Abstract
Deep-sea hydrothermal vents are one of the most unique and fascinating ecosystems on Earth. Although phylogenetic diversity of vent communities has been extensively examined, their physiological diversity is poorly understood. In this study, a GeoChip-based, high-throughput metagenomics technology revealed dramatic differences in microbial metabolic functions in a newly grown protochimney (inner section, Proto-I; outer section, Proto-O) and the outer section of a mature chimney (4143-1) at the Juan de Fuca Ridge. Very limited numbers of functional genes were detected in Proto-I (113 genes), whereas much higher numbers of genes were detected in Proto-O (504 genes) and 4143-1 (5,414 genes). Microbial functional genes/populations in Proto-O and Proto-I were substantially different (around 1% common genes), suggesting a rapid change in the microbial community composition during the growth of the chimney. Previously retrieved cbbL and cbbM genes involved in the Calvin Benson Bassham (CBB) cycle from deep-sea hydrothermal vents were predominant in Proto-O and 4143-1, whereas photosynthetic green-like cbbL genes were the major components in Proto-I. In addition, genes involved in methanogenesis, aerobic and anaerobic methane oxidation (e.g., ANME1 and ANME2), nitrification, denitrification, sulfate reduction, degradation of complex carbon substrates, and metal resistance were also detected. Clone libraries supported the GeoChip results but were less effective than the microarray in delineating microbial populations of low biomass. Overall, these results suggest that the hydrothermal microbial communities are metabolically and physiologically highly diverse, and the communities appear to be undergoing rapid dynamic succession and adaptation in response to the steep temperature and chemical gradients across the chimney.
Keywords: metagenomics, microarrays, chimney, deep sea, dynamic
Since the discovery of deep-sea (>2000 m) hydrothermal vents in 1977 (1), studies of the biological systems surrounding the vent environments have greatly expanded our knowledge of life forms on Earth. These environments provide unique models for understanding living strategies in extreme environments and for exploring questions regarding the origins and limits of life on this planet or the potential for extraterrestrial life (2, 3). The base of the deep-sea life pyramid in the vent environments consists of chemolithoautotrophic microorganisms, which are fueled by geochemical energy, such as H2S and H2. The vent chimney, formed by chemical interactions between the hot fluids and cold sea water, has steep chemical and thermal gradients, which provide a wide range of microhabitats for microorganisms (4, 5). Recent studies have demonstrated that microbial diversity varied from vent to vent and from days to years within the same vent field (6, 7). The unique mineralogical and chemical compositions of a chimney in the early stages of its formation may support a distinct pioneering microbial community (8, 9). By analyzing the conserved specific functional genes (such as nifH and mcrA), the metabolic diversity of some specific organisms in the mature chimneys was partially investigated (10, 11). However, because of a lack of appropriate detection technologies, the metabolic diversity and dynamics of whole-microbial communities in primitive and mature chimneys have not been examined.
Recently, high-throughput genomics technologies have shown the great potential to reveal the driving forces of evolution and how ecosystems originate in different geological settings (12). Among these advanced technologies, the microarray-based, high-throughput technologies, such as GeoChip (13), are enabling microbial ecologists to address complex ecological hypotheses at the community-wide scale (14). The GeoChip contains tens of thousands of functional gene markers so that many microbial populations and functional groups can be simultaneously detected at the whole community-wide scale. This unique capability provides incomparable insight into the spatial distribution patterns of many individual functional genes in the same sample sets. Here, we report the metabolic diversity and dynamics of the deep-sea hydrothermal vent chimneys in the Endeavour Segment of Juan de Fuca Ridge by using combined molecular approaches, including microarray hybridization, quantitative real-time PCR, 16S rRNA gene, and functional gene libraries. Our GeoChip study demonstrated that the hydrothermal microbial communities are metabolically and physiologically highly diverse, and the communities appear to be undergoing rapid dynamic succession and adaptation in response to the steep temperature and chemical gradients across the chimney.
Results and Discussion
Sample Description.
During the expedition of the Alvin/Atlantis to the Juan de Fuca Ridge in 2006, we placed a cone-shaped cap on the top of a small chimney (Alvin dive no. 4243), which was vigorously venting at about 310 °C (Fig. S1). The hot fluid vented directly through the central opening of the cap, and a protochimney was observed to form on the top of the cap a few days later. This protochimney was named Proto-O, and the anhydrite and sulfide minerals formed inside the cap were referred to as Proto-I. The Proto-O represented the newly formed porous chimney that has continuous interaction with the cold seawater, whereas the Proto-I may represent the more pristine sulfate and sulfide structures that lack intense interaction with sea water because of the protection of the cap. The protochimney was allowed to grow for 15 days before being collected for this study.
Proto-O and Proto-I are mainly composed of euhedral anhydrite (>90% in volume) by X-ray diffraction and microscopic observation of thin sections. Anhydrite is a highly porous mineral and reflects the first stage in sulfide chimney development (5). The mature chimney rock 4143-1 was from the outer part of a venting chimney from the Mothra Field and was characterized by a predominance of Zn sulfide minerals, such as wurtzite and sphalerite. These minerals commonly indicate the late stages of chimney development (5).
Quantitative PCR and Clone Libraries of 16S rRNA Genes in the Mature Chimney.
Proto-O and Proto-I had an extremely low biomass, as demonstrated by very low concentrations of DNA (5–10 ng/g). The mature chimney rock 4143-1, however, had DNA concentrations up to 200 ng/g. The microbial community structure in the chimney samples was examined by 16S rRNA gene analysis. Unfortunately, no PCR products were obtained from the Proto-I and Proto-O samples, even though several different pairs of PCR primers for Bacteria and Archaea were used. This may be due to the extremely low amounts of biomass in these 2 samples.
