Abstract
MicroRNA (miRNA) gene expression profiling has provided important insights into plant and animal biology. However, there has not been ample published work about pitfalls associated with technical parameters in miRNA gene expression profiling. One source of pertinent information about technical variables in gene expression profiling is the separate and more well-established literature regarding mRNA expression profiling. However, many aspects of miRNA biochemistry are unique. For example, the cellular processing and compartmentation of miRNAs, the differential stability of specific miRNAs, and aspects of global miRNA expression regulation require specific consideration. Additional possible sources of systematic bias in miRNA expression studies include the differential impact of pre-analytical variables, substrate specificity of nucleic acid processing enzymes used in labeling and amplification, and issues regarding new miRNA discovery and annotation. We conclude that greater focus on technical parameters is required to bolster the validity, reliability, and cultural credibility of miRNA gene expression profiling studies.
Keywords: microRNA, miRNA, microarray, profiling, technical
Introduction
Over the past 5 years, literally dozens of distinct miRNA gene expression profiling platforms (miGEPs) have been introduced. Studies using miGEPs have helped to establish that miRNA biology is fundamentally important in plants and animals with clinical implications for human diseases. Excellent prior reviews have described some of the important aspects of miRNA profiling (see for example [1–3]). However, research literature to improve quality control for miGEPs has not developed in parallel. Nor has there been commensurate published work about the effects of pre-analytical and other technical variables in miRNA gene expression profiling.
The purpose of this review is to describe some technical parameters that may be relevant to miRNA expression profiling. Unfortunately, there are many stages of a gene expression study where systematic bias can be introduced. The expression ‘garbage in, garbage out’ can be applied to gene expression profiling; however, in the context of high-throughput techniques, subtle bias can be more problematic than manifestly flawed data. This review is not oriented toward ‘solving’ technical problems. Instead, we wish to begin bringing important technical parameters to light because no variable will have the same significance for each miGEP. In order to not repeat the ‘learning curve’ of the mRNA profiling field, it seems advisable to focus on technical parameters for the sake of improving the validity and reliability of miRNA gene expression profiling studies.
Technical variables in gene expression profiling: lessons from mRNA-oriented gene expression profiling
General considerations
Relative to miRNA studies, mRNA profiling parameters have been assessed over a longer time, and with greater attention to technical details. Recent reviews of mRNA-related expression profiling have identified potential sources for systematic biases in expression profiling, along with strategies to overcome those potential pitfalls [4–6] (see table 1). The initial studies using high-throughput mRNA profiling microarrays were performed before the technical parameters for those studies were fully optimized. Hence an appreciation of technical barriers for mRNA gene expression profiling lagged behind the initial implementation of these techniques. This is noteworthy because some early mRNA gene expression profiling studies were plagued by flawed assumptions and non-reproducible results [7, 8]. These problems have been manifested in later years by some degree of skepticism about gene expression profiling results [9]. With these lessons in mind, it is important to begin addressing these issues soon in the context of miRNAs.
Table 1.
A sample of technical variables shown to play a role in mRNA expression studies
| Time between sample removal and RNA |
| isolation |
| RNA extraction efficiency |
| Amplification yield |
| Labeling yield |
| Hybridization efficiency |
| Dye bias |
| Fluorescence gain bias |
| cDNA array printing pins |
| Properties of probe plates |
| Inter-individual differences |
| Non-linear cross-talk effects |
| Pooling/non-pooling bias |
| Genome annotation irregularities |
| Using correct analytical tools |
Another general consideration for which mRNA profiling may provide insights for miRNA profiling is in the cross-comparisons between the results of different gene expression profiling platforms. In principle, for a given RNA sample, each gene expression profiling platform should ‘report’ the same repertoire of transcripts. However, this has not been shown necessarily to be the case (for an excellent review of this literature see [6]). For example, it has been shown that different cDNA-base mRNA profiling microarray techniques provide differing results, and these differences are greater when comparing the results between separate laboratories [6, 10–13]. Experimental results tend to vary even more when comparing between less-related technologies (e.g., microarrays and SAGE) [14–17]. This problem of reproducibility across platforms renders very difficult any biologically-relevant data mining of public access ‘gene expression’ databases [18]. Under the most ideal circumstances (within-lab studies performed in parallel), different gene expression platforms tend to have acceptable agreement as long as emphasis is placed on the direction of change, rather than the magnitude of change with regard to differentially expressed genes [19, 20].
There has been improvement in recent years in mRNA gene expression profiling—better experimental design has improved the replicability of expression profiling studies [6]—and this is largely because the field has focused during that time on technical parameters. A consensus seems to have emerged that regardless of the gene expression profiling platform, attention to study design is a key prerequisite to obtaining valid data [6, 7, 9, 21–23]. This focus has been reflected by a great deal of work, as well as large-scale international consortiums dedicated to quality control [11, 24]. The positive impact that these forward-thinking measures has had on mRNA expression profiling should have direct implications for researchers interested in miRNA expression profiling.
Pre-analytical variables
The pre-analytical variables that have been most carefully studied in mRNA expression studies regard the effect of fixation, tissue embedding, freezing and other forms of storage, post-mortem interval, RNA amplification, and tissue pH (see for example [25]). These mRNA-related studies will not be reviewed here, partly because there seems to be important differences between miRNAs and mRNAs in the importance of these parameters. It is also probable that the effects of particular technical parameters are distinct for different tissue types, so these results should be interpreted and generalized only with caution.
Relatively few mRNA gene expression studies have addressed systematically the effects of pre-analytical technical issues as the methods of cell lysis and of RNA isolation. However, the published studies indicate that standardizing and optimizing RNA isolation techniques, and the particular manner in which samples are handled, are vitally important to obtaining valid results in gene expression profiling studies [26–28]. Dell’Orto et al showed that the particular mRNA isolation technique chosen can impact significantly on the amount of variation within and between experiments, and that optimizing pre-analytical variables can help to better demonstrate more accurately which genes are differentially expressed [29]. The importance of RNA isolation in miRNA studies is discussed below.
Depending on the miGEP technique, processing samples after RNA extraction can involve many additional steps. For example, in order to label and/or amplify nucleic acids, catalysts are required. Purified proteins such as T4 RNA ligase, poly(A) polymerase, or others are utilized. These molecules are highly effective usually but are known to have bias with regard to donor and/or acceptor sequence specificity. To provide a few illustrations, labeling and amplification steps are predicted to be systematically biased because of the use of T4 RNA ligase [30, 31]. For example, the oligonucleoside electron acceptor C-C-C was ligated by T4 RNA ligase more than three times more efficiently in comparison to the acceptor sequence ending U-A-G [30]. There is also bias introduced during T7-based RNA amplification (aRNA) [32]. Thus it has been shown to be problematic to compare between the results of small-sample and large-sample experiments [33]. Random priming has also been used to label miRNAs, however, it has been pointed out that the nature of miRNAs (i.e., their short size) may results in nonrandom labeling [1]. The poly(A) polymerase (PAP) enzyme has been used for direct RNA dye labeling, with impressive results [1, 34], however, it should also be kept in mind that PAP may also show substrate sequence-related biases [35–37]. These data do not argue against the usefulness of the ligating, labeling or amplification techniques, which have been well established (in the context of miRNAs, see for example [38]). However, the potential biases should be known about and argue strongly for using identical methods in any single study.
