Abstract
Short-sequence fragments (‘DNA barcodes’) used widely for plant identification and inventorying remain to be applied to complex biological problems. Host–herbivore interactions are fundamental to coevolutionary relationships of a large proportion of species on the Earth, but their study is frequently hampered by limited or unreliable host records. Here we demonstrate that DNA barcodes can greatly improve this situation as they (i) provide a secure identification of host plant species and (ii) establish the authenticity of the trophic association. Host plants of leaf beetles (subfamily Chrysomelinae) from Australia were identified using the chloroplast trnL(UAA) intron as barcode amplified from beetle DNA extracts. Sequence similarity and phylogenetic analyses provided precise identifications of each host species at tribal, generic and specific levels, depending on the available database coverage in various plant lineages. The 76 species of Chrysomelinae included—more than 10 per cent of the known Australian fauna—feed on 13 plant families, with preference for Australian radiations of Myrtaceae (eucalypts) and Fabaceae (acacias). Phylogenetic analysis of beetles shows general conservation of host association but with rare host shifts between distant plant lineages, including a few cases where barcodes supported two phylogenetically distant host plants. The study demonstrates that plant barcoding is already feasible with the current publicly available data. By sequencing plant barcodes directly from DNA extractions made from herbivorous beetles, strong physical evidence for the host association is provided. Thus, molecular identification using short DNA fragments brings together the detection of species and the analysis of their interactions.
Keywords: herbivory, host plant, molecular identification, coevolution, trnL cpDNA
1. Introduction
DNA is surprisingly resilient to degradation when ingested by organisms with diet or prey. Molecules of hundreds of base pairs in length can be recovered easily despite extra-oral enzymatic digestion of predatory insects or after passage through the digestive tract of mammals (Symondson 2002). Thus, the presence of prey DNA in predator samples has been successfully exploited to identify prey species via PCR (Zaidi et al. 1999; Chen et al. 2000; Hoogendoorn & Heimpel 2001; Greenstone et al. 2007). Herbivory is the most widely observed feeding mode in insects (May 1988) and tight ecological links exist between herbivores and their food plants. Therefore, information on the precise feeding source is important for studies of ecology, speciation, coevolution and applied sciences. Host associations can be established by direct observations of feeding or by morphological or chemical studies of gut content (e.g. Isley & Alexander 1949; Post 2002), but require precise identification of plants, which may be complicated when reproductive parts or other diagnostic features are absent (Stewart 1967).
Large-scale genetic data acquisition in taxonomy (‘DNA barcoding’) now permits whole species inventories or assessment of species diversity, based on standardized short-sequence fragments. In plants, several ‘barcode’ loci have been proposed (e.g. Chase et al. 2007; Kress & Erickson 2007; Taberlet et al. 2007; Fazekas et al. 2008; Lahaye et al. 2008) for which representation in databases increases rapidly, improving the accuracy and speed of host plant identification. The wide availability of the cytochrome oxidase subunit 1 (cox1) marker fulfils a similar role in animals (Hebert et al. 2003). When used in comparative studies, e.g. for the analysis of host plant associations, the sequence fragments are used by linking them to a named species or DNA-based group to which ecological information from literature or field observations has been associated (Hebert et al. 2004). These groups and their host information provide the starting point for analysing coevolutionary relationships of plants and herbivores. A key aspect in these studies is the authentication of the feeding source, whereby the strongest evidence linking an individual to the food plant is provided through analysis of ingested host tissue (e.g. Isley & Alexander 1949; Fry et al. 1978). DNA-based approaches could greatly facilitate this step, in particular if the DNA of the food source could be obtained directly from the insect tissue.
Beetles account for nearly one quarter of the known animal species on the Earth; this species richness is closely linked to the diversity of their angiosperm food plants (Grimaldi & Engel 2005). The Chrysomelidae (leaf beetles) are a major herbivore group with some 35 000 species (Jolivet & Verma 2002). Among these, the subfamily Chrysomelinae represents some of the largest and most colourful groups and includes major agricultural pests, such as the Colorado potato beetle (Leptinotarsa). Both adults and larvae feed externally on leaves from a broad variety of angiosperm families, and most species are restricted in host range (Jolivet & Hawkeswood 1995; Pokon et al. 2005; Reid 2006). Their defensive compounds, the precursors of which are sequestered from plant tissue, make them an important group for coevolutionary analysis. Chrysomelinae are proportionally most diverse in Australia, with approximately 25 per cent of the world genera (42 recognized in the latest revision; Reid 2006) and approximately 750 species. Host plants of Chrysomelinae are well known in the North Temperate Zone to include mainly Asteraceae, Fabaceae, Lamiaceae, Plantaginaceae and Salicaceae (Jolivet & Hawkeswood 1995). In Australia, species within a genus are conservative in their food choice and those in several species-rich genera feed mainly on the equally diverse genera Acacia (Fabaceae) and Eucalyptus (Myrtaceae) with several hundred species each, but the details of host associations remain unknown (Reid 2006) and coevolutionary patterns have never been investigated. Here, we obtained host associations for a significant proportion of the Australian Chrysomelinae, in particular for those feeding on endemic Australian plant lineages. This information was obtained from the insect specimens themselves, by applying PCR amplifications with plant-specific primers directly to whole-insect DNA extractions. The ingested DNA constitutes a record of the individual's food plant(s).