PCR amplification of 16S rRNA gene could be achieved easily with the mature chimney 4143-1. The abundance of microorganisms in this sample was estimated to be 4.6 × 109 16S rRNA gene copies per gram (wet weight) for Bacteria, and about 104 copies per gram (wet weight) for Archaea by quantitative PCR, demonstrating that the detected bacterial abundance was significantly greater than Archaea in this sample. Dramatic variations in the distribution of microorganisms within chimneys have been documented from the outer part to the inner part of the chimney, with bacteria more abundant in the cooler, more oxygen-rich outer layer of the chimney, whereas hyperthermophilic Archaea were predominant near the hot interior (15, 16). Our data are in accordance with these previous observations, because 4143-1 was from the outer layer of a venting chimney. Clone libraries were constructed for both archaeal and bacterial 16S rRNA genes. A total of 84 and 55 clones were randomly selected from the bacterial and archaeal 16S rRNA gene library for restriction fragment length polymorphism (RFLP) analyses, respectively. Representative clones of each RFLP type were sequenced. Rarefaction analysis of the bacterial and archaeal 16S rRNA gene clones suggested a much higher diversity of bacteria than archaea, as shown by the curve slopes (Fig. S2D).
The diverse bacterial community in 4143-1 was composed of γ-, ε-, α-, and δ-Proteobacteria, Nitrospirae, Bacteroidetes, and Planctomycetes (Fig. S2 A and B). The γ-Proteobacteria dominated the bacterial community [≈54%, represented by 6 operational taxonomic units (OTUs)], with the majority of OTUs clustering with the symbiont sequences from Codakia orbicularis, Solemya terraeregina gill, and Ifremeria nautilei gill (Fig. S2B). The symbiont γ-Proteobacteria are known to be involved in sulfur oxidation, which provides energy for their host organisms (17). The ε-Proteobacteria were the second most abundant phylotype (≈23% represented by 5 OTUs). Some of the ε-Proteobacteria are also known to be involved in sulfur oxidation (18). A large proportion of the retrieved bacterial sequences had high similarity with symbiotic bacterial sequences, implying exchange between free-living and symbiotic bacteria of similar species. These results indicated that sulfur-oxidizing bacteria could dominate the bacterial community in 4143-1.
The archaeal community in 4143-1 was relatively simple and contained exclusively Euryarchaea (Fig. S2C). Unidentified Euryarchaea cluster (UEII; Fig. S2C, cluster I) and Thermococcus were predominant in the library (51% and 42%, respectively). The physiology and metabolic properties for most of the archaea in 4143-1 could not be determined, because they are distinct from the known species.
Overview of Functional Gene Diversity.
Because of the low quantity of DNA in the protochimney, whole-community genome amplification (19) was carefully performed to obtain enough DNA for microarray hybridization. On the GeoChip, there were a total of 8,371 genes from bacteria and 594 genes from archaea. After hybridization, there were 113 genes detected in Proto-I and 504 genes in Proto-O, whereas 5,414 functional genes were detected in 4143-1 (Table 1). In addition, the microbial diversity was found to be lowest in Proto-I, highest in 4143-1, and intermediate in Proto-O (Table 1). Proto-O and Proto-I had very different community compositions, as shown by the circa 1% overlapping genes between them. On the other hand, most of the genes detected in Proto-O (95%) could be found in 4143-1, suggesting that part of the microbial populations might have become stable as the chimney continued to grow. The low percentages of overlapping genes between Proto-I and Proto-O or 4143-1 indicated that a significantly different microbial community might have developed inside the cap. The fact that most of the genes detected in Proto-O were found in 4143-1 implies that common microbial populations existed in the 2 geographically different hydrothermal fields (the Main Endeavour Field vs. the Mothra Field).
Table 1.
The proportion of unique genes (in bold) in each sample and a matrix representation of the overlapping number of genes between samples
| Proto-O | Proto-I | 4143-1 | |
|---|---|---|---|
| Proto-O, Main Endeavour | 24 (4.8%) | 6 (1.2%, 5.3%) | 480 (95.2%, 8.8%) |
| Proto-I, Main Endeavour | 51 (45.13%) | 62 (54.9%, 1.1%) | |
| 4143-1, Mothra | 4,878 (90.1%) | ||
| Total no. of genes detected | 504 | 113 | 5,414 |
| Shannon Weaver's H′ | 5.93 | 5.18 | 6.16 |
| Shannon Weaver's evenness | 0.88 | 0.90 | 0.84 |
| Simpson's (1/D) | 45.1 | 28.7 | 48.5 |
When 2 different samples are compared, the italicized value indicates the number of overlapping genes between the 2 samples, and the 2 percentage values in parentheses indicate proportions of these genes in each individual sample (sample in row, sample in column).
Although the GeoChip contains probes from both Bacteria and Archaea, the total signal intensity of the detected probes could not be used directly to estimate the relative abundance of each in these samples, because 16 times more probes on the GeoChip are derived from Bacteria than Archaea. However, because the same arrays are used for all 3 samples under the same hybridization conditions, the relative proportions of the detected probes vs. the total probes on the arrays for both Bacteria and Archaea will stay roughly constant across these 3 samples if the ratios of Archaea to Bacteria are very similar across 3 samples and if the archaeal and bacterial probes on the arrays have similar power in detecting indigenous archaeal and bacterial populations in these samples. Thus, to further explore this idea of whether Bacteria are dominant in the matured chimney, the relative proportions of the detected archaeal or bacterial probe numbers vs. the total archaeal or bacterial probe numbers on the arrays were estimated (Fig. S3A). The relative proportions of the detected bacterial genes were substantially higher than those of the detected archaeal genes in 4143-1 and Proto-O, whereas they were very similar in Proto-I. These results suggested that the ratios of Archaea to Bacteria could be quite different among these 3 samples. Because the relative proportion of the detected bacterial genes was twice that of the detected archaeal genes in 4143-1, Bacteria could be dominant over Archaea in this sample, which is consistent with the 16S rRNA-based quantitative PCR results described above. It should be noted that if the archaeal and bacterial probes on the arrays do not have similar power in detecting indigenous archaeal and bacterial populations in these samples, it will be difficult to use such relative proportions to infer the relative abundance of Archaea and Bacteria. Direct experimental evidence using other approaches, such as lipid biomarker analysis, are needed.
