;1-GFP has been shown to be internalized in transgenic plants (Paciorek et al., 2005). In this case, an E3 Ub ligase localized to the plasma membrane may be involved in the internalization of PIP2;1-GFP. Ubiquitination and subsequent endocytosis and degradation of aquaporin-2 water channel were also reported in mammalian cells (Kamsteeg et al., 2006).
While our data demonstrate that Rma1H1 and PIP2;1 interact, and that Rma1H1 regulates PIP2;1 levels and localization in transgenic Arabidopsis, it is possible that the drought resistance of the 35S:Rma1H1 transgenic plants was due to other effects of Rma1H1. For example, ectopic expression of Rma1H1 may have resulted in ubiquitination of other PIP2 isoforms, leading to altered function of multiple aquaporins. There are at least eight homologous PIP2 isoforms in Arabidopsis. To further complicate matters, aquaporin isoforms physically interact with other isoforms, and heteromerization of aquaporins may modulate water channel activity in maize (Zea mays) (Fetter et al., 2004). In addition, we cannot rule out the possibility that other target proteins of Rma1H1 rather than aquaporins could be responsible for the drought-tolerant phenotype of 35S:Rma1H1 plants. Thus, further experiments are required to more precisely define the functional relationships between Rma1H1 and other target proteins. We are currently repeating yeast two-hybrid assay to identify possible substrates and/or protein partners of Rma1H1 and Rmas. In conclusion, our data argue that ubiquitination and subsequent degradation of PIP2;1 may play a critical role in the cellular mechanism underlying the tolerance to dehydration. These results will be applied to construct transgenic crop plants that are tolerant to drought stress.
METHODS
Plant Materials and Application of Various Abiotic Stresses
Dry seeds of hot pepper (Capsicum annuum cv Pukang) and Arabidopsis thaliana ecotype Columbia were grown, transformed, and treated as described previously (Cho et al., 2006b). Hot pepper plants were subjected to various abiotic stresses, such as drought, high salinity, and cold, as described by Kim et al. (2007) and Jun et al. (2008). The rma1 (line GT_5_68771), rma2 (Salk_136700), and rma3 (SAIL_218_G01) T-DNA insertion mutants were obtained from the ABRC (http://www.Arabidopsis.org).
Cloning of the Full-Length cDNA of Hot Pepper Rma1H1
To obtain a full-length cDNA of Rma1H1, pCa-DI6, a partial clone for Rma1H1, was used as a probe to screen the λ-uni-Zap II cDNA library constructed from water-stressed leaves of hot pepper plants (Park et al., 2003). The cDNAs containing putative Rma1H1 were subcloned into Bluescript SK plasmid by in vivo excision of pBluescript from pepper cDNA library vector according to the manufacturer's protocol (Stratagene). Restriction enzyme mapping and DNA sequencing analysis confirmed that one of the isolated clones (pRma1H1) represented a full-length Rma1H1 cDNA, including 5′- and 3′-untranslated regions. Deduced amino acid sequences of Rma1H1 and Rma homologs from other plant species and human were analyzed and aligned using ClustalW in Mega4 software (Tamura et al., 2007).
RNA Gel Blot and RT-PCR Analyses
Total RNAs of hot pepper and transgenic Arabidopsis plants were obtained as described by Cho et al. (2006a, 2008). Hot pepper total RNAs were separated by electrophoresis on a 1.0% agarose gel and blotted onto a nylon membrane filter (Amersham). The filter was hybridized to various 32P-labeled cDNA probes for Rma1H1, PINII, RCI, and LEAL1 under high stringency hybridization and washing conditions as described by Lee et al. (2004). The blot was washed and visualized by autoradiography at −80°C using Kodak XAR-5 film and an intensifying screen. For RT-PCR analysis of Arabidopsis RNAs, the first-strand cDNA synthesis and RT-PCR were performed as described previously (Joo et al., 2006). Primers used in RT-PCR are listed in Supplemental Table 1 online. The RT-PCR products were separated in a 1.0% agarose by electrophoresis.