Note about real-time “quantitative” PCR (RTQPCR)
In either low- or high-throughput format, RTQPCR is an important and widely-used platform for mature and precursor miRNA expression profiling. RTQPCR is a versatile, sensitive, and convenient method that provides a numerical read-out of DNA concentration in a sample. Messenger RNA quantification gives some insights about the validity and reliability of RTQPCR studies. Much of these data are impressive with regard to the reliability and validity of the data. However, when evaluated as a group, these studies may be biased toward reporting positive results. A number of experienced users have raised concerns about the technique. Even in the best of hands, high-throughput RTQPCR can give variable results in quantifying DNA as other techniques do, and care must be taken with controls and technical parameters [39–43]. Accordingly, when carefully compared with microarray and Northern blot results, RTQPCR results may provide very good but by no means perfect correlation in miRNA profiling [38, 44]. Hence despite its descriptive name, and the fact that RTQPCR has been repeatedly used as a validation technique of choice (see for example [45–47]), mRNA-related studies indicate that it is not necessarily appropriate to use RTQPCR data as an absolute ‘gold standard’. Like Northern blots and microarrays, RTQPCR is another good but imperfect tool for nucleic acid quantitation. This is not a criticism of the technique but simply a caveat based upon experience and supported by the pertinent literature. As concluded by Bustin and Nolan, “…real-time qPCR is a…powerful technique. But, like anything powerful, it needs to be treated with respect.” [40]
Technical issues in miRNA expression profiling
Relative to mRNAs, miRNAs are distinguished because of their relatively uniform and small size, their lack of poly(A) tails, and that they can be represented by very high cellular copy numbers [48]. Further, the overall mechanism(s) of miRNA function are distinct from mRNAs. Hence there are special aspects of miRNA biology that need to be taken into account by researchers interested in miRNA profiling.
MiRNA research is a fast-moving field and produces an evolving set of technical challenges for miRNA profiling
The study of miRNAs is still in its infancy. The field has grown explosively, with many new biological paradigms discovered. Researchers interested in miRNA expression profiling cannot ignore the technical implications of these novel data.
At the most basic level, the full complement of miRNAs expressed in most animals including humans has not been determined. As of December 2007, the Rfam registry (v. 10.1) has annotated 563 human miRNA genes, an increase from 475 miRNAs in v. 9.2. However, many of these are identical or nearly-identical paralogs (see http://microrna.sanger.ac.uk/ and Ref. [49]). It is important to note that certain miRNA assays, including some RTQPCR and some microarray methods, are highly specific to 3' ending positions of targeting miRNAs. The end-position specificity is considered to represent an advantage of those assays. However, the recent updates of sequence information, particularly in miRBase changes from v9.2 to v10.0, have led to changes in the ‘canonical’ end positions of many miRNAs. We have found that many different 3’ end configurations are represented a given miRNA; for example, Argonaute proteins-associated hsa-miR-451 has at least seven different 3’ end configurations in human blood [50]. These differences may indeed lead to different target selectivity for miRNAs. Hence it may be necessary to incorporate more flexibility into miRNA profiling assays, as was the case in the microarray studies by Berezikov et al [51].
In contrast to the official annotated miRNA count, some studies indicate that expressed humans miRNAs number well over a thousand. For example, a study of human and chimpanzee brains yielded 447 completely new miRNA genes (see [51–54]). There may be a diminishing return because the miRNAs described earlier were probably those with the highest expression (see Figure 2 and Ref [53]). However, the miRNAs that are expressed at low levels in a tissue may be very important to subpopulations of cells within those tissues (see for example [55]). Seeking new miRNAs is a challenge because many miRNAs may be expressed preferably during particular developmental timepoints, or within certain cells or tissues, anatomical subregions, or environmental conditions. In any case, many current miGEPs survey only several hundred different miRNAs. For this reason, future miRNA researchers may be compelled to perform the profiling experiments over again in order to incorporate the many new miRNAs.
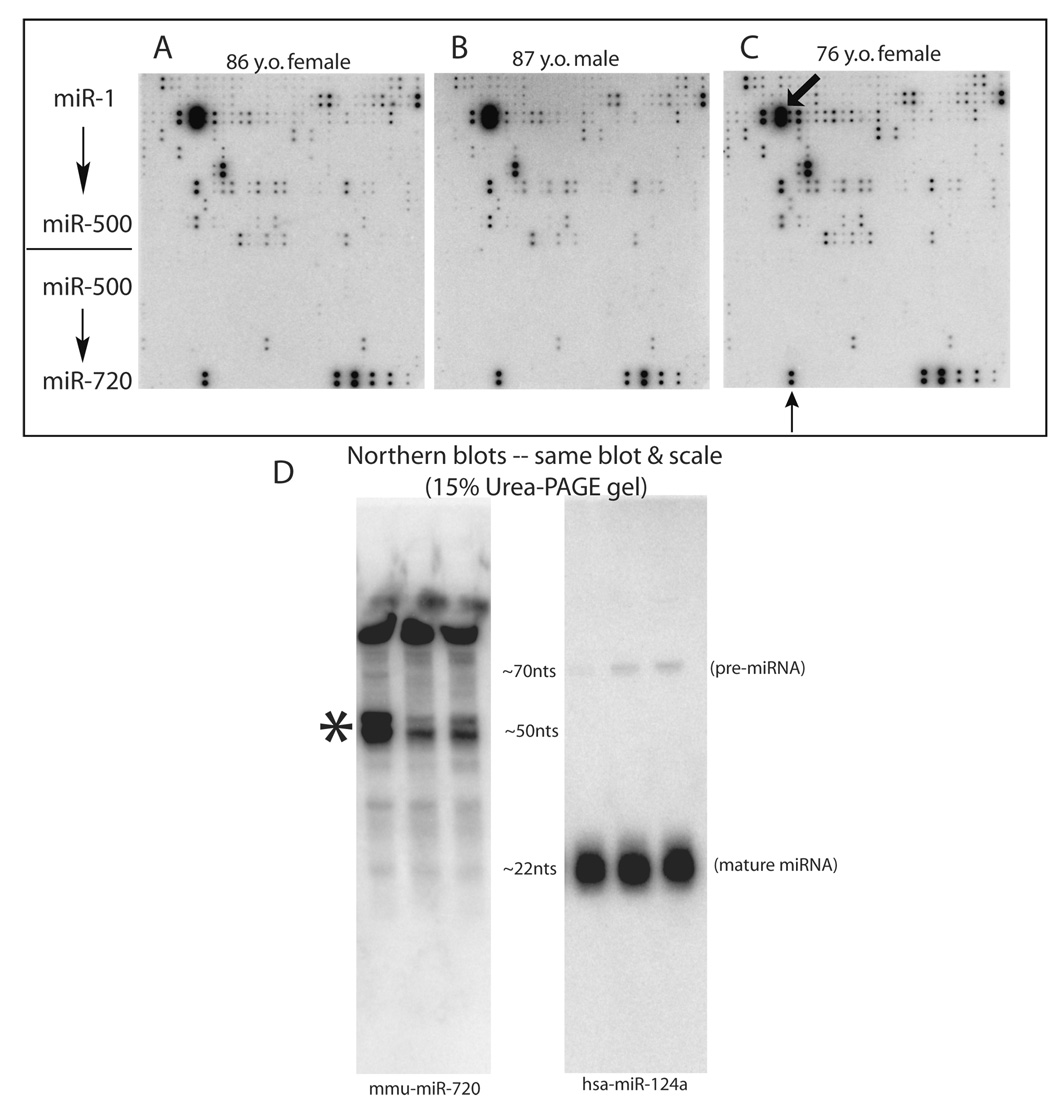
Figure 2.