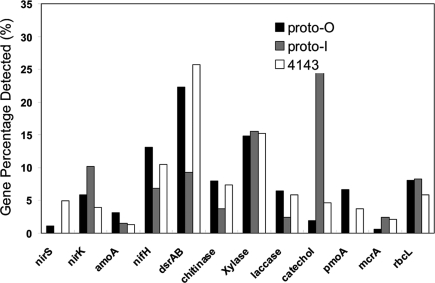
Among the genes detected, those involved in organic contaminant degradation were most abundant in all 3 samples, followed by genes involved in carbon degradation and metal resistance (Table S1). Proto-I contained significantly higher numbers of genes involved in carbon fixation and nitrification than the other 2 samples, whereas fewer genes for methane oxidation and sulfate reduction were present in Proto-I than in the other samples. The proportion of specific functional gene groups detected in the samples is presented in Fig. 1. The proportions of genes detected in Proto-O and 4143-1 showed similar variation, but were different from those detected in Proto-I. The metal-resistance genes detected are shown in Fig. S3B (discussed below). Because we primarily focus on carbon and nitrogen metabolisms of the chemoautotrophic communities in the chimney, genes for organic contaminant degradation and sulfate reduction will only be briefly mentioned in this paper.
Fig. 1.
Proportion of functional gene categories detected. The percentages were calculated by dividing the total signal intensity values of each gene group by total signal intensity values of all genes detected on the array.
Functional Community Involved in CO2 Fixation Revealed by GeoChip.
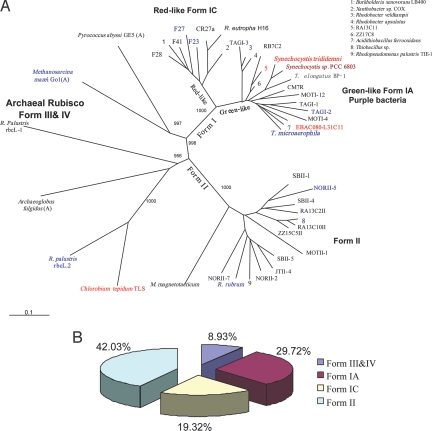
Chemolithoautotrophic microorganisms are known to be at the base of the pyramid of life because they provide food for the heterotrophic communities in the hydrothermal vents. There are, in total, 391 rbcL probes on the GeoChip encompassing both Form I and Form II RubisCO sequences and other RubisCO forms. Of the 391 rbcL probes, 10, 5, and 96 were detected in Proto-O, Proto-I, and 4143-1, respectively. The primary producers in Proto-O were similar to previously detected uncultured, deep-sea autotrophic bacteria, uncultured bacteria from groundwater, α-Proteobacterium Rhodobacter and Rhodospirillum, and Thioalkalispira microaerophila (Fig. S4A). The most dominant genes in Form I RubisCO observed here were similar to the clones from a hydrothermal vent chimney sample from TAG mound, Mid-Atlantic (20). These genes were green-like type IA, occupying about 38% of the chemosynthetic community. The most abundant genes in Form II RubisCO were similar to the clones from reducing sediment of cold seeps, symbiont bacteria of a tubeworm, and from groundwater samples (Fig. 2A, Fig. S4A, and Table S2).
Fig. 2.
Rubisco genes detected in the samples. (A) Phylogenetic tree based on the RubisCO large-subunit amino acid sequences obtained by GeoChip hybridization. Tree topography and evolutionary distance are given by a neighbor-joining method with Kimura distances. This tree is unrooted, with 1,000 replicates of bootstrapping. Bootstrap values are indicated only at major nodes of the tree. The designation of different colors is as follows: Red indicates unique sequences in Proto-I (4 genes), and dark red signifies the single sequence discovered in both Proto-I and 4143-1; blue designates the common sequences from both Proto-O and 4143-1 (10 genes); and black indicates the unique sequences found in 4143-1. All genes detected in Proto-I and Proto-O are listed here, but only the genes with signal intensities greater than 1 from 4143-1 were included in the tree. All genes found in Proto-O could be found in 4143-1. (Scale bar: 0.1 substitutions per site.) (B) Percentage of different types of rbcL genes detected in 4143-1.
