Abstract
Glutaredoxins (GRXs) have thus far been associated mainly with redox-regulated processes participating in stress responses. However, ROXY1, encoding a GRX, has recently been shown to regulate petal primorida initiation and further petal morphogenesis in Arabidopsis thaliana. ROXY1 belongs to a land plant-specific class of GRXs that has a CC-type active site motif, which deviates from ubiquitously occurring CPYC and CGFS GRXs. Expression studies of yellow fluorescent protein-ROXY1 fusion genes driven by the cauliflower mosaic virus 35S promoter reveal a nucleocytoplasmic distribution of ROXY1. We demonstrate that nuclear localization of ROXY1 is indispensable and thus crucial for its activity in flower development. Yeast two-hybrid screens identified TGA transcription factors as interacting proteins, which was confirmed by bimolecular fluorescence complementation experiments showing their nuclear interaction in planta. Overlapping expression patterns of ROXY1 and TGA genes during flower development demonstrate that ROXY1/TGA protein interactions can occur in vivo and support their biological relevance in petal development. Deletion analysis of ROXY1 demonstrates the importance of the C terminus for its functionality and for mediating ROXY1/TGA protein interactions. Phenotypic analysis of the roxy1-2 pan double mutant and an engineered chimeric repressor mutant from PERIANTHIA (PAN), a floral TGA gene, supports a dual role of ROXY1 in petal development. Together, our results show that the ROXY1 protein functions in the nucleus, likely by modifying PAN posttranslationally and thereby regulating its activity in petal primordia initiation. Additionally, ROXY1 affects later petal morphogenesis, probably by modulating other TGA factors that might act redundantly during differentiation of second whorl organs.
INTRODUCTION
Glutaredoxins (GRXs) are small ubiquitous glutathione-dependent oxidoreductases that play a crucial role in the response to oxidative stress (Fernandes and Holmgren, 2004; Buchanan and Balmer, 2005). According to the amino acid sequences at their active sites, plant GRXs fall into three groups, the CPYC, CGFS, and CC-type classes (Rouhier et al., 2004). The CPYC and CGFS classes are common to all prokaryotes and eukaryotes, whereas the CC-type class is specific for land plants (Lemaire, 2004; Rouhier et al., 2006; Xing et al., 2006). Out of the 31 GRX genes identified in Arabidopsis thaliana, 21 belong to the CC-type class. Comparisons of the GRX subfamily compositions in evolutionarily informative plant species revealed that only the size of the CC-type class expanded during the evolution of land plants, suggesting that these GRXs might have been recruited to participate in the evolution of more complex land plants (Xing et al., 2006). Despite well-established roles of plant thioredoxins in chloroplast and mitochondrial processes, seed development, and germination (Buchanan and Balmer, 2005), knowledge has been scarce about the biological functions and physiological roles of plant GRXs. Recently, we have identified and functionally characterized two CC-type GRX genes, ROXY1 and ROXY2, and demonstrated their function during Arabidopsis flower development (Xing et al., 2005; Xing and Zachgo, 2008). The roxy1 mutant initiates only 2.5 petal primordia on average instead of 4.0 and exhibits abnormalities during further petal morphogenesis (Xing et al., 2005). ROXY1, together with its closest homolog, ROXY2, acts redundantly during anther development and microspore formation (Xing and Zachgo, 2008). Another CC-type GRX gene, GRX480, has been demonstrated to participate in plant defense responses (Ndamukong et al., 2007).
A large body of evidence is accumulating for redox-regulated disulfide bridge formation and protein S-glutathionylation through conserved Cys residues that positively or negatively affect protein activities and, in the case of transcription factors, DNA binding and/or transcriptional activities (Sun and Oberley, 1996; Zheng and Storz, 2000; Heine et al., 2004; Dixon et al., 2005; Gallogly and Mieyal, 2007; Reichheld et al., 2007; Tada et al., 2008). The Arabidopsis transcription factor TGA1 relies on the reduction state of conserved Cys residues to enable its interaction with NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 (NPR1); this interaction is prevented by an intramolecular disulfide bridge in TGA1 (Després et al., 2003). NPR1 and TGA factors are required for salicylic acid (SA)–inducible GRX480 transcription. These proteins can form a ternary complex, implying that the GRX480 protein might be involved in the regulation of the redox state of unbound TGA factors or the TGA/NPR1 complexes (Ndamukong et al., 2007). Another member of the TGA gene family, PERIANTHIA (PAN), is involved in regulating the number of floral organ primorida formation in the first three Arabidopsis floral whorls, such that four sepals, four petals, and six stamens develop. The pan mutant reveals a pentamerous arrangement of floral organs in the first three whorls (Running and Meyerowitz, 1996; Chuang et al., 1999). Thus, PAN, compared with ROXY1, exerts an opposing effect on the initiation of petal primordia.
ROXY1 affects petal formation probably by mediating posttranslational modifications of target proteins. Intracellular localization studies demonstrate the importance of nuclear activity for this CC-type GRX. TGA proteins, including PAN, were identified as ROXY1 interactors in a yeast two-hybrid screen. Together with the data from in planta bimolecular fluorescence complementation (BiFC) interaction experiments and genetic studies, we demonstrate that nuclear interactions of ROXY1 with TGA factors are likely required for normal petal initiation and later petal morphogenesis.
RESULTS
Intracellular Localization of ROXY1
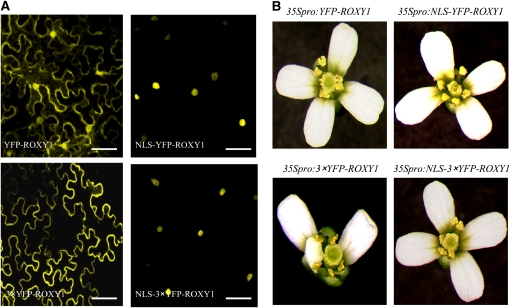
Information about intracellular localization of GRXs is scarce. For plant CPYC or CGFS GRXs, it has been reported that cytosolic and chloroplastic activities are implicated in the assembly and delivery of iron-sulfur clusters (Cheng et al., 2006; Rouhier et al., 2006; Bandyopadhyay et al., 2008). Given the function of ROXY1 in petal primordia initiation and morphogenesis, we were interested in determining where this land plant-specific CC-type GRX exerts its function within the cell. Yellow fluorescent protein (YFP) was fused to the N terminus of ROXY1. The construct was transiently expressed in tobacco (Nicotiana benthamiana) leaf epidermal cells under the control of the cauliflower mosaic virus (CaMV) 35S promoter and then monitored by capturing YFP fluorescence. The YFP-ROXY1 fusion protein was localized in the nucleus and also in the cytoplasm (Figure 1A). Availability of a characteristic petal phenotype in the roxy1-2 mutant, which produces only 2.5 instead of 4.0 petals on average, allowed us to determine the complementation capacity of this fusion protein in roxy1-2 mutant plants expressing the fusion gene. Eighty-nine percent (64/72) of the analyzed transgenic T1 plants formed four wild-type petals, proving that the nucleocytoplasmic expression of YFP-ROXY1 is capable of rescuing the petal phenotype of the roxy1-2 mutant (Figure 1B). This also demonstrates that N-terminal fusions to ROXY1 do not disrupt its function. To discriminate between nuclear and cytoplasmic contributions to the ROXY1 function, we generated fusion proteins of ROXY1 that are either localized in the nucleus or excluded from it and accumulate in the cytoplasm. A nuclear localization signal (NLS) derived from the SV40 large T antigen, which has been shown to be functional in plant cells (Hicks and Raikhel, 1993; Merkle et al., 1996), was fused to the N terminus of YFP-ROXY1, yielding NLS-YFP-ROXY1. This fusion protein was detectable exclusively in the nucleus (Figure 1A). Indeed, this nuclear localization is sufficient to mediate a wild-type-like ROXY1 activity that complements the roxy1-2 petal phenotype. Out of the 65 T1 transgenic plants examined, 54 plants (83%) developed wild-type petals (Figure 1B). Exclusive localization of the ROXY1 protein in the cytoplasm was achieved by cloning three YFP fragments (3×YFP) in-frame upstream of ROXY1, generating 3×YFP-ROXY1 (Figure 1A). Strikingly, the restricted localization to the cytoplasm disturbed the capability to complement the roxy1-2 mutant. Fifty-eight T1 plants were investigated and all showed the roxy1-2 mutant petal phenotype, with a reduced number of petals and the occurrence of later abnormalities, such as formation of smaller or folded petals (Figure 1B). However, functionality of this large fusion protein could be proven. The generation of an NLS-3×YFP-ROXY1 construct allowed us to redirect this ROXY1 fusion protein exclusively to the nucleus (Figure 1A) and as a consequence restored its potential to rescue the petal development of the roxy1-2 mutant. Among the analyzed 73 T1 transgenic lines, 56 lines (77%) formed wild-type petals (Figure 1B). Collectively, these results show that in contrast with other intracellular localization studies of CPYC and CGFS GRXs, nuclear activity of the CC-type GRX ROXY1 is required and sufficient to regulate normal petal development.