A–C. Informative miRNA array results referent to separate brain samples using a cDNA array after the method of Tang et al [130]. The RNA samples were extracted from gray matter of the superior and mid-temporal gyri from nondemented elderly autopsy subjects (age and sex indicated). RNA was then run on a denaturing (urea-PAGE) gel and the small RNA excised, then labeled with P-32 and hybridized to the cDNA array. Each array element is spotted in duplicate with miRNAs numbering from top (hsa-miR-1) to bottom (mmu-mir-720). The extreme bottom right spots are internal spiked-in controls of differing concentrations. The array elements corresponding to miR-124a are indicated with a diagonal arrow in 1C. Several observations follow from these results. Firstly, the “signature” of miRNA expression is quite consistent across all three arrays. Secondly, in this array that queries 554 different array elements representing all unique annotated miRNAs (circa spring 2007) found in humans and mice, ~100 miRNAs are expressed above a moderately-high threshold and several dozen would be considered highly-expressed. Thirdly, most but not all of the miRNAs detected in these brain samples were discovered prior to 2005, i.e. annotated before miR-500. An apparently conspicuous exception to this ‘rule’ is mmu-miR-720, which is indicated with the vertical arrow at the bottom of 1C. However, a follow-up Northern blot using a probe against mmu-miR-720 is shown in Figure 1D. For the Northern blot, the three different human brain samples were again used (total RNA isolated using Trizol LS) and run on a 15% urea-PAGE gel. Note that rather than the expected banding pattern of pre-miRNA and mature miRNA, the miR-720 Northern blot shows that the apparent miR-720 signal is probably not a conventional miRNA. An anomalous ~50nt RNA band is present (asterisk). Note for comparison the same blot using a probe for miR-124a which shows the expected pattern of pre-miRNA and mature miRNA. These results illustrate the importance of confirming array results with another technique, and shows the strengths of the Northern blot when faced with unexpected array data.
Another concern is that the exact biological definition of miRNAs will probably evolve further in the future. Some miRNAs may have special rules, for example, they may be processed differently from the current canonical guidelines [56], and unconventional miRNAs are derived from noncoding RNAs [57–59]). Viruses can either suppress or employ the miRNA pathways (see [60–63]). Another twist is that RNA editing, particularly A-to-I conversion, may dramatically alter miRNA function to the extent of generating a novel miRNA species and/or altering the degree to which mature miRNAs are produced from the pre-miRNA [64–67]. Furthermore, plant miRNAs and human piRNAs are methylated at the 3’ end [62, 68–73], which may add variables to profiling for 3’ end methylated miRNAs or small RNAs. In principle these biological ‘tricks’ would be detected by an optimal miRNA profiling platform.
Just as with the annotation of new miRNAs, the regulation of miRNA expression is a novel topic and no doubt many surprises are in store for researchers. A lurking uncertainty relevant to miGEP experiments is the relationship between the cellular levels of a miRNA in a biological sample, and that miRNA’s “biological activity”. Analogy can be drawn to messenger RNAs: mRNAs are templates for the production of polypeptides. However, gene expression is apparently regulated predominantly post-transcriptionally, so the levels of mRNAs in a biological sample tend to correlate poorly if at all with protein levels [74]. Likewise, we cannot query the percentage of miRNA molecules engaged in actively targeting mRNAs. This is a profound caveat to mRNA based ‘gene expression’ analyses. It is already known that miRNA function can be affected in cell culture without changing the ‘expression’ (i.e., transcription) of the miRNA [75]. There may also be regulation of miRNA activity at brain synapses via proteolytic processing of miRNA processing proteins [76]. Could the detected amount of a given miRNA correlate poorly in fact with the degree to which that miRNA is actively involved in regulating target mRNAs? Further work remains to be performed in this area.
Depending upon study design, the distinct characteristics of miRNA biochemistry would be predicted to impact upon still more aspects of miGEP studies. For example, considerable miRNA processing occurs in the cytoplasm, where pre-miRNAs are cleaved to produce ‘mature’ ~22nts miRNAs. It is possible that some miGEPs would have difficulty in differentiating between pre-miRNAs and miRNAs and this would affect the data readout [1]. Some miGEPs have intrinsic advantages in this area because they can detect the sizes and/or end sequences of small RNAs: miRNA cloning, PCR-based techniques, Northern blots, and RNA-primed array-based Klenow enzyme (RAKE) microarray techniques (see [51, 77–79]). Many microarray studies have been performed on RNA samples that have been enriched for small RNAs, or RNA samples that have been cut from PAGE gels, and thus should contain a high proportion of a given size range of RNAs. However, depending on the RNA isolation technique, the miGEP platform, and other experimental factors, it is possible that these techniques may introduce as well as eliminate some systematic biases. For example, whether or not one spins down the membranous portion of the cell lysate (which may include nucleoplasm) may strongly influence the relative amount of nuclear-enriched miRNAs.
While endonucleolytic processing of miRNAs may affect miGEP results, miRNAs’ heterogeneous spatial distribution is another feature that can influence experimental outcome. Animal miRNAs are generally considered to localize diffusely within the cytoplasm, within cytosolic P bodies, stress granules, and/or in association with polyribosomes [80–85]. It has been established that particular miRNAs tend to be targeted to different cellular compartments. This can even be true of closely paralogous miRNAs. For example, hsa-miR-29a has been shown to be targeted to the nucleus, whereas hsa-miR-29b is a predominantly cytoplasmic human miRNA [86]. These paralogous miRNAs differ only at one central nucleotide and in several other nucleotides at the 3’ end. In cultured rat neurons, it has been shown that a particular group of miRNAs is targeted to dendrites, whereas others are more concentrated in the neuronal soma [87]. These differences could in principle alter the results of miGEPS under some circumstances. It is possible for example that some cell lysis techniques would be relatively enriched for cytoplasmic RNA, and thus they would systematically exclude nuclear miRNAs such as miR-29a.
As with the spatial processing of cellular miRNAs, the temporal sequence of miRNA expression may be also subject to complex regulation. MiRNAs were discovered in the context of worm developmental stages [88–90], and they may play a role in cell fate determination [91–93]. Hence some miRNAs, within a developmental framework, may exhibit stable and fixed “on/off” expression pattern. The stability of miRNA expression is supported by the observation that some unmodified siRNAs are stable and functional for days[94, 95] or weeks[96] in cultured cells. However, new data appears to indicate that certain miRNAs may be a more dynamic source of biochemical regulation (see for example [85, 97–99]), and hence may exhibit differential stability kinetics and/or rapidly-changing compartmentation in animal cells. Published data are relatively scanty about rates and mechanism(s) of physiological miRNA turnover whether in vivo, in vitro, or ex vivo. However, these data will be highly relevant to miGEP studies. For example, miRNAs are apparently differentially stable during storage, i.e. some miRNAs are more unstable than others, as quantified using RTQPCR [100]. Surprisingly, this also extends to miRNAs isolated using Trizol and stored at −80°C [100]. These are exactly the sort of data that needs to be more known and better understood because there is obvious potential impact on the work of many researchers.