In Vitro Self-Ubiquitination and Immunoblot Analyses
The full-length Rma1H1 cDNA was amplified by PCR using primers (Rma1H1 Full F and Rma1H1 Full R in Supplemental Table 1 online). The PCR product was digested with EcoRI and then ligated into EcoRI-digested pMAL c2x vector (New England BioLabs). The mutant Rma1H1 clones (H58A, C61S, C89S, and K115R) were produced by site-directed mutagenesis using the QuikChange site-directed mutagenesis system (Stratagene). The mutagenic oligonucleotide primers for site-directed mutagenesis are presented in Supplemental Table 1 online. Recombinant MBP-Rma1H1 wild-type and mutant fusion proteins were expressed in Escherichia coli, purified by affinity chromatography using amylose resin (New England BioLabs), and used for in vitro self-ubiquitination assays as described (Cho et al., 2008). Purified MBP-Rma1H1 (500 ng) was incubated in 60 μL ubiquitination reaction buffer [50 mM Tris-HCl, pH 7.5, 2.5 mM MgCl2, 0.5 mM DTT, 4 mM ATP, 5 μg ubiquitin (Sigma-Aldrich), 100 ng Arabidopsis E1 (UBA1), and 100 ng Arabidopsis E2 (UBC8)] at 30°C for 1 h. The full-length Arabidopsis UBA1 (U21814) and UBC8 (DQ027022) clones were obtained from the ABRC. The UBA1 and UBC8 cDNAs were amplified by PCR using primers listed in the Supplemental Table 1 online, digested with EcoRI, and ligated into the pProEx Hta vector (Invitrogen) digested with EcoRI. (His)6-UBA1 and (His)6-UBC8 fusion proteins were purified by affinity chromatography using Ni-NTA superflow resin (Quiagen). Reaction products were separated by SDS-PAGE and subjected to immunoblot analysis using anti-MBP antibody (New England BioLabs) or anti-Ub antibody (Santa Cruz Biotechnology) as described previously (Lee et al., 2006).
Construction of 35S:Rma1H1 Transgenic Plants and Their Phenotypic Analysis
The full-length Rma1H1 and HA-Rma1H1 cDNAs were amplified by PCR using Rma1H1 BamH1 F and Rma1H1 SacI R primers (see Supplemental Table 1 online). PCR products were inserted into the pGEM T Easy vector (Promega) and confirmed by DNA sequencing. Digested BamH1-Rma1H1-SacI and BamH1-HA-Rma1H1-SacI inserts were ligated into the pBI121 binary vector (Clontech) digested with BamHI and SacI. Generation and phenotypic analysis of 35S:Rma1H1 and 35S:HA-Rma1H1 transgenic Arabidopsis plants were performed as described by Cho et al. (2008). Expression of Rma1H1 mRNA in independent transgenic lines was examined by RT-PCR using gene-specific primers for Rma1H1 (see Supplemental Table 1 online). Water loss measurements were conducted by the method described by Li et al. (2008) with the following modifications. Briefly, rosette leaves of wild-type and 35S:Rma1H1 plants, which had been grown under normal conditions for 3 weeks, were excised and dehydrated on Whatman 3MM filter paper at room temperature and ∼60% humidity under dim light. Water loss was determined as the percentage of fresh weight at zero time.
Construction of Plasmid DNAs
To generate 35S:PIP2;1-GFP, 35S:PIP2;1-mRFP and 35S:PIP2;1-HA, PIP2;1 were amplified by PCR using PIP2;1 XhoI F and PIP2;1 BamHI R primers (see Supplemental Table 1 online), digested with XhoI and BamHI, and inserted between the 35S cauliflower mosaic virus promoter and nos-terminator of the pUC-GFP, pUC-mRFP, and pUC-HA vectors, respectively, which were digested with XhoI and BamHI. The construction and structure of the pUC-GFP, pUC-mRFP, and pUC-HA vectors are described by Jin et al. (2001).
To generate 35S:HA-Rma1H1, Rma1H1 amplified by PCR using Rma1H1 SmaI F and Rma1H1 BglII R primers (see Supplemental Table 1 online) was digested with SmaI and BglII and ligated to an N-terminal HA tagging pUC-HA vector digested with SmaI and BglII. To generate 35S:GFP-Rma1H1, Rma1H1 amplified by PCR using Rma1H1 ClaI F and Rma1H1 XhoI R primers (see Supplemental Table 1 online) was digested with ClaI and XhoI and ligated to an N-terminal GFP tagging pUC-GFP vector. The constructs were confirmed by DNA sequencing.