In Proto-I, 5 rbcL genes were detected, among which 2 were from Cyanobacteria, 2 from uncultured Proteobacteria, and 1 from green sulfur bacteria. Based on signal intensity of the detected genes, rbcL genes from Cyanobacteria occupied 44% and green sulfur bacteria 34%, with the remaining being represented by the uncultured Proteobacteria (Fig. S4A and Table S2). The detection of cbbL genes closely related to phototrophic bacteria, including Cyanobacteria, has been documented for the dark deep-sea environment (20, 21). The similarity between the deep-sea chemoautotrophic OTUs and RubisCO from photosynthetic organisms may indicate the possible lateral transfer of RubisCO genes among deep-sea and surface water organisms (20). It should be noted that detection of rbcL genes in an organism does not necessarily mean that they are involved in the Calvin cycle. There are some reports demonstrating the presence of type III RubisCOs in some Archaea, but they lack the other genes in the Calvin cycle, and thus do not have the functional CBB pathway. Recently, it was found that the type III RubisCO in the archaeon Thermococcus kodakaraensis participates in adenosine 5-monophosphate (AMP) metabolism, distinct from the role of the classical RubisCOs in the CBB cycle (22). Previously, an obligate photosynthetic bacterial anaerobe, GSB1, was isolated from a deep-sea hydrothermal vent (23). GSB1 belongs to the green sulfur bacteria, and is related to Chlorobium and Prosthecochloris genera. The isolation of GSB1 from the vent implies that geothermal radiation and associated reduced S compounds could be sufficient to at least enhance the survival of GSB1 in the otherwise dark, oxygenated ocean depths. Here, the finding of abundant RubisCO genes of photosynthetic organisms likely supports the argument that unique photosynthetic organisms capable of harvesting geothermal radiation for growth may exist in the extremely light-deficient deep-sea hydrothermal vents; however, more direct experimental evidence is needed.
In 4143-1, Form I and Form II RubisCO genes coexisted and had comparable hybridization intensities (Fig. 2). This was verified by quantitative PCR analysis, which showed that the cbbL and cbbM gene copies were both about 105 copies per gram of the sample material. The main cbbL genes detected in 4143-1 by GeoChip belong to forms IA and IC groups (Fig. 2). Around 42% of the primary producers in the 4143-1 sample were related to deep-sea microorganisms detected previously (Fig. S4A and Table S2) (20).
Functional gene libraries were constructed for cbbL and cbbM to verify the GeoChip data. About 50 positive clones were randomly picked from each of the cbbL and cbbM libraries for RFLP analysis, respectively. After sequencing, 10 OTUs were obtained for cbbL and 7 for cbbM at a 3% cutoff. For cbbL, the clones grouped into 2 clusters (cluster I and cluster II; Fig. S4B), which belonged to the Form IA subfamily. No clones belonging to Form IB or Form IC were retrieved. Six OTUs in cluster I formed a distinct subgroup, which was closely related to the clones from the α- and γ-Proteobacteria. For the cbbM clones retrieved from 4143-1, the majority of the OTUs had the highest identity (85%) to Magnetospirillum gryphiswaldense MSR-1. The phylogenetic relationship of the cloned cbbM genes from 4143-1 with reference sequences is shown in Fig. S4B.
The cbbL and cbbM genes retrieved from 4143-1 by PCR amplification showed much lower diversity than those revealed by the GeoChip. Only dominant groups of sequences could be obtained by using the library construction method in our study. This may be due to the bias of the PCR primers, which preferentially target dominant groups. On the other hand, it also suggests that GeoChip hybridization might be more effective in detecting less-abundant genes. We designed specific PCR primers for several cbbL and cbbM genes, including Form IB cbbL, from uncultured, deep-sea autotrophic bacterium TAGI-2, and group III cbbM from aquifer environmental clone RA13C2II, which were detected by GeoChip hybridization but not by the library analysis. Specific PCR bands of the desired sizes for cbbL and cbbM genes were obtained, indicating that these functional genes were indeed present in the sample.
For the rTCA cycle, GeoChip only detected 2 aclB genes in 4143-1, which were related to uncultured episymbiont of Alvinella pompejana and Chlorobium limicola. This result suggested that the autotrophic microorganisms could predominantly use the CBB cycle in this environment. This is, to some extent, in agreement with our species diversity analysis demonstrating that γ-Proteobacteria using the CBB cycle dominated in 4143-1, which corroborated the observation of abundant cbbL genes from the microarray. On the other hand, the rare detection of aclB genes by GeoChip may suggest that the ε-Proteobacteria in this environment contain novel aclB genes for the rTCA cycle. This is supported by PCR amplification using aclB degenerate primers, which yielded a PCR band but not the expected aclB genes after sequencing. We have constructed a fosmid library for 4143-1, which will allow us to determine whether this sample contains novel aclB genes through library screening and sequencing.
Functional Community for Methane Cycling Revealed by GeoChip.
A characteristic of the thermal fluid from the Endeavour segment of the Juan de Fuca Ridge is its anomalously high CH4 concentration (24). The CH4 carbon isotopic compositions (13CH4 = ≈−55‰ vs. Pee-Dee Belemnite) of these fluids are the lightest yet found in a submarine system devoid of sediment (24). These data, in concert with the elevated CH4 concentrations, which are up to >900 times background seawater values, likely imply a robust subseafloor methanogen community within the Endeavour system (24). The communities in the 3 chimneys were analyzed by using functional genes targeting the methane cycle. The mcrA gene was used to detect both methanogenesis and anaerobic methane oxidation, whereas pmoA was used to track aerobic methane oxidation.
Diversity of mcrA genes.
Proto-O contained 2 mcrA genes, with one similar to Methanothermobacter thermoautotrophicus and the other to an uncultured euryarchaeote soil clone MCR-U3SP-12 from the Florida Everglades. Proto-I contained only 1 mcrA gene, which is closely related to the clone OS82 isolated from a landfill (Fig. S5 A and C).
Sample 4143-1, however, contained many more mcrA genes (Fig. S5 A and C). The dominant mcrA genes could be divided into 4 clusters belonging to Methanomicrobiales, Methanosarcinales, Methanobacteriales, and the anaerobic methane-oxidation ANME group (Fig. S5A). The mcrA gene clones in 4143-1 revealed a high diversity of methanogens and putative anaerobic methane oxidizers. The detection of clones belonging to the ANME1 and ANME2 groups implies that anoxic methane oxidation might occur in this hydrothermal vent chimney environment.