Figure 1.
Nuclear Localization of ROXY1 Is Required for Normal Petal Development.
Fusion proteins of ROXY1 engineered by N-terminally incorporating an NLS and/or three YFP fragments were used to examine contributions of cytoplasmic versus nuclear ROXY1 localization to petal development.
(A) Subcellular localization of ROXY1 fusion proteins transiently expressed in N. benthamiana leaves. Fluorescent images show different intracellular localizations of genetically engineered ROXY1 fusion proteins. Bars = 50 μm.
(B) Complementation analysis of ROXY1 fusion genes. Indicated ROXY1 fusion genes driven under the control of the CaMV 35S promoter were transformed in roxy1-2 mutants. At least 58 transgenic T1 plants were examined for each construct, and representative pictures of petal phenotypes are shown.
ROXY1 Interacts Differentially with TGA Factors in Yeast Two-Hybrid Assays
Intrigued by the finding that nuclear ROXY1 activity is crucial for flower development, we aimed to identify target proteins that interact with ROXY1. A yeast two-hybrid screen was conducted using ROXY1 as bait to isolate interaction partners from a prey library containing Arabidopsis cDNAs. Sequence analysis of interacting partners led to the identification of four cDNAs encoding TGA2, TGA3, TGA7, and PAN, respectively, which all belong to the family of TGA transcription factors for whose members nuclear activity has been demonstrated (Després et al., 2000, 2003; Johnson et al., 2003). Retransformation of these prey cDNAs into the GAL4 BD:ROXY1-expressing yeast strain AH109 further confirmed that truncated versions of TGA factors interacted with ROXY1. Moreover, in the absence of ROXY1 (empty bait vector), none of the transformants grew on selection medium. Similarly, the GAL4 BD:ROXY1-containing yeast strain did not grow on selection medium without TGA factors (empty prey vector).
The Arabidopsis genome contains 10 TGA genes (Jakoby et al., 2002), and eight of them (TGA1–TGA7 and PAN) have been further investigated (Running and Meyerowitz, 1996; Chuang et al., 1999; Zhou et al., 2000; Després et al., 2000). Full-length cDNAs for these eight TGA factors were cloned into pGADT7 behind the GAL AD domain and transformed into the GAL4 BD:ROXY1-expressing yeast strain. Prototrophic growth was observed when coexpressing ROXY1 with any of the TGA genes tested. Moreover, ROXY1 protein was found to interact differentially with TGA factors. ROXY1 showed a strong affinity for TGA3, TGA7, and PAN, an intermediate affinity for TGA2, and a weak affinity for TGA1 and TGA4-6, respectively (Table 1).
Table 1.
Interactions of Wild-Type or Mutagenized ROXY1 with TGA Factors Revealed by Yeast Two-Hybrid Screens
| Baita | Preyb | Interactionc |
|---|---|---|
| BD:ROXY1 | AD | − |
| BD | AD:PAN | − |
| BD | AD:TGA1-7 | − |
| BD:ROXY1 | AD:PAN | + + + |
| BD:ROXY1 | AD:TGA3,7 | + + + |
| BD:ROXY1 | AD:TGA2 | ++ |
| BD:ROXY1 | AD:TGA1,4-6 | + |
| BD:ROXY1ΔN1-38d | AD:PAN | + + + |
| BD:ROXY1ΔN1-38 | AD:TGA3,7 | + + + |
| BD:ROXY1ΔI85-98e | AD:PAN | + + + |
| BD:ROXY1ΔI85-98 | AD:TGA3,7 | + + + |
| BD:ROXY1ΔC129-136f | AD:PAN | − |
| BD:ROXY1ΔC129-136 | AD:TGA3,7 | − |
BD, GAL4 DNA binding domain.
AD, GAL4 activation domain.
+ + +, strong growth; + +, intermediate growth; +, weak growth; −, no growth.
ΔN1-38, deletion of the N-terminal extension and the αN.
ΔI85-98, deletion of the intervening region.
ΔC129-136, C-terminal deletion.
Analysis of ROXY1 and TGA Interactions in Planta
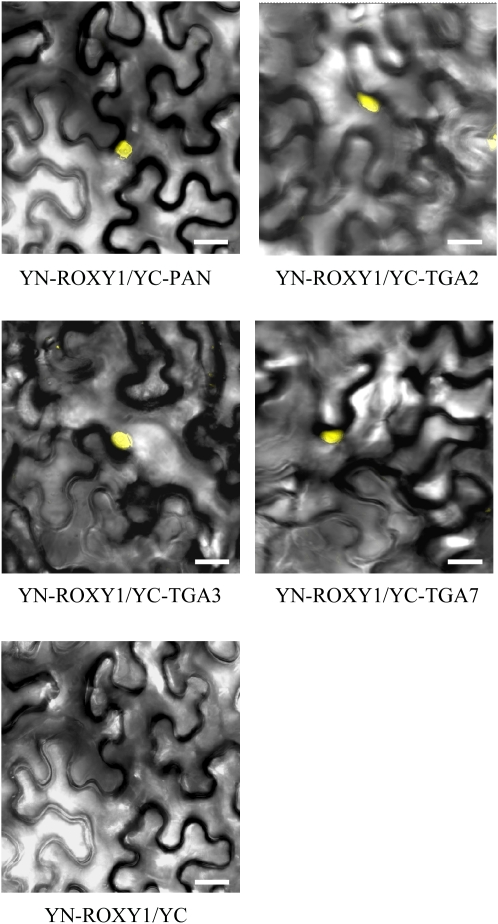
To further investigate the interactions of ROXY1 with TGA factors in planta, the BiFC technique was employed. For BiFC experiments, the N terminus of YFP (YN) was cloned upstream of ROXY1. The C terminus of YFP (YC) was fused N- and C-terminally in-frame to TGA2, TGA3, TGA7, and PAN, individually. By conducting Agrobacterium tumefaciens coinfiltration, constructs were pairwise transiently expressed in abaxial epidermal cells of tobacco (N. benthamiana) leaves (Voinnet et al., 2003). Reconstitution of YFP fluorescence was examined by confocal microscopy 3 to 5 d after transient coexpression of protein pairs. Yellow fluorescence in the nucleus was detected for coexpression of ROXY1 and PAN, TGA2, TGA3, or TGA7, regardless of the two tested fusion orientations for PAN, TGA2, TGA3, or TGA7 (Figure 2; see Supplemental Table 1 online). No yellow fluorescence was observed when coexpressing ROXY1 and each of the other TGA factors that showed only a weak interaction in the yeast two-hybrid experiments. As negative controls, coexpression of nonfused YN with nonfused YC or one of the fusion proteins with nonfused YN or nonfused YC failed to reconstitute a fluorescent YFP chromophore (Figure 2). The in planta nuclear interaction of ROXY1 with TGA2/3/7 and PAN, together with the finding that the nuclear activity of ROXY1 is crucial for its function, suggests that this CC-type GRX might modify TGA factors posttranslationally in the nucleus.
Figure 2.
ROXY1 Interacts with TGA Factors in the Nuclei of Transiently Transformed N. benthamiana Leaves.
The BiFC technique was employed to investigate ROXY1 interactions with TGA factors in planta. The N terminus of YFP (YN) was cloned in-frame upstream of ROXY1. The C terminus of YFP (YC) was cloned in-frame upstream of TGA2, TGA3, TGA7, and PAN individually. Constructs were pairwise transiently expressed in N. benthamiana leaves. Reconstitution of YFP fluorescence was examined by confocal microscopy 3 to 5 d after transient coexpression of protein pairs, and representative pictures are shown. Yellow fluorescence in the nucleus was detected for interactions of ROXY1 with PAN, TGA2, TGA3, and TGA7, respectively. As a negative control, coexpression of YN-ROXY1 with nonfused YC failed to reconstitute a fluorescent YFP chromophore. Bars = 50 μm.