Clinical/co-morbidity variables
Human tissue is an important source of RNA for gene expression profiling. Even in relatively tightly-controlled experimental contexts (surgical pathology biopsy specimens, biopsies from volunteers, or autopsy series from a closely-followed clinical cohort), there are challenges to adequate controls in miRNA and mRNA studies alike (see [25]). This is because there are a wide variety of conditions that can introduce experimental confounds and/or biological variability. Some of these are presented in Table 2, revised from [25]. Relatively little work has been performed to probe systematically the importance of premortem clinical features and co-morbidities, partly because expression profiling is expensive, the variables are so numerous, and grappling with obstacles of clinical documentation alone is a daunting task.
Table 2.
Clinical variables of potential relevance in miGEP studies on human tissues
| Demographics | Age |
| Sex | |
| Race | |
| Genetic factors | Congenital diseases |
| SNPs | |
| Non-SNP polymorphisms | |
| Mosaicism | |
| 'Background' comorbidity (chronic) | Sophistication of clinical documentation |
| Substance abuse/cigarettes | |
| Prescription drugs/supplements | |
| Metabolic disease | |
| Degenerative processes | |
| Major psychiatric disorder(s) | |
| Inflammatory/infectious disease | |
| Prior sugery/transplant | |
| Radiation therapy | |
| Body habitus/nutritional status | |
| Toxic/environmental exposures | |
| Organ function: kidney, liver, lungs, etc. | |
| Acute condition | (see above) |
| Vascular status (blood pressure problem/shock) | |
| External support--respirator, dialysis | |
| Metabolic perturbations | |
| Acute treatment incl chemotherapy, morphine, etc. | |
| State of consciousness incl sleep/wake | |
| Trauma | |
| Related to death/surgery | Single or multi-factorial |
| Biopsy/autopsy confirmed via pathology | |
| Anesthesia | |
| Preanalytical variables | Post-mortem interval |
| Interval prior to freezing/fixative | |
| Tissue pH | |
| Expertise/number of tissue handler(s) | |
| Sophistication of documentation | |
| Detailed information regarding anatomy |
Some potential confounds may be unexpectedly relevant to human miRNA expression studies. For example, circadian rhythms are apparently important in cell culture [101–103] as well as in vivo. Since miRNAs have been shown to be differentially expressed in different circadian stages [97, 104], it seems likely that some of the miGEP results will simply result from differences in sleep-wake cycle or, biorhythms that can result from various stimuli [103]. Additionally, the importance of particular anesthesia modalities and pain stimuli cause changes in mammalian mRNA gene expression [105, 106], and these data would also be interesting to correlate with miRNA expression studies. As more studies accumulate it will become easier to filter these variables. However, for now these potential confounds should be kept in mind.
Tissue processing
Important variables in tissue processing include tissue procurement, fixation, embedding, and RNA extraction method. Tissue fixation and embedding are technical parameters where the unique characteristics of miRNAs have a strong impact on gene expression profiling. This is evidently explained both by the shortness of the molecules as well as protection from degradation by intimate RNA-protein interactions. Other factors may be related—miRNA sequences may have evolved to elude RNA nucleases. Whereas mRNA tends to be labile in fixed and/or embedded tissue [25], a number of studies have shown robust correlations between miRNA profiling results in fresh versus in formalin fixed paraffin-embedded (FFPE) tissue [78, 107–110]. Perhaps surprisingly, biologically-relevant profiling data can be obtained from FFPE tissue that had been stored embedded at room temperature for years [107, 108]. However, it has been noted that RNA isolation from FFPE tissue is far less efficient than from fresh-frozen tissue, possibly because of fixative-induced RNA-protein cross-links [110], and the subtle biases of these reactions have yet to be entirely elucidated. Furthermore, it is possible that some miRNAs are relatively less likely to be ‘protected’ via RNA-protein interactions, and these would be predicted to be more affected by FFPE processing. Or more globally, during stress [75, 111], the overall miRNA-protein pattern may be altered, which may in turn change the degree to which miRNAs are protected from fixative cross-linking.
After the tissue sample is obtained, there are many different techniques that can be used to extract or isolate RNA. Considering the importance to the overall field, relatively little research has been performed to probe the impact of the particular RNA isolation techniques on miRNA expression. Different techniques rely on phenol/chloroform extraction, denaturing polyacrylamide gel electrophoresis (PAGE) with small RNA extraction, and/or column-based small RNA enrichment. The companion article shows that in human brain isolation for miRNA microarrays, the different RNA isolation techniques produce results with nontrivial differences [112]. Since we do not have a current “gold standard”, we do not know which is technically superior. However, we can certainly recommend that one should never compare samples against each other that have been isolated using different techniques. It has been noted that PAGE involves a loss of RNA, although it is unknown whether this can lead to a systematic bias in miRNA repertoire [1]. Another variable in RNA extraction that goes without saying, but which is no doubt important, is the expertise of the individual(s) who perform the experiment. Tissue dissection and RNA extraction can be a challenge to even experienced individuals. Whether or not an experienced person performs the RNA extraction, it should be kept in mind that even slight deviations from protocol can induce relatively large changes in experimental outcome.
Validation (alternative method, in situ hybridization)
Validating the results of miGEP studies is vitally important. As for any high-throughput and data rich technique, some testable false-positives are to be expected. It is necessary to know if the differential expression revealed by one platform is also shown using another expression platform, as this cannot be taken for granted. There is currently no acknowledged ‘gold standard’ for assessing the concentration of miRNAs in a sample. For example, Northern blots and RTQPCR have been shown to be very good at quantifying miRNAs, however each can be problematic in some circumstances [40, 78].
Validation experiments should also extend the tissue-level results (most miGEPs) to the cellular and subcellular levels, via in situ hybridization (ISH). A number of studies have shown robust ISH results with miRNAs in a number of different organisms (see [107, 113–116]). The insights from ISH can be vitally important, partly because almost every tissue comprises a variety of cell types and it is important to know which cells express the miRNA(s) of interest. For example, in human brain tumors, tissue-level miRNA microarray showed that miR-124 expression was decreased in oligodendrogliomas, whereas the expression of miR-9 was increased [107]. In situ hybridization revealed that the observed miR-124 decrease was accurate but not biologically relevant because miR-124 is specific to neurons and neurons are present in lower density in tissue samples that include brain tumors. By contrast, ISH showed that miR-9 expression was more robust in tumor cells than normal glial cells, and thus represent a better candidate for further study in the clinicobiological behavior of brain tumors [107]. We also found ISH to be an imperative complement to miRNA expression profiling in the study of Alzheimer’s disease where the cellular localization of miR-107 was important [117]. Another example in which ISH is helpful is in understanding the subcellular distribution of miRNAs. This principle is illustrated in myocytes where miR- 206 was shown to colocalize with ribosomes and within cell nucleoli [118]. Finally, ISH can be used to detect biologically important differences in the distribution of pre-miRNA and mature miRNA species [119]. Thus, ISH data provides a strongly complementary tool in shifting from high-throughput miGEP experiments to the more focused analyses of individual miRNAs.