2. Material and methods
(a) Data collection
The provenance of specimens used in this study is presented in the electronic supplementary material (figs S1 and S2). DNA was extracted from whole beetles by soaking the specimens in extraction buffer using the DNeasy Blood and Tissue Extraction kit (Qiagen, West Sussex, UK). After extraction, beetle specimens were mounted and retained as vouchers (Australian Museum, Sydney; IBE, Barcelona). The DNA was used as template for PCR amplification of the plastid trnL intron using the plant-specific primers c A49325 (5′CGAAATCGGTAGACGCTACG) and d B49863 (5′GGGGATAGAGGGACTTGAAC; Taberlet et al. 1991). PCR conditions used 0.2 μM of each primer and 3.5 mM MgCl2 using a touchdown protocol of 16 cycles with decreasing annealing temperature from 60 to 43°C (60 s), and 27 cycles at 42°C (60 s). Denaturation (94°C) and elongation (72°C) lasted 30 and 60 s, respectively. PCR products were visualized on 1.5 per cent agarose gel electrophoresis and subsequently purified using MSB Spin PCRapace (Invitek, Berlin, Germany). A few products showing multiple bands were cloned using the TOPO-TA Cloning kit (Invitrogen, Carlsbad, CA, USA). Inserts were amplified using vector primers, and those showing different lengths in agarose gels were selected for sequencing. In all other cases, PCR products were sequenced directly using the same primers as above and the BigDye Terminator Cycle Sequencing kit (Applied Biosystems, Foster City, CA, USA). Chromatograms showing double peaks were also interpreted as evidence for wider trophic range and the original sample was also subjected to cloning and screening of different size classes. In addition, all DNA extractions were used for amplification of animal cox1 (C1-J-2183 and TL2-N-3014; Simon et al. 1994) and nuclear elongation factor 1-alpha (EF1a), using two primers sets (efs149 and efa1043; Normark et al. 1999), and customized internal primers for Chrysomelinae (efs303-chrysomelinae 5′CACAGAGATTTCATCAAGAAC and efa923-chrysomelinae 5′CGTTCTTAACGTTGAAACCAA). PCR conditions used a standard protocol with annealing ranging between 45 and 50°C in the case of cox1 and the same conditions as described for the amplification of the trnL intron for EF1a. Sequences were edited and contigs were assembled using BioEdit v. 7 (Hall 1999). trnL sequences have been deposited at the EBI under accession numbers FM160425–FM160505, and beetle sequences under FM209215–FM209246 (cox1) and FM209248-FM209279 (EF1a).
(b) Phylogenetic reconstruction
Each plant trnL sequence was compared against the GenBank nr database using the BLASTn algorithm and default search parameters (Altschul et al. 1990). The 500 best hits were arranged according to the ‘total score’, a recommended strategy for this marker where a hypervariable region is flanked by areas of high similarity. This measure of similarity considers the sum of scores for different fragments in which a query sequence can be potentially fragmented as part of the heuristic BLAST search, as opposed to the score of a single fragment (‘maximum score’). The top 100 hits were retrieved and both the query sequence and three gymnosperm outgroups (Cycas siamensis: AY651841; Gingko biloba: AY145323; and Pseudotsuga menziensii: AF327589) were added to the matrix. Sequence alignment was done with MAFFT 5 and the iterative search strategy L-INS-i using default parameters (Katoh et al. 2005). Aligned matrices were used for Bayesian reconstruction of phylogenetic relationships using MrBayes v. 3.1.2 (Ronquist & Huelsenbeck 2003) with a GTR+I+G model (Rodríguez et al. 1990), as recommended by ModelTest v. 3.7 (Posada & Crandall 1998). Trees were obtained after two independent runs of four MCMC chains each for one million generations, sampling trees every 100th generation. After discarding trees in the pre-stationary phase, the tree topology and posterior probability (PP) for each clade were obtained as the strict consensus tree. Phylogenetic analysis of combined beetle cox1 and EF1a sequences (trivially aligned manually) was also based on Bayesian inference rooting the trees with Macrolema marginata and Richmondia olliffi (Chrysomelidae: Spilopyrinae).
3. Results
(a) Characterization of diet trnL and its usage as barcode
Among possible plant barcodes (ITS, trnH-psbA, rbcL, trnL, rpoC1, rpoB, matK; Taberlet et al. 2007; Fazekas et al. 2008), rbcL and trnL stood out as best candidates for identification based on their high representation in sequence databases. A fragment corresponding to the trnL intron of the chloroplast genome produced very reliable PCR amplifications and was our marker of choice. We obtained trnL sequences ranging between 313 and 581 bp for 78 beetle specimens of 76 species from sclerophyll and rainforest biomes across Australia, representing 24 genera. Most samples produced sequence reads of high quality after direct sequencing of the PCR products. Where mixed sequence reads required cloning and sequencing of multiple clones, two divergent trnL sequences were obtained in three of seven cases (table S1 in the electronic supplementary material), resulting in 81 different trnL intron sequences.
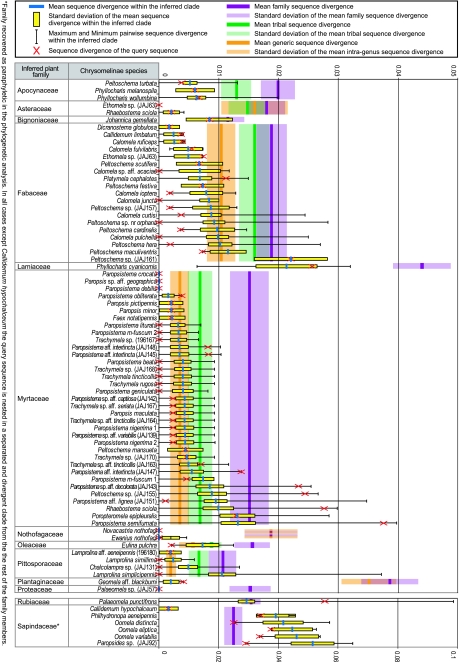
For taxonomic identifications, the trnL intron sequences were associated with existing data using phylogenetic analysis and measures of similarity. The precision of the identification differed greatly dependent on the taxonomic coverage in particular groups, as sequence divergence to the closest GenBank hit ranged from 0 to 5.5 per cent (figure 1). Phylogenetic analysis of each trnL sequence together with the respective 100 GenBank top hits placed the query sequences within clades at various hierarchical levels. Membership in a particular taxon was determined by establishing the smallest group of sequences encompassing the query that was supported by a PP≥0.7; using the taxonomic assignment common to all of these GenBank entries (e.g. at the level of genus, tribe or family) identified the focal sequence as a member of a certain taxon. Depending on the botanical group, we could infer associations in most cases to tribes, groups of related genera, genera, subgenera and even species (table 1; fig. S1 in the electronic supplementary material). The tribe and genus levels were inferred confidently for 82.7 and 51.0 per cent of the taxa, respectively. These percentages are conservative because they do not include cases where the focal sequence is well supported as sister group to a single representative of a tribe or a genus, i.e. consistent with a particular position within those groups. The phylogenetic reconstruction was always sufficient to narrow down the host plant hypothesis to very few species using floristic data for the particular area where the beetles were collected (fig. S1 in the electronic supplementary material).