Deletion Analysis of ROXY1
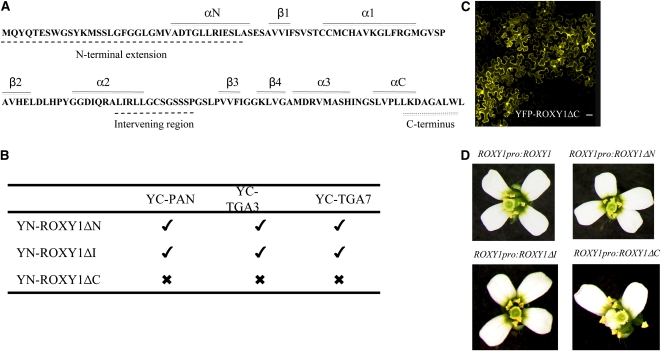
All GRXs share a conserved β1-α1-β2-α2-β3-β4-α3 thioredoxin fold (Holmgren et al., 1975; Rouhier et al., 2002; D'Ambrosio et al., 2003). In addition, all Arabidopsis CC-type GRXs possess a predicted α-helix at the N terminus (αN), a predicted α-helix at the C-terminus (αC), and a special C-terminal extension (Figure 3A). In comparison with other CC-type GRXs, ROXY1 also has a specific 28–amino acid N-terminal extension and an additional 15–amino acid intervening region between α2 and β3, which are also present only in its closest homolog ROXY2 (Figure 3A).
Figure 3.
The C terminus of ROXY1 Is Crucial for Interaction with TGA Factors and Complementation of roxy1-2 Mutants.
(A) The amino acid sequence of ROXY1 is shown using the one-letter code. The ROXY1-specific N-terminal extension, intervening region, and C terminus are highlighted with dashed lines below the sequence. Amino acid residues that form α-helices (αN, α1-3, and αC) and β-sheets (β1-4) are indicated by solid lines above the sequence.
(B) Interactions between three mutated ROXY1 versions (ΔN1-38, ΔI85-98, and ΔC129-136) and PAN, TGA3, and TGA7 in the nuclei of transiently transformed N. benthamiana leaves using BiFC. Ticks indicate that YFP fluorescence was reconstituted by an interaction, whereas crosses denote the inability to detect an interaction.
(C) Nucleocytoplasmic localization of the transiently expressed YFP-ROXY1ΔC fusion protein in N. benthamiana leaves was determined by confocal laser scanning microscopy, proving that the C-terminal deletion of ROXY1 does not affect its intracellular localization. Bar = 50 μm.
(D) Complementation analysis of the three mutant versions of ROXY1. Wild-type and the three mutagenized ROXY1 genes driven by endogenous ROXY1 regulatory sequences were transformed into roxy1-2 mutant plants. Petal phenotypes of transgenic T1 plants were examined, and representative phenotypes are shown.
To determine the effects of these specific regions on the ROXY1 activity, three deletion mutants were constructed: ROXY1ΔN1-38, in which the N-terminal extension including the αN helix was deleted, ROXY1ΔI85-98, lacking the intervening region, and ROXY1ΔC129-136, where the C-terminal eight amino acids were removed. Interactions of these mutagenized proteins with TGA3, TGA7, and PAN were tested in yeast two-hybrid and BiFC experiments (Table 1, Figure 3B). ROXY1ΔN1-38 and ROXY1ΔI85-98 still revealed a strong interaction affinity for the tested TGA factors in the yeast experiments (Table 1). These results are consistent with a recent report that the N terminus of the Arabidopsis CC-type GRX480 protein does not contribute to the interactions with various TGA factors (Ndamukong et al., 2007). However, ROXY1ΔC129-136 failed to interact with any of the tested TGA factors. These findings were confirmed by in planta BiFC experiments. ROXY1ΔN1-38, ROXY1ΔI85-98, and ROXY1ΔC129-136 each were cloned downstream of YN. YC was fused N-terminally to TGA3, TGA7, and PAN individually. Yellow fluorescence reconstitution was observed in the nuclei of tobacco leaf cells when ROXY1ΔN1-38 and ROXY1ΔI85-98 were coexpressed with any of the examined TGA factors (Figure 3B). Although ROXY1ΔC129-136 still showed a nucleocytoplasmic expression (Figure 3C), it lost the ability to interact with these TGA factors in transient expression experiments (Figure 3B), demonstrating the importance of the ROXY1 C terminus for mediating interactions with TGA factors in planta. Finally, the three mutated versions of ROXY1 were introduced into roxy1-2 mutants and expressed under the control of the endogenous regulatory sequences of ROXY1 (Xing et al., 2005). The construct ROXY1pro:ROXY1 served as a positive control and rescued the petal phenotype of roxy1-2 mutants in all analyzed 62 ROXY1pro:ROXY1 T1 plants (Figure 3D). Similarly, all analyzed T1 lines expressing ROXY1ΔN1-38 (72 T1 plants examined) and ROXY1ΔI85-98 (68 T1 plants examined) complemented the roxy1-2 mutant (Figure 3D). By contrast, none of the investigated 57 ROXY1pro:ROXY1ΔC129-136 T1 plants produced wild-type flowers (Figure 3D). These experiments prove that the N-terminal extension and the intervening region are dispensable, whereas the CC-type-specific C terminus is required for interactions with TGA factors and for the ROXY1 protein to exert its function during petal development.
Comparisons of the C Termini for the Arabidopsis CC-Type GRXs
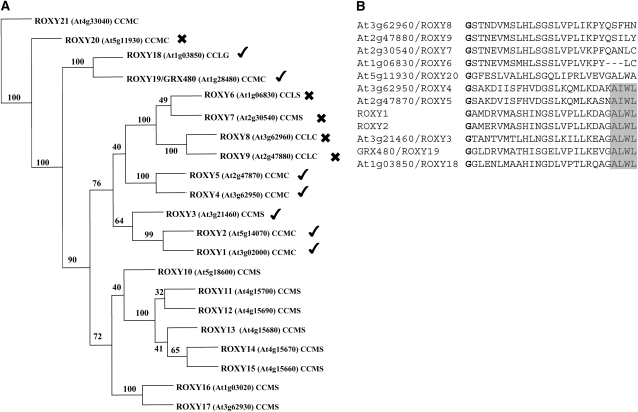
Given the importance of the ROXY1 C terminus, CC-type GRXs with a different composition of the C-terminal extension were further analyzed (Figures 4A and 4B). To this end, a phylogenetic tree was constructed for all CC-type GRXs, including ROXY1 and ROXY2, the first two characterized members of this GRX type (Xing et al., 2005; Xing and Zachgo, 2008). All other CC-type GRXs were named successively ROXY3 to ROXY21, according to decreasing protein sequence similarities with ROXY1 (Figure 4A). Complementation analysis of roxy1-2 mutants was conducted for selected GRX genes driven under the control of ROXY1 endogenous promoter sequences. Besides ROXY1 and ROXY2, serving as positive controls, known to complement the roxy1-2 mutant (Xing and Zachgo, 2008), seven closely related CC-type GRXs (ROXY3 to 9) were tested and three more distantly related ones (ROXY18 yo 20) were included (Figure 4A). This selection comprised at least one GRX as a representative for the different CC-type active site motifs, the CCMC, CCMS, CCLC, and CCLG, respectively. Petal phenotypes of roxy1-2 mutants were rescued in seven out of the 12 tested CC-type GRX genes (60 T1 plants per construct examined; Figure 4A). Alignment of the C-terminal amino acids for the 12 CC-type GRXs (Figure 4B) reveals that all complementing GRXs share a short motif, AL/IWL, located at the end of the C terminus. One CC-type GRX, ROXY20, contains a similar ALWA motif at the C terminus but did not show a ROXY1-like activity, which might be due to a Leu to Ala exchange in the last position (Figures 4A and 4B). These findings further highlight the importance of the ROXY1 C terminus, particularly the presence of an ALWL motif, for its functionality.
Figure 4.
Comparisons of the C termini for Arabidopsis CC-Type GRXs.
(A) Phylogenetic tree of Arabidopsis CC-type GRXs. Numbers at the branches indicate the bootstrap values for 1000 replicates. The alignment used to generate this tree is available as Supplemental Data Set 1 online. Except for the characterized ROXY1/2, all other CC-type GRXs were named successively as ROXY3 to ROXY21, according to decreasing protein sequence similarities with ROXY1. Respective Arabidopsis Genome Initiative codes and active site motifs are indicated, and the previously described GRX480 is denoted. Ticks indicate CC-type GRXs that were able to complement roxy1-2 mutants, and crosses denote those that did not exhibit a ROXY1-like activity and failed to complement the mutant. Expression of CC-type GRXs was driven under the control of ROXY1 regulatory sequences. Unmarked ROXYs were not tested in this study.
(B) Alignment of C-terminal extensions of CC-type GRXs chosen for complementation experiments. Bold letters indicate a conserved Gly, representing the putative glutathione binding sites (G110 in ROXY1). The C-terminal four amino acids A(L/I)WL indicated by a gray box are conserved for all tested CC-type GRXs that could rescue the roxy1-2 mutant petal phenotype.