A clone library of mcrA was constructed for 4143-1. Phylogenetic analysis revealed that the PCR-derived mcrA clones of 4143-1 fall into Methanomicrobiales and Methanosarcinales (Fig. S5A). No ANME-related groups were detected by the PCR library analysis. Comparison of the mcrA genes detected on the GeoChip and mcrA clones from the PCR library further suggested that GeoChip could be a more sensitive gene detection method.
Diversity of pmoA sequences.
Aerobic methanotrophs are a unique group of methylotrophic bacteria that use methane as their sole carbon and energy source. Based on cell morphology, ultrastructure, phylogeny, and metabolic pathways, methanotrophs can be divided into 2 taxonomic groups, type I and type II. Type I methanotrophs belong to the γ-Proteobacteria, whereas type II methanotrophs are in the α-Proteobacteria.
Several pmoA sequences were found in Proto-O, but none were detected in Proto-I (Fig. S5 B and D). The absence of pmoA sequences in Proto-I indicates that no aerobic methane oxidation had taken place, which is consistent with the fact that the hydrothermal fluid has no or limited oxygen. Aerobic methane oxidation can take place after seawater mixes with the fluid, bringing oxygen into the environment. Most of the pmoA sequences from Proto-O fell into the type I thermophilic methanotrophs (cluster A) and cluster B (Fig. S5B).
The main methanotrophic bacteria in 4143-1 contained both type I and II methanotrophs (Fig. S5 B and D). Based on GeoChip signal intensities, type I methanotrophs were predominant and accounted for about 54% of all pmoA probes on the array. Type II methanotrophs accounted for about 22%, whereas the remaining 24% were from unknown species. Among the type I methanotrophs, 3 clusters (A, B, and C) could be observed, with cluster A sequences predominating (64%). Cluster A consisted of sequences from a thermophilic methanotroph strain HB and pmoA clones from deep-sea hydrothermal vents (11). Cluster B contained clones from organic soil and Lake Washington sediment. Cluster C contained clones from a Movile cave (Fig. S5B).
Functional Community for Nitrogen Cycle Revealed by GeoChip.
Diversity of nifH sequences.
Dissolved dinitrogen gas (N2) is abundant in deep seawater and in hot hydrothermal vent fluids. Nitrogen isotope ratios (15N/14N) of vent animals are much lower than the ratios of deep-sea organic nitrogen, ammonium, and nitrate, but are similar to those of deep oceanic N2 and marine biota associated with nitrogen fixation (25). Recently, a thermophilic methanogen capable of nitrogen fixation at 92 °C was isolated from the hydrothermal vent environment (26). All data indicate that biological nitrogen fixation is an important process of nitrogen cycling in the vent environments. Biological nitrogen fixation uses the nitrogenase enzyme complex encoded by nifHDK to reduce dissolved N2 to ammonium (NH4+). nifH encodes the iron-containing protein and is highly conserved among various microorganisms. All nifH genes fall into 4 clusters: cluster I includes standard molybdenum nitrogenases from Cyanobacteria and Proteobacteria (α, β, γ), as well as nvfH from γ-Proteobacteria; cluster II includes methanogen nitrogenases and bacterial anfH; cluster III is composed of nitrogenases from diverse anaerobic bacteria, such as Clostridia and δ-Proteobacterial sulfate reducers; cluster IV includes nitrogenases from methanogens (10).
In this study, results from GeoChip showed an increasing number of nifH genes across samples: 5 in Proto-I, 20 in Proto-O, and 58 in 4143-1. Cluster III nifH genes were dominant in all samples (Fig. S6). Among the 5 nifH genes detected from Proto-I, 4 of them fell in nifH cluster III, and one in cluster IV. This is a good reflection of the strict anaerobic environment of Proto-I. The nifH genes in Proto-O fell into clusters I, III, and IV, with cluster III being predominant (around 70%). The main nifH genes in Proto-O were from unidentified or uncultured bacteria retrieved from different environments.
Sample 4143-1, again, contained significantly more nifH genes than Proto-O and Proto-I, and they were distributed among all 4 clusters. Cluster III nifH genes were dominant (59.9%), followed by cluster I sequences (27.6%). The archaeal nifH genes distributed in clusters II and IV only constituted a small proportion of the total nifH genes. This is consistent with the quantification results of microbial 16S rRNA genes, which showed that bacteria were predominant in the sample.
Genes involved in nitrification and denitrification.
Nitrification and denitrification have been observed in the hydrothermal environments (27, 28). The GeoChip contains extensive probes targeting genes involved in nitrification, such as amo (ammonia monooxygenase), and nitrogen metabolism, such as gdh (glutamate dehydragenase), nasA (assimilatory nitrate reductase), nar (nitrate reductase), nir (nitrite reductase), norB (nitric oxide reductase), and nosZ (nitrous oxide reductase). Most of these essential genes involved in nitrification and denitrification were detected in all 3 samples. However, the genes detected from Proto-I and Proto-O were completely different, indicating different microorganisms might be involved in these processes. In 4143-1, the majority of the amoA sequences were from Nitrosomonas and uncultivated β-Proteobacteria. Crenarchaeotal amoA genes were also detected (Fig. S7A). The presence of archaeal amoA genes was confirmed by positive PCR amplification using archaeal amoA-specific primers; however, PCR amplification of bacterial amoA genes failed. The nitrifying archaea may be derived from mixing of seawater with the vent fluid or from thermophilic nitrifying archaea in the chimney, because thermophilic and moderately thermophilic nitrifying archaea have been enriched from hot spring environments (29–31). Most of the nir sequences detected were from uncultured organisms from various environments (Fig. S7 B and C). Our results provide the genetic evidence for possible existence of nitrification and denitrification in deep-sea hydrothermal vent chimneys. This is consistent with the observation of the relatively high ammonia concentrations in some chimney environments in Juan de Fuca Ridge (24), suggesting that nitrification could be an important process in deep-sea hydrothermal vents.