Figure 1.
Identification of host plants against GenBank entries. trnL intron sequences obtained from beetle tissue were subjected to phylogenetic analysis together with their respective GenBank top hits. In each case, sequence divergence was estimated by pairwise comparisons with all sequences in this clade; minimum, mean and standard deviation measures (p-distance) are given here. Details on each clade are in fig. S1 in the electronic supplementary material. Similarity levels in each individual case are compared with the mean trnL divergences and standard deviations for all sequences available on GenBank for the identified family, shown by coloured bands to illustrate divergences at the level of the entire family and the average divergence for tribes and genera within each family.
Table 1.
Summary of trophic inferences for 76 species (n) of Australian Chrysomelinae. (Both angiosperm family and most inclusive supported clade (PP≥0.70) for each inference are given. The trees used for the inferences are in fig. S1 in the electronic supplementary material.)
| beetle genus | n | plant family | support (PP) | most inclusive plant group | support (PP) | no. |
|---|---|---|---|---|---|---|
| Callidemum | 2 | Sapindaceae | 0.71 | sister to Dodonaea viscosa | 0.97 | 1 |
| Fabaceae | 1.00 | Acacia | 0.83 | 2 | ||
| Calomela | 9 | Fabaceae | 0.86–1.00 | Acacia | 0.73–0.97 | 3–11 |
| Chalcolampra | 1 | Pittosporaceaea | 1.00 | Billardiera, Marianthus and Cheiranthera | 0.93 | 12 |
| Dicranosterna | 1 | Fabaceae | 1.00 | Acacia | 0.95 | 13 |
| Ethomela | 2b | Asteraceaea,c | 0.85–1.00 | Asteraceaec | — | 14 |
| Craspedia | 1.00 | 15 | ||||
| Fabaceaea | 1.00 | Acacia | 0.88 | 16 | ||
| Eulina | 1 | Oleaceae | 0.80 | Ligustruma | 0.97 | 17 |
| Ewanius | 1 | Nothofagaceae | 1.00 | Nothofagus subgenus Lophozonia | 1.00 | 18 |
| Faex | 1 | Myrtaceae | 1.00 | tribe Leptospermeae sister to Leptospermum scoparium | 0.90 | 19 |
| Geomela | 1 | Plantaginaceaea | 1.00 | Plantago subgenus Plantago section Mesembrynia | 0.98 | 20 |
| Johannica | 1 | Bignoniaceaec | — | sister to Pandorea | 0.89 | 21 |
| Lamprolina | 3 | Pittosporaceae | 1.00 | Bursaria sister to B. spinosa | 0.71 | 22 |
| Pittosporum | 0.95 | 23 | ||||
| Pittosporaceae | — | 24 | ||||
| Novacastria | 1 | Nothofagaceae | 1.00 | Nothofagus subgenus Lophozonia sister to N. moorei | 0.98 | 25 |
| Oomela | 3 | Sapindaceaec | — | Allophylus, Serjania and ?Aesculus | 0.75–0.96 | 26–28 |
| Palaeomela | 2 | Proteaceaea | 1.00 | sister to Orites lancifolia | 1.00 | 29 |
| Rubiaceaea | 1.00 | Rubioideae | 1.00 | 30 | ||
| Paropsides | 1 | Sapindaceaec | — | Allophylus, Serjania and ?Aesculus | 0.96 | 31 |
| Paropsis | 4 | Myrtaceae | 1.00 | Angophora and Corymbia | 1.00 | 32 |
| Eucalyptus | 1.00 | 33 | ||||
| sister to Leptospermum scoparium | 1.00 | 34–35 | ||||
| Paropsisterna | 16 | Myrtaceae | 1.00 | Angophora and Corymbia | 1.00 | 36 |
| sister to Angophora and Corymbia | 1.00 | 37 | ||||
| basal and close to tribe Leptospermeae | 1.00 | 38–41 | ||||
| basal and divergent to tribe Leptospermeae | 0.93–1.00 | 42–43 | ||||
| Eucalyptus | 0.88–1.00 | 44–52 | ||||
| Kunzea | 1.00 | 53 | ||||
| Peltoschema | 11b | Apocynaceaea | 1.00 | Artia, Parsonsia and Prestonia | 0.84 | 54 |
| Asteraceaea,c | 0.95 | Asteraceae | — | 55 | ||
| Fabaceae | 0.93–1.00 | Acacia | 0.76–1.00 | 56–62 | ||
| Daviesiaa | 1.00 | 63 | ||||
| Myrtaceae | 1.00 | basal and divergent to tribe Leptospermeaea | 0.93 | 64 | ||
| tribe Leptospermeae sister to Leptospermum scoparium | 0.89 | 65 | ||||
| Philhydronopa | 1 | Sapindaceaec | — | Allophylus and Serjania | 0.73 | 66 |
| Phyllocharis | 3 | Apocynaceaea | 1.00 | Artia, Parsonsia and Prestonia | 0.93 | 67 |
| sister to Parsonsia eucalyptophylla | 0.70 | 68 | ||||
| Lamiaceaea,c | — | subfamily Teucrioideae sister to genus Ajuga | 1.00 | 69 | ||
| Platymela | 1 | Fabaceaea | 0.94 | Acacia | 0.79 | 70 |
| Poropteromela | 1 | Myrtaceae | 0.97 | Myrtaceae | — | 71 |
| Rhaebosterna | 1b | Asteraceaea,c | — | Asteraceae | — | 72 |
| Myrtaceae | 1.00 | basal and divergent to tribe Leptospermeae | 1.00 | 73 | ||
| Trachymela | 8 | Myrtaceae | 1.00 | Eucalyptus | 1.00 | 74–81 |
New trophic link.