Overlapping Floral Expression of ROXY1 and TGA Genes
To investigate whether ROXY1 and TGA genes are expressed in overlapping domains, a prerequisite for an in vivo interaction, we conducted in situ hybridization experiments for ROXY1, PAN, and TGA2 during different floral stages (Figure 5). ROXY1 is expressed in regions where floral buds and floral organ primordia are formed (Figures 5A and 5D), confirming earlier results (Xing et al., 2005). Generally, both PAN (Figures 5B and 5E) and TGA2 (Figures 5C and 5F) exhibit a broader expression pattern that includes the region of ROXY1 expression. In young flower buds, PAN and TGA2 transcripts are localized in the floral meristem (Figures 5B and 5C). For PAN, a signal is also detected in the inflorescence apical meristem (Figure 5B), confirming earlier protein localization data (Chuang et al., 1999). ROXY1 continues to be expressed in a few cells in younger developing floral organs, like petals and stamens (Figure 5D). Here, PAN (Figure 5E) and TGA2 (Figure 5F) show hybridization signals, although weaker, throughout petals, stamens, and carpels in flowers of the same developmental stages. Expression of ROXY1 (Figure 5G), PAN (Figure 5H), and TGA2 (Figure 5I) can be detected at later stages of flower development at the inner surface of carpels, where ovules will be formed. In summary, ROXY1, PAN, and TGA2 are coexpressed in floral tissues, including petals, demonstrating that ROXY1/TGA protein interactions can occur in planta.
Figure 5.
Overlapping Expression Domains of ROXY1, PAN, and TGA2 in Floral Tissues.
ROXY1, PAN, and TGA2 antisense probes were hybridized to longitudinal sections through wild-type Arabidopsis flowers. c, carpel; fl, young flower; st, stamen; pe, petal. Bars = 50 μm.
(A) ROXY1 is expressed (blue color) in the inflorescence apex at positions where future floral primordia will be formed. Expression in young flower buds indicates the initiation of sepal and stamen primordia.
(B) and (C) PAN (B) and TGA2 (C) expression in young flowers is less distinct than that of ROXY1. PAN mRNA is detected in several cell layers of the inflorescence apical meristem and the upper half of a young flower bud. Likewise, TGA2 is expressed in cells of the upper floral dome.
(D) to (F) In older flowers, after the initiation of floral organs, ROXY1 shows a distinct expression in young petal and stamen primordia (D), whereas PAN (E) and TGA2 (F) expression patterns are broader and their mRNAs are located in all cells of developing floral organs.
(G) to (I) Later in flower development, ROXY1 (G), PAN (H), and TGA2 (I) mRNAs are localized to the inner carpel surface and delineate the area where ovule primordia will be initiated. Asterisks indicate inflorescence apices in (A) and (B). The arrow in (G) points to the ROXY1 expression domain.
Genetic Interaction of ROXY1 and PAN
Currently, PAN is the sole TGA gene in Arabidopsis with a known function specifically associated with flower development (Chuang et al., 1999). To further test the genetic relationship between ROXY1 and PAN, double mutants between roxy1-2 and pan were generated. Whereas the petal number is reduced in the roxy1-2 mutant and later petal morphogenesis also is disturbed (Figures 6A and 6B), floral organ numbers in the pan mutant are increased in whorls 1 and 2 and decreased in whorl 3, such that a pentamerous pattern is formed (Figure 6C), confirming data obtained from other pan alleles (Running and Meyerowitz, 1996; Chuang et al., 1999). In roxy1-2 pan double mutants, similar to the pan single mutant, flowers develop a pentamerous phenotype and are composed of 4.8 ± 0.6 sepals, 4.7 ± 0.5 petals, and 4.4 ± 0.7 stamens (400 flowers from 20- to 40-d-old plants were examined; values are means ± se) in the first three whorls, respectively (Figures 6D to 6F). Thus, PAN is epistatic to ROXY1 with respect to its function of regulating petal primorida initiation. However, pentamerous double mutant petals also display an abnormal later petal morphogenesis, such as the formation of smaller or folded petals (Figures 6D to 6F). This effect on later stages of petal development is also observed in roxy1-2 single mutants. Therefore, ROXY1 likely exerts an additional function that acts independently of PAN.
Figure 6.
Interaction between ROXY1 and PAN.
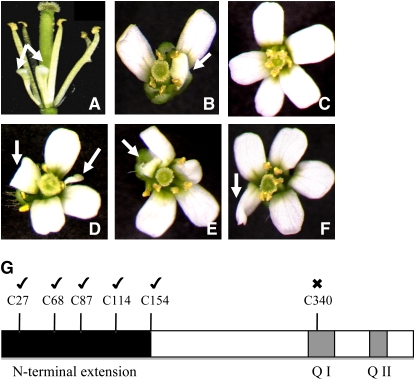
(A) and (B) show typical petal phenotypes of roxy1-2 mutants, where petals remain often smaller or folded. The roxy1-2 pan double mutant (D) to (F) develops pentamerous flowers resembling those of the pan single mutant (C). Additionally, later petal morphogenesis is affected in double mutants that also form smaller (D) or folded petals in (D) to (F), thus resembling later roxy1-2 petal differentiation.
(G) C340 is essential for PAN functionality during flower development. Schematic representation of the PAN protein indicates the positions of its six Cys residues (top), a specific N-terminal extension (black box), and two conserved Gln-rich regions (Q I and Q II, gray boxes). C27, C68, C87, C114, and C154 and C340 were individually mutagenized into Ser residues. For complementation experiments, the pan mutant plants were transformed with wild-type and mutated versions of PAN genes driven under the control of the CaMV 35S promoter. Flower phenotypes of transgenic T1 plants carrying each construct were examined. Ticks or crosses above Cys residues denote if mutagenized PAN proteins were still able to rescue the pan mutant phenotype (tick) or failed to exhibit a wild type-like PAN protein activity (cross). Arrows indicate altered petal morphogenesis, such as smaller or folded petals.
Mutagenesis of PAN Cys Residues
As a GRX, ROXY1 likely modifies Cys residues in target proteins posttranslationally. As our data show physical and genetic interactions of PAN and ROXY1, we further tested the importance of different Cys residues in the PAN protein that might represent target sites for modification. Out of the six Cys residues present in PAN, five of them, C27, C68, C87, C114, and C154, are located in a PAN-specific N-terminal extension (Figure 6G). The sixth Cys, C340, is localized in one of the two conserved Gln-rich regions that may function as transactivation domains (Figure 6G; Després et al., 2003). All Cys residues were individually mutagenized and converted into Ser residues. Wild-type and the six mutated PAN cDNAs were put under the control of the CaMV 35S promoter and introduced into the pan mutant, and the complementation capacity of mutagenized proteins was analyzed in T1 transformants. The wild-type and five N-terminally mutated PAN proteins (C27S, C68S, C87S, C114S, and C154S) were able to complement the pan mutant (50 T1 plants examined for each construct, all producing wild-type flowers; Figure 6G). However, the PAN protein with a mutagenized C340 failed to exert a wild-type-like activity (50 T1 plants examined, all forming pan mutant flowers; Figure 6G). These data show that the conserved C340 is crucial for PAN activity in flower development and this Cys might therefore represent a target for a posttranslational modification by ROXY1.
The Chimeric PAN Repressor Disturbs Petal Development
TGA factors are known to act redundantly in the regulation of pathogen response (PR) gene expression and disease resistance in Arabidopsis (Zhang et al., 2003; Kesarwani et al., 2007). The floral expression of PAN and TGA2 raises the possibility that TGA factors might act redundantly during flower development. The chimeric repressor gene-silencing technology (CRES-T) was employed to overcome experimental limitations caused by functional redundancy of transcription factor families (Hiratsu et al., 2003; Koyama et al., 2007; Heyl et al., 2008). PAN was fused to the EAR-motif repression domain (SRDX) that has been shown to dominantly repress the target gene expression of transcription factors and was expressed under the control of the CaMV 35S promoter (35Spro:PANSRDX) in wild-type plants. Ectopic expression of PANSRDX induced severe defects during flower development (500 flowers from 50 T1 transgenic plants and 10 flowers per T1 plant examined; Figure 7), whereas vegetative development was not affected. Floral phenotypes of 35Spro:PANSRDX T1 transgenic plants could be classified into four groups: tetramerous, wild-type flowers developing four normal petals (10.6%, Figure 7A), pentamerous flowers with five normal sepals, petals, and stamens (19.0%; Figure 7B), tetramerous flowers forming four petals with normal, small, or folded shapes (19.6%; Figures 7C and 7D), and pentamerous flowers with five petals that are normal, small, or folded and five normal sepals and stamens (50.8%; Figures 7E and 7F). A similar phenotype distribution was observed when we expressed PANSRDX in wild-type plants under the control of a 2.5-kb-long 5′-upstream region of the PAN gene (see Supplemental Table 2 online). Overall, the majority of flowers in the transgenic 35Spro:PANSRDX and PANpro:PANSRDX plants thus resembled those of the roxy1-2 pan double mutant, which also showed a pentamerous second whorl pattern and abnormalities in later petal morphogenesis. The pentamerous pan-like floral phenotype is likely caused by specific repression of PAN target genes by the CRES-T approach. However, the additional phenotype, an altered petal morphogenesis, cannot solely be attributed to a reduced PAN activity. This phenotype might be caused by the chimeric PAN repressor having also silenced activities of other TGA factors, which participate redundantly in the regulation of petal morphogenesis.