Genes for Metal Resistance Revealed by GeoChip.
The deep-sea hydrothermal vent environment is rich in various heavy metals. Microorganisms are known to respond or adapt to potentially toxic levels of iron through multiple strategies, including dissimilatory and assimilatory metal oxidation and reduction, as well as metal transport (32). Much more is known regarding dissimilatory reactions than assimilatory reactions, although most studies have focused on mesophilic microbes. Many microorganisms isolated from hydrothermal environments are metal-resistant. Generally, metal-resistance mechanisms of microorganisms from hydrothermal vents are similar to those of mesophilic organisms isolated from other environments; however, novel mechanisms for metal reduction in hyperthermophilic archaea have been observed (33). It would be difficult to detect novel metal-resistance mechanisms by using GeoChip, because only known sequences are covered by the array. However, this study allows a careful survey and comparison of genes putatively involved in metal resistance from hydrothermal vent environments at the community level.
In Proto-I, most of the detected metal resistance genes were for cadmium, tellurium, aluminum, and chromium resistance (Fig. S3B). In Proto-O and 4143-1, the majority of resistance genes were for arsenic and mercury, with the remaining being for chromium, tellurium, and other metals (Fig. S3B). The data imply that microorganisms in Proto-I may be generally resistant to chromium and tellurium but more sensitive to arsenic and mercury.
Other Main Genes Revealed by GeoChip.
Large numbers of genes detected by the GeoChip also included those involved in sulfate reduction and carbon/organic contaminant degradation. Sulfate reduction is thought to be one of the important energy sources for microbial systems at hydrothermal vents (34). Only 3 dsrB genes were detected in Proto-I, which are related to those found in Desulfotomaculum geothermicum, a thermophilic Firmicutes originally isolated from geothermal ground water; Syntrophobacter fumaroxidans, an H2-producing syntroph of δ-Proteobacteria; and an unidentified clone from groundwater at a uranium mill tailings site. The dsr genes detected in Proto-O and 4143-1 were highly diverse and were related to those in Desulfobacteraceae, Desulfovibrionaceae, Peptococcaceae, Desulfobulbacaea, Syntrophobacteraceae, and unidentified clusters.
Little is known about carbon and organic contaminant degradation and their contribution to the vent ecosystems. GeoChip analysis revealed that carbon degradation genes are highly abundant in these vent systems, suggesting that they may play important roles in carbon cycling and metabolism in the geothermal environment. However, further studies are needed in terms of carbon cycling processes and their associated microbial communities. It should be noted that many genes classified in the category of organic contaminant degradation (e.g., aromatic degradation genes) are also important in the degradation of various carbon polymers in nature. Thus, the detection of these types of genes does not necessarily indicate the existence of such contaminants in the vent systems.
Summary.
In this study, GeoChip hybridization, clone libraries, and quantitative PCR were integrated to examine the abundance and metabolic diversity of microbial populations in the Juan de Fuca hydrothermal vent chimneys. Microbial populations detected by the GeoChip were significantly higher than estimates based on gene clone library analyses but were consistent with the observation of a vast diversity of microorganisms in the hydrothermal vents revealed by extensive (hundreds of thousands of sequences) sequencing analysis (35). Significant variability of functional genes related to carbon, sulfur, and nitrogen metabolism was observed during the development of vent chimneys. This may be due to the changing vent structure and chemical composition. By 16S rRNA gene library analysis, bacteria affiliated within γ- and ε-Proteobacteria groups were dominant in 4143-1. Quantitative PCR analysis of 16S rRNA genes suggested that Bacteria could be more abundant than Archaea in 4143-1. The primary producers in the 3 samples could primarily use the CBB cycle. In particular, photosynthetic green-like cbbL genes were the major components in Proto-I, whereas previously retrieved cbbL and cbbM genes from deep-sea hydrothermal vents were predominant in Proto-O and 4143-1. Clone libraries using the 16S rRNA and functional genes (mcrA, cbbL, cbbM) supported the GeoChip results but were less effective than the microarray technology in delineating the microbial community structure of low-biomass communities. Our study revealed extremely rich and diverse metabolic reservoirs of the microbial community in the hydrothermal vent chimney environment, which has not been fully recognized before. Furthermore, our data showed the great potential of high-throughput microarray technology in understanding ecosystem dynamics in the complex hydrothermal vent environments.
Materials and Methods
Samples from the sulfide chimneys were collected by the submersible Alvin supported by the R/V Atlantis in 2005 and 2006 at the Endeavour Segment of the Juan de Fuca Ridge, located ≈300 km west of Vancouver Island, Canada. During a cruise in 2006, a stainless steel cap was deployed on top of a chimney that was vigorously venting at the Maine Endeavour field (Fig. S1). The fluid temperature was higher than 300 °C. After 15 days of deployment, samples were collected from the chimney formed on top of the cap (Proto-O; Fig. S1) and the sulfate deposit accumulated on the surface of the inner cap (Proto-I). An outer part of a mature sulfide chimney (4143-1) collected in 2005 from the Mothra Field, which was venting at about 316 °C at the time of sampling, was also used for comparison. Precautions were taken during sampling and handling of the chimney samples to preserve their integrity for microbiological analyses. The chimneys were stored at −20 °C as soon as collected, kept on dry ice during transportation, and stored at −80 °C until further analysis.
The methods of molecular manipulations, including DNA isolation, amplification, labeling, and microarray hybridization; construction of 16S rRNA, mcrA, cbbL, and cbbM gene clone libraries; and quantitative PCR are essentially based on established methods described previously (19, 20, 36–40). Sequences obtained for bacterial and archaeal 16S rRNA, mcrA, cbbL, and cbbM genes were deposited in the GenBank database under accession numbers FJ640793–FJ640842. The details are provided as SI Methods.