Bioinformatics and normalization
A primary challenge to miGEP studies is to identify miRNA expression changes between samples that are valid and replicable. Although statistical tools are important in any high-throughput technology, including miGEPs, the statistical tasks to be performed are different. For most tissues the number of different miRNAs that are highly-expressed is fairly modest: usually less than a hundred miRNAs are even moderately expressed in a given tissue. Thus the same emphases that are placed on data management for a very-high throughput mRNA experiment, in which >100-fold times more transcripts are present, may not be as important to miRNA studies. MiRNA microarray experiments may align closer with large-scale Northern blotting than with expression arrays. Furthermore, in our experience, even sensible-seeming measures such as spiked-in controls must be performed mindful of their own potential for introducing, rather than dampening, data variability. In any case, the experiments must be performed considering the underlying biochemistry and the limitations inherent to each profiling platform.
Global miRNA expression patterns are thought to change dramatically in response to Drosha and histone deacetylase levels, cell division status, neoplastic transformation, developmental stage(s), circadian rhythms, cellular stress, and other factors (see for example [97, 104, 111, 116, 120–124]). Hence the assumptions—common to many mRNA expression profiling experiments—that overall RNA transcription is ‘constant’, and that a low percentage of individual transcripts are changed under different test conditions, are not applicable to miRNA studies. Some informatics, statistical, and normalization issues in miGEPs has been previously discussed [1]. In short, many researchers employ median scaling normalization (with the caveat that the results are probably not biologically relevant because global miRNA expression is changeable) and other means of stabilizing variance with a combination of technical replicates, logarithmic transformation, with or without spiked-in positive and negative controls [2, 125–128]. Innovative approaches have been employed when reconciling different expression platforms [129]. However, more work can be performed in order to reconcile miGEP profiling data analyses with the specifics of miRNA biology.
Conclusion
In high-throughput miGEP studies, the saying applies: “the devil is in the details”. Unfortunately, the details for every system (every profiling platform, cell type, disease, and experimental design) will be distinct. The impact of each technical parameter will hence be different – however subtly – for each of the profiling platforms. This argues for the need of more work on the topic of technical parameters in miGEP studies. Yet such papers are not necessarily considered high-yield in careerist or grantsmanship senses. There is not a high premium placed on working out details in a given experimental system, much less comparing different systems in a rigorous way. This problem has no quick fix. Rather, the field must acknowledge the importance of quality control and implementing measures to ensure that miGEP results can be considered valid and reliable. The first steps of this process involve the identification of the different areas of miGEP experiments that can contribute to systematic bias and/or error. These data begin to provide researchers with the necessary parameters for designing and implementing rigorous and valid future experiments that can be compared to each other across the many different miRNA profiling platforms.
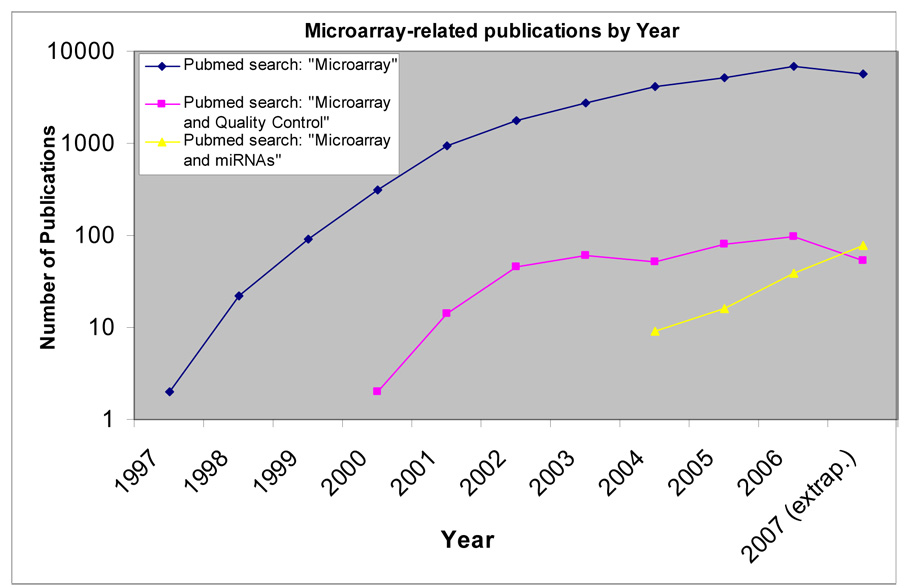
Figure 1.
A chart of the numbers of available papers referent to microarrays, as defined by searches on the Pubmed database. Papers about mRNA microarrays increased dramatically beginning in 1997, followed by a later increase in the number of papers cross-referenced by the search terms “microarray” and “quality control”. Papers returned by the search for the terms “microarray” and “miRNAs” appear to be increasing considerably since first appearing in 2004.
Acknowledgements
We thank Ms. Willa Huang for technical help. Funding was provided through NIH K08 NS050110 and NIH P30 49008400 (PTN), and the Kentucky Tobacco Research and Development Center (KTRDC), the USDA-NRI grants 2006-35301-17115 and 2006-35100-17433, the NSF MCB-0718029 (Subaward No. S-00000260), and the NIH 5 R03 AI 068934-02 (GT).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References Cited
- 1.Davison TS, Johnson CD, Andruss BF. Analyzing micro-RNA expression using microarrays. Methods Enzymol. 2006;411:14–34. doi: 10.1016/S0076-6879(06)11002-2. [DOI] [PubMed] [Google Scholar]
- 2.Garzon R, et al. MicroRNA expression and function in cancer. Trends Mol Med. 2006;12(12):580–587. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 3.Krichevsky AM. MicroRNA profiling: from dark matter to white matter, or identifying new players in neurobiology. ScientificWorldJournal. 2007;7:155–166. doi: 10.1100/tsw.2007.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kreil DP, Russell RR. There is no silver bullet--a guide to low-level data transforms and normalisation methods for microarray data. Brief Bioinform. 2005;6(1):86–97. doi: 10.1093/bib/6.1.86. [DOI] [PubMed] [Google Scholar]
- 5.Tinker AV, Boussioutas A, Bowtell DD. The challenges of gene expression microarrays for the study of human cancer. Cancer Cell. 2006;9(5):333–339. doi: 10.1016/j.ccr.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Yauk CL, Berndt ML. Review of the literature examining the correlation among DNA microarray technologies. Environ Mol Mutagen. 2007 doi: 10.1002/em.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller RA, Galecki A, Shmookler-Reis RJ. Interpretation, design, and analysis of gene array expression experiments. J Gerontol A Biol Sci Med Sci. 2001;56(2):B52–B57. doi: 10.1093/gerona/56.2.b52. [DOI] [PubMed] [Google Scholar]
- 8.Marshall E. Getting the noise out of gene arrays. Science. 2004;306(5696):630–631. doi: 10.1126/science.306.5696.630. [DOI] [PubMed] [Google Scholar]
- 9.Blalock EM, et al. Harnessing the power of gene microarrays for the study of brain aging and Alzheimer's disease: statistical reliability and functional correlation. Ageing Res Rev. 2005;4(4):481–512. doi: 10.1016/j.arr.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 10.Wang H, et al. A study of inter-lab and inter-platform agreement of DNA microarray data. BMC Genomics. 2005;6(1):71. doi: 10.1186/1471-2164-6-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi L, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24(9):1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuo WP, et al. A sequence-oriented comparison of gene expression measurements across different hybridization-based technologies. Nat Biotechnol. 2006;24(7):832–840. doi: 10.1038/nbt1217. [DOI] [PubMed] [Google Scholar]