Figure 7.
Petal Phenotypes Induced by the Chimeric PAN Repressor.
To overcome functional redundancy of TGA factors, PAN was fused to the EAR-motif repression domain (SRDX). Ectopic expression of PANSRDX induced severe defects during flower development (500 flowers, 10 flowers from 50 T1 plants, were examined). Floral phenotypes of 35Spro:PANSRDX T1 transgenic plants grouping into four distinct classes are shown.
(A) Tetramerous wild-type-like flowers with four normal petals (10.6%).
(B) Pentamerous flowers with five normal petals resembling those of the single pan mutant (19.0%).
(C) and (D) Tetramerous flowers with normal, smaller, or folded petals resembled those of the single roxy1-2 mutant (19.6%). The majority of analyzed flowers was pentamerous (50.8%) and developed normal but also smaller (E) or folded petals (F), thus resembling those of the roxy1-2 pan double mutant. Abnormal petals, such as smaller or folded ones, are marked by arrows.
DISCUSSION
GRXs are small oxidoreductases that reduce disulfide bonds reversibly in response to redox alterations. Unexpectedly, ROXY1, a land plant-specific Arabidopsis CC-type GRX, was found to regulate petal development (Xing et al., 2005). Therefore, the identification of ROXY1-interacting proteins is of importance to understand the molecular mechanism by which ROXY1 exerts its function in petal development. Our data show that the ROXY1 protein acts in the nucleus, where it likely modulates activities of TGA factors and thereby contributes to normal petal development.
ROXY1 Interacts with TGA Transcription Factors
Yeast two-hybrid assays were used to isolate ROXY1 interaction partners and led to the identification of four TGA proteins: TGA2, TGA3, TGA7, and PAN. Further screening showed that ROXY1 interacts differentially with the eight members of the TGA family of basic leucine zipper (bZIP) transcription regulators: ROXY1 shows a strong affinity for TGA3, TGA7, and PAN, an intermediate affinity for TGA2, and a weak affinity for TGA1 and TGA4-6. Furthermore, in planta BiFC experiments revealed nuclear interactions of ROXY1 with the TGA factors that exhibit strong and intermediate binding affinities for ROXY1 in the yeast two-hybrid experiments. In situ hybridization experiments for the selected TGA genes, PAN and TGA2, revealed overlapping mRNA expression patterns with ROXY1 during flower development and thus demonstrate that ROXY1/TGA protein interactions can occur in vivo. The identification of TGA factors as ROXY1 interaction partners might be biologically relevant as two Cys residues of TGA1, C260 and C266, were previously shown to be reduced in vivo after SA treatment (Després et al., 2003). The specific interactions of ROXY1 with TGA factors suggest that ROXY1 could be involved in posttranslational modifications of TGA factor activities and thereby various TGA factor-mediated cellular processes. This notion is further supported by the finding that the SA-inducible CC-type ROXY19/GRX480 protein also interacts with several TGA factors and plays a role in the SA/jasmonic acid (JA) crosstalk by repressing JA-responsive PDF1.2 transcription (Ndamukong et al., 2007).
The analyzed specific N-terminal and intervening regions of the ROXY1 protein are neither required for interactions with TGA factors nor for its functionality in petal development. Contrarily, the specific ROXY1 C terminus is crucial for the ROXY1/TGA protein interactions and the ROXY1 function in petal development. ROXY1ΔC129-136 failed to interact with TGA factors in yeast two-hybrid and BiFC experiments and could not rescue the roxy1-2 petal phenotype, although the C-terminal deletion of ROXY1 does not affect its intracellular localization. So far, ROXY2, the closest ROXY1 homolog, sharing >70% amino acid identity with ROXY1, has been shown to exert ROXY1-like activities (Xing and Zachgo, 2008). The capability of other closely (ROXY3-6) and more distantly related (ROXY18-20) Arabidopsis CC-type GRXs to complement the mutant was analyzed. We show that ROXY3-5 and ROXY18/19 can exert ROXY1-like activities if they are expressed under the control of ROXY1 regulatory sequences. These experiments allowed the identification of a conserved AI/LWL C-terminal motif present in the seven (out of 12 investigated) GRXs that can functionally replace the ROXY1 protein. All analyzed CC-type GRXs sharing this AI/LWL motif therefore seem to be able to interact with ROXY1 target proteins and exert similar posttranslational protein modifications. The importance of the C terminus is further highlighted by the observation that the closest ROXY1/2 homologs from rice (Oryza sativa), Os ROXY1/2, also share this AI/LWL motif and can complement the roxy1-2 mutant (Wang et al., 2008). ROXY4, one of the GRXs that we found to complement roxy1-2, has been recently shown to participate in GA signaling and is a target gene upregulated by DELLA (Hou et al., 2008). However, if expressed under the control of ROXY1 endogenous regulatory sequences, it exerts a ROXY1-like function in petal development. Together, our data emphasize the importance of expression regulation as a crucial mechanism by which specificity is conferred to the function of CC-type GRXs sharing conserved C-terminal extensions.
Nuclear Activity of ROXY1 Is Required for Petal Development
Plant GRXs can be targeted to multiple subcellular compartments as predicted by bioinformatic approaches (Lemaire, 2004; Rouhier et al., 2004), and cytosolic or chloroplastic activities of several CPYC and CGFS GRXs have been analyzed. Cytosolic poplar (Populus trichocarpa) CPYC GRXC1, chloroplast-localized CGFS GRXS14 from Arabidopsis, and poplar and GRXS16 from Arabidopsis and poplar exist as dimeric inactive holoproteins containing a subunit-bridging [2Fe-2S] cluster and are implicated in the assembly and delivery of iron-sulfur clusters (Cheng et al., 2006; Rouhier et al., 2007; Bandyopadhyay et al., 2008). Fe-S clusters have been proposed to function as redox sensors for the activation of human GRX2 in the response to oxidative stress (Lillig et al., 2005), suggesting the possible involvement of plant GRXs in oxidative stress sensing or iron-sulfur assembly. Pteris vittata GRX5, an arsenate-activated chloroplast-localized CGFS GRX, regulates cellular arsenite levels and arsenic resistance (Sundaram et al., 2008). The CaMV 35S promoter-driven expression of YFP-ROXY1 fusion genes shows a nucleocytoplasmic distribution of ROXY1. To discriminate between nuclear and cytoplasmic contributions of ROXY1 to petal development, exclusive localization of ROXY1 proteins in the nucleus was obtained by fusing a NLS to ROXY1. Complementation experiments proved the functionality of this redirected fusion protein. On the contrary, the fusion of 3×YFP to ROXY1 caused a cytoplasmic protein localization that failed to rescue the roxy1-2 petal phenotype, although we show that the functionality of this fusion protein was not disturbed by the addition of 3xYFP. This demonstrates a nuclear activity of a plant GRX. Given the observed interaction with TGA factors, it is therefore likely that ROXY1 acts as an oxidoreductase in the nucleus, where it might posttranslationally modify transcription factors.
Dual Role of ROXY1 in Petal Initiation and Morphogenesis
It has been demonstrated for different organisms that GRXs are able to modulate transcription factor activities and thereby participate in the regulation of various cellular processes. In yeast, the CGFS GRX3 and GRX4 interact with the transcription factor Sc AFT1, which induces the transcription of iron regulon genes in iron-deficient yeast (Ojeda et al., 2006). Overexpression of PICOT, a human GRX, inhibits transactivation activities of AP-1 and NF-κB (Witte et al., 2000).