Transient Expression, Treatment with MG 132, in Vivo Targeting of Reporter Protein, and Microscopy
For transient expression, plasmids were introduced by polyethylene glycol–mediated transformation into Arabidopsis protoplasts prepared from leaf tissues (Jin et al., 2001; Lee et al., 2002). Expression of fusion constructs was monitored after transformation and images were captured with a cooled CCD camera and a Zeiss Axioplan fluorescence microscope. Fractionations of the soluble and membrane-bound proteins and subsequent immunoblot analysis were performed as described by Jin et al. (2001). Treatment of protoplasts and transgenic Arabidopsis plants with MG132 was performed as described previously (Cho et al., 2006a). For immunohistochemistry, protoplasts were placed onto poly-L-lysine–coated glass slides and fixed with 3% paraformaldehyde in buffer (10 mM HEPES, pH 7.4, 154 mM NaCl, 125 mM CaCl2, 2.5 mM maltose, and 5 mM KCl) for 1 h at room temperature (Park et al., 2005). Fixed cells were incubated with rat monoclonal anti-HA antibody (Applied Biological Materials) at 4°C overnight and washed with TSW buffer (10 mM Tris, pH 7.4, 0.9% [w/v] NaCl, 0.25% [w/v] gelatin, 0.02% [w/v] SDS, and 0.1% [v/v] Triton X-100) three times. Subsequently, cells were incubated with TRITC-conjugated anti-rat IgG (Zymed). Images were captured as described above.
For confocal laser scanning microscopy, roots were examined with a Zeiss LSM 510 META laser scanning confocal microscope using a C-APOCHROMAT (×40/1.2w numerical aperture water immersion) lens in multitrack mode. Excitation/emission wavelengths were 488/505 to 530 nm for GFP. Transmitted light reference images were captured using differential interference contrast optics and argon laser illumination at 488 nm. Images are presented as stacks of neighboring sections. Data were processed using Adobe Photoshop (Adobe Systems) software, and images are presented in pseudocolor (Jin et al., 2001).
Yeast Two-Hybrid Assay
The full-length Rma1H1 and PIP2;1 cDNAs, or N- and C-terminal deletion constructs of Rma1H1, were amplified by PCR using primers listed in the Supplemental Table 1 online, digested with EcoRI, and ligated into EcoRI-digested pGBKT7 and pGADT7 vectors (Clontech; Matchmaker3), respectively. These constructs and empty vector controls were transformed into yeast strain AH109. Yeast cells were plated onto SD/-His/-Trp/-Leu medium including 10 mM 3-amino-1,2,4-triazole and allowed to grow for 5 d at 30°C. To ensure the nuclear localization of Rma1H1 and PIP2;1, yeast proteins were fractionated into nuclear and cytosolic fractions. Transformed yeast cells were grown on YPAD medium (1% [w/v] Bacto yeast extract, 2% [w/v] Bacto peptone, 80 mg/L adenine hemisulfate, and 4% [w/v] glucose) to mid-log phase and harvested by centrifugation. Pellets were resuspended in zymolyase buffer (50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 M sorbitol, and 30 mM DTT) and treated with 200 units/mL zymolyase. Spheroplasts were collected, resuspended in 1 mL of cold Ficoll buffer (18% Ficoll-400, 10 mM Tris-HCl, pH 7.5, 20 mM KCl, 5 mM MgCl2, 3 mM DTT, 1 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride [PMSF]), and homogenized with a mortar and pestle. To remove cell debris and unlysed spheroplasts, the homogenate was centrifuged (3000g) at 4°C for 10 min. The supernatant was then recentrifuged (20,000g) at 4°C for 20 min. This final supernatant was collected as a cytosolic fraction. The pellet was resuspended with storage buffer (20 mM Tris-HCl, pH 7.5, 0.1 mM EDTA, 10% glycerol, 100 mM KCl, 1 mM DTT, and 1 mM PMSF), mixed with an equal volume of 66% Percoll, and centrifuged (18,000g) at 4°C for 35 min. The band was collected as a nuclear fraction.