Specimens contributing two sequences (after cloning).
Paraphyletic.
The quality of identifications was also assessed with measures of sequence similarity. Pairwise sequence divergence of the query with the closest available GenBank sequence was compared with the mean divergences within the taxonomic group to which the sequence was assigned at the genus, tribe and family level (figure 1). In most cases the divergence between query and top GenBank hit was lower than the mean divergence at the genus or tribe level, demonstrating the great precision of most identifications. However, this type of analysis is only useful if taxonomic coverage of the predicted hierarchical group is largely uniform in the database, as divergences may be biased if only closely related species are represented (depressing the average divergence), while the query sequence may be highly divergent to the sampled species. This explains high variance in sequence divergence in the all-against-all comparisons and very high divergence of the query from its closest relative in some cases in our analysis (figure 1). Underlying taxonomy (but also taxonomic decisions of sequence data submitters) are an important source of bias as divergence estimates of non-natural groups can inflate both divergence and variance measures (e.g. Sapindaceae in figure 1). In general, these analyses confirmed the results from the phylogenetic identifications, showing that levels of sequence divergences with the query fell within the ranges observed in the groups to which a sequence had been associated.
(b) Barcoding an ecological association
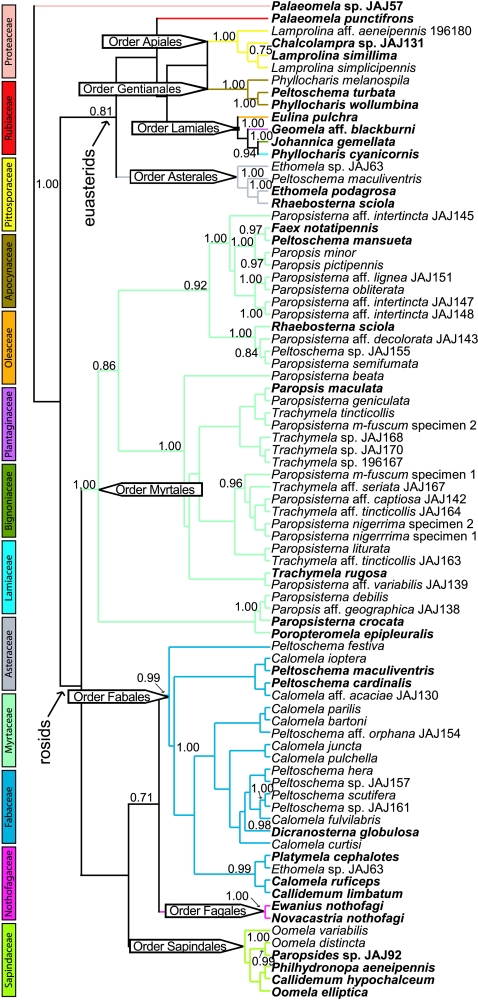
When trnL intron sequences were used for tree-building without inclusion of GenBank data, major clades in this tree corresponded well to families and genera when labelled based on their taxonomy assignment from top GenBank hits, and the tree was generally consistent with current plant systematics (figure 2). This procedure for summarizing the data confirmed that the identification obtained for each case (table 1; fig. S1 in the electronic supplementary material) was reliable. In total, the trnL intron sequences amplified from the beetle specimens were assigned to 13 plant families, with the greatest representation in Fabaceae and Myrtaceae, the dominant botanical features of the Australian landscape and among the known hosts of species in various Chrysomelinae genera (Reid 2006). Almost 40 per cent of associations represented new host plant records, either for genera with unknown host plants (Geomela on Plantaginaceae and Palaeomela on Proteaceae and Rubiaceae) or expanding their known range of hosts (Chalcolampra, Ethomela, Eulina, Peltoschema, Phyllocharis, Platymela and Rhaebosterna). In instances where two divergent trnL sequences were retrieved from a single individual, we found them to belong to different plant families in all but one case. Multiple food choice was also deduced by examining more than one species of the genus from different geographical sources (table 1; fig. S1 in the electronic supplementary material).
Figure 2.
Bayesian phylogenetic tree of trnL intron sequences obtained from beetle tissue. Terminals are given as the taxa of Chrysomelinae yielding each sequence; those in bold represent individuals used for reconstructing the genus-level phylogeny of the beetles (see figure 3). The topology is broadly congruent with plant systematics and major plant lineages are labelled. Only PP≥0.70 are shown.
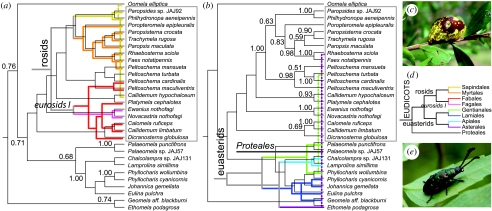
Conservatism of associations and potential coevolutionary patterns were examined by establishing relationships of the major chrysomeline lineages (figure 3). One representative for each of the 24 genera was selected at random for phylogenetic analysis, plus an additional representative of any genus (Callidemum, Phyllocharis, Peltoschema and Paleomela) that was found on a second order of plants (as identified in the trnL intron tree; figure 2), for a total of 30 taxa. The resulting tree showed a clade uniting all representatives of the tribe Gonioctenini, which was included in a basal grade representing the Phyllocharitini, in which Oomela and the sister pair Ethomela and Geomela were the basal lineages. Host use was conservative, with major lineages of beetles generally limited to particular groups of plants (figure 3). For instance, most members of the monophyletic paropsines (genera Faex, Paropsis, Paropsisterna, Poropteromela, Rhaebosterna and Trachymela) were associated with Myrtaceae, the sister pair Ewanius nothofagi and Novacastria nothofagi with Nothofagus (Fagales), and many other representatives of the same tribe Gonioctenini (genera Callidemum, Calomela, Dicranosterna, Peltoschema and Platymela) were found on Acacia (Fabales). With only two exceptions, the Gonioctenini were associated with four orders of the rosid angiosperms. In turn, the Phyllocharitini, with the exceptions of Oomela elliptica on the order Sapindales and an undescribed species of Paleomela on the eudicot primitive order Proteales, were associated with four orders of the euasterid angiosperms.