TGA factors can bind to a TGAC/G motif in the PDF1.2 promoter and the SA-inducible ROXY19/GRX480 interacts with TGA factors and represses JA-responsive PDF1.2 transcription (Ndamukong et al., 2007). Many TGA factors regulate stress responses, such as PR gene expression, redundantly. PAN represents a thus far exceptional TGA gene, as it participates in flower development and regulates the initiation of Arabidopsis floral organ primordia. In the first three floral whorls of the pan mutant, five organ primordia are initiated, giving rise to the formation of a pentamerous flower (Running and Meyerowitz, 1996; Chuang et al., 1999). Contrarily, in the roxy1-2 mutant, a reduced petal number is initiated, only 2.5 petals are formed on average, and additionally later petal morphogenesis is disturbed. Double mutant analysis revealed that roxy1-2 pan plants develop pentamerous flowers, indicating that PAN is epistatic to ROXY1 with respect to its function in the regulation of petal primordia initiation. Moreover, pentamerous petals of double mutants also exhibit abnormal morphogenesis, such as the formation of smaller or folded petals, resembling petal abnormalities observed in roxy1-2 single mutants. This phenotype implicates an additional function for ROXY1, where the ROXY1 protein can act independently of PAN and likely modifies additional proteins involved in later petal morphogenesis. CRES-T has been used to repress target gene expression of transcription activators fused to the EAR-motif repression domain (SRDX), thereby overcoming functional redundancy of transcription activators (Hiratsu et al., 2003; Koyama et al., 2007; Heyl et al., 2008). Functionality of the CRES-T was supported by the observation that ∼19.0% of flowers in T1 transgenic plants formed pan-like pentamerous flowers, most likely due to successful silencing of PAN target gene expression. This indicates that PAN probably activates genes that prevent petal primorida initiation. Surprisingly, the majority of T1 transgenic flowers (50.8%) resembled those of the roxy1-2 pan double mutant, where, in addition to the formation of pentamerous flowers, further abnormalities occurred during later petal morphogenesis. These later petal defects were also observed in 19.6% of examined flowers that did not form a pentamerous second whorl. Thus, in these plants, likely only TGA factors redundantly acting during later petal morphogenesis were silenced, and the CRES-T thus seemed to have uncoupled the dual function of TGA factors in petal development. To summarize, overlapping floral expression of ROXY1, PAN, and TGA2 yeast two-hybrid, BiFC, and genetic interaction data all together indicate that besides PAN, other TGA genes seem to participate in petal development, namely during later stages of petal differentiation.
In a previous study, the existence of two different forms of the PAN protein, along with the near absence of a phenotype in plants ectopically expressing PAN as well as the contrast between the broad expression of PAN and the specificity of its mutant phenotype, suggested that the PAN activity might be posttranslationally regulated (Chuang et al., 1999). Mutagenesis of six different PAN Cys residues followed by complementation experiments demonstrates that C340, present in one of the two Gln-rich regions that may function as transactivation domains and conserved in several TGA factors, is essential for PAN function. Thus, C340 likely represents a target site for modification by ROXY1. In the corresponding putative transactivation domain of TGA1, two critical Cys residues that form an intramolecular disulfide bridge and thereby preclude the interaction with NPR1 were identified (Després et al., 2003). TGA factors thus seem to contain crucial Cys residues that represent putative target sites for posttranslational modifications affecting their DNA binding and/or transcriptional activity.
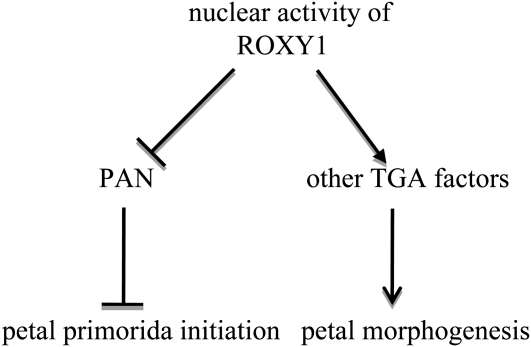
Based on our genetic, mRNA expression, and protein interaction data, a model is proposed to explain the dual role of a nuclear ROXY1 activity in early petal primordia initiation and later petal morphogenesis (Figure 8). Given the opposing effects of PAN and ROXY1 on petal initiation, where PAN decreases and ROXY1 enhances the number of initiated petal primordia, ROXY1 likely inhibits PAN activity in this pathway. By contrast, during later petal development, ROXY1 seems to exert a positive effect on the activities of other TGA factors to allow normal petal morphogenesis to occur. Interestingly, TGA factors that function both in floral organ development and in defense responses seem to be targets of CC-type GRXs. This modification mechanism might thus represent an important, conserved function of land plant-specific GRXs that has been integrated in different processes during the evolution of more complex land plants.
Figure 8.
ROXY1 Activities during Petal Development.
Intracellular localization studies coupled with complementation data prove that the nuclear activity of ROXY1 is required for petal development. Genetic and protein interaction studies unraveled two distinct ROXY1 functions during petal organogenesis. At the early stage of petal development, ROXY1 regulates the number of petal primordia probably by negatively regulating the PAN activity. PAN suppresses a pentamerous flower patterning and reduces the petal organ number such that a tetramerous second whorl is formed. Later in petal development, ROXY1 likely positively regulates other TGA factors, which seem to act redundantly to govern normal petal morphogenesis. Arrows and hatchets indicate positive and negative interactions, respectively.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana mutants and wild-type plants are all Columbia ecotypes. Arabidopsis seeds were sown in soil and treated at 4°C for a week. Both tobacco (Nicotiana benthamiana) and Arabidopsis plants were grown in the greenhouse under controlled environmental conditions (21 to 23°C, 100 μmol photons m−2 s−1, 60% relative humidity, 16 h light/8 h dark) and irrigated from below.
Yeast Two-Hybrid Assays
To identify target proteins that interact with ROXY1, the intronless ROXY1-coding sequence was PCR amplified from Arabidopsis genomic DNA and fused in-frame as bait to the DNA binding domain of GAL4 (GAL4 BD) in the bait vector pGBKT7 by EcoRI and BamHI restriction sites. The mere expression of this chimeric construct was not sufficient to activate marker genes in Y187 or AH109, thus enabling its use in screening ROXY1 interaction partners. An Arabidopsis whole-plant cDNA library fused to the coding sequence of the GAL4 transcriptional activation domain (GAL4 AD) of the prey vector pGADT7 (a gift from Hans Sommer, MPIZ) was transformed into the yeast strain AH109, serving as prey to identify ROXY1 interaction partners on plates containing SD/-Trp-His-Leu-Ade and 3-amino-1,2,4-triazole (3-AT). After mating, yeast cells were plated on selection medium as above, colonies were picked, and the corresponding cDNA inserts isolated. To examine whether all eight TGA factors interact with ROXY1, full-length cDNAs were PCR amplified, cloned into pGADT7, and introduced into the yeast strain AH109 using a recombination strategy as detailed in Supplemental Methods online. After cotransformation, yeast cells were plated on selection medium containing SD/-Trp-His-Leu-Ade and 3-amino-1,2,4-triazole (3-AT) and then incubated in a growth chamber at 28°C for 2 to 5 d. To examine whether the N terminus, the intervening region, or the C terminus is crucial for ROXY1/TGA protein interactions, three mutated versions of ROXY1 (ROXY1ΔN1-38, the N-terminal extension including the αN helix was deleted; ROXY1ΔI85-98, the intervening region were removed; and ROXY1ΔC129-136, lacking the C-terminal eight amino acids) were constructed and fused in-frame as bait to the DNA binding domain of GAL4 (GAL4 BD) in the bait vector pGBKT7 by EcoRI and BamHI restriction sites, respectively. Primers used for cloning are listed in Supplemental Table 3 online.
Transient Expression Assay
Leaves from 3- to 4-week-old N. benthamiana plants were transformed by infiltration using a 1-mL syringe of Agrobacterium tumefaciens cells harboring appropriate plasmids. A. tumefaciens strain GV3101 (pMP90RG) was used to examine intracellular localization of the YFP-ROXY1fusion proteins. YFP was fused to the N terminus of ROXY1, generating YFP-ROXY1. An NLS derived from the SV40 large T antigen (Hicks and Raikhel, 1993; Merkle et al., 1996) was fused to the N terminus of YFP-ROXY1, yielding NLS-YFP-ROXY1. Cloning three YFP fragments (3×YFP) in-frame upstream of ROXY1 generated 3×YFP-ROXY1. The generation of a NLS-3×YFP-ROXY1 construct was achieved by N-terminally fusing the same NLS to 3×YFP-ROXY1. ROXY1 fusion genes encoding these ROXY1 fusion proteins were introduced into the pBAR-35S expression vector by XmaI and XbaI restriction sites (Xing et al., 2005) and transiently expressed in tobacco leaf epidermal cells under the control of the CaMV 35S promoter and then monitored by capturing YFP fluorescence. To further confirm whether wild-type and three mutagenized ROXY1 proteins (ROXY1ΔN1-38, ROXY1ΔI85-98, and ROXY1ΔC129-136) as shown above interact in planta with TGA factors, the BiFC technique was applied. GATEWAY-compatible vectors, pE-SPYNE, pE-SPYCE, and pSYC (Walter et al., 2004) were used to generate expression vectors using a GATEWAY cloning strategy (www.invitrogen.com), where fusion genes are under the control of the CaMV 35S promoter. The N terminus of YFP (YN) was cloned upstream of wild-type and three mutagenized ROXY1 proteins in the pE-SPYNE vector. The C terminus of YFP (YC) was in-frame fused N-terminally for pE-SPYCE and C-terminally for pSYC to TGA2, TGA3, TGA7, and PAN individually. Constructs were transformed into A. tumefaciens strain GV3101 (pMP90RK). To avoid cosuppression, A. tumefaciens strain GV3101 (pMP90RG) carrying the p19 viral silencing suppressor gene was always included in the infiltration mixture (Voinnet et al., 2003). Infiltrated tobacco leaves stayed attached to the plants, and after 3 to 5 d, leaves were examined for reconstitution of YFP fluorescence and images captured with a Leica TCS SP2 AOBS confocal laser scanning microscope. Coexpression of nonfused YN with nonfused YC or one of the fusion proteins with nonfused YN or nonfused YC was used as a negative control. Primers used for cloning are listed in Supplemental Table 3 online.