In Vitro Pull-Down Assay
The full-length Rma1H1 and PIP2;1 cDNAs were amplified by PCR using Rma1H1 Full F and Rma1H1 R primers and PIP2;1 F and PIP2;1 R primers (see Supplemental Table 1 online), respectively, and digested with EcoRI restriction enzyme. The EcoRI-digested Rma1H1 was inserted into the HA-tagging pProEx Hta-HA vector, while the EcoRI-digested PIP2;1 was ligated into the pMAL c2x vector. HA-Rma1H1 and MBP-PIP2;1 fusion proteins were purified as described by Cho et al. (2008) with the following modifications. An overnight culture of transformed E. coli DH5-α cells containing pHA-Rma1H1-UC and pMBP-PIP2;1-UC plasmids, respectively, were diluted 1:50 in Luria-Bertani medium (10 g Bactotrypton, 5 g yeast extract, and 10 g NaCl per liter) containing 100 μg/L ampicillin and grown for 2 h at 37°C before addition of isopropyl β-d-thiogalactopyranoside to 0.3 mM. After a further 2 h of incubation, cells were pelleted and resuspended in Tris-buffered saline, pH 7.4, containing 1 mM EDTA and 1 mM phenylmethylsulfonyl fluoride (PMSF). Cells were lysed on ice by sonication and centrifuged at 9000g for 30 min at 4°C to remove insoluble materials. The resulting supernatant was applied to an amylase- or HA-affinity column (New England BioLabs) equilibrated with column buffer (20 mM Tris-HCl, pH 7.4, 1 mM EDTA, and 200 mM NaCl). After washing of the column, the fusion proteins were eluted with the same buffer containing 0.5% (w/v) SDS. For in vitro pull-down assays, the bacterially expressed HA-Rma1H1 and MBP-PIP2;1 were coincubated in 30 μL amylose resin (New England BioLabs), washed extensively, and eluted using 10 mM maltose as described previously (Cho et al., 2006a). The eluted protein was resolved by 12.5% SDS-PAGE, transferred to a PVDF membrane, and subjected to immunoblot analysis using anti-HA antibody or anti-MBP antibody (New England BioLabs) as described previously Cho et al. (2006a).
In Vivo Ubiquitination Analyses
In vivo ubiquitination analysis was performed as described by Cho et al. (2008) with the following modifications. Light-grown 2-week-old, intact whole seedlings of wild-type and T3 transgenic plants (35S:PIP2;1-GFP and 35S:PIP2;1-GFP/35S:Rma1H1) were pretreated for 4 h with 10 μM MG132. A total of 30 seedlings for each sample were ground in protein extraction buffer containing 50 mM Tris, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 10% glycerol, and 1× complete protease inhibitor cocktail (Roche). Total proteins (100 μg) were prepared, separated on an 8% SDS-PAGE, and visualized by staining with the Ponceau S solution. The crude extracts were coincubated with anti-GFP antibody and 40 μL protein A-Sepharose (GE Healthcare) and washed three times extensively with IP buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 1 mM MgCl2, and 1× complete protease inhibitor cocktail). The precipitated samples were washed four times with IP buffer and eluted with 0.1 M glycine buffer, pH 2.7. Each sample was separated by SDS-PAGE and subjected to immunoblot analysis using anti-GFP or anti-Ub antibody. The relative intensities of ubiquitinated smear bands of PIP2;1-GFP in 35S:PIP2;1-GFP/35S:Rma1H1 DT plants were determined using MultiGauge version 3.1 software (Fuji Photo Film) and normalized to 1.00 for the ubiquitinated bands of PIP2;1-GFP in 35S:PIP2;1-GFP plants.
Construction of rma3-Knockout/Rma1Rma2-RNAi-Knockdown Plants
The Rma1 and Rma2 cDNAs encompassing 300 bp of the C terminus were PCR amplified in the sense and antisense directions using primers listed in the Supplemental Table 1 online and ligated into the pKANNIBAL vector (http://www.pi.csiro.au/RNAi/vectors.htm) in the XhoI/KpnI sites (XhoI-Rma1Rma2-KpnI) and the XbaI/ClaI sites (XbaI-Rma1Rma2-Cla1), respectively. The plant expression cassette including the NotI-35S:Rma1Rma2-hairpin:OCS-NotI construct was moved from the pKANNIBAL vector into the binary pART27 vector (http://www.pi.csiro.au/RNAi/vectors.htm) digested with NotI. The plasmid was transformed into Agrobacterium tumefaciens strain GV3101 and then used for the transformation into the rma3 knockout mutant plants as described by Cho et al. (2008). Primers used in genotyping PCR and RT-PCR are listed in Supplemental Table 1 online.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: hot pepper Ca-DI6 (BAA28598), Rma1H1 (AY513612), and LEAL1 (AF543310); Arabidopsis PIP2;1 (P43286), Rma1 (At4g03510), Rma2 (At4g28270), Rma3 (At4g27470), UBA1 (U21814), and UBC8 (DQ027022); poplar Pta-Ring protein (AAN05420); rice RING protein (Os4g44820); and human Hs-Rma1 (Q99942).