Figure 3.
Phylogeny of hosts and herbivores. The tree shows the relationships of beetle genera from combined cox1 and EF1a sequences. Each genus is represented by one species, with further species added where a genus was feeding on more than one plant family (terminals in boldface in figure 2). Inferred host associations are plotted on the tree for (a) the Gonioctenini and (b) the Phyllocharitini chrysomelines showing a high degree of conservatism with few host shifts (dashed lines) between major plant lineages. (d) Simplified eudicot phylogeny for relevant plant orders, consistent with the tree obtained here from trnL intron sequences; redrawn from Soltis et al. 2005). Beetles: (c) Paropsis maculata Marsham on Myrtaceae; (e) Johannica gemellata (Westwood) on Bignoniaceae (photographs: J. A. Jurado).
Four beetle genera were found associated with two host plant families drawn from different orders or even major eudicot lineages (in one case, Callidemum, already recorded in the literature; Reid 2006). Remarkably, the genus Peltoschema was associated with four different host plants in three plant orders of rosids and euasterids (table 1; figures 2 and 3). The beetle genera occurring on multiple host groups were monophyletic in all cases except Callidemum, indicating that genus designation largely matches phylogenetic inferences from the two gene fragments used here. Generally, incidences of multiple host groups therefore represent host shifts across large phyletic distances. It remains to be tested as to what degree intra-generic lineages have undergone host shifts within a given order of plants.
4. Discussion
PCR amplification from DNA extractions of herbivorous beetles readily produced plant-derived trnL intron sequences that permitted a reliable authentication of the feeding source. The study shows that recently developed procedures (Matheson et al. 2008) that require time-consuming gut content preparations can be greatly simplified by amplifying directly from the whole-specimen DNA extraction. It is not clear as to whether the procedure simply co-purifies the gut content or the plant DNA is preserved in the insect haemolymph and other tissue, and for how long after feeding. The nearly uniform success of the amplification suggests that the precise conditions under which specimens were collected are not critical. Contamination with plant material externally from contact with plants or pollen, rather than ingested tissue, may also be amplified, but can be avoided through cleaning of specimens, while pollen in angiosperms very rarely bears chloroplasts used for amplification here (Zhang et al. 2003). The authenticity of plant DNA was also established for nine Chrysomelinae species of particularly well-known and narrow food plant associations. In all of these, trnL amplified from whole-insect DNA produced correct inferences of host association (table 2; fig. S2 in the electronic supplementary material). This observation greatly increases our confidence in the inference of host associations using this method.
Table 2.
Host identification from trnL intron in species of Chrysomelinae with well-established existing host records. (The trees used for the inferences are in fig. S2 in the electronic supplementary material.)
| beetle species | source | plant source | food plant (literature) | phylogenetic inference | no. |
|---|---|---|---|---|---|
| Araucanomela wellingtonensis (Bechyné et Bechyné) | Llanquihue, Chile | Nothofagus betuloides | Nothofagaceae (Nothofagus betuloides) | Nothofagaceae (1), Nothofagus gr. nitida (0.98) | 1 |
| Chrysolina americana (Linnaeus) | Granada, Spain | Rosmarinus officinalis | Lamiaceae (Rosmarinus, Lavandula) | Lamiaceae-Mentheae (1), Rosmarinus officinalis (0.99) | 2 |
| Chrysolina quadrigemina (Suffrian) | Bragança, Portugal | — | Clusiaceae (Hypericum) | Clusiaceae (1), Hypericum sp. (1) | 3 |
| Chrysolina viridana (Küster) | Granada, Spain | Mentha sp. | Lamiaceae (Mentha, Salvia) | Lamiaceae-Mentheae (1), Mentha gr. spicata-longifolia (0.74) | 4 |
| Chrysomela collaris Linnaeus | Altai, Siberia | — | Salicaceae (Salix, Populus) | Salicaceae (1), Salix (11 spp.; 0.79) | 5 |
| Gonioctena variabilis (Olivier) | Albacete, Spain | Genista scorpius | Fabaceae (Genista, Retama, Sarothamnus) | Fabaceae-Genisteae (1), Genista scorpius (0.99) | 6 |
| Leptinotarsa decemlineata (Say) | Granada, Spain | Solanum tuberosum | Solanaceae (Solanum, Lycopersicon) | Solanaceae (1), Solanum-Lycopersicon (1) | 7 |
| Phratora vitellinae (Linnaeus) | Lleida, Spain | Salix bicolor | Salicaceae (Salix, Populus) | Salicaceae (1), Salix gr. alba-pentandra (0.99) | 8 |
| Plagiodera versicolora (Laicharting) | Ourense, Spain | Salix sp. | Salicaceae (Salix, Populus) | Salicaceae (1), Salix gr. alba-pentandra (0.99) | 9 |
Correct identification of host plants greatly depends on the taxonomic representation in sequence databases. For example, host plant genera were well sampled at the species level or even population level for our test set of nine species with known hosts (table 2), which were mainly from the well-studied Palaearctic region. This permitted straightforward identifications of hosts at the species level in all cases, with the exception of Hypericum (Clusiaceae), which was less well represented in GenBank. Other plant families are clearly under-represented, given the number of described species. For example, only 14 and 15 of approximately 1000 and 800 described Australian species of Acacia and Eucalyptus, respectively, are represented in GenBank by trnL intron sequences. This affects the phylogenetic host inference when constructing the data matrix, as the assessment against distantly related taxa may lead to inconsistent relationships due to long-branch attraction. However, our supplementary approach of measuring the divergence with the closest relative in the context of all pairwise divergences in the wider taxonomic group provides an additional level of confidence. An inference can also be narrowed down by extrapolation from floristic catalogues from the insect's area of origin. This made it possible in most cases to hypothesize a single plant species, or group of related species, as possible host (see fig. S1 in the electronic supplementary material). Ultimately, however, only denser taxon sampling in the reference database can solve the problem of potentially spurious identification. The inverse problem of high-sequence homogeneity and a lack of discriminatory power may be circumvented by increasing the number of reference taxa obtained from GenBank for the phylogenetic analysis. We set the matrix size to 100, which provided sufficient taxa to yield resolution, while not compromising the speed of analysis.