Phylogenetic Analysis
Protein sequences for all CC-type Arabidopsis GRXs (ROXY1-21 accession numbers are shown in Supplemental Data Set 1 online) were aligned using the ClustalW program with default settings (http://www.ebi.ac.uk/clustalw) and adjusted manually (see Supplemental Data Set 1 online). A neighbor-joining tree was constructed with the program package PHYLIP (Phylogeny Inference Package, v3.573c, Department of Genetics, University of Washington, Seattle) and visualized with Treeview software (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html). Bootstrap analysis was performed with 1000 replications and obtained bootstrap values are indicated.
Construction of roxy1-2 pan Double Mutants
To obtain roxy1-2 pan double mutants, roxy1-2 was crossed with a homozygous pan line (SALK_057190, carrying a T-DNA insertion in the DNA sequence encoding the second exon of PAN). All F1 plants developed wild-type flowers. A segregating F2 population was genotyped by PCR, and 20 roxy1-2 pan double mutants were identified and phenotypically analyzed. Flower phenotypes were documented using a binocular microscope LEICA MZ-FLIII, and images were made using a digital camera (KY-F70). Primers used for PCR genotyping are listed in Supplemental Table 3 online.
Complementation Experiments
To test whether three deleted versions of ROXY1 (ΔN1-38, ΔI85-98, and ΔC129-136) or the 12 selected CC-type GRXs exert a ROXY1-like activity and complement roxy1-2 mutants, respective cDNAs were cloned in pGSA1252 (www.chromdb.org) by XbaI restriction sites, upstream of a NOS terminator and downstream of a 3.6-kb ROXY1 promoter fragment known to confer its endogenous expression (Xing et al., 2005). Secondary structure prediction for ROXY1 allowing the identification of ROXY1-specific regions was conducted using the Protein Sequence Analysis tool (http://bmerc-www.bu.edu/psa). As described below, intracellular ROXY1 localization studies were performed using the pBAR-35S vector (XmaI and XbaI restriction sites) by driving the ROXY1 expression under the control of the CaMV 35S promoter, which has been demonstrated to complement the roxy1-2 mutant (Xing et al., 2005). Nuclear localization of YFP-ROXY1 was obtained by fusing an NLS derived from the SV40 large T antigen to its N terminus. Cytoplasmic ROXY1 localization was achieved by adding three YFP fragments to the N terminus and further addition of an NLS to this construct redirected the protein to the nucleus and thereby restored its functionality in the roxy1-2 complementation test. Individual substitutions of all PAN Cys residues by Ser residues were performed by a PCR-based mutagenesis approach using paired mutagenic oligomers, and mutated versions of PAN were cloned into pBAR-35S by XmaI and XbaI restriction sites. As described previously by Hiratsu et al. (2003), the chimeric PAN-SRDX repressors were generated by fusing the SRDX-coding sequence to the C terminus of the PAN coding region, introduced by XmaI and XbaI restriction sites into pBAR-35S for ectopic expression under the control of the CaMV 35S promoter and into pBAR-A for expression in the genomic context of PAN using a 7020-bp genomic fragment. All these constructs were introduced into A. tumefaciens strain GV3101 (pMP90RG), and transformation of wild-type plants or mutants was performed with the floral dip method (Clough and Bent, 1998). T1 transgenic plants were obtained by spraying with a 0.2% (v/v) BASTA solution. For complementation analyses of roxy1-2 mutants, petal phenotypes of at least 58 T1 transgenic plants were examined. For complementation experiments of pan mutants and phenotypic analysis of PAN-SRDX transgenic plants, floral organ phenotypes were analyzed in at least 50 T1 transgenic plants. Primers used for cloning are listed in Supplemental Table 3 online.
mRNA in Situ Hybridization
Reverse primers containing the T7 polymerase binding site (5′-TAATACGACTCACTATAGGG-3′) and corresponding forward primers were used to prepare PCR templates for the production of PAN and TGA2 antisense probes using cDNAs of PAN and TGA2 (Zachgo, 2002). ROXY1 probe preparation was conducted as described (Xing et al., 2005). Digoxigenin-labeled riboprobes were prepared using DIG RNA labeling mix and T7 Polymerase (Roche). Hybridization and detection were performed as described previously (Zachgo, 2002). Primers used are listed in Supplemental Table 3 online.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: NM180942 (TGA1), NM120777 (TGA2), NM102057 (TGA3), NM121041 (TGA4), NM120778 (TGA5), NM202564 (TGA6), NM106441 (TGA7), AK118286 (PAN), AY910752 (ROXY1), EU332351 (ROXY2), and FJ611903-611921 (ROXY3-21).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. ROXY1 Interacts with TGA Factors in the Nuclei of Transiently Transformed Nicotiana benthamiana Leaves When the C Terminus of YFP Was in-Frame Fused C-Terminally to TGA2, TGA3, TGA7, and PAN Individually.
Supplemental Table 2. Floral Phenotypes Induced by Expression of the Chimeric PAN Repressor (PANSRDX) under the Control of the Endogeneous Regulatory Sequence of PAN (PANpro:PANSRDX).
Supplemental Table 3. Primers Used in This Study.
Supplemental Methods. The Recombination Strategy Used for Cloning Coding Sequences of PAN and TGAs into the pGADT7-rec Yeast Expression Vector.
Supplemental Data Set 1. Protein Sequence Alignment of All CC-Type GRXs in Arabidopsis.
Supplementary Material
Acknowledgments
We thank Ralph Panstruga (MPIZ Köln) for BiFC vectors, Elmon Schmelzer (MPIZ Köln) for assistance with confocal microscopy, and Hans Sommer (MPIZ) for the Arabidopsis yeast two-hybrid library. Seeds for the pan mutant line SALK_057190 were obtained from Jan Lohmann (University of Heidelberg). S.Z. is grateful to Heinz Saedler (MPIZ Köln) for supporting our ROXY work. This work was supported by grants from the Deutsche Forschungsgemeinschaft to S.Z. (SFB 635 and ZA259/4).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Sabine Zachgo (zachgo@biologie.uni-osnbrueck.de).
Online version contains Web-only data.