Author Contributions
I. Hwang and W.T. Kim designed research and wrote the paper, and therefore contributed equally to this article. H.K. Lee, S.K. Cho, O. Son, and Z. Xu performed research. H.K. Lee, S.K. Cho, I. Hwang, and W.T. Kim analyzed data.
Supplemental Data
The following materials are available in the online version of the article.
Supplemental Figure 1. Localization of GXK to the ER.
Supplemental Figure 2. Solubilization of PIP2;1-mRFP by Urea-Containing Denaturation Buffer.
Supplemental Figure 3. Expression of HA-Rma1 Protein in 35S:HA-Rma1 Arabidopsis Transgenic Plants.
Supplemental Figure 4. Molecular Characterization of the rma1, rma2, and rma3 Mutant Lines.
Supplemental Figure 5. Root Growth Patterns of Wild-Type and 35S:Rma1H1 Transgenic Plants in Response to Salt Treatment.
Supplemental Table 1. Primers for PCR, Cloning, and Construction of Transgenic Plants.
Supplementary Material
Acknowledgments
This work was supported in part by grants from the Plant Diversity Research Center (21st Century Frontier Research Program funded by the Ministry of Education, Science, and Technology) and the BioGreen 21 Program (funded by Rural Development Administration) to W.T.K. and by grants from the Creative Research Program of the Ministry of Education, Science, and Technology and the Agricultural R&D Promotion Center (ARPC) of the Ministry of Agriculture, Forestry, and Foods to I.H. S.K.C. and H.K.L. were recipients of Brain Korea 21 (BK21) graduate student scholarship.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Woo Taek Kim (wtkim@yonsei.ac.kr).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Abrami, L., Kunz, B., Iacovache, I., and van der Goot, F.G. (2008). Palmitoylation and ubiquitination regulate exit of the Wnt signaling protein LRP6 from the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 105 5384–5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharon, R., Shahak, Y., Wininger, S., Bendov, R., Kapulnik, Y., and Galili, G. (2003). Overexpression of a plasma membrane aquaporin in transgenic tobacco improves plant vigor under favorable growth conditions but not under drought or salt stress. Plant Cell 15 439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandersson, E., Fraysse, L., Sjovall-Larsen, S., Gustavsson, S., Fellert, M., Karlsson, M., Johanson, U., and Kjellbom, P. (2005). Whole gene family expression and drought stress regulation of aquaporins. Plant Mol. Biol. 59 469–484. [DOI] [PubMed] [Google Scholar]
- Benghezal, M., Wasteneys, G.O., and Jones, D.A. (2000). The C-terminal dilysine motif confers endoplasmic reticulum localization to type I membrane proteins in plants. Plant Cell 12 1179–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert, H.J., Gong, Q., Li, P., and Ma, S. (2006). Unraveling abiotic stress tolerance mechanisms - Getting genomics going. Curr. Opin. Plant Biol. 9 180–188. [DOI] [PubMed] [Google Scholar]
- Boursiac, Y., Boudet, J., Postaire, O., Luu, D.-T., Tournaire-Roux, C., and Maurel, C. (2008). Stimulus-induced downregulation of root water transport involves reactive oxygen species-activated cell signalling and plasma membrane intrinsic protein internalization. Plant J. 56 207–218. [DOI] [PubMed] [Google Scholar]
- Boursiac, Y., Chen, S., Luu, D.-T., Sorieul, M., van den Dries, N., and Maurel, C. (2005). Early effects of salinity on water transport in Arabidopsis roots. Molecular and cellular features of aquaporin expression. Plant Physiol. 139 790–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer, J.S. (1982). Plant productivity and environment. Science 218 443–448. [DOI] [PubMed] [Google Scholar]
- Bray, E.A. (1997). Plant responses to water deficit. Trends Plant Sci. 2 48–54. [Google Scholar]