Further limitations arise from using only a single marker. First, the paucity of representation for some botanical groups in the database may depend on which marker is used for the analysis. Here we opted for the trnL intron as the most widely represented marker in GenBank with 71 855 entries (5 August 2008). This marker was shown to provide a suitable level of resolution for identifications (Taberlet et al. 2007) and in our hands produced more robust amplifications than other potential barcode markers such as rbcL and matK (Bradley et al. 2007; Matheson et al. 2008). Second, lineage-specific evolutionary dynamics may compromise the resolving power of the trnL intron across plant groups. While highly discriminative in most cases, trnL alone may not recover some well-established plant taxa as monophyletic (e.g. family Bignonaceae, tribe Astereae) or show no structure for constituent members (e.g. Mimosoideae). However, we found that even in these cases, a query sequence will be placed correctly, provided there is at least one fairly close relative in the database. There is also the possibility that a higher taxon as currently defined may not constitute a monophyletic group (e.g. Acacia; Maslin et al. 2003), but this is not a problem for phylogenetic approaches to identification, as the query will still group with its closest relatives. Third, at a finer level of resolution, the identification will be limited by the lack of variation in trnL at the species level. Multiple barcode sequences can overcome this problem in most cases due to their combined greater resolution (Fazekas et al. 2008). Finally, using only a single marker may increase the susceptibility of the study to inaccurate identification, incorrect labelling or poor sequence quality in the reference database (e.g. Korning et al. 1996). In the course of this study, we found several examples of erroneous taxonomic assignments (e.g. Sapindaceae identified as Cypripedium, Cypripedioideae; Apocynaceae labelled as Sesamum, Pedaliaceae; one case of names switched between Pittosporum and Cheiranthera, both Pittosporaceae; suspicious generic assignment for Aesculus x carnea), and of sequencing artefacts (e.g. Tragopogon spp., Acacia usumatensis) and chimeras (e.g. Pentaphylax euryoides). Problems introduced by these sequences were only apparent after careful inspection of trees revealing suspicious relationships, and required phylogenetic re-evaluation after removing problematic sequence data.
All of the above would argue for the use of additional markers, and to improve precision and confidence of host plant identification. Similar arguments could be made for the use of cox1 as a single marker for barcoding in animals, in particular where coevolutionary studies are attempted. This is not a problem in principle, as the DNA extractions could be used for additional PCR (e.g. EF1a; figure 3). While the precision of the analysis would improve, this may come at the cost of fewer analyses and ultimately fewer host records that could establish precise host ranges. Sampling more individuals will also fill out the trees with additional species of both herbivores and plants, and will improve the quality of reference databases and the precision of identifications.
In this first application of the method, we obtained a highly diverse array of trophic behaviours for Australian Chrysomelinae. Our implementation of the test to Australian Chrysomelinae shows that we can reach a reliable identification to plant family in every case and very frequently the inference is possible at lower taxonomic levels as well. The tectonic isolation of Australia (ca 25 Mya) and its long-term aridity have favoured the unique sclerophyll biomes dominated by a few plant lineages, including eucalypts and acacias, that are predominant features of the Australian landscape (Crisp et al. 2004). Some species-rich chrysomeline genera including Calomela, Trachymela or Paropsisterna show a strict association with Acacia and Eucalyptus (or Leptospermum), respectively (Reid 2006). Other genera, including Paropsides or Peltoschema, show wide host ranges. The latter genus is reported here for the first time as polyphagous, with a trophic amplitude that may support its hypothesized non-monophyly (Reid & Ślipiński 2001; Reid 2006). Curiously, secondary hosts in individuals of Peltoschema, Rhaebosterna and Ethomela were from Asteraceae, the host of many Chrysomelinae in the Northern Hemisphere. These are unexpected host records that could represent either true polyphagy, accidental uptake or an opportunistic use of plant tissue for hydration. The evolution of host use in Australian Chrysomelinae is broadly congruent with that of their feeding sources at deep evolutionary scales, suggestive of coevolutionary patterns (figure 3). Nevertheless, host shifts or wide host spectra are also evident.
Our analysis not only shows the details of ecological associations for a dominant herbivore group, but also offers the basis for their evolutionary interpretation. Routine application of this procedure to herbivorous insects can solve a range of ecological and evolutionary questions about host plant associations with great reliability. We also explored the use of a smaller fragment (10–143 bp) nested within trnL, the so-called P6 loop (primers g and h; Taberlet et al. 2007), which seems to provide useful inferences at least at family level (data not shown). The use of this smaller, yet informative, marker could broaden the suitability of this technique for samples where an even higher degree of DNA degradation is expected (starved or even dry collection specimens).
The future implementation of this method will benefit from the growing taxonomic coverage in databases and regional genetic botanical inventories, improved methods for match analysis that overcome the limitations of BLAST, and the use of multiple marker systems to refine the identification of hosts. These developments will further increase the value of our demonstration that host plant DNA can be amplified with great reliability from a DNA sample extracted from herbivorous beetles. The procedure extends the use of DNA barcoding methods to species identification and coevolutionary relationships of trophic interactions.
Acknowledgments
We are indebted to J. A. Rosselló (Institut Cavanilles, València), A. Cardoso and J. Castresana (IBE, CSIC, Barcelona) and J. Pons (IMEDEA, CSIC, Esporles) for useful discussion. A. Muñoz (Palma de Mallorca), D. de Little (Hobart), A. Sundholm and R. de Keyzer (Sydney) helped with collecting. T. Houston and B. Hanich (Western Australian Museum, Perth) gave logistic support. The Departments of Conservation and Land Management of Western Australia and of Primary Industries, Water and Environment of Tasmania provided the required collecting permits. The editor and three reviewers made useful comments towards the final version of this paper. This work has been supported by the Spanish Ministry of Education with an FPI doctorate studentship and short-stay programme to J.A.J.R., as well as projects REN2003-03667 and CGL2006-08810 to E.P., the CSIC Intramural Project 200730I014 to J.G.-Z. and Australian Biological Resources Study funding to C.A.M.R.