References
- Bandyopadhyay, S., Gama, F., Molina-Navarro, M.M., Gualberto, J.M., Claxton, R., Naik, S.G., Huynh, B.H., Herrero, E., Jacquot, J.P., Johnson, M.K., and Rouhier, N. (2008). Chloroplast monothiol glutaredoxins as scaffold proteins for the assembly and delivery of [2Fe-2S] clusters. EMBO J. 27 1122–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan, B.B., and Balmer, Y. (2005). Redox regulation: A broadening horizon. Annu. Rev. Plant Biol. 56 187–220. [DOI] [PubMed] [Google Scholar]
- Cheng, N.H., Liu, J.Z., Brock, A., Nelson, R.S., and Hirschi, K.D. (2006). AtGRXcp, an Arabidopsis chloroplastic glutaredoxin, is critical for protection against protein oxidative damage. J. Biol. Chem. 281 26280–26288. [DOI] [PubMed] [Google Scholar]
- Chuang, C.F., Running, M.P., Williams, R.W., and Meyerowitz, E.M. (1999). The PERIANTHIA gene encodes a bZIP protein involved in the determination of floral organ number in Arabidopsis thaliana. Genes Dev. 13 334–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- D'Ambrosio, K., Kauffmann, B., Rouhier, N., Benedetti, E., Jacquot, J.P., Aubry, A., and Corbier, C. (2003). Crystallization and preliminary X-ray studies of the glutaredoxin from poplar in complex with glutathione. Acta Crystallogr. D Biol. Crystallogr. 59 1043–1045. [DOI] [PubMed] [Google Scholar]
- Després, C., Chubak, C., Rochon, A., Clark, R., Bethune, T., Desveaux, D., and Fobert, P.R. (2003). The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell 15 2181–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després, C., DeLong, C., Glaze, S., Liu, E., and Fobert, P.R. (2000). The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12 279–290. [PMC free article] [PubMed] [Google Scholar]
- Dixon, D.P., Skipsey, M., Grundy, N.M., and Edwards, R. (2005). Stress-induced protein S-glutathionylation in Arabidopsis. Plant Physiol. 138 2233–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes, A.P., and Holmgren, A. (2004). Glutaredoxins: Glutathione-dependent redox enzymes with functions far beyond a simple thioredoxin backup system. Antioxid. Redox Signal. 6 63–74. [DOI] [PubMed] [Google Scholar]
- Gallogly, M.M., and Mieyal, J.J. (2007). Mechanisms of reversible protein glutathionylation in redox signaling and oxidative stress. Curr. Opin. Pharmacol. 7 381–391. [DOI] [PubMed] [Google Scholar]
- Heine, G.F., Hernandez, J.M., and Grotewold, E. (2004). Two cysteines in plant R2R3 MYB domains participate in redox-dependent DNA binding. J. Biol. Chem. 279 37878–37885. [DOI] [PubMed] [Google Scholar]
- Heyl, A., Ramireddy, E., Brenner, W.G., Riefler, M., Allemeersch, J., and Schmulling, T. (2008). The transcriptional repressor ARR1-SRDX suppresses pleiotropic cytokinin activities in Arabidopsis. Plant Physiol. 147 1380–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks, G.R., and Raikhel, N.V. (1993). Specific binding of nuclear localization sequences to plant nuclei. Plant Cell 5 983–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu, K., Matsui, K., Koyama, T., and Ohme-Takagi, M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34 733–739. [DOI] [PubMed] [Google Scholar]
- Holmgren, A., Soberberg, B.O., Eklund, H., and Branden, C.I. (1975). Three-dimensional structure of Escherichia coli thioredoxin-S2 to 2.8 angstrom resolution. Proc. Natl. Acad. Sci. USA 72 2305–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, X.L., Hu, W.W., Shen, L.S., Lee, L.Y.C., Tao, Z., Han, J.H., and Yu, H. (2008). Global identification of DELLA target genes during Arabidopsis flower development. Plant Physiol. 147 1126–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby, M., Weisshaar, B., Droge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., and Parcy, F. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 1 106–111. [DOI] [PubMed] [Google Scholar]
- Johnson, C., Boden, E., and Arias, J. (2003). Salicyclic acid and NPR1 induce the recruitment of trans-activating TGA factors to a defense gene promoter in Arabidopsis. Plant Cell 15 1846–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesarwani, M., Yoo, J., and Dong, X. (2007). Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis. Plant Physiol. 144 336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama, T., Furutani, M., Masao, T., and Ohme-Takagi, M. (2007). TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19 473–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire, S.D. (2004). The glutaredoxin family in oxygenic photosynthetic organisms. Photosynth. Res. 79 305–318. [DOI] [PubMed] [Google Scholar]
- Lillig, C.H., Berndt, C., Vergnolle, O., Lönn, M.E., Hudemann, C., Bill, E., and Holmgren, A. (2005). Characterization of human glutaredoxin 2 as iron–sulfur protein: A possible role as redox sensor. Proc. Natl. Acad. Sci. USA 102 8168–8173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle, T., Leclerc, D., Marshallsay, C., and Nagy, F. (1996). A plant in vitro system for the nuclear import of proteins. Plant J. 10 1177–1186. [DOI] [PubMed] [Google Scholar]
- Ndamukong, I., Abdallat, A.A., Thurow, C., Fode, B., Zander, M., Weigel, R., and Gatz, C. (2007). SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J. 50 128–139. [DOI] [PubMed] [Google Scholar]
- Ojeda, L., Keller, G., Muhlenhoff, U., Rutherford, J.C., Lill, R., and Winge, D.R. (2006). Role of glutaredoxin-3 and glutaredoxin-4 in the iron regulation of the Aft1 transcriptional activator in Saccharomyces cerevisiae. J. Biol. Chem. 281 17661–17669. [DOI] [PubMed] [Google Scholar]
- Reichheld, J.P., Khafif, M., Riondet, C., Droux, M., Bonnard, G., and Meyer, Y. (2007). Inactivation of thioredoxin reductases reveals a complex interplay between thioredoxin and glutathione pathways in Arabidopsis development. Plant Cell 19 1851–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhier, N., Couturier, J., and Jacquot, J.P. (2006). Genome-wide analysis of plant glutaredoxin systems. J. Exp. Bot. 57 1685–1696. [DOI] [PubMed] [Google Scholar]
- Rouhier, N., Gelhaye, E., and Jacquot, J.P. (2002). Exploring the active site of plant glutaredoxin by site-directed mutagenesis. FEBS Lett. 511 145–149. [DOI] [PubMed] [Google Scholar]
- Rouhier, N., Gelhaye, E., and Jacquot, J.P. (2004). Plant glutaredoxins: still mysterious reducing systems. Cell. Mol. Life Sci. 61 1266–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhier, N., et al. (2007). Functional, structural, and spectroscopic characterization of a glutathione-ligated [2Fe-2S] cluster in poplar glutaredoxin C1. Proc. Natl. Acad. Sci. USA 104 7379–7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Running, M.P., and Meyerowitz, E.M. (1996). Mutations in the PERIANTHIA gene of Arabidopsis specifically alter floral organ number and initiation pattern. Development 122 1261–1269. [DOI] [PubMed] [Google Scholar]
- Sun, Y., and Oberley, L.W. (1996). Redox regulation of transcriptional activators. Free Radic. Biol. Med. 21 335–348. [DOI] [PubMed] [Google Scholar]
- Sundaram, S., Rathinasabapathi, B., Ma, L.Q., and Rosen, B.P. (2008). An arsenate-activated glutaredoxin from the arsenic hyperaccumulator fern Pteris vittata L. regulates intracellular arsenite. J. Biol. Chem. 283 6095–6101. [DOI] [PubMed] [Google Scholar]
- Tada, Y., Spoel, S.H., Pajerowska-Mukhtar, K., Mou, Z., Song, J., Wang, C., Zuo, J., and Dong, X. (2008). Plant immunity requires conformational changes of NPR1 via S-nitrosylatio and thioredoxins. Science 321 952–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet, O., Rivas, S., Mestre, P., and Baulcombe, D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33 949–956. [DOI] [PubMed] [Google Scholar]
- Walter, M., Chaban, C., Schütze, K., Batistic, O., Weckermann, K., Näke, C., Blazevic, D., Grefen, C., Schumacher, K., Oecking, C., Harter, K., and Kudla, J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40 428–438. [DOI] [PubMed] [Google Scholar]
- Wang, Z., Xing, S., Birkenbihl, R.P., and Zachgo, S. (2008). Conserved functions of Arabidopsis and rice CC-type glutaredoxins in flower development and pathogen response. Mol. Plant, in press. [DOI] [PubMed]
- Witte, S., Villalba, M., Bi, K., Liu, Y., Isakov, N., and Altman, A. (2000). Inhibition of the c- Jun N-terminal kinase/AP-1 and NF-kappaB pathways by PICOT, a novel protein kinase C-interacting protein with a thioredoxin homology domain. J. Biol. Chem. 275 1902–1909. [DOI] [PubMed] [Google Scholar]
- Xing, S., Lauri, A., and Zachgo, S. (2006). Redox regulation and flower development: a novel function for glutaredoxins. Plant Biol. 8 547–555. [DOI] [PubMed] [Google Scholar]
- Xing, S., Rosso, M.G., and Zachgo, S. (2005). ROXY1, a member of the plant glutaredoxin family, is required for petal development in Arabidopsis thaliana. Development 132 1555–1565. [DOI] [PubMed] [Google Scholar]
- Xing, S., and Zachgo, S. (2008). ROXY1 and ROXY2, two Arabidopsis glutaredoxin genes, are required for anther development. Plant J. 53 790–801. [DOI] [PubMed] [Google Scholar]
- Zachgo, S. (2002). In situ hybridization. In Molecular Plant Biology, P.M. Gilartin and C. Blower, eds (Oxford, UK: Oxford University Press), pp. 41–63.
- Zhang, Y.L., Tessaro, M.J., Lassner, M., and Li, X. (2003). Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 15 2647–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, M., and Storz, G. (2000). Redox sensing by prokaryotic transcription factors. Biochem. Pharmacol. 59 1–6. [DOI] [PubMed] [Google Scholar]
- Zhou, J.M., Trifa, Y., Silva, H., Pontier, D., Lam, E., Shah, J., and Klessig, D.F. (2000). NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol. Plant Microbe Interact. 13 191–202. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.