- 13.Yauk CL, et al. Comprehensive comparison of six microarray technologies. Nucleic Acids Res. 2004;32(15):e124. doi: 10.1093/nar/gnh123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffith OL, et al. Assessment and integration of publicly available SAGE, cDNA microarray, and oligonucleotide microarray expression data for global coexpression analyses. Genomics. 2005;86(4):476–488. doi: 10.1016/j.ygeno.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 15.Kavsan VM, et al. Comparison of microarray and sage techniques in gene expression analysis of human glioblastoma. Tsitol Genet. 2007;41(1):36–55. [PubMed] [Google Scholar]
- 16.Kim HL. Comparison of oligonucleotide-microarray and serial analysis of gene expression (SAGE) in transcript profiling analysis of megakaryocytes derived from CD34+ cells. Exp Mol Med. 2003;35(5):460–466. doi: 10.1038/emm.2003.60. [DOI] [PubMed] [Google Scholar]
- 17.van Ruissen F, et al. Evaluation of the similarity of gene expression data estimated with SAGE and Affymetrix GeneChips. BMC Genomics. 2005;6:91. doi: 10.1186/1471-2164-6-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li S, et al. Too much data, but little inter-changeability: a lesson learned from mining public data on tissue specificity of gene expression. Biol Direct. 2006;1:33. doi: 10.1186/1745-6150-1-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petersen D, et al. Three microarray platforms: an analysis of their concordance in profiling gene expression. BMC Genomics. 2005;6(1):63. doi: 10.1186/1471-2164-6-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shippy R, et al. Performance evaluation of commercial short-oligonucleotide microarrays and the impact of noise in making cross-platform correlations. BMC Genomics. 2004;5(1):61. doi: 10.1186/1471-2164-5-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shippy R, et al. Using RNA sample titrations to assess microarray platform performance and normalization techniques. Nat Biotechnol. 2006;24(9):1123–1131. doi: 10.1038/nbt1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yauk CL, et al. Novel design and controls for focused DNA microarrays: applications in quality assurance/control and normalization for the Health Canada ToxArray. BMC Genomics. 2006;7:266. doi: 10.1186/1471-2164-7-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo L, et al. Rat toxicogenomic study reveals analytical consistency across microarray platforms. Nat Biotechnol. 2006;24(9):1162–1169. doi: 10.1038/nbt1238. [DOI] [PubMed] [Google Scholar]
- 24.Strauss E. Arrays of hope. Cell. 2006;127(4):657–659. doi: 10.1016/j.cell.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 25.Van Deerlin VM, Gill LH, Nelson PT. Optimizing gene expression analysis in archival brain tissue. Neurochem Res. 2002;27(10):993–1003. doi: 10.1023/a:1020996519419. [DOI] [PubMed] [Google Scholar]
- 26.Debey S, et al. Comparison of different isolation techniques prior gene expression profiling of blood derived cells: impact on physiological responses, on overall expression and the role of different cell types. Pharmacogenomics J. 2004;4(3):193–207. doi: 10.1038/sj.tpj.6500240. [DOI] [PubMed] [Google Scholar]
- 27.Breit S, et al. Impact of pre-analytical handling on bone marrow mRNA gene expression. Br J Haematol. 2004;126(2):231–243. doi: 10.1111/j.1365-2141.2004.05017.x. [DOI] [PubMed] [Google Scholar]
- 28.Kim SJ, et al. Effects of storage, RNA extraction, genechip type, and donor sex on gene expression profiling of human whole blood. Clin Chem. 2007;53(6):1038–1045. doi: 10.1373/clinchem.2006.078436. [DOI] [PubMed] [Google Scholar]
- 29.Campo Dell'orto M, et al. New data on robustness of gene expression signatures in leukemia: comparison of three distinct total RNA preparation procedures. BMC Genomics. 2007;8(1):188. doi: 10.1186/1471-2164-8-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohtsuka E, et al. Joining of synthetic ribotrinucleotides with defined sequences catalyzed by T4 RNA ligase. Eur J Biochem. 1977;81(2):285–291. doi: 10.1111/j.1432-1033.1977.tb11950.x. [DOI] [PubMed] [Google Scholar]
- 31.Romaniuk E, et al. The effect of acceptor oligoribonucleotide sequence on the T4 RNA ligase reaction. Eur J Biochem. 1982;125(3):639–643. doi: 10.1111/j.1432-1033.1982.tb06730.x. [DOI] [PubMed] [Google Scholar]
- 32.Li Y, et al. Systematic comparison of the fidelity of aRNA, mRNA and T-RNA on gene expression profiling using cDNA microarray. J Biotechnol. 2004;107(1):19–28. doi: 10.1016/j.jbiotec.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 33.Wilson CL, et al. Amplification protocols introduce systematic but reproducible errors into gene expression studies. Biotechniques. 2004;36(3):498–506. doi: 10.2144/04363RN05. [DOI] [PubMed] [Google Scholar]
- 34.Shingara J, et al. An optimized isolation and labeling platform for accurate microRNA expression profiling. Rna. 2005;11(9):1461–1470. doi: 10.1261/rna.2610405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin G, et al. Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) polymerase. J Mol Biol. 2004;341(4):911–925. doi: 10.1016/j.jmb.2004.06.047. [DOI] [PubMed] [Google Scholar]
- 36.Yehudai-Resheff S, Schuster G. Characterization of the E.coli poly(A) polymerase: nucleotide specificity, RNA-binding affinities and RNA structure dependence. Nucleic Acids Res. 2000;28(5):1139–1144. doi: 10.1093/nar/28.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weiss EA, Gilmartin GM, Nevins JR. Poly(A) site efficiency reflects the stability of complex formation involving the downstream element. Embo J. 1991;10(1):215–219. doi: 10.1002/j.1460-2075.1991.tb07938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mattie MD, et al. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bustin SA. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol. 2002;29(1):23–39. doi: 10.1677/jme.0.0290023. [DOI] [PubMed] [Google Scholar]
- 40.Bustin SA, Nolan T. Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J Biomol Tech. 2004;15(3):155–166. [PMC free article] [PubMed] [Google Scholar]
- 41.Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nat Protoc. 2006;1(3):1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- 42.Stahlberg A, et al. Properties of the reverse transcription reaction in mRNA quantification. Clin Chem. 2004;50(3):509–515. doi: 10.1373/clinchem.2003.026161. [DOI] [PubMed] [Google Scholar]
- 43.Burns MJ, et al. Standardisation of data from real-time quantitative PCR methods - evaluation of outliers and comparison of calibration curves. BMC Biotechnol. 2005;5:31. doi: 10.1186/1472-6750-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beuvink I, et al. A novel microarray approach reveals new tissue-specific signatures of known and predicted mammalian microRNAs. Nucleic Acids Res. 2007;35(7):e52. doi: 10.1093/nar/gkl1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barczak A, et al. Spotted long oligonucleotide arrays for human gene expression analysis. Genome Res. 2003;13(7):1775–1785. doi: 10.1101/gr.1048803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee JK, et al. Comparing cDNA and oligonucleotide array data: concordance of gene expression across platforms for the NCI-60 cancer cells. Genome Biol. 2003;4(12):R82. doi: 10.1186/gb-2003-4-12-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li J, Pankratz M, Johnson JA. Differential gene expression patterns revealed by oligonucleotide versus long cDNA arrays. Toxicol Sci. 2002;69(2):383–390. doi: 10.1093/toxsci/69.2.383. [DOI] [PubMed] [Google Scholar]