- Cho, S.K., Chung, H.S., Ryu, M.Y., Park, M.J., Lee, M.M., Bahk, Y.-Y., Kim, J., Pai, H.S., and Kim, W.T. (2006. a). Heterologous expression and molecular and cellular characterization of CaPUB1 encoding a hot pepper U-box E3 ubiquitin ligase homolog. Plant Physiol. 142 1664–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, S.K., Kim, J.E., Park, J.-A., Eom, T.J., and Kim, W.T. (2006. b). Constitutive expression of abiotic stress-inducible hot pepper CaXTH3, which encodes a xyloglucan endotransglucosylase/hydrolase homolog, improves drought and salt tolerance in transgenic Arabidopsis plants. FEBS Lett. 580 3136–3144. [DOI] [PubMed] [Google Scholar]
- Cho, S.K., Ryu, M.Y., Song, C., Kwak, J.M., and Kim, W.T. (2008). Arabidopsis PUB22 and PUB23 are homologous U-box E3 ubiquitin ligases that play combinatory roles in response to drought stress. Plant Cell 20 1899–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushman, J.C., and Bohnert, H.J. (2000). Genomic approaches to plant stress tolerance. Curr. Opin. Plant Biol. 3 117–124. [DOI] [PubMed] [Google Scholar]
- Dong, C.-H., Agarwal, M., Zhang, Y., Xie, Q., and Zhu, J.-K. (2006). The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. USA 103 8281–8286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher, K., and Callis, J. (2007). Ubiquitin, hormones and biotic stress in plants. Ann. Bot. (Lond.) 99 787–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetter, K., Van Wilder, V., Moshelion, M., and Chaumont, F. (2004). Interaction between plasma membrane aquaporins modulate their water channel activity. Plant Cell 16 215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, A.E., Shachar-Hill, B., and Shachar-Hill, Y. (2004). What are aquaporins for? J. Membr. Biol. 197 1–32. [DOI] [PubMed] [Google Scholar]
- Hong, J.-P., and Kim, W.T. (2005). Isolation and functional characterization of the Ca-DREBLP1 gene encoding a dehydration-responsive element binding-factor-like protein 1 in hot pepper (Capsicum annuum L. cv Pukang). Planta 220 875–888. [DOI] [PubMed] [Google Scholar]
- Jang, J.Y., Kim, D.G., Kim, Y.O., Kim, J.S., and Kang, H. (2004). An expression analysis of a gene family encoding plasma membrane aquaporins in response to abiotic stresses in Arabidopsis thaliana. Plant Mol. Biol. 54 713–725. [DOI] [PubMed] [Google Scholar]
- Javot, H., Lauvergeat, V., Santoni, V., Martin-Laurent, F., Guclu, J., Vinh, J., Heyes, J., Franck, K.I., Schaffner, A.R., Bouchez, D., and Maurel, C. (2003). Role of a single aquaporin isoform in root water uptake. Plant Cell 15 509–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, J.B., Kim, Y.A., Kim, S.J., Lee, S.H., Kim, D.H., Cheong, G.W., and Hwang, I. (2001). A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell 13 1511–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson, U., Karlsson, M., Johansson, I., Gustavsson, S., Sjovall, S., Fraysse, L., Weig, A.R., and Kjellbom, P. (2001). The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiol. 126 1358–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo, S., Seo, Y.S., Kim, S.M., Hong, D.K., Park, K.Y., and Kim, W.T. (2006). Brassinosteroid-induction of AtACS4 encoding an auxin-responsive 1-aminocyclopropane-1-carboxylate synthase 4 in Arabidopsis seedlings. Physiol. Plant. 126 592–604. [Google Scholar]
- Jun, S.-S., Choi, H.J., Lee, H.Y., and Hong, Y.-N. (2008). Differential protection of photosynthetic capacity in trehalose- and LEA protein-producing transgenic plants under abiotic stresses. J. Plant Biol. 51 327–336. [Google Scholar]
- Kamsteeg, E.-J., Hendriks, G., Boone, M., Konings, I.B.M., Oorschot, V., van der Sluijs, P., Klumperman, J., and Deen, P.M.T. (2006). Short-chain ubiquitination mediates the regulated endocytosis of the aquaporin-2 water channel. Proc. Natl. Acad. Sci. USA 103 18344–18349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D.H., Eu, Y.-J., Yoo, C.M., Kim, Y.W., Pih, K.T., Jin, J.B., Kim, S.J., Stenmark, H., and Hwang, I. (2001). Trafficking of phosphatidylinositol 3-phosphate from the trans-Golgi network to the lumen of the central vacuole in plant cells. Plant Cell 13 287–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K.-A., Hwang, E.-W., Park, S.-C., Jeong, M.-J., Byun, M.-O., and Kwon, H.-B. (2007). Transgenic tobacco expressing a ring domain-containing protein of Capsicum annuum confers improved cold tolerance. J. Plant Biol. 50 44–49. [Google Scholar]