Supplementary Material
Cloning strategy
Bayesian phylogenetic trees based on chloroplast trnL intron under a GTR + I + G evolutionary model and sequence alignment obtained with MAFTT 5 using the L-INS-I iterative search strategy. Outgroup sequences have been removed secondarily as well as posterior probability (PP) values equal or lower than 0.15. Terminals of the most inclusive clade containing the query sequence supported with a PP value equal or higher than 0.7 appear enclosed in a box. Tree branches have been colored according to botanical gropus following the taxonomic information attached to each sequence in the nucleotide sequence public database. Each tree appears associated to the available information used to elaborate the trophic inference: collecting locality, field observations (i.e. plant on which the beetle was found), literature feeding records, a summary of our phylogenetic inference, the regional and local flora related with the inference that have been cited in the study area and a concluding field with the most plausible trophic hypothesis based on this information
Bayesian phylogenetic trees based on chloroplast trnL intron under a GTR + I + G evolutionary model and sequence alignment obtained with MAFTT 5 using the L-INS-I iterative search strategy. Outgroup sequences have been removed secondarily as well as posterior probability (PP) values equal or lower than 0.15. Terminals of the most inclusive clade containing the query sequence supported with a PP value equal or higher than 0.7 appear enclosed in a box. Tree branches have been colored according to botanical gropus following the taxonomic information attached to each sequence in the nucleotide sequence public database
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. doi:10.1006/jmbi.1990.9999 [DOI] [PubMed] [Google Scholar]
- Bradley B.J., Stiller M., Doran-Sheehy D.M., Harris T., Chapman C.A., Vigilant L., Poinar H. Plant DNA sequences from feces: potential means for assessing diets of wild primates. Am. J. Primatol. 2007;69:699–705. doi: 10.1002/ajp.20384. doi:10.1002/ajp.20384 [DOI] [PubMed] [Google Scholar]
- Chase M.W., et al. A proposal for a standardised protocol to barcode all land plants. Taxon. 2007;56:295–299. [Google Scholar]
- Chen Y., Giles K.L., Payton M.E., Greenstone M.H. Identifying key cereal aphid predators by molecular gut analysis. Mol. Ecol. 2000;9:1887–1898. doi: 10.1046/j.1365-294x.2000.01100.x. doi:10.1046/j.1365-294x.2000.01100.x [DOI] [PubMed] [Google Scholar]
- Crisp M., Cook L., Steane D. Radiation of the Australian flora: what can comparisons of molecular phylogenies across multiple taxa tell us about the evolution of diversity in present-day communities? Phil. Trans. R. Soc. B. 2004;359:1551–1571. doi: 10.1098/rstb.2004.1528. doi:10.1098/rstb.2004.1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazekas A.J., Burgess K.S., Kesanakurti P.R., Graham S.W., Newmaster S.G., Husband B.C., Percy D.M., Hajibabaei M., Barrett S.C.H. Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well. PLoS ONE. 2008;3:e2882. doi: 10.1371/journal.pone.0002802. doi:10.1371/journal.pone.0002802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry B., Joern A., Parker P.L. Grasshopper food web analysis: use of carbon isotope ratios to examine feeding relationships among terrestrial herbivores. Ecology. 1978;59:498–506. doi:10.2307/1936580 [Google Scholar]
- Greenstone M.H., Rowley D.L., Weber D.C., Payton M.E., Hawthorne D.J. Feeding mode and prey detectability half-lives in molecular gut-content analysis: an example with two predators of the Colorado potato beetle. Bull. Entomol. Res. 2007;97:201–209. doi: 10.1017/S000748530700497X. doi:10.1017/S000748530700497X [DOI] [PubMed] [Google Scholar]
- Grimaldi D., Engel M.S. Cambridge University Press; Cambridge, UK: 2005. Evolution of the insects. [Google Scholar]
- Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- Hebert P.D.N., Cywinska A., Ball S.L., de Waard J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. doi:10.1098/rspb.2002.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert P.D.N., Penton E.H., Burns J.M., Janzen D.H., Hallwachs W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc. Natl Acad. Sci. USA. 2004;41:14 812–14 817. doi: 10.1073/pnas.0406166101. doi:10.1073/pnas.0406166101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogendoorn M., Heimpel G.E. PCR-based gut content analysis of insect predators: using ribosomal ITS-1 fragments from prey to estimate predation frequency. Mol. Ecol. 2001;10:2059–2067. doi: 10.1046/j.1365-294x.2001.01316.x. doi:10.1046/j.1365-294X.2001.01316.x [DOI] [PubMed] [Google Scholar]
- Isley F.B., Alexander G. Analysis of insect food habits by crop examination. Science. 1949;109:115–116. doi: 10.1126/science.109.2823.115. doi:10.1126/science.109.2823.115 [DOI] [PubMed] [Google Scholar]
- Jolivet P., Hawkeswood T.J. Backhuys Publishers; Leiden, Germany: 1995. Host-plants of the Chrysomelidae of the world. [Google Scholar]
- Jolivet P., Verma K.K. Intercept Publishers; Andover, UK: 2002. Biology of leaf beetles. [Google Scholar]
- Katoh K., Kuma K.-i., Toh H., Miyata T. MAFFT v. 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. doi:10.1093/nar/gki198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korning P.G., Hebsgaard S.M., Rouzé P., Brunak S. Cleaning the GenBank Arabidopsis thaliana data set. Nucleic Acids Res. 1996;24:316–320. doi: 10.1093/nar/24.2.316. doi:10.1093/nar/24.2.316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress W.J., Erickson D.L. A two locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE. 2007;2:e508. doi: 10.1371/journal.pone.0000508. doi:10.1371/journal.pone.0000508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahaye R., et al. DNA barcoding the floras of biodiversity spots. Proc. Natl Acad. Sci. USA. 2008;105:2923–2928. doi: 10.1073/pnas.0709936105. doi:10.1073/pnas.0709936105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslin B.R., Miller J.T., Seigler D.S. Overview of the generic status of Acacia (Leguminosae: Mimosoideae) Aust. Syst. Bot. 2003;16:1–18. doi:10.1071/SB02008 [Google Scholar]