- 48.Lim LP, et al. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17(8):991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32(Database issue):D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nelson PT, et al. A novel monoclonal antibody against human Argonaute proteins reveals unexpected characteristics of miRNAs in human blood cells. RNA. 2007;13(10):1787–1792. doi: 10.1261/rna.646007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berezikov E, et al. Many novel mammalian microRNA candidates identified by extensive cloning and RAKE analysis. Genome Res. 2006;16(10):1289–1298. doi: 10.1101/gr.5159906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bentwich I, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37(7):766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 53.Berezikov E, et al. Diversity of microRNAs in human and chimpanzee brain. Nat Genet. 2006;38(12):1375–1377. doi: 10.1038/ng1914. [DOI] [PubMed] [Google Scholar]
- 54.Cummins JM, et al. The colorectal microRNAome. Proc Natl Acad Sci U S A. 2006;103(10):3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johnston RJ, Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426(6968):845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- 56.Ambros V, et al. A uniform system for microRNA annotation. Rna. 2003;9(3):277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim HK, et al. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol. 2006;174(5):677–687. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCarthy JJ, Esser KA, Andrade FH. MicroRNA-206 is overexpressed in the diaphragm but not the hindlimb muscle of mdx mouse. Am J Physiol Cell Physiol. 2007;293(1):C451–C457. doi: 10.1152/ajpcell.00077.2007. [DOI] [PubMed] [Google Scholar]
- 59.Rao PK, et al. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci U S A. 2006;103(23):8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chapman EJ, et al. Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 2004;18(10):1179–1186. doi: 10.1101/gad.1201204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pfeffer S, et al. Identification of virus-encoded microRNAs. Science. 2004;304(5671):734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 62.Xie Z, et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2(5):E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xie Z, Kasschau KD, Carrington JC. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr Biol. 2003;13(9):784–789. doi: 10.1016/s0960-9822(03)00281-1. [DOI] [PubMed] [Google Scholar]
- 64.Blow MJ, et al. RNA editing of human microRNAs. Genome Biol. 2006;7(4):R27. doi: 10.1186/gb-2006-7-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kawahara Y, et al. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315(5815):1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Luciano DJ, et al. RNA editing of a miRNA precursor. Rna. 2004;10(8):1174–1177. doi: 10.1261/rna.7350304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kawahara Y, et al. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep. 2007;8(8):763–769. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kirino Y, Mourelatos Z. The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. RNA. 2007;13(9):1397–1401. doi: 10.1261/rna.659307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kirino Y, Mourelatos Z. Mouse Piwi-interacting RNAs are 2'-O-methylated at their 3' termini. Nat Struct Mol Biol. 2007;14(4):347–348. doi: 10.1038/nsmb1218. [DOI] [PubMed] [Google Scholar]
- 70.Ohara T, et al. The 3' termini of mouse Piwi-interacting RNAs are 2'-O-methylated. Nat Struct Mol Biol. 2007;14(4):349–350. doi: 10.1038/nsmb1220. [DOI] [PubMed] [Google Scholar]
- 71.Ronemus M, Martienssen R. RNA interference: methylation mystery. Nature. 2005;433(7025):472–473. doi: 10.1038/433472a. [DOI] [PubMed] [Google Scholar]
- 72.Seitz H, et al. Imprinted small RNA genes. Biol Chem. 2004;385(10):905–911. doi: 10.1515/BC.2004.118. [DOI] [PubMed] [Google Scholar]
- 73.Yu B, et al. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307(5711):932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nelson PT, Keller JN. RNA in brain disease: no longer just "the messenger in the middle". J Neuropathol Exp Neurol. 2007;66(6):461–468. doi: 10.1097/01.jnen.0000240474.27791.f3. [DOI] [PubMed] [Google Scholar]
- 75.Bhattacharyya SN, et al. Stress-induced reversal of microRNA repression and mRNA P-body localization in human cells. Cold Spring Harb Symp Quant Biol. 2006;71:513–521. doi: 10.1101/sqb.2006.71.038. [DOI] [PubMed] [Google Scholar]
- 76.Lugli G, et al. Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J Neurochem. 2005;94(4):896–905. doi: 10.1111/j.1471-4159.2005.03224.x. [DOI] [PubMed] [Google Scholar]
- 77.Krichevsky AM, et al. A microRNA array reveals extensive regulation of microRNAs during brain development. Rna. 2003;9(10):1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nelson PT, et al. Microarray-based, high-throughput gene expression profiling of microRNAs. Nat Methods. 2004;1(2):155–161. doi: 10.1038/nmeth717. [DOI] [PubMed] [Google Scholar]
- 79.Schmittgen TD, et al. A high-throughput method to monitor the expression of microRNA precursors. Nucleic Acids Res. 2004;32(4):e43. doi: 10.1093/nar/gnh040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chan SP, Slack FJ. microRNA-mediated silencing inside P-bodies. RNA Biol. 2006;3(3):97–100. doi: 10.4161/rna.3.3.3499. [DOI] [PubMed] [Google Scholar]
- 81.Kim J, et al. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc Natl Acad Sci U S A. 2004;101(1):360–365. doi: 10.1073/pnas.2333854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nelson PT, Hatzigeorgiou AG, Mourelatos Z. miRNP:mRNA association in polyribosomes in a human neuronal cell line. Rna. 2004;10(3):387–394. doi: 10.1261/rna.5181104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu J, et al. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7(7):719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Leung AK, Sharp PA. Function and localization of microRNAs in mammalian cells. Cold Spring Harb Symp Quant Biol. 2006;71:29–38. doi: 10.1101/sqb.2006.71.049. [DOI] [PubMed] [Google Scholar]
- 85.Leung AK, Calabrese JM, Sharp PA. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc Natl Acad Sci U S A. 2006;103(48):18125–18130. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315(5808):97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 87.Kye MJ, et al. Somatodendritic microRNAs identified by laser capture and multiplex RT-PCR. Rna. 2007 doi: 10.1261/rna.480407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107(7):823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- 89.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294(5543):862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 90.Pasquinelli AE, Ruvkun G. Control of developmental timing by micrornas and their targets. Annu Rev Cell Dev Biol. 2002;18:495–513. doi: 10.1146/annurev.cellbio.18.012502.105832. [DOI] [PubMed] [Google Scholar]
- 91.Hobert O. Common logic of transcription factor and microRNA action. Trends Biochem Sci. 2004;29(9):462–468. doi: 10.1016/j.tibs.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 92.Schier AF, Giraldez AJ. MicroRNA function and mechanism: insights from zebra fish. Cold Spring Harb Symp Quant Biol. 2006;71:195–203. doi: 10.1101/sqb.2006.71.055. [DOI] [PubMed] [Google Scholar]