- Ko, J.-H., Yang, S.H., and Han, K.-H. (2006). Upregulation of an Arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. Plant J. 47 343–355. [DOI] [PubMed] [Google Scholar]
- Kölling, R., and Hollenberg, C.P. (1994). The ABC-transporter Ste6 accumulates in the plasma membrane in a ubiquitinated form in endocytosis mutants. EMBO J. 13 3261–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft, E., Stone, S.L., Ma, L., Su, N., Gao, Y., Lau, O.-S., Deng, X.-W., and Callis, J. (2005). Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol. 139 1597–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H.J., Xiong, L.M., Gong, Z.Z., Ishitani, M., Stevenson, B., and Zhu, J.K. (2001). The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes Dev. 15 912–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.-H., Deng, X.W., and Kim, W.T. (2006). Possible role of light in the maintenance of EIN3/EIL1 stability in Arabidopsis seedlings. Biochem. Biophys. Res. Commun. 350 484–491. [DOI] [PubMed] [Google Scholar]
- Lee, J.-H., Hong, J.-P., Oh, S.-K., Lee, S., Choi, D., and Kim, W.T. (2004). The ethylene-responsive factor like protein 1 (CaERFLP1) of hot pepper (Capsicum annuum L.) interacts in vitro with both GCC and DRE/CRT sequences with different binding affinities: possible biological roles of CaERFLP1 in response to pathogen infection and high salinity conditions in transgenic tobacco plants. Plant Mol. Biol. 55 61–81. [DOI] [PubMed] [Google Scholar]
- Lee, M.H., Min, M.K., Lee, Y.J., Jin, J.B., Shin, D.H., Kim, D.H., Lee, K.H., and Hwang, I. (2002). ADP-ribosylation factor 1 of Arabidopsis plays a critical role in intracellular trafficking and maintenance of endoplasmic reticulum morphology in Arabidopsis. Plant Physiol. 129 1507–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W.-X., Oono, Y., Zhu, J., He, X.-J., Wu, J.-M., Iida, K., Lu, X.-Y., Cui, X., Jin, H., and Zhu, J.-K. (2008). The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 20 2238–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda, N., and Nakano, A. (1998). RMA1, an Arabidopsis thaliana gene whose cDNA suppresses the yeast sec15 mutation, encodes a novel protein with a RING finger motif and a membrane anchor. Plant Cell Physiol. 39 545–554. [DOI] [PubMed] [Google Scholar]
- Maurel, C. (2007). Plant aquaporins: Novel functions and regulation properties. FEBS Lett. 581 2227–2236. [DOI] [PubMed] [Google Scholar]
- Maurel, C., Javot, H., Lauvergeat, V., Gerbeau, P., Tournaire, C., Santoni, V., and Heyes, J. (2002). Molecular physiology of aquaporins in plants. Int. Rev. Cytol. 215 105–148. [DOI] [PubMed] [Google Scholar]
- Moon, J., Parry, G., and Estelle, M. (2004). The ubiquitin-proteasome pathway and plant development. Plant Cell 16 3181–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay, D., and Riezman, H. (2007). Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315 201–205. [DOI] [PubMed] [Google Scholar]
- Oono, Y., et al. (2003). Monitoring expression profiles of Arabidopsis gene expression during rehydration process after dehydration using ca. 7000 full-length cDNA microarray. Plant J. 34 868–887. [DOI] [PubMed] [Google Scholar]
- Paciorek, T., Zazímalová, E., Ruthardt, N., Petrásek, J., Stierhof, Y.D., Kleine-Vehn, J., Morris, D.A., Emans, N., Jürgens, G., Geldner, N., and Friml, J. (2005). Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435 1251–1256. [DOI] [PubMed] [Google Scholar]