- Matheson C.D., Muller G.C., Junnila A., Vernon K., Hausmann A., Miller M.A., Greenblatt C., Schlein Y. A PCR method for detection of plant meals from the guts of insects. Org. Div. Evol. 2008;7:294–303. doi:10.1016/j.ode.2006.09.002 [Google Scholar]
- May R.M. How many species are there on earth? Science. 1988;241:1441–1449. doi: 10.1126/science.241.4872.1441. doi:10.1126/science.241.4872.1441 [DOI] [PubMed] [Google Scholar]
- Normark B.B., Jordal B.H., Farrell B.D. Origin of a haplodiploid beetle lineage. Proc. R. Soc. B. 1999;226:2253–2259. doi:10.1098/rspb.1999.0916 [Google Scholar]
- Pokon R., Novotny V., Samuelson G.A. Host specialization and species richness of root-feeding chrysomelid larvae (Chrysomelidae, Coleoptera) in a New Guinea rain forest. J. Trop. Ecol. 2005;21:595–604. doi:10.1017/S0266467405002567 [Google Scholar]
- Posada D., Crandall K.A. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. doi:10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Post D.M. Using stable isotopes to estimate trophic position: models, methods, and assumptions. Ecology. 2002;83:703–718. doi:10.1890/0012-9658(2002)083[0703:USITET]2.0.CO;2 [Google Scholar]
- Reid C.A.M. A taxonomic revision of the Australian Chrysomelinae, with a key to the genera (Coleoptera: Chrysomelidae) Zootaxa. 2006;1292:1–119. [Google Scholar]
- Reid C.A.M., Ślipiński S.A. Peltoschema Reitter, a hitherto unrecognized Chrysomelinae: redescription and systematic placement (Coleoptera: Chrysomelidae) Coleopt. Bull. 2001;55:330–337. doi:10.1649/0010-065X(2001)055[0330:PRAHUC]2.0.CO;2 [Google Scholar]
- Rodríguez F., Oliver J.L., Marín A., Medina J.R. The general stochastic model of nucleotide substitution. J. Theor. Biol. 1990;142:485–501. doi: 10.1016/s0022-5193(05)80104-3. doi:10.1016/S0022-5193(05)80104-3 [DOI] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. doi:10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Simon C., Frati F., Beckenbach A., Crespi B., Liu H., Flook P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and compilation of conserved polymerase chain reaction primers. Ann. Entomol. Soc. Am. 1994;87:651–701. [Google Scholar]
- Soltis D.E., Soltis P.S., Endress P.K., Chase M.W. Sinauer Associates; Sunderland, MA: 2005. Phylogeny and evolution of angiosperms. [Google Scholar]
- Stewart D.R.M. Analysis of plant epidermis in faeces: a technique for studying the food preferences of grazing herbivores. J. Appl. Ecol. 1967;4:83–111. doi:10.2307/2401411 [Google Scholar]
- Symondson W.O.C. Molecular identification prey in predator diets. Mol. Ecol. 2002;11:627–641. doi: 10.1046/j.1365-294x.2002.01471.x. doi:10.1046/j.1365-294X.2002.01471.x [DOI] [PubMed] [Google Scholar]
- Taberlet P., Gielly L., Pautou G., Bouvet J. Universal primers for amplification of three noncoding regions of chloroplast DNA. Plant Mol. Biol. 1991;17:1105–1109. doi: 10.1007/BF00037152. doi:10.1007/BF00037152 [DOI] [PubMed] [Google Scholar]
- Taberlet P., et al. Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Res. 2007;35:e14. doi: 10.1093/nar/gkl938. doi:10.1093/nar/gkl938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi R.H., Jaal Z., Hawkes N.J., Hemingway J., Symondson W.O.C. Can the detection of prey DNA amongst the gut contents of invertebrate predators provide a new technique for quantifying predation in the field? Mol. Ecol. 1999;8:2081–2088. doi: 10.1046/j.1365-294x.1999.00823.x. doi:10.1046/j.1365-294x.1999.00823.x [DOI] [PubMed] [Google Scholar]
- Zhang Q., Liu Y., Sodmergen Examination of the cytoplasmatic DNA in male reproductive cells to determine the potential for cytoplasmatic inheritance in 295 angiosperm species. Plant Cell. Physiol. 2003;44:941–951. doi: 10.1093/pcp/pcg121. doi:10.1093/pcp/pcg121 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cloning strategy
Bayesian phylogenetic trees based on chloroplast trnL intron under a GTR + I + G evolutionary model and sequence alignment obtained with MAFTT 5 using the L-INS-I iterative search strategy. Outgroup sequences have been removed secondarily as well as posterior probability (PP) values equal or lower than 0.15. Terminals of the most inclusive clade containing the query sequence supported with a PP value equal or higher than 0.7 appear enclosed in a box. Tree branches have been colored according to botanical gropus following the taxonomic information attached to each sequence in the nucleotide sequence public database. Each tree appears associated to the available information used to elaborate the trophic inference: collecting locality, field observations (i.e. plant on which the beetle was found), literature feeding records, a summary of our phylogenetic inference, the regional and local flora related with the inference that have been cited in the study area and a concluding field with the most plausible trophic hypothesis based on this information
Bayesian phylogenetic trees based on chloroplast trnL intron under a GTR + I + G evolutionary model and sequence alignment obtained with MAFTT 5 using the L-INS-I iterative search strategy. Outgroup sequences have been removed secondarily as well as posterior probability (PP) values equal or lower than 0.15. Terminals of the most inclusive clade containing the query sequence supported with a PP value equal or higher than 0.7 appear enclosed in a box. Tree branches have been colored according to botanical gropus following the taxonomic information attached to each sequence in the nucleotide sequence public database