- 93.Wilfred BR, Wang WX, Nelson PT. Energizing miRNA research: A review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Mol Genet Metab. 2007;91(3):209–217. doi: 10.1016/j.ymgme.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15(2):188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Holen T, et al. Positional effects of short interfering RNAs targeting the human coagulation trigger Tissue Factor. Nucleic Acids Res. 2002;30(8):1757–1766. doi: 10.1093/nar/30.8.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Omi K, Tokunaga K, Hohjoh H. Long-lasting RNAi activity in mammalian neurons. FEBS Lett. 2004;558(1–3):89–95. doi: 10.1016/S0014-5793(04)00017-1. [DOI] [PubMed] [Google Scholar]
- 97.Cheng HY, et al. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54(5):813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Poy MN, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432(7014):226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 99.Schratt GM, et al. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439(7074):283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 100.Bravo V, et al. Instability of miRNA and cDNAs derivatives in RNA preparations. Biochem Biophys Res Commun. 2007;353(4):1052–1055. doi: 10.1016/j.bbrc.2006.12.135. [DOI] [PubMed] [Google Scholar]
- 101.Ralph MR, Joyner AL, Lehman MN. Culture and transplantation of the mammalian circadian pacemaker. J Biol Rhythms. 1993;(8 Suppl):S83–S87. [PubMed] [Google Scholar]
- 102.Ren D, Miller JD. Primary cell culture of suprachiasmatic nucleus. Brain Res Bull. 2003;61(5):547–553. doi: 10.1016/s0361-9230(03)00193-x. [DOI] [PubMed] [Google Scholar]
- 103.Schibler U. Circadian time keeping: the daily ups and downs of genes, cells, and organisms. Prog Brain Res. 2006;153:271–282. doi: 10.1016/S0079-6123(06)53016-X. [DOI] [PubMed] [Google Scholar]
- 104.Xu S, et al. MicroRNA transcriptome of mouse retina and identification of a sensory organ specific miRNA cluster. J Biol Chem. 2007 doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
- 105.Culley DJ, et al. Altered hippocampal gene expression 2 days after general anesthesia in rats. Eur J Pharmacol. 2006;549(1–3):71–78. doi: 10.1016/j.ejphar.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 106.Ko J, et al. cDNA microarray analysis of the differential gene expression in the neuropathic pain and electroacupuncture treatment models. J Biochem Mol Biol. 2002;35(4):420–427. doi: 10.5483/bmbrep.2002.35.4.420. [DOI] [PubMed] [Google Scholar]
- 107.Nelson PT, et al. RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain. Rna. 2006;12(2):187–191. doi: 10.1261/rna.2258506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lawrie CH, et al. Microrna expression distinguishes between germinal center B cell-like and activated B cell-like subtypes of diffuse large B cell lymphoma. Int J Cancer. 2007;121(5):1156–1161. doi: 10.1002/ijc.22800. [DOI] [PubMed] [Google Scholar]
- 109.Wang H, Ach RA, Curry B. Direct and sensitive miRNA profiling from low-input total RNA. Rna. 2007;13(1):151–159. doi: 10.1261/rna.234507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li J, et al. Comparison of miRNA expression patterns using total RNA extracted from matched samples of formalin-fixed paraffin-embedded (FFPE) cells and snap frozen cells. BMC Biotechnol. 2007;7:36. doi: 10.1186/1472-6750-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Marsit CJ, Eddy K, Kelsey KT. MicroRNA responses to cellular stress. Cancer Res. 2006;66(22):10843–10848. doi: 10.1158/0008-5472.CAN-06-1894. [DOI] [PubMed] [Google Scholar]
- 112.Wang WX, et al. Focus on RNA isolation: Obtaining RNA for microRNA (miRNA) expression profiling analyses of neural tissue. Biochim Biophys Acta. 2008 doi: 10.1016/j.bbagrm.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.De Pietri Tonelli D, et al. Single-cell detection of microRNAs in developing vertebrate embryos after acute administration of a dual-fluorescence reporter/sensor plasmid. Biotechniques. 2006;41(6):727–732. doi: 10.2144/000112296. [DOI] [PubMed] [Google Scholar]
- 114.Obernosterer G, Martinez J, Alenius M. Locked nucleic acid-based in situ detection of microRNAs in mouse tissue sections. Nat Protoc. 2007;2(6):1508–1514. doi: 10.1038/nprot.2007.153. [DOI] [PubMed] [Google Scholar]
- 115.Wheeler G, et al. In situ detection of animal and plant microRNAs. DNA Cell Biol. 2007;26(4):251–255. doi: 10.1089/dna.2006.0538. [DOI] [PubMed] [Google Scholar]
- 116.Wienholds E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309(5732):310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 117.Wang WX, et al. The expression of microRNA miR-107 decreases early in Alzheimer's disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008;28(5):1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Politz JC, Zhang F, Pederson T. MicroRNA-206 colocalizes with ribosome-rich regions in both the nucleolus and cytoplasm of rat myogenic cells. Proc Natl Acad Sci U S A. 2006;103(50):18957–18962. doi: 10.1073/pnas.0609466103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Obernosterer G, et al. Post-transcriptional regulation of microRNA expression. RNA. 2006;12(7):1161–1167. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Muralidhar B, et al. Global microRNA profiles in cervical squamous cell carcinoma depend on Drosha expression levels. J Pathol. 2007;212(4):368–377. doi: 10.1002/path.2179. [DOI] [PubMed] [Google Scholar]
- 121.Mishima Y, et al. Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr Biol. 2006;16(21):2135–2142. doi: 10.1016/j.cub.2006.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shalgi R, et al. Global and Local Architecture of the Mammalian microRNA-Transcription Factor Regulatory Network. PLoS Comput Biol. 2007;3(7):e131. doi: 10.1371/journal.pcbi.0030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang T, et al. A micro-RNA signature associated with race, tumor size, and target gene activity in human uterine leiomyomas. Genes Chromosomes Cancer. 2007;46(4):336–347. doi: 10.1002/gcc.20415. [DOI] [PubMed] [Google Scholar]
- 124.Scott GK, et al. Rapid alteration of microRNA levels by histone deacetylase inhibition. Cancer Res. 2006;66(3):1277–1281. doi: 10.1158/0008-5472.CAN-05-3632. [DOI] [PubMed] [Google Scholar]
- 125.Calin GA, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353(17):1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 126.Garzon R, et al. MicroRNA fingerprints during human megakaryocytopoiesis. Proc Natl Acad Sci U S A. 2006;103(13):5078–5083. doi: 10.1073/pnas.0600587103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Barad O, et al. MicroRNA expression detected by oligonucleotide microarrays: system establishment and expression profiling in human tissues. Genome Res. 2004;14(12):2486–2494. doi: 10.1101/gr.2845604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 130.Tang X, et al. A simple array platform for microRNA analysis and its application in mouse tissues. RNA. 2007;13(10):1803–1822. doi: 10.1261/rna.498607. [DOI] [PMC free article] [PubMed] [Google Scholar]