- Park, J.A., Cho, S.K., Kim, J.E., Chung, H.S., Hong, J.-P., Hwang, B., Hong, C.B., and Kim, W.T. (2003). Isolation of cDNAs differentially expressed in response to drought stress and characterization of the Ca-LEAL1 gene encoding a new family of atypical LEA-like protein homologue in hot pepper (Capsicum annuum L. cv. Pukang). Plant Sci. 165 471–481. [Google Scholar]
- Park, M., Lee, D., Lee, G.J., and Hwang, I. (2005). AtRMR1 functions as a cargo receptor for protein trafficking to the protein storage vacuole. J. Cell Biol. 170 757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart, C.M., and Eddins, M.J. (2004). Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta 1695 55–72. [DOI] [PubMed] [Google Scholar]
- Preston, G.M., Carroll, T.P., Guggino, W.B., and Agre, P. (1992). Appearance of water channels in Xenopus oocytes expressing red blood cells CHIP28 protein. Science 256 385–387. [DOI] [PubMed] [Google Scholar]
- Qin, F., et al. (2008). Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20 1693–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, Y.S., Kim, E.Y., Mang, H.G., and Kim, W.T. (2008). Heterologous expression and biochemical and cellular characterization of CaPLA1 encoding a hot pepper phospholipase A1 homolog. Plant J. 53 895–908. [DOI] [PubMed] [Google Scholar]
- Shinozaki, K., and Yamaguchi-Shinozaki, K. (2007). Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58 221–227. [DOI] [PubMed] [Google Scholar]
- Siefritz, F., Tyree, M.T., Lovisolo, C., Schubert, A., and Kaldenhoff, R. (2002). PIP1 plasma membrane aquaporins in tobacco: From cellular effects to function in plants. Plant Cell 14 869–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle, J., and Vierstra, R.D. (2004). The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55 555–590. [DOI] [PubMed] [Google Scholar]
- Stone, S.L., Hauksdottir, H., Troy, A., Herschleb, J., Kraft, E., and Callis, J. (2005). Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 137 13–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24 1596–1599. [DOI] [PubMed] [Google Scholar]
- Tyerman, S.D., Bohnert, H., Maurel, C., Steudle, E., and Smith, J.A. (1999). Plant aquaporins: Their molecular biology, biophysics and significance for plant water relations. J. Exp. Bot. 50 1055–1071. [Google Scholar]
- Tyerman, S.D., Niemietz, C.M., and Bramley, H. (2002). Plant aquaporins: Multifunctional water and solute channels with expanding roles. Plant Cell Environ. 25 173–194. [DOI] [PubMed] [Google Scholar]
- Vera-Estrella, R., Barkla, B.J., Bohnert, H.J., and Pantoja, O. (2004). Novel regulation of aquaporins during osmotic stress. Plant Physiol. 135 2318–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra, R.D. (2003). The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 8 135–142. [DOI] [PubMed] [Google Scholar]
- Vij, S., and Tyagi, A.K. (2007). Emerging trends in the functional genomics of the abiotic stress response in crop plants. Plant Biotechnol. J. 5 361–380. [DOI] [PubMed] [Google Scholar]
- Younger, J.M., Chen, L., Ren, H.Y., Rosser, M.F., Turnbull, E.L., Fan, C.Y., Patterson, C., and Cyr, D.M. (2006). Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell 126 571–582. [DOI] [PubMed] [Google Scholar]
- Zelazny, E., Borst, J.W., Muylaert, M., Batoko, H., Hemminga, M.A., and Chaumont, F. (2007). FRET imaging in living maize cells reveals that plasma membrane aquaporins interact to regulate their subcellular localization. Proc. Natl. Acad. Sci. USA 104 12359–12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Yang, C., Li, Y., Zheng, N., Chen, H., Zhao, Q., Gao, T., Guo, H., and Xie, Q. (2007). SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell 19 1912–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J.K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53 247–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.