Supplementary Material
Acknowledgments.
We thank Maurice A. Tivey (Woods Hole Oceanographic Institution, Woods Hole, MA), Marvin D. Lilley (University of Washington, Seattle, WA), Deborah Kelly (University of Washington, Seattle, WA), Kang Ding (University of Minnesota, Minneapolis, MN), and all of the crew members from R/V Atlantis/DSV Alvin for their efforts and help in sample collection. This study was supported by China Ocean Mineral Resources R&D Association Grant DYXM-115-02-2-01), by National Natural Science Foundation of China Grants 40532011, 40403004, and 40473032, and by the U.S. Department of Energy under the Genomics: GTL program through the Virtual Institute of Microbial Stress and Survival (http://vimss.lbl.gov), Office of Biological and Environmental Research, Office of Science.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810418106/DCSupplemental.
References
- 1.Francheteau J, et al. Massive deep-sea sulfide ore deposits discovered on the East Pacific Rise. Nature. 1979;277:523–528. [Google Scholar]
- 2.Zierenberg RA, Adams MW, Arp AJ. Life in extreme environments: Hydrothermal vents. Proc Natl Acad Sci USA. 2000;97:12961–12962. doi: 10.1073/pnas.210395997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deming JW, Baross JA. Deep-sea smokers: Windows to a subsurface biosphere? Geochim Cosmochim Acta. 1993;57:3219–3230. doi: 10.1016/0016-7037(93)90535-5. [DOI] [PubMed] [Google Scholar]
- 4.Kelley DS, Baross JA, Delaney JR. Volcanoes, fluids, and life at mid-ocean ridge spreading centers. Annu Rev Earth Planet Sci. 2002;30:385–491. [Google Scholar]
- 5.Kristall B, Kelly DS, Hannington MD, Delaney JR. Growth history of a diffusely venting sulfide structure from the Juan de Fuca ridge: A petrological and geochemical study. Geochem Geophys Geosyst. 2006 Jul 4; doi: 10.1029/2005GC001166. [DOI] [Google Scholar]
- 6.Huber JA, Butterfield DA, Baross JA. Temporal changes in archaeal diversity and chemistry in a mid-ocean ridge subseafloor habitat. Appl Environ Microbiol. 2002;68:1585–1594. doi: 10.1128/AEM.68.4.1585-1594.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nercessian O, Reysenbach AL, Prieur D, Jeanthon C. Archaeal diversity associated with in situ samplers deployed on hydrothermal vents on the East Pacific Rise (13 degrees N) Environ Microbiol. 2003;5:492–502. doi: 10.1046/j.1462-2920.2003.00437.x. [DOI] [PubMed] [Google Scholar]
- 8.McCliment EA, et al. Colonization of nascent, deep-sea hydrothermal vents by a novel Archaeal and Nanoarchaeal assemblage. Environ Microbiol. 2006;8:114–125. doi: 10.1111/j.1462-2920.2005.00874.x. [DOI] [PubMed] [Google Scholar]
- 9.Page A, Tivey MK, Stakes DS, Reysenbach AL. Temporal and spatial archaeal colonization of hydrothermal vent deposits. Environ Microbiol. 2008;10:874–884. doi: 10.1111/j.1462-2920.2007.01505.x. [DOI] [PubMed] [Google Scholar]
- 10.Mehta MP, Butterfield DA, Baross JA. Phylogenetic diversity of nitrogenase (nifH) genes in deep-sea and hydrothermal vent environments of the Juan de Fuca Ridge. Appl Environ Microbiol. 2003;69:960–970. doi: 10.1128/AEM.69.2.960-970.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nercessian O, Bienvenu N, Moreira D, Prieur D, Jeanthon C. Diversity of functional genes of methanogens, methanotrophs and sulfate reducers in deep-sea hydrothermal environments. Environ Microbiol. 2005;7:118–132. doi: 10.1111/j.1462-2920.2004.00672.x. [DOI] [PubMed] [Google Scholar]
- 12.Reysenbach AL, Shock E. Merging genomes with geochemistry in hydrothermal ecosystems. Science. 2002;296:1077–1082. doi: 10.1126/science.1072483. [DOI] [PubMed] [Google Scholar]
- 13.He Z, et al. GeoChip: A comprehensive microarray for investigating biogeochemical, ecological and environmental processes. Isme J. 2007;1:67–77. doi: 10.1038/ismej.2007.2. [DOI] [PubMed] [Google Scholar]
- 14.Zhou J, Kang S, Schadt CW, Garten CTJ. Spatial scaling of functional gene diversity across various microbial taxa. Proc Natl Acad Sci USA. 2008;105:7768–7773. doi: 10.1073/pnas.0709016105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harmsen H, Prieur D, Jeanthon C. Distribution of microorganisms in deep-sea hydrothermal vent chimneys investigated by whole-cell hybridization and enrichment culture of thermophilic subpopulations. Appl Environ Microbiol. 1997;63:2876–2883. doi: 10.1128/aem.63.7.2876-2883.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schrenk MO, Kelley DS, Delaney JR, Baross JA. Incidence and diversity of microorganisms within the walls of an active deep-sea sulfide chimney. Appl Environ Microbiol. 2003;69:3580–3592. doi: 10.1128/AEM.69.6.3580-3592.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duperron S, Laurent MC, Gaill F, Gros O. Sulphur-oxidizing extracellular bacteria in the gills of Mytilidae associated with wood falls. FEMS Microbiol Ecol. 2008;63:338–349. doi: 10.1111/j.1574-6941.2008.00438.x. [DOI] [PubMed] [Google Scholar]
- 18.Campbell BJ, Engle AS, Porter ML, Takai K. The versatile epsilon-Proteobacteria: Key players in sulphidic habitats. Nat Rev Microbiol. 2006;4:458–468. doi: 10.1038/nrmicro1414. [DOI] [PubMed] [Google Scholar]
- 19.Wu L, Liu X, Schadt CW, Zhou J. Microarray-based analysis of subnanogram quantities of microbial community DNAs by using whole-community genome amplification. Appl Environ Microbiol. 2006;72:4931–4941. doi: 10.1128/AEM.02738-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elsaied H, Naganuma T. Phylogenetic diversity of ribulose-1,5-bisphosphate carboxylase/oxygenase large-subunit genes from deep-sea microorganisms. Appl Environ Microbiol. 2001;67:1751–1765. doi: 10.1128/AEM.67.4.1751-1765.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elsaied HE, Kimura H, Naganuma T. Composition of archaeal, bacterial, and eukaryal RuBisCO genotypes in three Western Pacific arc hydrothermal vent systems. Extremophiles. 2007;11:191–202. doi: 10.1007/s00792-006-0025-2. [DOI] [PubMed] [Google Scholar]
- 22.Sato T, Atomi H, Imanaka T. Archaeal type III RuBisCOs function in a pathway for AMP metabolism. Science. 2007;315:1003–1006. doi: 10.1126/science.1135999. [DOI] [PubMed] [Google Scholar]
- 23.Beatty JT, et al. An obligately photosynthetic bacterial anaerobe from a deep-sea hydrothermal vent. Proc Natl Acad Sci USA. 2005;102:9306–9310. doi: 10.1073/pnas.0503674102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lilley MD, et al. Anomalous CH4 and NH4+ concentrations at an unsedimented mid-ocean-ridge hydrothermal system. Nature. 1993;364:45–47. [Google Scholar]
- 25.Rau GH. Low 15N/14N in hydrothermal vent animals: Ecological implications. Nature. 1981;289:484–485. [Google Scholar]
- 26.Mehta MP, Baross JA. Nitrogen fixation at 92 degrees C by a hydrothermal vent archaeon. Science. 2006;314:1783–1786. doi: 10.1126/science.1134772. [DOI] [PubMed] [Google Scholar]
- 27.Jannasch HW. In: Hydrothermal Processes at Seafloor Spreading Centers. Rona PA, Bostrom K, Laubier L, Smith KL, editors. New York: Plenum; 1984. pp. 677–709. [Google Scholar]
- 28.Lilley M, Baross JA, Gordon LI. In: Hydrothermal Processes at Seafloor Spreading Centers. Rona PA, Bostrom K, Laubier L, Smith KL, editors. New York: Plenum; 1984. pp. 411–449. [Google Scholar]
- 29.Hatzenpichler R, et al. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci USA. 2008;105:2134–2139. doi: 10.1073/pnas.0708857105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de la Torre JR, Walker CB, Ingalls AE, Konneke M, Stahl DA. Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ Microbiol. 2008;10:810–818. doi: 10.1111/j.1462-2920.2007.01506.x. [DOI] [PubMed] [Google Scholar]
- 31.Zhang CL, et al. Global occurrence of archaeal amoA genes in terrestrial hot springs. Appl Environ Microbiol. 2008;74:6417–6426. doi: 10.1128/AEM.00843-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holden JF, Adams MW. Microbe-metal interactions in marine hydrothermal environments. Curr Opin Chem Biol. 2003;7:160–165. doi: 10.1016/s1367-5931(03)00026-7. [DOI] [PubMed] [Google Scholar]
- 33.Childers SE, Lovley DR. Differences in Fe(III) reduction in the hyperthermophilic archaeon, Pyrobaculum islandicum, versus mesophilic Fe(III)-reducing bacteria. FEMS Microbiol Lett. 2001;195:253–258. doi: 10.1111/j.1574-6968.2001.tb10529.x. [DOI] [PubMed] [Google Scholar]
- 34.Nakagawa T, Nakagawa S, Inagaki F, Takai K, Horikoshi K. Phylogenetic diversity of sulfate-reducing prokaryotes in active deep-sea hydrothermal vent chimney structures. FEMS Microbiol Lett. 2004;232:145–152. doi: 10.1016/S0378-1097(04)00044-8. [DOI] [PubMed] [Google Scholar]
- 35.Huber JA, et al. Microbial population structures in the deep marine biosphere. Science. 2007;318:97–100. doi: 10.1126/science.1146689. [DOI] [PubMed] [Google Scholar]
- 36.Zhou J, Bruns MA, Tiedje JM. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yergeau E, Kang S, He Z, Zhou J, Kowalchuk GA. Functional microarray analysis of nitrogen and carbon cycling genes across an Antarctic latitudinal transect. Isme J. 2007;1:163–179. doi: 10.1038/ismej.2007.24. [DOI] [PubMed] [Google Scholar]
- 38.Wu L, et al. Development and evaluation of microarray-based whole-genome hybridization for detection of microorganisms within the context of environmental applications. Environ Sci Technol. 2004;38:6775–6782. doi: 10.1021/es049508i. [DOI] [PubMed] [Google Scholar]
- 39.Hales BA, et al. Isolation and identification of methanogen-specific DNA from blanket bog peat by PCR amplification and sequence analysis. Appl Environ Microbiol. 1996;62:668–675. doi: 10.1128/aem.62.2.668-675.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Campbell BJ, Stein JL, Cary SC. Evidence of chemolithoautotrophy in the bacterial community associated with Alvinella pompejana, a hydrothermal vent polychaete. Appl Environ Microbiol. 2003;69:5070–5078. doi: 10.1128/AEM.69.9.5070-5078.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.